Abstract
We present a prototype for conducting rapid, inexpensive and point-of-care-compatible nucleic acid amplification tests (NAATs) for tuberculosis (TB). The fluorescent isothermal paper-and-plastic NAAT (FLIPP-NAAT) uses paper-based loop mediated isothermal amplification (LAMP) for DNA detection. The cost of materials required to build a 12-test-zone device is $0.88 and the cost of reagents per reaction is $0.43. An inexpensive imaging platform enables filter-free fluorescence detection of amplified DNA using a cell-phone camera. FLIPP-NAAT can be operated by an untrained user and only requires a regular laboratory incubator as ancillary equipment. All reagents can be dry-stored in the device, facilitating storage and transportation without cold chains. The device design is modular and the assay demonstrated high specificity to Mycobacterium tuberculosis (Mtb), analytical sensitivity of the order of 10 copies of Mtb gDNA, and tolerance to complex samples. The clinical sensitivity and specificity of sputum-based FLIPP NAAT tests were 100% (zero false negatives) and 68.75% (five false positives), respectively (N = 30), using Xpert MTB/RIF assay as the reference standard. FLIPP-NAAT has the potential to provide affordable and accessible molecular diagnostics for TB in low- and middle-income countries, when used in conjunction with an appropriate sample preparation technique. Although demonstrated for the detection of TB, FLIPP-NAAT is a platform technology for amplification of any nucleic acid sequence.
Subject terms: Genetic testing, Biomedical engineering
Introduction
Tuberculosis (TB) is the deadliest infectious disease today, having claimed over 1.6 million lives worldwide in 20171. A lack of rapid and inexpensive TB diagnostic tests has been a major roadblock in managing the disease. Because the disease primarily affects people from poor economic backgrounds, it is imperative to provide access to state-of-the-art rapid TB diagnostic technologies to these populations. Culture tests, the gold standard for definitive TB diagnosis, take a long time (6 to 8 weeks) and are associated with other challenges2. Nucleic acid amplification tests (NAATs) are the new gold standards for rapid TB diagnosis.
The WHO-endorsed GeneXpert (Xpert MTB/RIF assay) provides an unprecedented level of automation of the multiple steps involved in a NAAT and enables a minimally-trained user to conduct a NAAT, but the cost of this instrument remains prohibitively high in many settings3. Another PCR-based test from a joint venture of Molbio diagnostics and Bigtec labs (Bangalore, India)4 consists of two units -Trueprep Mag for sample preparation and Truelab Uno for DNA amplification. This device consists of one additional step compared to the Xpert MTB/RIF assay because purified DNA generated by the Trueprep Mag must be manually transferred into the Truelab Uno unit. Compared to Xpert MTB/RIF assay, this test is only slightly lower in terms of user-friendliness, but moderately lower in terms of instrument-cost. On the low end of the cost spectrum, there are generic PCR machines like the Open qPCR (CHAI, Santa Clara, CA), not specifically designed for TB NAATs, and are only suitable for research labs for operation by skilled users. Instrument costs have also been driven down by isothermal nucleic acid amplification methods5,6 replacing PCR, a strategy which has recently gained significant interest for POC diagnostics. A commercially available TB testing kit and instrument from Eiken (Tokyo, Japan), based on an isothermal NAAT, have been endorsed by the WHO7. While the instrument cost is low, conducting the assay involves multiple careful pipetting steps that must be conducted by trained users. Hence, there continues to be a tradeoff between the cost of TB NAATs and their user-friendliness and ultimately, ease-of-use is the most critical parameter in making a NAAT amenable to use at the point-of-care. Therefore, there exists significant room for technological innovation to provide easy-to-use, yet affordable solutions for TB-NAATs.
Paper-based isothermal NAATs have recently emerged as popular platforms for low-cost nucleic acid detection. These platforms have the potential to fill the abovementioned gap8,9 and the opportunities and challenges in the area of paper-based NAATs have recently been extensively reviewed10,11. While paper-based NAATs have been demonstrated for the detection of various infectious diseases11, there has been only one report of their use for TB detection. Shetty et al.12 used helicase-dependent amplification (HDA) in chromatography paper for detection of TB. The target DNA was an 84-base pair fragment from the insertion sequence 6110, amplified using PCR. A disk of chromatography paper containing dried HDA reagents was rehydrated with a solution of this synthetic target, sealed in a polyethylene pouch, and kept on a heater to conduct HDA. The detection was based on end-point fluorescence using a fluorescence microscope.
In this study, we present a new paper-based device for conducting isothermal NAATs at the point-of-care and demonstrate its utility in sensitive and specific detection of TB, directly from whole Mycobacterium tuberculosis (Mtb) genomic DNA (gDNA). We also present a very low-cost imaging box for filter-free end-point fluorescence detection of the amplified DNA using a cell phone. To the best of our knowledge, this is the first demonstration of a paper-based NAAT for TB starting from gDNA as the template. Compared to existing paper-based NAATs for other infectious diseases, our device presents several improvements in terms of ability to multiplex, user experience, and cost reduction. The prototype called fluorescent isothermal paper-and-plastic NAAT (FLIPP-NAAT) uses loop mediated isothermal amplification (LAMP) for DNA amplification and the only ancillary equipment required for testing is a laboratory incubator. The design of the FLIPP-NAAT is ‘modular’ and the number of testing zones can easily be increased by increasing the number of modules. FLIPP-NAAT is sensitive (10 copies, analytical sensitivity) and specific towards Mtb gDNA and can detect DNA directly from viscous sputum. TB testing from 30 clinical samples demonstrated a clinical sensitivity of 100% and clinical specificity of 68.75%. By providing a very low-cost and user-friendly means for DNA amplification, FLIPP NAAT represents a significant step towards filling the technological gap in TB-NAATs. In combination with an appropriate DNA extraction procedure, it promises to be an essential tool for conducting NAATs in places like rural India.
Materials and Methods
Materials for fabrication
Standard 17 glass fiber (referred to as paper pads henceforth) was acquired from GE Healthcare Life Sciences (Bangalore, India). A double-sided pressure sensitive adhesive (PSA; 3MTM 9731) was used to secure the different layers of the device together. Transparent acrylic sheets (2.8 mm-thick), black acrylic sheets (2 mm-thick) and transparency sheets (0.07 mm) were acquired locally. All materials were cut using a 50 W CO2 laser in a VLS 3.60 laser engraver (Universal Laser Systems, Scottsdale, AZ). All designs were created in AutoCAD (Autodesk, San Rafael, CA).
DNA amplification in tube
Mtb gDNA (strain H37Rv) was obtained from BEI resources (Manassas, VA, USA) and quantified using a Nanodrop 2000 (ThermoFisher Scinetific). DNA amplification was performed using loop-mediated isothermal amplification (LAMP) targeting a region of the hspX gene of Mtb using six primers as described by Bi et al.13 (primer sequences are provided in Supplementary Table S1). Initial standardization of the LAMP reaction was done for a 25 µl reaction with the following composition: 1.6 µM forward inner primer (FIP) and backward inner primer (BIP), 0.2 µM forward outer primer (F3) and backward outer primer (B3), 1.2 µM forward loop primer (FLP) and backward loop primer (BLP), 1X LAMP reaction buffer, 8 mM MgSO4, 0.9 M Betaine (B0300-1VL, Sigma-Aldrich), 2.5 µl dNTP mix (10 mM each, N0447S, New England Biolabs (NEB)), 8 units of Bst 2.0 WarmStart® DNA polymerase (M0538S, NEB), 1 µl of the target DNA, and DEPC water to make up the final volume to 25 µl. Specificity of the assay was tested by replacing the Mtb gDNA with equal volume of Escherichia coli gDNA, Mycobacterium smegmatis gDNA, or plasmid DNA containing inserts for Hepatitis C and Dengue virus. Tolerance to human gDNA was also tested by mixing 10–104 starting copies of human gDNA (G3041, Promega) with 1000 and 100 starting copies of Mtb gDNA. A range of temperatures starting from 56 °C to 65 °C was tested to find the optimum reaction temperature. For real-time LAMP reactions, four different fluorescent dyes - SYBR green I (Sigma-Aldrich, S9430), SYBR safe (Invitrogen, P/NS33102), Evagreen (Biotium, 31019) and SYBR gold (Invitrogen, S11494) were tested using a Quant Studio 3 (Applied Biosystems).
DNA amplification in FLIPP-NAAT
The reaction was ported from tube to paper by making the entire reaction mix in a tube along with the template and adding it to paper pads through sample addition holes. The device was then sealed and kept in an incubator set to the optimum reaction temperature for a specified period, following which, the reaction was terminated by removing the device from the incubator. Over the course of working on this device, the reaction volume was reduced to 12.5 µl maintaining the concentrations of all reagents.
Detection of LAMP products using gel electrophoresis
LAMP products were run in 2% agarose gels (115V) stained with ethidium bromide and the gels were imaged in a custom-made gel imager. In cases where DNA amplification was done in FLIPP-NAAT, the device was carefully pried open using a blade and fluid from within the paper pads was eluted by centrifugation at 10,000 rpm for 1 minute before conducting electrophoresis. Target specific amplification was confirmed by digesting the LAMP amplicons with the restriction enzyme AatII. DNA ladders (O’ Gene Ruler; Low range and 100 bp) were obtained from ThermoFisher Scientific.
Fluorescence detection of LAMP products in FLIPP-NAAT
Two fluorescent DNA intercalating dyes, SYBR Green I (Sigma- Aldrich, S9430) and SYBR Safe (Invitrogen, P/NS33102) were tested for end-point detection of LAMP amplicons. On completion of the reaction, the device was removed from the incubator, top flap was peeled, DNA intercalating fluorescent dye (5 µl, 50X) was added to the reaction pads through the top holes, and top flap was placed back to seal the device. The device was then placed inside a custom-made imaging station such that bottom transparent acrylic faced the source of excitation and the camera. The imaging station (20 cm × 22 cm) was a cuboid without top cover, made from black acrylic, and consisted of an inclined platform to fix the phone. A UV torch was used as the excitation source and fluorescent images were captured using a smartphone (Redmi 5, 64 GB Phone) with the HDR option turned off. The imaging set-up was checked for uniformity of field by adding 12.5 µl of a well-mixed LAMP amplicons and fluorescent dye mix to each of the 12 paper pads and imaging it within the box. The images were analyzed using ImageJ in the green channel and two-tailed Student’s t-tests were used for statistical comparison.
DNA amplification from dry-stored reagents in FLIPP-NAAT
Circular paper pads were pre-treated by dipping in a 1% bovine serum albumin (BSA) and 0.1% tween20 solution for two hours, following which the excess fluid was removed by placing the pads on Kimwipes. These pre-treated paper pads were dried at 37 °C for at least 24 hours and stored at 37 °C until further use. For dry storage of LAMP reagents in paper pads, 12.5 µl of the LAMP mix containing an optimum trehalose concentration and water replacing the template was made in tubes and added to the paper pads. The paper pads were immediately put under liquid nitrogen and placed in a lyophilizer (Labconco, 7670560) set to −45 °C and 0.01mBar for 12 hours. After removal from lyophilizer, FLIPP-NAAT was assembled using these paper pads and LAMP reactions were conducted within 2 hours. Paper pads were rehydrated with 9 µl of a template solution (1 µl target DNA + 8 µl of water), the device sealed, and put inside an incubator for the reaction.
Detection of TB from complex samples
Mock sputum was prepared by intermittently dissolving 2 g of methyl cellulose (Sigma-Aldrich, M0262) in 90 ml of autoclaved distilled water with constant stirring at 80 °C. This solution was then stored at 42 °C overnight to ensure complete dissolution of methyl cellulose14. Following this, egg yolk emulsion (Sigma-Aldrich, 17148) was added intermittently with constant stirring at room temperature to make a final solution containing 10% egg yolk emulsion. The viscosity of this 1X solution of mock sputum was measured using a cone and plate rheometer (MCR 301 Anton Paar, Graz, Austria). The reaction pads containing dried reagents were then rehydrated with 9 µl of different dilutions (1X, 2X and 5X) of mock sputum, each of which were spiked with 100 copies of Mtb genomic DNA. The compatibility of DNA amplification in FLIPP-NAAT was also tested with conventional methods of DNA extraction using methods described under Supplementary Methods.
Testing of clinical samples in FLIPP-NAAT
A set of 30 clinical sputum samples obtained at Christian Medical College (CMC), Vellore, India were tested using sputum smear microscopy, Xpert MTB/RIF, and FLIPP-NAAT after all methods were approved by the CMC Institutional Review Board Ethics Committee. All standard operating procedures (SOPs) were in accordance with the RNTCP (Revised national tuberculosis control program) and C&DST (culture and drug susceptibility testing) guidelines. The set of clinical samples was diverse to include samples that were smear microscopy negative, smear microscopy positive and positive for other respiratory diseases but not TB. The Xpert MTB/RIF negative samples were culture positive at CMC Vellore for other microorganisms such as Viridans group of Streptococci, Pseudomonas, Klebsiella, Candida tropicalis, and Candida glabrata. Sputum smear microscopy and Cepheid Xpert MTB/RIF tests were conducted at CMC as part of routine clinical diagnostic procedures. DNA was extracted from the samples at CMC using a QIAamp DNA mini kit (Qiagen, 51304). 200 µl of native sputum sample was added to a Qiagen column and the DNA was extracted in a final volume of 100 µl. Extracted and purified DNA from the 30 clinical samples was then tested in FLIPP-NAAT in a blind study at IISc Bangalore, where only the sample identity numbers were known. The reaction was set-up as explained in the section on ‘DNA amplification in tube’, with two modifications.
First, the total volume of the reaction was reduced to 11.5 µl from 12.5 µl to ensure that the paper pads were not filled to capacity to completely prevent leakage issues (if any). Second, 3.5 µl of template DNA was added instead of 1 µl, eliminating additional water that was being added to fill the remaining volume. 3.5 µl of 100copies/µl solution of purified Mtb gDNA or water was added for positive and negative controls, respectively. After addition of the reaction mix into paper pads, the protocols followed for conducting the reaction and fluorescence imaging were as explained earlier. The following rules were used for interpreting test results: (i) the threshold intensity was set to be µN + 3σN, where µN is the mean intensity from the three negative controls present in each device, and σN is the standard deviation of mean intensities across these negative controls, (ii) the test would be considered valid only if all the three positive controls in the device showed amplification (mean intensity >100) while none of the three negative controls showed amplification (mean intensity <100), (iii) Any sample with intensity greater than the threshold was considered as a positive while a sample with mean intensity below the threshold was considered as a negative, (iv) Each sample was tested in duplicates and a positive test result for at least one replicate was interpreted as a TB-positive test, (v) To address the challenges faced while testing of smear-negative samples, each smear negative sample was tested in six replicates and a positive test result for at least one replicate was interpreted as a TB-positive test.
Results
FLIPP NAAT: Design, fabrication, and user experience
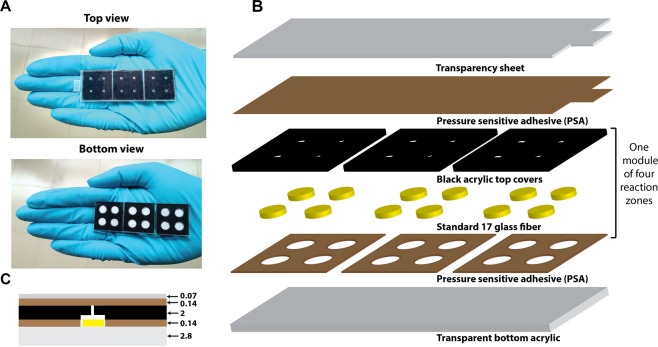
The top and bottom view of a 12-reaction-zone FLIPP NAAT device is shown in Fig. 1A and a schematic of the layer-by-layer assembly is shown in Fig. 1B. The device consists of a 7.7 cm × 2.6 cm rectangular base constructed from 2.8 mm thick transparent acrylic. Three 2.5 cm × 2.5 cm PSA sheets, each having four equidistant circular holes (6.75 mm diameter), were adhered on the base with a gap of 1 mm between them. Each 2.5 cm × 2.5 cm PSA represented one section/module of the device (Fig. 1B). In each module, four circular reaction pads (6.3 mm diameter) made from Standard 17 were placed within the 6.75 mm circular boundaries defined by the PSA sheets, such that the pads directly contacted the acrylic base. The top covers for each section were constructed from 2 mm thick black acrylic. For each module, a 2.5 cm × 2.5 cm acrylic cover comprised of four circles (6.75 mm diameter), engraved to a depth of ~600 um, such that they aligned with the holes in the PSA (Fig. 1C). The engraved regions provided tightly closed reaction zones for each reaction pad. For addition of reagents, 2 mm × 2 mm square openings were cut in the top cover at the center of each reaction zone. This assembly was pressurized using a vice to ensure that the bottom and top acrylic sections stick well to each other. The top seal of the device consisted of a combination of PSA and a transparency sheet cut to match the dimensions of the acrylic base, with an additional flap to ease peeling off. After addition of reagents, the top seal was pressed hard manually to ensure a good seal.
Figure 1.
FLIPP-NAAT design. (A) Top and bottom view of the fabricated device (B) Schematic of the layer-by-layer assembly of the device. (C) Schematic of front view of device demonstrating the position of paper (yellow) between the bottom acrylic (grey) and the engraved top acrylic (black). The color of the materials has been kept same as in (B) and thickness of each layer is marked in millimeters.
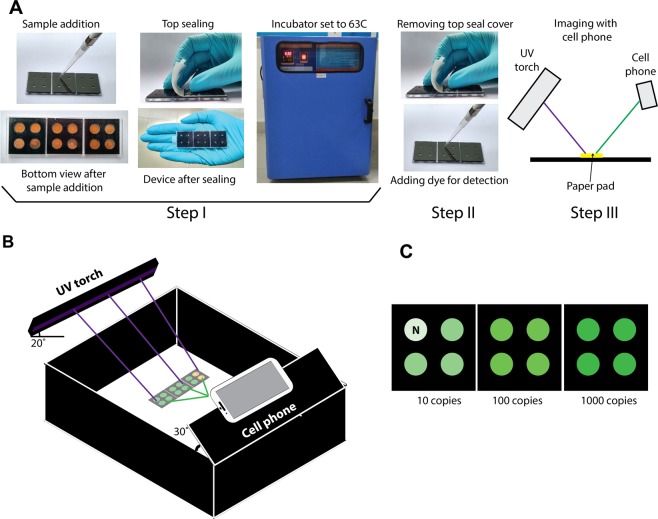
Conducting a NAAT in FLIPP-NAAT involves a simple three-step user experience (Fig. 2A). The first step involves addition of the samples to the device, putting the top seal, and placing the device in an incubator (Step I; Fig. 2A). The second step involves removing the device from the incubator and adding a DNA-intercalating fluorescent dye (Step II; Fig. 2A). The last step involves capturing an image of test results using a cell phone camera (Step III; Fig. 2A). Figure 2B shows the schematic of the imaging box built in-house, demonstrating positions of the UV torch and the cell phone. A schematic of expected results shows green fluorescence in the presence of 10, 100, and 1000 copies of target DNA (Fig. 2C) and lack of green fluorescence in the absence of target (N; Fig. 2C). In one implementation, each module can be used for testing an independent sample in duplicates along with a positive and a negative control and hence 3 samples can be tested on the device at a time. In an alternate implementation, a 3-module device can be used for conducting as many as 6 replicates of a single sample along with 3 positive and 3 negative controls, to increase the probability of detection in samples with low bacillary load.
Figure 2.
User experience of conducting a NAAT in FLIPP-NAAT. (A) User steps involved in operating FLIPP-NAAT. Step I: Sample addition to FLIPP-NAAT, sealing the device and placing it in an incubator set to the reaction temperature. Step II: Removing FLIPP-NAAT from the incubator and adding the DNA intercalating dye. Step III: A UV torch is used for excitation and results are captured using a cell phone camera. (B) Schematic of an inexpensive imaging set-up for end-point fluorescence detection. (C) Schematic of expected TB test results from FLIPP-NAAT. In one implementation, green fluorescence intensity increases with increasing number of target molecules.
Analytical sensitivity and specificity of the LAMP assay
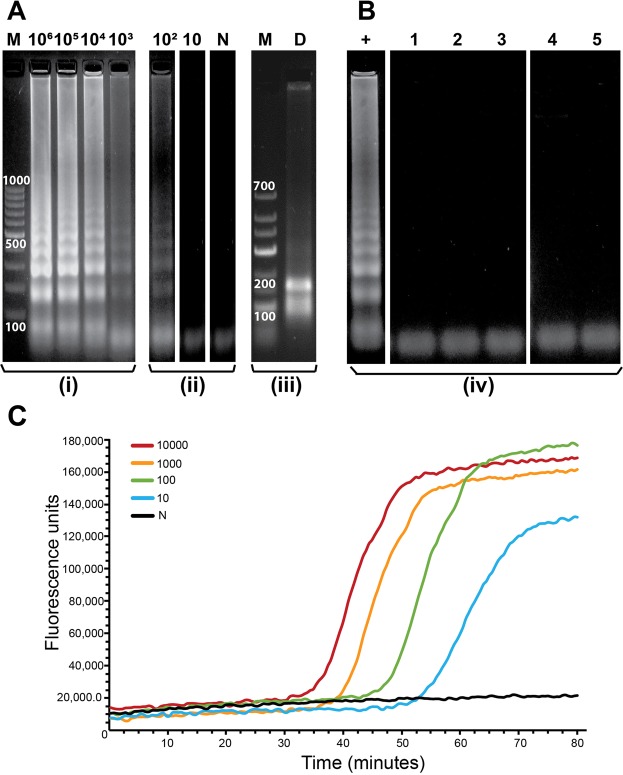
The limit of detection (LOD) of the LAMP assay in tube, using gel electrophoresis as the detection method, was determined to be 100 starting copies of Mtb genomic DNA for a 40-minute reaction. Figure 3A shows the results of gel electrophoresis conducted on products of LAMP reactions starting from 106 copies, serially diluted 10x, down to 10 copies. A ladder-like pattern of amplified DNA products, typical of LAMP reactions, was observed for reactions starting from 106 to 102 copies, but not for 10 copies. The negative control (lane N; Fig. 3A) showed no amplification. Digestion of LAMP amplicons was theoretically expected to produce bands of size 100 and 160 bp, which were observed on the gel (lane D; Fig. 3A), confirming target-specific amplification. The analytical specificity of the reaction was tested against five different non-Mtb targets and no amplification was observed for any of these (Fig. 3B). Additionally, Bi et al.13 had tested the analytical specificity of this assay against 22 species of non-tuberculous mycobacteria, 7 non-mycobacterial species that present Mtb-like symptoms and 50 Mtb clinical isolates. This was a crucial factor for selection of this assay for use in FLIPP-NAAT. The LAMP assay was also found to be tolerant to high concentrations of human gDNA for a reaction time of 80 minutes. Target-specific amplification was observed for the positive controls even in the presence of varying concentrations of human gDNA while the negative controls containing only human gDNA showed no amplification (see Supplementary Fig. S1).
Figure 3.
LAMP in tube. (A) Gel electrophoresis analysis of products of LAMP reactions conducted in tube with varying starting copy numbers of Mtb gDNA (106 copies up to 10 copies). The numbers on top of the lanes indicate the starting copy number of the template. N- No template control, M- DNA marker and D- digested LAMP amplicons. Reaction time was 40 minutes. (B) Specificity of the LAMP assay tested against non-Mtb targets. +: Positive control (Mtb gDNA), 1: Human gDNA, 2: E. coli gDNA, 3: Mycobacterium smegmatis gDNA, 4: plasmid with hepatitis C insert, 5: plasmid with dengue insert. (C) Real-time amplification curves for LAMP in tube. Reaction time was 80 minutes. (i–iv) denote lanes from different gels. Full-length gels are presented in Supplementary Fig. S9 (i), (ii), (iii) and (iv).
Real-time LAMP reactions
Out of the four DNA-intercalating dyes tested, only SYBR Safe worked with this LAMP assay for real-time detection and the concentration of the dye in the LAMP mix was optimized to be 3X (where 1X is the manufacturer-recommended concentration for use in agarose gels). At concentrations below 3X, we observed that the signal to noise ratio was very low and concentrations above 3X adversely affected the reaction kinetics, delaying the lift-off times. Real-time curves using the optimum dye concentration revealed that the limit of detection of the assay could be improved by increasing reaction time. Real-time amplification curves for 10, 102, 103, and 104 copies lifted off at approximately 52, 44, 38, and 34 minutes, respectively (Fig. 3C). At 80 minutes, real-time curves for all starting copy numbers saturated (Fig. 3C). For all further experiments, the reaction time was accordingly increased to 80 minutes to enable low copy number detection. The LOD in tube improved to 10 copies of Mtb gDNA with the 80-minute reaction.
Porting the LAMP assay to paper
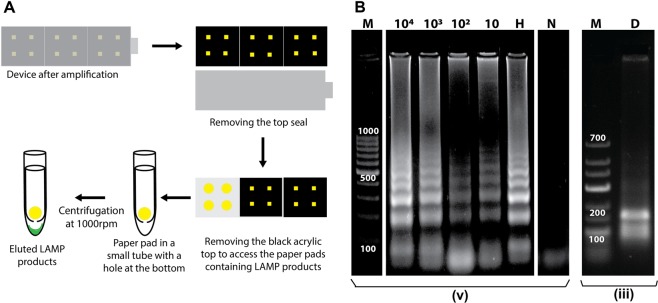
Before porting the assay into paper, its sensitivity to temperature was tested and the assay was found to be very sensitive to temperature. Amplification was observed only at temperatures of 61, 62 and 63 °C (see Supplementary Fig. S2), with maximum efficiency at 63 °C, which was chosen as the operating temperature. For the initial trials of porting the reaction to paper, amplicons were analyzed using gel electrophoresis to confirm that the amplification was target specific. Figure 4A demonstrates the method used for extracting the LAMP amplicons from the paper pads after the LAMP reaction. Figure 4B shows the gel results for LOD of 10 starting copies of Mtb gDNA in paper and the digestion of the amplicons confirmed target-specific amplification (D; Fig. 4B). Since sputum samples also carry human gDNA, the tolerance of the assay to foreign DNA was tested by adding 1000 copies of (15 ng) human gDNA along with 1000 copies of Mtb gDNA to the reaction. Target-specific amplification was still achieved (H; Fig. 4B) with no interference observed due to presence of human genomic DNA.
Figure 4.
LAMP amplification in paper. (A) Schematic of the method used for retrieving the LAMP amplicons from the paper pads. (B) Gel electrophoresis analysis of products of LAMP reactions conducted in paper starting from varying number of starting Mtb gDNA copies (104 up to 10). The numbers on top of the wells indicate the starting copy number of the template. H- 1000 copies of human gDNA were added to the reaction mix along with 1000 copies of Mtb gDNA; N- No template control; M- DNA marker; and D- digested LAMP amplicons. The reaction time was 40 minutes. Larger spaces between sections of lanes in a gel image indicate lanes from different gels and smaller spaces represent lanes from the same gel. Full-length gels are presented in Supplementary Fig. S9 (iii) and (v).
DNA amplification and fluorescence detection in FLIPP-NAAT
Based on real-time LAMP curves, an 80 minute-reaction time was chosen for FLIPP-NAAT to enable detection at low copy numbers. Because fluorescence imaging is a crucial part of testing in FLIPP-NAAT, an economical imaging set-up (Fig. 2B) was designed and fabricated out of black acrylic to avoid any background fluorescence. The angle of the excitation source (20°) was optimized to maximize sample excitation and minimize reflection of the UV light from the device surface, which would otherwise be captured by the camera leading to background noise. The angle of the phone camera (30 ) was optimized to minimize capturing reflected UV light from the device surface. Out of the two fluorescent dyes tested, SYBR Green I was chosen for end-point detection. Though SYBR Safe was more specific to double stranded DNA and generated very little background for negative controls, its excitation peak lay in the UV range and it was very difficult to find an inexpensive material transmissive to UV. SYBR Green I interacts non-specifically with single stranded DNA which led to some background fluorescence in negative controls due to the presence of high concentration of LAMP primers. But because target-specific LAMP reactions produced large amounts of amplicons, the positives were distinguishable from the negatives using SYBR Green I.
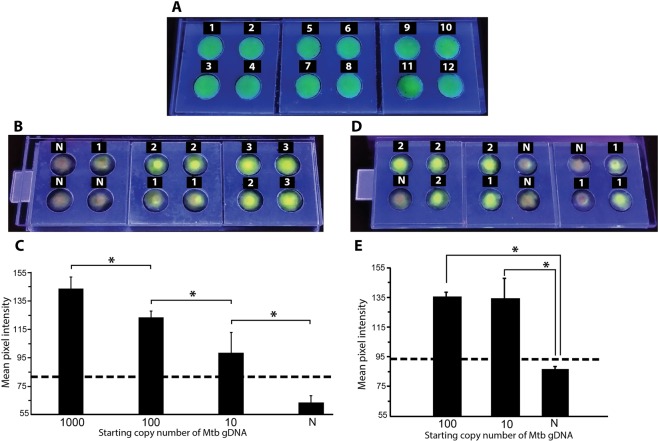
The uniformity of field for fluorescence imaging in FLIPP-NAAT was first tested (Fig. 5A) to ensure that the fluorescence was independent of the reaction zone’s location in the device. The mean green fluorescence intensity across 12 reaction zones had a standard deviation of ±9 intensity values and coefficient of variation of 5.4% (Fig. 5A). Hence, we did not find any significant difference in the mean intensities across locations. TB testing was then performed in FLIPP-NAAT with varying starting copy numbers of Mtb gDNA. The NTCs appeared slightly orange (N; Fig. 5B) as SYBR green I is orange in its native state while pads containing target DNA appeared green due to the DNA-dye complex formation (Fig. 5B). The intensity of green fluorescence increased with increasing starting copy numbers (Fig. 5B). The mean intensity for reaction zones containing 1000 starting copies of Mtb gDNA was statistically higher than 100 copies, for 100 copies was statistically higher than 10 copies, and for 10 copies was statistically higher than negative controls that did not contain Mtb gDNA (P < 0.05; N = 3; Fig. 5C). The threshold for a positive test (µN + 3σN) is represented by the dashed line (Fig. 5C). The mean intensity for all samples containing Mtb gDNA was greater than the threshold value, enabling successful distinction from samples not containing Mtb gDNA. The tolerance to presence of human gDNA was also tested in FLIPP-NAAT. All the negative controls containing only human gDNA showed no amplification while samples containing both Mtb gDNA and human gDNA showed amplification (Fig. 5D,E). One dropout was observed for reactions set-up with 10 starting copies of Mtb gDNA. A green band pass filter (510–550 nm) was tested for reducing background fluorescence but was found to produce no significant improvement in the ability to distinguish between a positive and a negative test (see Supplementary Fig. S3).
Figure 5.
LAMP in FLIPP-NAAT and end-point fluorescence detection. (A) Testing uniformity of field in FLIPP-NAAT. Numbers on each reaction zone mark the 12 replicates. The mean intensity for signals from the 12 reaction zones was 160 with a coefficient of variation of only 5.4%. (B) Cell phone image of FLIPP-NAAT after 5 minutes of addition of SYBR Green I dye for fluorescence detection. Numbers on top of each reaction zone indicate log10 of Mtb gDNA starting copy number. N – no template control. (C) Mean pixel intensity of green color for the different starting copy numbers. The mean intensity of each starting copy number was statistically different from that of others (*P < 0.05; N = 3). (D) LAMP reactions in FLIPP-NAAT in presence of 3.1 * 103 copies of human gDNA. Numbers on top of each reaction zone indicate log10 of Mtb gDNA starting copy number. N – no template control containing only human gDNA. (E) Mean pixel intensity of green color for the different starting copy numbers. The mean intensity of 100 and 10 copy number was statistically different from that of the negative controls (*P < 0.05; N = 3). The one drop out for 10 copies (second reaction zone from the right in the bottom row, Panel D) was not included for the intensity analysis as it was a false negative. The dashed line represents the threshold value of (µN + 3σN). Samples with intensity below the threshold were considered as TB negative while samples with green color intensity above the threshold were considered as TB positive.
Performance of FLIPP-NAAT containing dried LAMP reagents
LAMP reaction mixes were dry-stored in reaction pads within FLIPP-NAAT. Because trehalose is a well-known enzyme cryopreserving agent, a range of trehalose concentrations up to a maximum of 10% w/v were first tested for compatibility with the assay. No significant loss in amplification was observed at any concentration (see Supplementary Fig. S4). Ten per cent trehalose was therefore selected for dry-storage of LAMP reagents. Although the reaction volume for fresh reagents was set to 12.5 µl, we found that 12.5 µl was excessive for rehydration of paper pads containing lyophilized reagents, potentially because of residual glycerol in the lyophilized pads. In addition, when rehydrated with 12.5 µl, there was poor mixing of the amplicon solution with the fluorescent dye (added after completion of the reaction) because the pads were filled beyond capacity. Reducing the rehydration volume to 9 µl resolved this issue and allowed better mixing of the dye with amplicons. Bright green color was observed in reaction zones with 1000, 100 and 10 Mtb gDNA starting copies (3, 2 and 1 respectively, Fig. 6A) while the negative controls appeared predominantly orange (N; Fig. 6A). The mean fluorescence intensity for different copy numbers for positives were significantly distinguishable from the negative controls (P < 0.05; N = 3; Fig. 6B) and all the positives were above the threshold value (as indicated by the dashed line, Fig. 6B).
Figure 6.
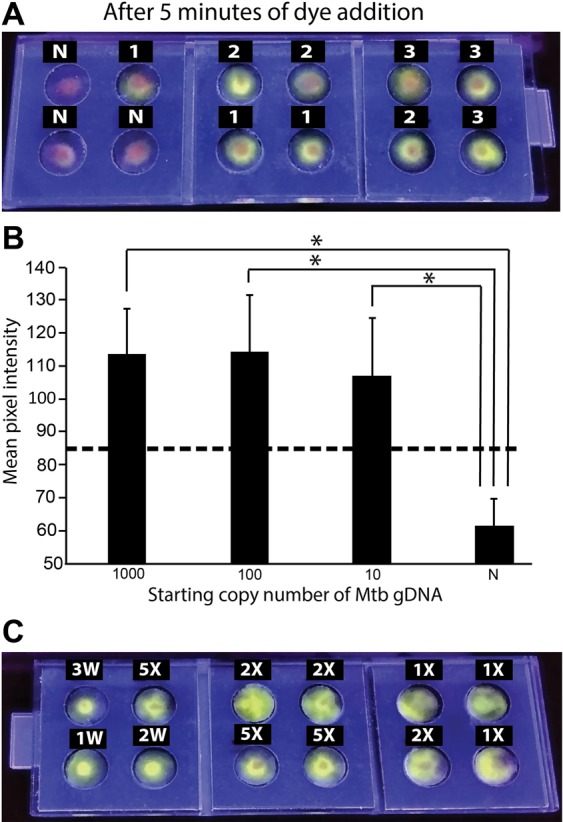
LAMP in FLIPP-NAAT from dry-stored reagents. (A) End-point imaging of the FLIPP-NAAT device after 5 minutes of addition of SYBR green I dye. Numbers on top of each reaction zone indicate log10 of starting copy number. N – no template control. (B) Mean pixel intensity of the green color from all starting copy numbers was statistically higher (*P < 0.05; N = 3) from that of no template control. The dashed line represents the threshold value of (µN + 3σN). Samples with intensity below the threshold were considered as TB negative while samples with green color intensity above the threshold were considered as TB positive. (C) LAMP in FLIPP-NAAT from dry-stored reagents and rehydration with mock sputum. Each reaction zone had 100 copies of Mtb gDNA as the starting template. The markings on top of each of the reaction zone indicate the dilution of mock sputum used for rehydration. 3 W, 2 W, 1 W – reaction set-up with template suspended in water containing 1000, 100 and 10 copies of Mtb gDNA respectively, as positive controls. All reactions worked despite the presence of the complex sputum matrix.
Detection of TB from complex samples
Since sputum is a viscous fluid, it is important that DNA amplification techniques used in TB-NAATs be compliant with viscous sputum. To test robustness in the presence of sputum, FLIPP-NAAT test zones with dried reagents were rehydrated with different dilutions (1X, 2X and 5X) of mock sputum spiked with 100 copies of Mtb gDNA. The viscosity of 1X mock sputum was measured to be 350 cP. FLIPP NAAT successfully amplified DNA within undiluted (1X) and diluted (2X and 5X) mock sputum (Fig. 6C). Three positive controls with 1000, 100 and 10 starting copies of Mtb gDNA in water (3 W, 2 W and 1 W, respectively; Fig. 6C) were run on the same device. No significant reduction was observed in the intensity of green color from zones rehydrated with mock sputum as compared to zones rehydrated with water. As the next step, the compatibility of FLIPP NAAT with commonly used bacterial lysis and DNA extraction methods was tested; successful amplification was observed from DNA extracted using both a commercially available Qiagen kit and a crude culture boiling and centrifugation method (see Supplementary Methods and Supplementary Fig. S5).
Testing of clinical samples in FLIPP-NAAT
The results for TB testing of 30 clinical samples in a blind study in FLIPP-NAAT were interpreted according to the rules described in the materials and methods section. A representative result of testing three samples (in duplicates) in FLIPP-NAAT is shown in Fig. 7A. The validity of the test was confirmed by no amplification for negative controls (1_N, 2_N and 3_N; Fig. 7A) and amplification for positive controls (1_P, 2_P and 3_P; Fig. 7A). The emergence of bright green color spots for both the duplicates of Samples 1 and 3 (1_I, 1_II and 3_I, 3_II; Fig. 7A) indicated a positive test while absence of the bright green spot for both duplicates of Sample 2 (2_I and 2_II; Fig. 7A) indicated a negative test. These results were interpreted by image analysis (Fig. 7B) where the green color intensity for sample 1 and sample 3 was above the threshold (µN + 3σN) while that for sample 2 was below the threshold. The end-point images for all the devices used for testing of clinical samples were analyzed in the same fashion (see Supplementary Figs S6–S8 and Table S2). When each sample was tested in duplicates, 21 samples reported TB positive and 9 samples reported TB negative with FLIPP-NAAT. As it was a blind study, Xpert MTB/RIF assay results were received after FLIPP-NAAT testing and were found to have 19 true positives and 11 true negatives. When compared to Xpert MTB/RIF assay, FLIPP-NAAT reported 5 false positives (Samples 11,20, 27, 28 and 30) and 3 false negatives (Samples 21, 24 and 25) resulting in a clinical sensitivity of 86.36% and clinical specificity of 68.75%. It was further observed that the clinical sensitivity of FLIPP-NAAT was 100% for smear-positive samples but dropped to 86.36% when smear-negative samples were included in the analysis. This loss in clinical sensitivity was addressed by retesting the smear-negative TB-positive samples by conducting six replicates of each sample (instead of duplicates) in a 3-module FLIPP NAAT device. Incorporation of more replicates increased the clinical sensitivity for the entire set of 30 samples to 100% while the clinical specificity remained 68.75%. To investigate the imperfect specificity, the cases of false positives were analyzed further. Amplicons from these FLIPP NAAT test zones were loaded on agarose gels, before and after restriction enzyme digestion (see Supplementary Figs S7 and S8A,B). The gel patterns for the false positive samples were very different from true positives (see Supplementary Fig. S8A) and these samples did not undergo enzyme digestion (see Supplementary Fig. S8B), confirming that the amplification was not specific to Mtb. The final positive and negative predictive values for the set of 30 clinical samples was 79.16 and 100%, respectively. FLIPP-NAAT, smear microscopy, and Xpert MTB/RIF results for the 30 clinical samples have been compiled in Table 1.
Figure 7.
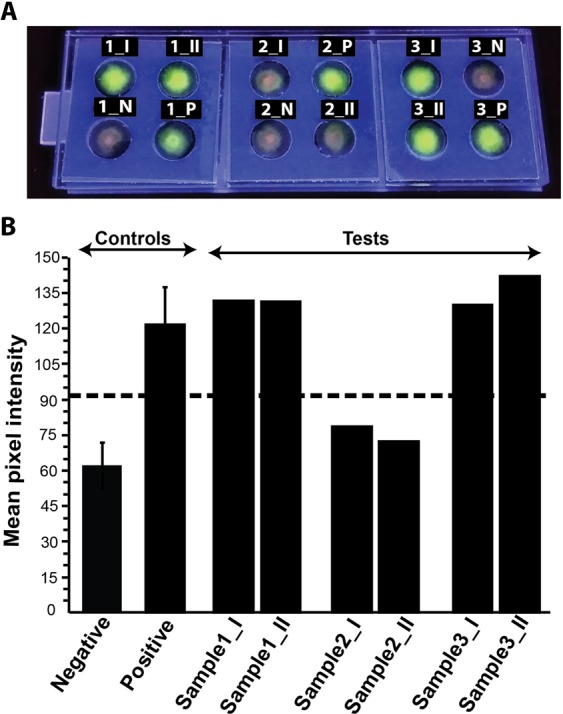
TB testing for clinical samples in FLIPP-NAAT. (A) Three clinical samples were tested per device with a negative and positive control in each module. Each sample was run in duplicates. Numbers on top of each zone represent the sample number and suffix ‘I’ and ‘II’ represent the duplicates. N – negative control and P – positive control. (B) Mean pixel intensity of green color from all the six blind clinical samples, negative and positive controls. The dashed line represents the threshold value of (µN + 3σN). Samples with intensity below the threshold were considered as TB negative while samples with green color intensity above the threshold were considered as TB positive.
Table 1.
Comparison of FLIPP-NAAT with smear microscopy and Xpert MTB/RIF.
| Study No. | Smear | Xpert MTB/RIF assay | VL/L/M/H | FLIPP-NAAT |
|---|---|---|---|---|
| 1 | 3+ | Mtb+ | H | Mtb+ |
| 2 | No AFB | Negative | N/A | Negative |
| 3 | 2+ | Mtb+ | H | Mtb+ |
| 4 | 3+ | Mtb+ | H | Mtb+ |
| 5 | No AFB | Negative | N/A | Negative |
| 6 | No AFB | Negative | N/A | Negative |
| 7 | 2+/3+ | Mtb+ | H | Mtb+ |
| 8 | 3+ | Mtb+ | M | Mtb+ |
| 9 | Sc | Mtb+ | M | Mtb+ |
| 10 | 2+ | Mtb+ | L | Mtb+ |
| 11 | No AFB | Negative | N/A | Mtb+ |
| 12 | 2+ | Mtb+ | M | Mtb+ |
| 13 | 1+ | Mtb+ | M | Mtb+ |
| 14 | 3+ | Mtb+ | M | Mtb+ |
| 15 | No AFB | Negative | N/A | Negative |
| 16 | 2+ | Mtb+ | L | Mtb+ |
| 17 | 1+ | Mtb+ | L | Mtb+ |
| 18 | 3+ | Mtb+ | M | Mtb+ |
| 19 | 1+ | Mtb+ | M | Mtb+ |
| 20 | No AFB | Negative | N/A | Mtb+ |
| 21 | No AFB | Mtb+ | L | Mtb+ |
| 22 | No AFB | Mtb+ | VL | Mtb+ |
| 23 | No AFB | Mtb+ | L | Mtb+ |
| 24 | No AFB | Mtb+ | VL | Mtb+ |
| 25 | No AFB | Mtb+ | VL | Mtb+ |
| 26 | No AFB | Negative | N/A | Negative |
| 27 | No AFB | Negative | N/A | Mtb+ |
| 28 | No AFB | Negative | N/A | Mtb+ |
| 29 | No AFB | Negative | N/A | Negative |
| 30 | No AFB | Negative | N/A | Mtb+ |
VL: Very low, L: Low, M: Medium, H: High.
Discussion
Despite some challenges that surfaced during testing of clinical samples, the results of FLIPP-NAAT testing are encouraging. The following sections elaborate upon the issues related to clinical sensitivity and specificity.
Clinical sensitivity
When all samples were tested in duplicates, the sensitivity for smear-positive samples was 100%, but the pooled sensitivity including smear-negative samples was 86.36%. This loss is attributed to the low bacterial load present in smear-negative samples leading to stochasticity in the presence of target DNA in the 3.5 µl of sample added to the device. This limitation can be addressed in two ways. First, sample preparation could be improved to suit the needs of FLIPP NAAT. The generic Qiagen extraction kit used in this work recommended that DNA be extracted in 100 µl final volume, only a small fraction of which could be tested in a single FLIPP-NAAT test zone. Alternate sample preparation techniques that elute DNA in smaller volumes will improve the clinical sensitivity of FLIPP NAAT. Nevertheless, resetting the clinical sensitivity back to 100% by increasing the number of test replicates demonstrates the ability of FLIPP-NAAT to detect TB from a wide range of samples having varying loads of Mtb (as indicated in Table 1). The second method of potentially enhancing sensitivity is to conduct FLIPP NAAT testing from dried reagents, which will enable using up to 9 µl of sample per test zone, ~2.5x higher than the volume accommodated in the current tests. Preliminary results of successful amplification from dried reagents are presented in this work. Even though the above-mentioned challenges exist, the current performance of FLIPP-NAAT with clinical samples substantiates its potential to be a higher-sensitivity replacement for smear microscopy.
Clinical specificity
The clinical specificity of FLIPP-NAAT could be improved further. Loss in specificity ensued from the use of a sequence-independent DNA intercalating fluorescent dye. The finding that all false positive reactions generated Mtb-independent amplicons reveals a path to recover the lost specificity by introducing sequence-specific detection probes in FLIPP-NAAT, which will be the topic of our future work.
Conventionally, NAATs have been conducted in tubes placed inside a thermal cycler. Tubes have an associated form factor and require specially designed heating blocks and instruments. A heated and heavy top cover is also required to prevent the tubes from opening due to generation of vapors at high temperature and to ensure that the vapors do not condense. Lack of proper sealing could lead to carry-over contamination which is highly undesirable for diagnostic devices. Fluorescence imaging in tubes is challenging – multiple internal reflections make it difficult to differentiate positives from negatives and to provide quantitative results. Paper, on the other hand, is flat and does not require any particular form factor-accommodating infrastructure. Fluorescence imaging is more reliable as shown in the results for fluorescence imaging in FLIPP-NAAT. Compared to tubes, FLIPP-NAAT also presents a mixing advantage. Conducting NAATs from dry stored reagents in tubes requires vortexing to mix the rehydrated reagents followed by centrifuging to push the contents to the bottom of the tube. On the other hand, FLIPP-NAAT is designed such that mixing is automatic on rehydration, reducing the requirement of ancillary equipment, making it more point-of-care compatible. While several isothermal nucleic acid amplification techniques exist, we chose LAMP for FLIPP-NAAT because of its multiple distinctive characteristics (see Supplementary Note S1).
There have recently been three reports on paper-based NAAT devices, designs of which appear similar to the eye as that of FLIPP NAAT15–17. However, the fabrication techniques, working principles, flexibility in device design, imaging set-up and costs involved in making FLIPP NAAT are strikingly different (see Supplementary Note S2). One of the most desirable features of FLIPP NAAT is that the material cost of making a 12-reaction-zone device is only $0.88 and the cost of reagents required per reaction zone is $0.43 (see Supplementary Tables S3 and S4). Modular design is a powerful feature of FLIPP-NAAT and using the current definition of a module, devices may be designed to contain 4 N reaction zones (where N = 1, 2, 3…), without any major modification in fabrication methods. The device design has been optimized (see Supplementary Note S3) as a simple layer-by-layer assembly of paper pads, plastics, and adhesives, and therefore it is compatible with mass manufacturing using injection molded plastic components.
Although this report focuses on TB diagnosis, FLIPP-NAAT is a platform technology for conducting NAATs by minimally trained users. FLIPP-NAAT is amenable to the detection of any DNA or RNA target from any sample type, provided it is appended to an appropriate sample-preparation technique. Because paper can be used to conduct many types of isothermal amplification techniques11, FLIPP-NAAT is not restricted to LAMP and can accommodate other isothermal amplification chemistries. Given its semi-quantitative fluorescence detection capabilities, FLIPP-NAAT can potentially be used for detection of fold-changes in gene expression. While it is a tool designed to be compatible with limited resource settings, it will also find applications in healthcare settings like clinics, hospitals, and beyond healthcare, in classrooms and teaching laboratories. It is a tool to make molecular diagnostics accessible, fast, easy, and affordable for all.
Supplementary information
Acknowledgements
This work has been funded by the Department of Biotechnology (DBT), Government of India, in the form of an Innovative Young Biotechnologist award (BT/010/IYBA/2016/07) to B.J.T., by the Bill & Melinda Gates Foundation in the form of a Grand Challenges Exploration award (OPP1182249) to B.J.T., by an extramural research grant from the Science and Engineering Research Board, India (EMR/2016/006029) to B.J.T., by the Indian Institute of Science in the form of a generous startup grant to B.J.T. and by the Ministry of Human Resource Development (MHRD), India, in the form of graduate student support to N.K. Collaboration with CMC Vellore for clinical trials was facilitated by a Margdarshi Fellowship awarded to Sandhya S. Visweswariah by the Wellcome Trust-DBT India Alliance (IA/M/16/1/502606). We acknowledge Prof. Deepak Saini (IISc) and Mr. Krishna Kumar Singh for providing H37Ra cultures, and Ms Andrea D’souza and Mr. Tabish Khan for help with viscosity measurements. The following reagent was obtained through BEI resources, NIAID, NIH: Genomic DNA from Mycobacterium tuberculosis, strain H37Rv, NR-48669.
Author contributions
B.J.T. contributed towards conception and design of the work, and analysis and interpretation of data. N.K. performed all experiments and contributed towards design of the work, and analysis and interpretation of data. J.S.M. collected clinical samples, obtained routine clinical TB-diagnostic results and helped with analysis and interpretation of data. B.J.T. and N.K. together wrote the main manuscript text and prepared figures.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51873-8.
References
- 1.World Health Organization. Global Tuberculosis Report 2017. World Health Organization, WHO/HTM/TB/2017.23 (2017).
- 2.Nema V. Tuberculosis diagnostics: Challenges and opportunities. Lung India. 2012;29:259. doi: 10.4103/0970-2113.99112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdurrahman ST, et al. The hidden costs of installing xpert machines in a Tuberculosis [TB] high-burden country: Experiences from Nigeria. Pan Afr. Med. J. 2014;18:277. doi: 10.11604/pamj.2014.18.277.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diagnostics, M. Truelab Uno, http://www.molbiodiagnostics.com/molbio_order.html Available at, http://www.molbiodiagnostics.com/molbio_order.html.
- 5.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 6.Yan L, et al. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014;10:970. doi: 10.1039/c3mb70304e. [DOI] [PubMed] [Google Scholar]
- 7.The use of loop-mediated isothermal amplifi cation (TB-LAMP) for the diagnosis of pulmonary tuberculosis Loop-Mediated Isothermal Amplification Dna Tuberculosis Tb-Lamp The Strategy End TB. World Heal. Organ. 52, doi:9789241511186 (2016).
- 8.Reboud Julien, Xu Gaolian, Garrett Alice, Adriko Moses, Yang Zhugen, Tukahebwa Edridah M., Rowell Candia, Cooper Jonathan M. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proceedings of the National Academy of Sciences. 2019;116(11):4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafleur Lisa K., Bishop Joshua D., Heiniger Erin K., Gallagher Ryan P., Wheeler Maxwell D., Kauffman Peter, Zhang Xiaohong, Kline Enos C., Buser Joshua R., Kumar Sujatha, Byrnes Samantha A., Vermeulen Nicolaas M. J., Scarr Noah K., Belousov Yevgeniy, Mahoney Walt, Toley Bhushan J., Ladd Paula D., Lutz Barry R., Yager Paul. A rapid, instrument-free, sample-to-result nucleic acid amplification test. Lab on a Chip. 2016;16(19):3777–3787. doi: 10.1039/C6LC00677A. [DOI] [PubMed] [Google Scholar]
- 10.Magro L, et al. Paper Microfluidics for Nucleic Acids Amplification Testing (NAAT) Lab Chip. 2017;17:2347–2371. doi: 10.1039/C7LC00013H. [DOI] [PubMed] [Google Scholar]
- 11.Kaur Navjot, Toley Bhushan J. Paper-based nucleic acid amplification tests for point-of-care diagnostics. The Analyst. 2018;143(10):2213–2234. doi: 10.1039/C7AN01943B. [DOI] [PubMed] [Google Scholar]
- 12.Shetty P, Ghosh D, Singh M, Tripathi A, Paul D. Rapid amplification of Mycobacterium tuberculosis DNA on a paper substrate. RSC Adv. 2016;6:56205–56212. doi: 10.1039/C6RA07529K. [DOI] [Google Scholar]
- 13.Bi, A., Nakajima, C., Fukushima, Y., Tamaru, A. & Sugawara, I. Short Communication A Rapid Loop-Mediated Isothermal Amplification Assay Targeting hspX for the Detection of Mycobacterium tuberculosis Complex. 247–251 (2012). [DOI] [PubMed]
- 14.I. N. de Kantor et al. Laboratory Services In Tuberculosis Control Organization And Management Part I. Glob. Tuberc. Program. World Heal. Organ (1998).
- 15.Seok Y, et al. A Paper-Based Device for Performing Loop-Mediated Isothermal Amplification with Real-Time Simultaneous Detection of Multiple DNA Targets. Theranostics. 2017;7:2220–2230. doi: 10.7150/thno.18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn H, Batule BS, Seok Y, Kim MG. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal. Chem. 2018;90:10211–10216. doi: 10.1021/acs.analchem.8b01309. [DOI] [PubMed] [Google Scholar]
- 17.Trinh TND, Lee NY. A rapid and eco-friendly isothermal amplification microdevice for multiplex detection of foodborne pathogens. Lab Chip. 2018;18:2369–2377. doi: 10.1039/C8LC00424B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).