Abstract
Aurantiadioic acids A (1) and B (2), two new furan-containing polyketides, and aurantoic acid A (3), a new natural product, were isolated from the liquid fermentation of the sika deer dung-derived actinomycete Actinocorallia aurantiaca. The structures of the new compounds were established by extensive spectroscopic methods, including 1D & 2D NMR, HRESIMS spectroscopic analysis. The absolute configuration of 3 was assigned by comparison of the specific optical rotations with the reported derivatives. Biological activity evaluations suggested that compounds 1–3 showed weak inhibition on NO production in the murine monocytic RAW 264.7 macrophages with IC50 values of 35.8, 41.8, 45.2 μM, respectively. Compound 3 showed weak inhibition on influenza A virus (A/PuertoRico/8/1934, H1N1) with an EC50 value of 35.9 μM, and a selective index higher than 13.3.
Electronic supplementary material
The online version of this article (10.1007/s13659-019-00217-0) contains supplementary material, which is available to authorized users.
Keywords: Actinomycete, Actinocorallia aurantiaca, Polyketides, Anti-NO activity, Antiviral activity
Introduction
The actinomycetous secondary metabolites, which have attracted great attention from natural product research community in past decades, are considered to be a promising reservoir of new bioactive natural products for drug discovery [1]. The examples of secondary metabolites from actinomycetes, such as streptomycin, actinomycin, tetracycline, rifamycin, vancomycin and mitomycin, etc., have great influence on the treatment of human diseases. The gut microbes from insects have emerged to be fruitful resources for drug leads in recent years [2–4]. However, the gut actinomycetes associated with wild animals have long been underexplored for their potential in drug discovery [5].
In this study, we examined the secondary metabolites of the actinomycete Actinocorallia aurantiaca which was isolated from the feces of sika deer. The actinomycete A. aurantiaca belongs to the family Thermomonosporaceae, and it has never been chemically investigated. Herein, we report the isolation, structural elucidation, and anti-NO activity of three compounds from the cultures of A. aurantiaca.
Results and Discussion
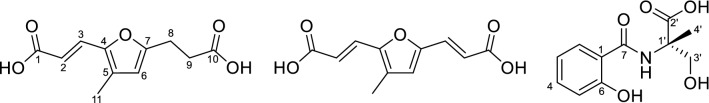
Compound 1 was obtained as a yellow oil. It had a molecular formula of C11H12O5 as determined by (+)-HRESIMS analysis with the protonated ion peak at m/z 225.07576 [M+H]+ (calcd for C11H13O5, 225.07575), corresponding to six degrees of unsaturation. The 1H NMR spectroscopic data of 1 (Table 1) showed the presence of one methyl singlet at δH 2.11 (CH3-11), and two trans-olefinic methines at δH 6.07 (d, J = 15.4 Hz, H-2), 7.44 (d, J = 15.4 Hz, H-3), and an olefinic methine singlet at δH 6.12 (s, H-2). The 13C NMR and DEPT spectroscopic data (Table 1) presented eleven carbons ascribable to one methyl, two methylenes, three methines, and five quaternary carbons (two carbonyl groups). The spectroscopic features of the chemical shifts at δC 112.2, 129.2, 147.2, and 158.8 implied the presence of a trisubstituted furan ring in compound 1. Exhaustive analysis of the 2D NMR spectra furnished the establishment of the structure of 1. The 1H–1H COSY correlations allowed the connection of C-2–C-3, and C-8–C-9. The HMBC correlations from Me-11 to C-4 (δC 147.2), C-5 (δC 129.2), and C-6 (δC 112.2) indicated the methyl connected to C-5. Furthermore, the trans-olefinic protons (H-2, H-3) correlated to a carboxylic group at δC 171.2 (C-1), and C-4 in the HMBC spectrum, indicative of the connection of C-1–C-2–C-3–C-4. Besides, the HMBC correlations from two methylene protons (H-8, H-9) to C-7 and C-10 enabled the connection of C-7–C-8–C-9–C-10 (Fig. 2). Therefore, compound 1 was established to be a furan derivative with two carboxylic groups (Fig. 1), and was given the name aurantiadioic acid A.
Table 1.
1H NMR and 13C NMR spectroscopic data for compounds 1–3 (CD3OD, δ in ppm)
| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δ C | δ H | δ C | δ H | δ C | δ H | |
| 1 | 171.2, C | 170.8, C | 118.3, C | |||
| 2 | 113.4, CH | 6.07, d (15.4) | 117.8, CH | 6.32, d (15.6) | 130.1, CH | 7.83, d (8.0) |
| 3 | 130.7, CH | 7.44, d (15.4) | 129.5, CH | 7.47, d (15.6) | 120.4, CH | 6.91, t (4.4) |
| 4 | 147.2, C | 150.2, C | 134.8, CH | 7.36, t (7.7) | ||
| 5 | 129.2, C | 129.4, C | 118.3, CH | 6.90, d (5.7) | ||
| 6 | 112.2, CH | 6.12, s | 120.2, CH | 6.72, s | 160.4, C | |
| 7 | 158.8, C | 153.2, C | 169.6, C | |||
| 8 | 24.7, CH2 | 2.94, t (7.3) | 131.5, CH | 7.36, d (15.8) | ||
| 9 | 33.0, CH2 | 2.65, t (7.3) | 119.7, CH | 6.39, d (15.8) | ||
| 10 | 176.1, C | 170.5, C | ||||
| 11 | 10.3, CH3 | 2.11, s | 10.1, CH3 | 2.17, s | ||
| 1′ | 62.9, C | |||||
| 2′ | 177.0, C | |||||
| 3′ | 66.1, CH2 |
3.96, d (11.0) 4.03, d (11.0) |
||||
| 4′ | 20.6, CH3 | 1.60, s | ||||
1H NMR were measured at 600 MHz; 13C NMR were measured at 150 MHz
Fig. 2.
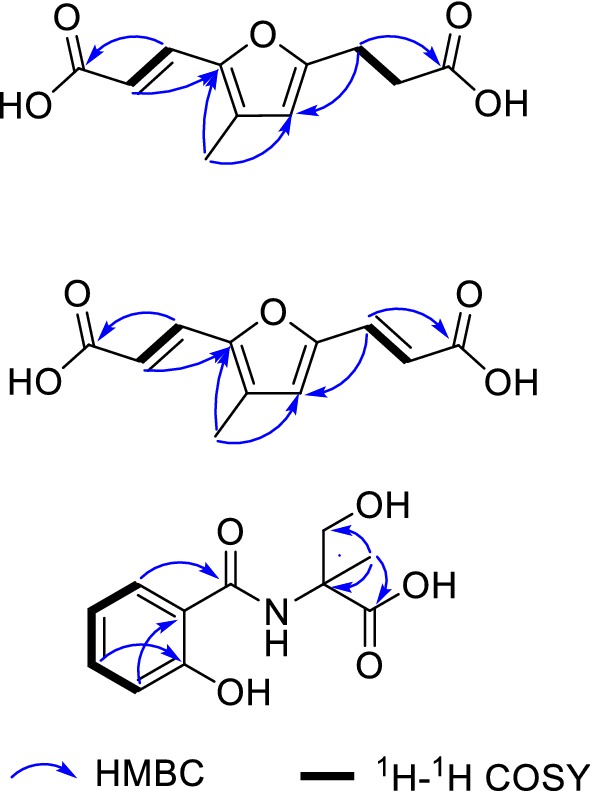
Characteristic mutual HMBC (blue arrow) and 1H–1H COSY correlations for compounds 1–3
Fig. 1.
Chemical structures of compounds 1–3
Compound 2 was obtained as a yellow oil. The molecular formula of C11H10O5 was determined by the (+)-HRESIMS protonated ion peak at m/z 223.06007 [M+H]+, and sodium-adduct ion peak at m/z 245.04192 [M+Na]+, corresponding to seven degrees of unsaturation. The 1H and 13C NMR spectroscopic data of 2 (Table 1) showed highly similarities to those of 1, indicating that it was a congener of 1. The presence of an additional carbon double bond at δC 119.7 (C-9), 131.5 (C-8) of 2 compared to those of 1 was assigned by the HMBC correlations from the trans-double bond protons H-8 and H-9 to C-7 (δC 153.2) and C-10 (δC 170.5) (Fig. 2). Thus, the structure of 2 was established as shown in Fig. 1, and was trivially named as aurantiadioic acid B.
The yellow oil compound 3, possessed the molecular formula of C11H13O5N as determined by the (+)-HRESIMS sodium-adduct ion peak at m/z 262.06842 [M+Na]+ (calcd for C11H13O5NNa, 262.06914), indicating six indices of hydrogen deficiency. The 1H NMR spectroscopic data of 3 (Table 1) showed the presence of an ortho-substituted benzene ring at δH 7.83 (J = 8.0 Hz, H-2), 6.90 (J = 5.7 Hz, H-5), δH 6.91(J = 4.4 Hz, H-3), and 7.36 (J = 7.7 Hz, H-4), which also confirmed by the 1H–1H COSY correlations of H-2/H-3/H-4/H-5. The HMBC correlations from H-2 to a carbonyl at δC 169.6 (C-7) indicated the attachment of a carbonyl at C-1. The down-field chemical shifts of C-6 (δC 160.4) suggested the presence of a hydroxy substituent. Furthermore, the methyl singlet at δH 1.60 showed HMBC correlations to the hydroxymethyl at δC 66.1 (C-3′), the quaternary carbon at δC 62.9 (C-1′), and the carboxylic group at δC 177.0 (C-2′) suggested the presence of an isolated unit assembled by C-1′ to C-4′. The nitrogen atom was assigned between C-7 and C-1′ based on the chemical shifts of C-7 and C-1′ to satisfy the element composition of the molecular formula. Thus, compound 3 was established as shown in Fig. 1. However, although this compound was recorded in SciFinder database, but there was no literature information available. We herein reported the chemical shifts, and first origin organism of this compound.
When examining the structure of 3, it was possibly generated by dehydration of an anthranilic acid and an unusual amino acid 2-methylserine. Since compound 3 harbored a sole chiral center, it was subjected to the chiral-phase HPLC analysis to investigate the optical purity. As depicted in Fig. 3, the analysis result suggested that it presented in enantiomerically pure form. Thus, the absolute configuration of 3 was assigned as 1′S according to the specific optical rotatory data ([α] + 17.2) compared with the reported specific optical rotatory values for (−)-2-methyl-d-serine (2R, [α] − 6.0) and (+)-2-methyl-l-serine (2S, [α] + 6.0) [6].
Fig. 3.
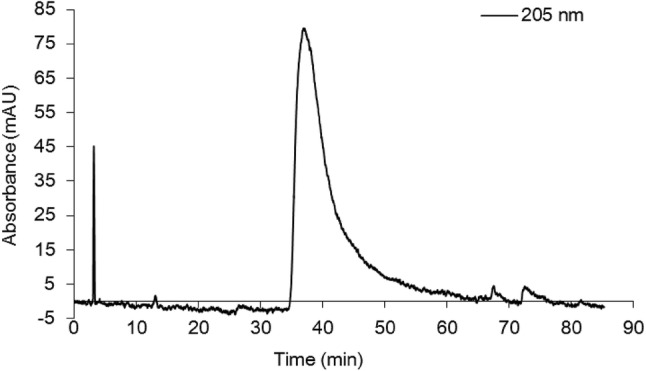
Chiral-phase HPLC analysis of 3
Compounds 1–3 were evaluated for their inhibition against NO production in murine monocytic RAW 264.7 macrophages. As a result, compounds 1–3 displayed weak inhibitory activity with IC50 values of 35.8, 41.8, 45.2 μM, respectively. The IC50 value for the positive control PDTC (ammonium pyrrolidine dithiocarbamate) was 15.3 μM.
Compound 3 was further screened for inhibitory activity against the influenza virus strain A/PuertoRico/8/1934 (H1N1). The result suggested that 3 displayed weak inhibition on the virus A/PuertoRico/8/1934 with an EC50 value of 35.9 μM, and a selective index higher than 13.3.
Experimental
General Experimental Procedures
Optical rotations were obtained on an Autopol IV Automatic Polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA). UV spectra were recorded on a Hitachi UH5300 UV–Vis spectrophotometer (Hitachi, Ltd., Tokyo, Japan). An IRTracer-100 Fourier transform infrared spectrophotometer (Shimazu Corporation, Kyoto, Japan) was used for scanning IR spectroscopy using KBr pellets. 1D and 2D NMR spectra were obtained on Bruker Ascend 600 MHz spectrometers (Bruker Corporation, Karlsruhe, Germany). HRESIMS were recorded on a Q Exactive HF Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Sephadex LH-20 (Amersham Biosciences, Uppsala, Sweden) and silica gel (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China) were used for column chromatography (CC). Medium pressure liquid chromatography (MPLC) was performed on a Büchi Sepacore System equipped with pump manager C-615, pump modules C-605 and fraction collector C-660 (Büchi Labortechnik AG, Flawil, Switzerland), and columns packed with Chromatorex C-18 (dimensions 450 mm × i.d. 14 mm, particle size: 40–75 μm, Fuji Silysia Chemical Ltd., Kasugai, Japan). Chiral-phase HPLC analysis were conducted on an Agilent 1260 Infinity II liquid chromatography system with a Diacel Chiralpak AD-H column (i.d. 4.6 mm × 250 mm, 1 mL min−1), eluting with n-hexane–isopropanol 90:10. Preparative high performance liquid chromatography (prep-HPLC) were performed on an Agilent 1260 Infinity II liquid chromatography system equipped with a Zorbax SB-C18 column (particle size 5 μm, dimension 150 mm × i.d. 9.4 mm, flow rate 5 mL min−1, respectively) and a DAD detector (Agilent Technologies, Santa Clara, CA, US).
Actinomycete Material
The strain was identified as Actinocorallia aurantiaca by Prof. Shen Qin of Yunnan University. A voucher strain (YIM 111109) was deposited at the School of Pharmaceutical Sciences, South-Central University for Nationalities, China.
The actinomycete A. aurantiaca strain was cultured in 50 500-mL Erlenmeyer flasks with the liquid culture medium consist of glucose 20 g, peptone 2 g, yeast extract 2 g, soluble starch 5 g, K2HPO4 0.5 g, MgSO4 0.5 g, NaCl 4 g, CaCO3 2 g in 1 L of deionized water, the pH was adjusted to 7.8 before autoclaving. All flasks were incubated at 25 °C and shaking at 150 rpm for 25 days.
Extraction and Isolation
The total liquid culture (20 L) was evaporated to 5 L, then extracted with EtOAc for four times to obtain a total extract 19.2 g. The crude extract was eluted on MPLC with a stepwise gradient of MeOH/H2O (0–100%) to afford eight fractions (A–H).
Fraction H (3.2 g) was applied to silica gel column chromatography eluting with petroleum ether/acetone (5:1–2:1) to give ten subfractions (H1–H10). Subfraction H3 was purified by prep-HPLC (MeCN/H2O = 3:97 → 23:77, 5 mL min−1, 25 min) to yield compounds 2 (3.1 mg, tR = 15.1 min) and 3 (8.2 mg, tR = 20.3 min). Fraction G (2.1 g) was subjected to Sephadex LH-20 (acetone) to furnish five subfractions (G1–G3). Subfraction G2 was purified on prep-HPLC (MeCN/H2O = 15:85, isocratic, 5 mL min−1, 25 min) to yield compound 1 (10.3 mg, tR = 12.5 min).
Spectroscopic Data Of Compounds
Aurantiadioic Acid A (1)
Yellow oil; UV (MeOH) λmax (log ε) 215.0 (2.66); IR (KBr) νmax 3338, 2943, 2381, 1454, 1114, 1031 cm−1; 1H NMR (600 MHz, CD3OD) and 13C NMR (150 MHz, CD3OD) data: Table 1; HRESIMS m/z 225.07576 [M + H]+ (calcd for C11H13O5, 225.07575).
Aurantiadioic Acid B (2)
Yellow oil; UV (MeOH) λmax (log ε) 325.0 (4.57); IR (KBr) νmax 3338, 2943, 2831, 1452, 1114, 1031 cm−1; 1H NMR (600 MHz, CD3OD) and 13C NMR (150 MHz, CD3OD) data: Table 1; HRESIMS m/z 223.06007 [M+H]+, 245.04192 [M+Na]+ (calcd for C11H11O5, 223.06065, C11H10O5Na, 245.04259).
Aurantoic Acid A (3)
Yellow oil; [α] − 16.9 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 235.0 (3.24); IR(KBr) νmax 3342, 2943, 2831, 1452, 1148, 1031; 1H NMR (600 MHz, CD3OD) and 13C NMR (150 MHz, CD3OD) data: Table 1; HRESIMS m/z 262.06842 [M+Na]+ (calcd for C11H13O5NNa, 262.06914).
Nitric Oxide Inhibitory Assay
The procedures of nitric oxide inhibitory assay were similar with those in previously reported literature [7]. Moreover, PDTC (ammonium pyrrolidine dithiocarbamate) was used as positive control in this research.
Viral Replication Inhibition Assay
The viral replication inhibition assay against the influenza virus strain A/PuertoRico/8/1934 (H1N1) was performed with the procedures that similar with those reported in the literature [8].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant No. 81773590). The authors thank Analytical & Measuring Centre, South-Central University for Nationalities, for the NMR measurements.
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interests.
Contributor Information
He-Ping Chen, Email: chenhp@mail.scuec.edu.cn.
Ji-Kai Liu, Email: jkliu@mail.kib.ac.cn.
References
- 1.Genilloud O. Nat. Prod. Rep. 2017;34:1203–1232. doi: 10.1039/C7NP00026J. [DOI] [PubMed] [Google Scholar]
- 2.Guo ZK, Jiao RH, Dai HF, Mei WL, Tan RX, Ge HM. Chem. Biodivers. 2013;10:296–302. doi: 10.1002/cbdv.201200224. [DOI] [PubMed] [Google Scholar]
- 3.Guo ZK, Wang R, Chen FX, Liu TM. Fitoterapia. 2018;127:25–28. doi: 10.1016/j.fitote.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Kellner K, Ishak HD, Linksvayer TA, Mueller UG, Microbiol FEMS. FEMS Microbiol. Ecol. 2015;91:fiv073. doi: 10.1093/femsec/fiv073. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZX, Shen Q, Xu LH, Chen HP, Sun H, Huang R, Li ZH, Feng T, Liu JK. Nat. Prod. Bioprospect. 2018;8:91–95. doi: 10.1007/s13659-018-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang HF, Ma GH, Yang SB, Han RG, Xu PF. Tetrahedron: Asymmetry. 2008;19:1630–1635. doi: 10.1016/j.tetasy.2008.06.033. [DOI] [Google Scholar]
- 7.Chen HP, Zhao ZZ, Li ZH, Huang Y, Zhang SB, Tang Y, Yao JN, Chen L, Isaka M, Feng T, Liu JK. J. Agric. Food Chem. 2018;66:3146–3154. doi: 10.1021/acs.jafc.8b00287. [DOI] [PubMed] [Google Scholar]
- 8.An L, Liu R, Tang W, Wu JG, Chen X. Antivir. Res. 2014;109:54–63. doi: 10.1016/j.antiviral.2014.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.