Abstract
Drug-resistant tuberculosis (TB) is a serious global problem, and pathogen factors involved in the transmission of isoniazid (INH)-resistant TB have not been fully investigated. We performed whole genome sequencing of 332 clinical Mycobacterium tuberculosis (Mtb) isolates collected from patients newly diagnosed with smear-positive pulmonary TB in Hanoi, Vietnam. Using a bacterial genome-wide approach based on linear mixed models, we investigated the associations between 31-bp k-mers and clustered strains harboring katG-S315T, a major INH-resistance mutation in the present cohort and in the second panel previously published in South Africa. Five statistically significant genes, namely, PPE18/19, gid, emrB, Rv1588c, and pncA, were shared by the two panels. We further identified variants of the genes responsible for these k-mers, which are relevant to the spread of INH-resistant strains. Phylogenetic convergence test showed that variants relevant to PPE46/47-like chimeric genes were significantly associated with the same phenotype in Hanoi. The associations were further confirmed after adjustment for the confounders. These findings suggest that genomic variations of the pathogen facilitate the expansion of INH-resistance TB, at least in part, and our study provides a new insight into the mechanisms by which drug-resistant Mtb maintains fitness and spreads in Asia and Africa.
Subject terms: Bacterial genes, Tuberculosis
Introduction
In 2017, 558,000 tuberculosis (TB) cases of multidrug resistance (MDR) or rifampicin (RMP) resistance were estimated globally1. Resistance to anti-TB drugs increases the burden of TB because the treatment of drug-resistant TB is generally prolonged and costly, while the outcome is relatively poor2,3. Isoniazid (INH) resistance without concurrent RMP resistance accounts for 7.1% of the new TB cases in the world1. The spread of INH resistance serves as a reservoir of more combined drug resistance4,5; empirical treatment after rapid genetic assessment of RMP resistance alone increases the future risk for developing MDR-TB or extensively drug-resistant (XDR)-TB6,7. INH-containing preventive therapy is also affected heavily when the INH-resistance rate is high in a target population.
Extensive studies have been performed to identify the genetic mutations responsible for Mycobacterium tuberculosis (Mtb) drug resistance, and comprehensive lists of drug-resistance-conferring mutations have been provided8,9. However, unidentified genetic variants, including compensatory mutations, may also assist or facilitate the transmission of drug-resistant TB without reducing the fitness of the bacilli10. Investigation of this mechanism would contribute to the effective control of drug-resistant TB, and the accumulation of molecular epidemiological data in many areas around the world would deepen the understanding of the dynamics.
Vietnam is one of the 30 TB high-burden countries, with approximately 124,000 incidence cases reported in 20171. According to the national drug resistance survey conducted in 2011, among the new cases, resistance to any anti-TB drug accounted for 32.7% of the cases, and the proportion of INH-resistant TB reached 18.9%11. In our previous study cohort of 489 newly diagnosed patients in a city area, INH resistance was observed in 28.2% of the patients12, while RMP resistance remained in 4.9% of the patients. The predominant genetic mutations katG-S315T for INH and rpoB-S450L for RMP that confer drug resistance in Vietnam are similar to those reported in other Asian countries13–15. However, pathogen factors correlated with the transmission of INH-resistant TB have not been fully investigated.
Whole genome sequencing (WGS) has been recently used globally, offering new opportunities in the management of drug-resistant TB, since it can provide a huge amount of information, including genetic variants that are relevant to drug resistance throughout the genome16. WGS data also offer critical insights into the dynamics of TB endemics, transmission route, and the evolutionary patterns of genomic mutations10,17–19. Recently, bacterial genome-wide association studies (GWAS) controlling for population structure have also been performed for identifying the genes or genetic variants relevant to the TB phenotype, including drug resistance, by analyzing all single-nucleotide polymorphisms, small and large insertions/deletions (indels), or k-mers obtained from massive short-read data from next-generation sequencers (NGS)20,21.
In this study, we investigated drug resistance-conferring mutations carried by the clinical Mtb isolates from patients newly diagnosed with smear-positive pulmonary TB in Hanoi, Vietnam, by using WGS with a bacterial GWAS approach incorporating linear mixed models (LMMs). We then identified the genetic variants that may be relevant to the success in an extensive spread of INH-resistant strains, in reference to a previously published cohort study in KwaZulu-Natal, South Africa, as the second panel19.
Results
Prevalence of known drug resistance-conferring mutations
Among the 332 Hanoi samples analyzed with WGS, known mutations, which have been registered in the TBProfiler’s mutation database9,22, accounted for 80 (90.9%) of the 88 isolates with phenotypic INH resistance, 12 (92.3%) of the 13 isolates with RMP resistance, 63 (73.3%) of the 86 isolates with streptomycin (SM) resistance, 5 (100.0%) of the 5 isolates with ethambutol resistance, and 5 (55.6%) of the 9 isolates with pyrazinamide (PZA) resistance (Table 1). The most prevalent drug-resistance-conferring mutations were katG-S315T (26.2%) to INH, rpsL-K43R (13.3%) to SM, and rpoB-S450L (2.7%) to RMP. Mutations conferring resistance to the second-line drugs were rrs-A514C (4.5%) to amikacin and those in the fabG1-promoter (2.7%) to ethionamide. Others were rare mutations such as ethR-F110L to ethionamide, and gyrA-A90V and gyrA-D94G to fluoroquinolone (0.6%, 0.3%, and 0.6%, respectively). Among the 87 strains harboring katG-S315T mutations, 54 (62.1%) had at least one other drug-resistance-conferring mutation. Co-occurrence of katG-S315T and rpsL-K43R with or without other known mutations was observed most frequently (31 of the 87 isolates; 35.6%).
Table 1.
Frequencies of known mutations conferring resistance to first-line drugs among all isolates, phenotypically resistant isolates, and susceptible isolates in the Hanoi sample set (N = 332).
| Drug resistance-conferring mutations | Study population n (%) |
Phenotypically resistant isolates n (%) |
Phenotypically susceptible isolates n (%) |
|---|---|---|---|
| Isoniazid | N = 332 | N = 88 | N = 244 |
| (−) | 233 (70.2) | 8 (9.1) | 225 (92.2) |
| katG (S315T) | 87 (26.2) | 75 (85.3) | 12 (4.9) |
| Rv1482c-fabG1 (C-15T) | 7 (2.1) | 3 (3.4) | 4 (1.6) |
| Rv1482c-fabG1 (C-15T), inhA (S94A) | 1 (0.3) | 1 (1.1) | 0 (0.0) |
| Rv1482c-fabG1 (C-15T), katG (S315N) | 1 (0.3) | 1 (1.1) | 0 (0.0) |
| katG (G234R) | 1 (0.3) | 0 (0.0) | 1 (0.4) |
| katG (S315G) | 1 (0.3) | 0 (0.0) | 1 (0.4) |
| katG (W191R) | 1 (0.3) | 0 (0.0) | 1 (0.4) |
| Streptomycin | N = 332 | N = 86 | N = 246 |
| (−) | 259 (78.0) | 23 (26.6) | 236 (96.0) |
| rpsL (K43R) | 44 (13.3) | 42 (48.8) | 2 (0.8) |
| rrs (A514C) | 14 (4.2) | 11 (12.8) | 3 (1.2) |
| rpsL (K88R) | 6 (1.8) | 6 (7.0) | 0 (0.0) |
| gid (L16R), rrs (A514C) | 1 (0.3) | 1 (1.2) | 0 (0.0) |
| rpsL (K43R), rrs (A1401G) | 1 (0.3) | 1 (1.2) | 0 (0.0) |
| rrs (A907T) | 1 (0.3) | 1 (1.2) | 0 (0.0) |
| rrs (C517T) | 2 (0.6) | 1 (1.2) | 1 (0.4) |
| rrs (T1239C) | 1 (0.3) | 0 (0.0) | 1 (0.4) |
| rpsL (K88T) | 1 (0.3) | 0 (0.0) | 1 (0.4) |
| gid (L16R) | 2 (0.6) | 0 (0.0) | 2 (0.8) |
| Rifampicin | N = 332 | N = 13 | N = 319 |
| (−) | 310 (93.4) | 1 (7.7) | 309 (96.9) |
| rpoB (S450L) | 9 (2.7) | 4 (30.8) | 5 (1.6) |
| rpoB (H445D) | 4 (1.2) | 4 (30.8) | 0 (0.0) |
| rpoB (H445L) | 2 (0.6) | 2 (15.4) | 0 (0.0) |
| rpoB (K446Q) | 1 (0.3) | 1 (7.7) | 0 (0.0) |
| rpoB (Q432K) | 1 (0.3) | 1 (7.7) | 0 (0.0) |
| rpoB (L452P) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| rposB (S450L), rpoB (E761D) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| rpoB (L430P) | 3 (0.9) | 0 (0.0) | 3 (0.9) |
| Ethambutol | N = 332 | N = 5 | N = 327 |
| (−) | 303 (91.3) | 0 (0.0) | 303 (92.7) |
| embB (M306V) | 6 (1.8) | 3 (60.0) | 3 (0.9) |
| embB (L370R) | 13 (3.9) | 1 (20.0) | 12 (3.7) |
| embC-embA (C-12T) | 1 (0.3) | 1 (20.0) | 0 (0.0) |
| embB (D354A) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| embB (G406A) | 2 (0.6) | 0 (0.0) | 2 (0.6) |
| embB (G406D) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| embB (M306I) | 4 (1.2) | 0 (0.0) | 4 (1.2) |
| embB (Q497R) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Pyrazinamide | N = 332 | N = 9 | N = 323 |
| (−) | 319 (96.1) | 4 (44.4) | 315 (97.5) |
| pncA (Q141P) | 3 (0.9) | 2 (22.2) | 1 (0.3) |
| pncA (L159R) | 1 (0.3) | 1 (11.1) | 0 (0.0) |
| pncA (S59P) | 1 (0.3) | 1 (11.1) | 0 (0.0) |
| pncA-Rv2044c (T-11C) | 3 (0.9) | 1 (11.1) | 2 (0.6) |
| pncA (A46V) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| pncA (I31S) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| pncA (Q10P) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| pncA (Q10R) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| pncA (S104R) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
Other than SNVs, in one isolate, a 353-bp deletion was found in the pncA region after the screening of zero or low-depth areas, which also covered the pncA promoter and a part of the Rv2044c nearby. One sample had an in-frame 6-bp deletion (M434-D435) in the rpoB gene; this strain was not resistant to RMP. Eight isolates harboring 1-bp deletion in the gid gene, and two other isolates carrying 1-bp deletion of the ethA gene were also found (Supplementary Fig. S1).
Mtb lineages/sublineages, drug-resistance-conferring mutations, and genetic clusters
The lineage (L)2 East Asian—mostly the Beijing strains—possessed the mutations that conferred drug resistance most frequently (52.6%); L1 (Indo-Oceanic; 23.2%) carried less and then L4 (Euro-American; 18.3%) the least (P < 0.0001) in Hanoi. The proportion of the above drug resistance was the highest in ancient Beijing strains (59.8%), followed by modern Beijing strains (40.9%), and then by non-Beijing strains (20.8%) (P < 0.0001).
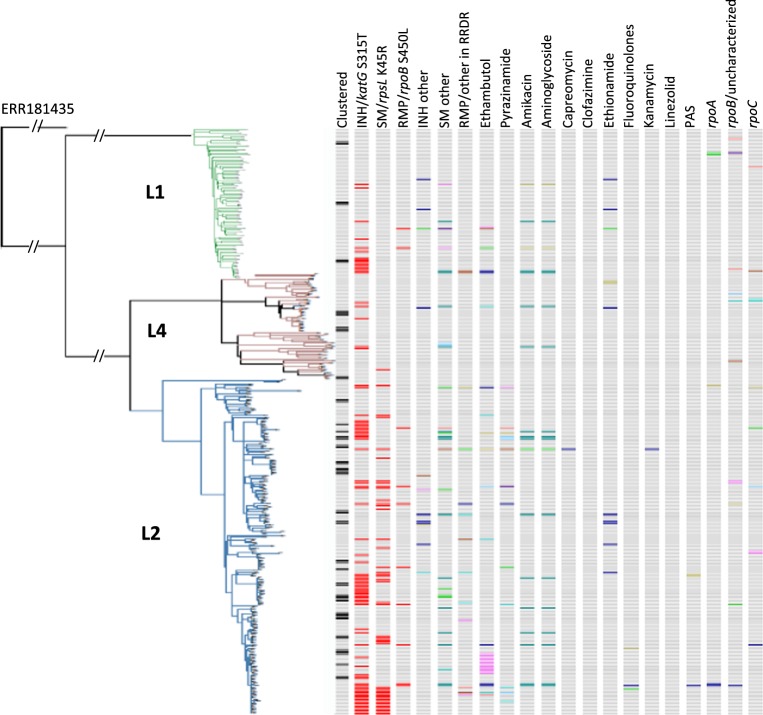
Figure 1 shows the distribution of the strains harboring mutations of interest. Of note, katG-S315T, known as a major INH-resistance mutation in the world23, was reported with a frequency of 26.2% in Hanoi, which accounted for 85.3% of the primary INH-resistance, and it was distributed unevenly among the strains like scattered islands. This S315T mutation, regardless of whether it occurred alone or in combination with other mutations, was more frequently observed in the L2 and L1 strains than in L4 (P < 0.0001) and also more in ancient Beijing than in modern Beijing and non-Beijing strains (59.0%, 40.9%, and 19.4%, respectively, P < 0.001). katG-S315T was often carried by ancient Beijing strains, whereas clustered strains defined by pairwise SNV differences of <6 alone were observed more frequently among modern Beijing strains than among ancient and non-Beijing strains (24.2%, 18.9%, and 9.7%, respectively, P = 0.016).
Figure 1.
Distribution of drug resistant-relevant mutations and their presence in correlation with Mtb lineages and clustering. Clustered: defined by pairwise SNV difference <6 SNVs. Different colors indicate different mutations.
Framework of bacterial GWAS analysis
To investigate the pathogen factors involved in the wide spread of INH-resistant strains in Hanoi, we used a combination of a representative INH resistance-conferring mutation katG-S315T (S315T[+]) with genetic cluster (cluster[+]) <6 SNV differences as a “phenotype” or objective variable for GWAS. katG-S315T and SNV-based genetic clusters alone served as phenotypes for comparison. Initially 31-bp k-mers throughout the genome were set as the “genotype” or explanatory variables, because the presence or absence of such k-mers provides clues for identifying the SNVs, indels, or even structural variants without a standard reference sequence for mapping reads20. When candidate genes were obtained from the k-mer analysis, individual variants, including SNVs and indels, were further analyzed for confirmation using other variant-based platforms.
Association between phenotypes and 31-bp k-mers throughout the genome
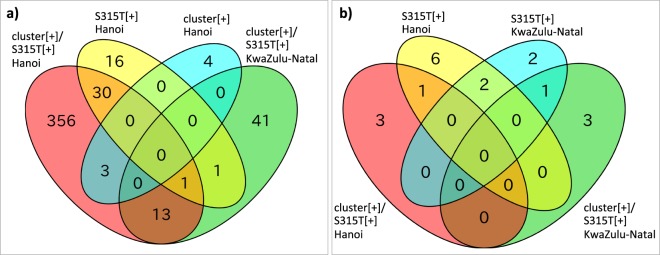
When k-mer GWAS was performed for our Hanoi samples, the number of genes harboring significant k-mers associated with cluster[+] alone and S315T[+] alone were 7 and 48, respectively, whereas the combined phenotype, cluster[+]/S315T[+] extracted 403 statistically significant genes, controlling for the population structure using LMM (Table 2). For comparison, 337 samples from the South African KwaZulu-Natal study were analyzed in a similar way as for the combined phenotype (Table 2), and 14 genes were eventually extracted in common (Fig. 2a). Of these, eight (Rv1148c, mmpL6, PE_PGRS21, PE_PGRS53, pks12, rpoB, Rv2090, and wag22) were excluded from further analysis because the presence or absence of k-mers appeared only in one sample of either case or control group. PE_PGRS10 was further excluded since the presence of k-mers was not further confirmed by BLAST search for read sequences from both Hanoi and KwaZulu-Natal samples.
Table 2.
Genes harboring k-mers significantly associated with different phenotypes (from (1) to (6)) in Hanoi and KwaZulu-Natal studies.
| Phenotype | Gene name | Number of genes | |||||
|---|---|---|---|---|---|---|---|
| (1) | (2) | (3) | (4) | (5) | (6) | ||
| No | Yes | Yes | Yes | Yes | Yes | gid | 1 |
| No | Yes | No | Yes | No | Yes | rrs | 1 |
| No | Yes | Yes | No | Yes | No | katG | 1 |
| No | No | Yes | Yes | Yes | Yes | PPE18, Rv1148c | 2 |
| Yes | No | Yes | No | No | No | ephB, glpK, mce1A | 3 |
| No | No | Yes | No | No | Yes | emrB, mmpL6, Rv1588c | 3 |
| No | No | Yes | Yes | No | Yes | PE_PGRS10, PE_PGRS21, PE_PGRS53, pks12, pncA, rpoB, Rv2090, wag22 | 8 |
| No | No | No | Yes | No | Yes | atsD, fabG1, gyrA, helZ, murF, PE_PGRS52, Rv2000, Rv2161c, Rv3471c, ubiA | 10 |
| No | No | No | Yes | Yes | Yes | ctpB, drrA, ethA, fhaA, PPE19, PPE60, Rv0094c, Rv1945, Rv2019, Rv2141c, Rv3921c | 11 |
| No | Yes | Yes | No | No | No | accD2, eccC3, eccC4, fadE7, fas, glfT1, icd1, lepB, lprN, mbtJ, mce1R, menA, mez, PE_PGRS42, PPE30, PPE4, PPE54, rbsK, Rv1106c, Rv1147, Rv1215c, Rv1747, Rv2005c, Rv2566, Rv3085, Rv3401, Rv3677c, yfiH, zwf1 | 29 |
| No | No | No | No | Yes | No | fadD5, Rv0336, Rv0348 | 3 |
| Yes | No | No | No | No | No | atsA, gltA2, PE_PGRS12, Rv3863 | 4 |
| No | Yes | No | No | No | No | mycP2, rpsL, Rv0012, Rv0219, Rv0893c, Rv1217c, Rv1443c, Rv1515c, Rv1516c, Rv2033c, Rv2277c, Rv2672, Rv2825c, Rv3177, Rv3618, yjcE | 16 |
| No | No | No | No | No | Yes | cysK1, espI, glmS, hsaA, lprQ, PE23, pks15, pks7, pmmB, rhlE, Rv0121c, Rv1073, Rv1639c, Rv2219, Rv3217c, Rv3254, Rv3660c, Rv3707c, thiL, vapB34 | 20 |
| No | No | Yes | No | No | No | 35kd_ag, accA3, adhE1, aglA, aldA, alkB, alr, amiB2, amiC, aprA, aroA, aroG, arsA, arsB2, bfrA, bfrB, blaC, cdh, cobN, cut1, cyp139, cysA2, cysD, deoC, devS, dinF, dinG, dinX, dnaB, dnaE2, dnaG, dnaK, dxs2, eccA3, eccB2, eccC2, eccD5, efp, egtE, eno, erg3, espH, esxJ, esxK, esxM, etgB, fadB3, fadB4, fadD15, fadD18, fadD22, fadD24, fadE18, fadE25, fadE34, fbiC, fecB, folC, frdD, fusA1, gatB, gcvB, glgA, gnd2, groEL2, guaB1, guaB2, hemB, hemY, hisH, hupB, hycQ, iniC, ispD, kshA, lepA, leuA, lipD, lipP, lppA, lppH, lppZ, lpqH, lprA, ltp1, mbtE, mce3B, mce3C, mcr7, menC, metH, mmpL1, mmpL11, mmpL5, mmsA, mmuM, moeB1, murE, mutT3, mymT, narG, nlhH, nrp, oppB, oxyR, papA2, papA5, PE_PGRS1, PE_PGRS13, PE_PGRS16, PE_PGRS17, PE_PGRS19, PE_PGRS2, PE_PGRS22, PE_PGRS23, PE_PGRS24, PE_PGRS25, PE_PGRS26, PE_PGRS27, PE_PGRS30, PE_PGRS31, PE_PGRS32, PE_PGRS33, PE_PGRS38, PE_PGRS4, PE_PGRS43, PE_PGRS44, PE_PGRS45, PE_PGRS46, PE_PGRS47, PE_PGRS48, PE_PGRS50, PE_PGRS54, PE_PGRS55, PE_PGRS56, PE_PGRS57, PE_PGRS6, PE_PGRS62, PE_PGRS7, PE_PGRS9, PE1, pepC, pepD, pepR, php, pknB, pknJ, pknK, pknL, pks13, pks9, pncB2, pntAb, ppa, PPE11, PPE12, PPE15, PPE21, PPE24, PPE34, PPE42, PPE46, PPE55, PPE56, PPE57, PPE59, PPE6, PPE8, ppm1, ppnK, ppsA, ppsC, proV, prpD, psd, purH, purL, purT, recC, relE, rfe, rimJ, rpfA, rpsD, Rv0057, Rv0060, Rv0074, Rv0095c, Rv0111, Rv0133, Rv0149, Rv0178, Rv0180c, Rv0188, Rv0192, Rv0193c, Rv0203, Rv0218, Rv0238, Rv0257, Rv0278c, Rv0302, Rv0323c, Rv0339c, Rv0367c, Rv0378, Rv0386, Rv0412c, Rv0457c, Rv0474, Rv0477, Rv0492c, Rv0537c, Rv0538, Rv0575c, Rv0584, Rv0647c, Rv0650, Rv0658c, Rv0686, Rv0696, Rv0740, Rv0790c, Rv0842, Rv0845, Rv0874c, Rv0876c, Rv0921, Rv0939, Rv0996, Rv1004c, Rv1024, Rv1043c, Rv1045, Rv1047, Rv1048c, Rv1050, Rv1069c, Rv1085c, Rv1178, Rv1225c, Rv1273c, Rv1313c, Rv1318c, Rv1372, Rv1395, Rv1425, Rv1455, Rv1488, Rv1498A, Rv1499, Rv1526c, Rv1610, Rv1667c, Rv1683, Rv1702c, Rv1707, Rv1714, Rv1725c, Rv1726, Rv1728c, Rv1729c, Rv1739c, Rv1749c, Rv1815, Rv1825, Rv1828, Rv1831, Rv1835c, Rv1879, Rv1895, Rv1941, Rv1999c, Rv2017, Rv2023A, Rv2024c, Rv2025c, Rv2038c, Rv2074, Rv2134c, Rv2143, Rv2164c, Rv2177c, Rv2190c, Rv2219A, Rv2228c, Rv2254c, Rv2256a, Rv2286c, Rv2314c, Rv2319c, Rv2327, Rv2424c, Rv2456c, Rv2532c, Rv2556c, Rv2567, Rv2621c, Rv2630, Rv2638, Rv2656c, Rv2668, Rv2669, Rv2729c, Rv2733c, Rv2792c, Rv2798c, Rv2851c, Rv2864c, Rv2893, Rv2897c, Rv2951c, Rv2974c, Rv2994, Rv3027c, Rv3060c, Rv3064c, Rv3074, Rv3104c, Rv3169, Rv3193c, Rv3201c, Rv3208, Rv3216, Rv3218, Rv3230c, Rv3263, Rv3365c, Rv3400, Rv3439c, Rv3549c, Rv3668c, Rv3689, Rv3717, Rv3728, Rv3729, Rv3807c, Rv3856c, Rv3896c, Rv3898c, sirA, smc, tal, TB9.4, tgs2, thiD, tig, treX, trxB1, uspC, uvrA, vapB36, vapC29, whiA, yrbE3A, yrbE3B | 356 |
Phenotype (1): clustered strains (cluster[+]) (pairwise SNV difference between two isolates is no more than five SNVs) in Hanoi’s study cohort; (2): strains harboring katG-S315 T mutation (S315 T[+]) in Hanoi’s cohort; (3): cluster[+]/S315T[+] in Hanoi’s cohort; (4): cluster[+] in KwaZulu-Natal’s cohort; (5) S315T[+] in KwaZulu-Natal’s cohort; (6): cluster[+]/S315T[+] in KwaZulu-Natal’s cohort; SNV: single nucleotide variant.
The genes in bold are associated with cluster[+]/S315T[+], and shared between Hanoi and KwaZulu-Natal study population.
Figure 2.
Venn diagram showing the number of genes significantly associated with the given phenotypes and shared by the two study cohorts. (a) results from k-mer-based GWAS; (b) results from phyOverlap; cluster[+]: clustered (pairwise SNV difference between two isolates is no more than five SNVs); S315T[+]: harboring katG-S315T mutation; Hanoi: Hanoi’s study cohort; KwaZulu-Natal: KwaZulu-Natal’s study cohort.
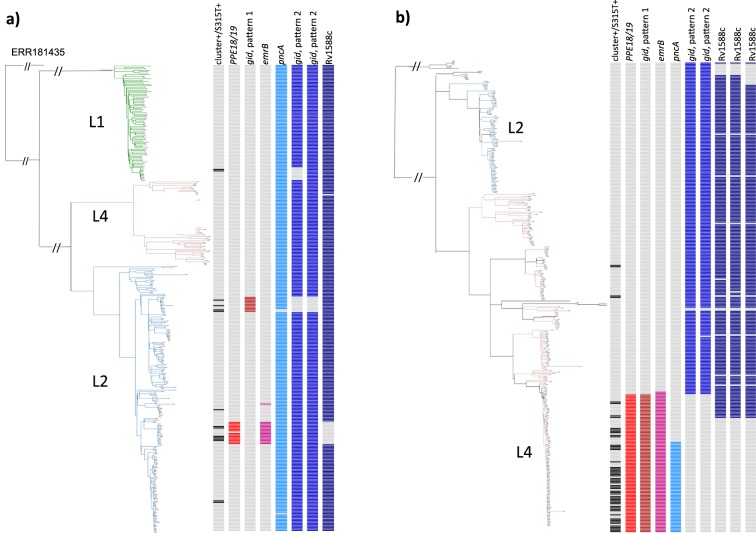
The remaining five genes, namely, PPE18, gid, emrB, Rv1588c, and pncA, were confirmed by BLAST search and were aligned to the H37Rv reference sequence (Supplementary Fig. S2) and nominated as real candidates. In the Hanoi study population, significant k-mers included 36 k-mers from the PPE18 gene (all had the same P value = 1.840E-09), 33 k-mers from gid (the best P value = 5.213E-08), 31 from emrB (8.579E-08), 4 from Rv1588c (8.579E-08), and 31 from the pncA gene (4.437E-07) (Supplementary Table S1), when 6.208E-06 was applied as the threshold of statistical significance after Bonferroni correction. For the KwaZulu-Natal study population, within the same set of genes, the best P values obtained from 5, 101, 31, 12, and 29 k-mers were 2.998E-08, 2.998E-08, 7.802E-07, 2.453E-06, and 4.802E-11, respectively, with the threshold of significance after Bonferroni correction as 4.591E-06 (Supplementary Table S1). Positive associations with the phenotype were mainly observed in L2 of Hanoi’s samples and L4 of KwaZulu-Natal’s samples (Fig. 3).
Figure 3.
Distribution of k-mers derived from variant (or wild) types of five genes (PPE18/19, gid, emrB, Rv1588c, and pncA) that showed positive or negative associations with clustered strains carrying katG-S315T in the phylogenetic trees of Hanoi (a) and KwaZulu-Natal (b) study population.
Next, we attempted to confirm significant k-mers by using a DBGWAS approach to identify all the relevant sequences as “unitigs” (Table 3). Consequently, one significant unitig indicating merged k-mers was identified in PPE18, three were identified in gid, two in emrB, and two in Rv1588c (the best q value in each gene was 5.562E-07, 3.676E-05, 1.704E-05, and 1.704E-05, respectively) for the Hanoi samples (Supplementary Table S2, Fig. S3). For the KwaZulu-Natal samples, one significant unitig was found in PPE18, seven in gid, two in emrB, and one in pncA (the best q value in each gene was 4.856E-10, 7.943E-09, 7.943E-09, and 7.639E-12, respectively) (Supplementary Table S2).
Table 3.
Genes with k-mers significantly associated with clustered strains harboring katG-S315T mutation (cluster[+]/S315T[+]) in both Hanoi and KwaZulu-Natal study cohorts, further investigated by using the DBGWAS platform and variant/deletion-based search.
| Gene name | Sample panel | K-mer-based GWAS | Strongest P value | DBGWAS* | Strongest P value | Variant-based GWAS/deletion-based GWAS/BLAST search | Strongest P value | Conclusion on variant(s) corresponding to significant k-mer(s) |
|---|---|---|---|---|---|---|---|---|
| PPE18/19 | Hanoi | 36 k-mers, SNV at 1339644 and 1339649 | 1.840E-09 | 1 unitig, SNVs at 1339644 and 1339649 | 5.56188E-07 | SNVs at 1339644 (E99A) and 1339649 (A101T) found by variant-based GWAS | 1.941E-09 | Significant SNVs E99A and A101T |
| PPE18/19 | KwaZulu-Natal | 5 k-mers, SNV at 1340231 | 2.998E-08 | 1 unitig**, SNV at 1532777 | 4.8561E-10 | SNV at 1532777 (Q286R) detected by BLAST search. By variant-based GWAS, SNV was not called properly | NA | SNV Q286R |
| gid | Hanoi | 33 k-mers, SNV at 4407686 | 5.213E-08 | 3 unitgs, SNV at 4407686 | 3.676E-05 | SNV at 4407686 (E173*) fount by variant-based GWAS | 2.991E-06 | Significant SNV E173* |
| gid | KwaZulu-Natal | 101 k-mers | 7.802E-07 | 7 unitigs, no SNV | 7.943E-09 | Large deletions (from 120 bp to 675 bp) | 2.178E-04 | Large deletions |
| emrB | Hanoi | 31 k-mers, SNV at 876918 | 8.579E-08 | 2 unitigs, SNV at 876918 | 1.704E-05 | SNV at 876918 (F508S) found by variant-based GWAS | 1.507E-08 | Significant SNV F508S |
| emrB | KwaZulu-Natal | 31 k-mers, SNV at 877058 | 7.802E-07 | 2 unitigs**, SNV at 877058 | 7.943E-09 | SNV at 877058 (I461I) found by variant-based GWAS | 1.077E-06 | Significant SNV I461I |
| Rv1588c | Hanoi | 4 k-mers without SNV (neg.***) | 8.579E-08 | 2 unitigs**, no SNV | 1.704E-05 | SNV at 1789735 (P34P) found by variant-based GWAS and BLAST search | 1.511E-08 | Significant SNV P34P |
| Rv1588c | KwaZulu-Natal | 12 k-mers without SNV (neg.***) | 2.453E-06 | 0 unitig | NA | No SNV inside the corresponding k-mers | NA | None |
| pncA | Hanoi | 31 k-mers without SNV (neg.***) | 4.437E-07 | 0 unitig (pncA not found) | NA | SNV at 2288820 (Q141P) found by variant-based GWAS | 4.919E-07 | Significant SNV Q141P |
| pncA | KwaZulu-Natal | 29 k-mers, SNV at 2288785 and 2288788 | 4.802E-11 | 1 unitig, G insertion at 2288785 | 7.639E-12 | SNV at 2288785 (T153fs) found by variant-based GWAS | 6.569E-11 | Significant SNV T153fs |
cluster[+]: clustered (pairwise SNV difference between two isolates is no more than five SNVs); S315T[+]: harboring katG-S315T mutation; Hanoi: Hanoi’s study cohort; KwaZulu-Natal: KwaZulu-Natal’s study cohort; GWAS: genome wide association study; SNV: single nucleotide variant.
*Only significant unitigs and their SNVs are shown.
**Found by sequence search.
***Showing negative association with Cluster[+]/S315T[+] by logistic regression analyses
All k-mers annotated with PPE18 in both study sets were mapped to either PPE18, 19, or 60. Nucleotide sequences within these three PPE genes were hardly distinguishable from each other (Supplementary Fig. S4); BWA-MEM mapping or simple BLAST search to the reference could not specify the exact PPE gene that each 31-bp k-mer belonged to. Nevertheless, the analysis of de novo assembled contigs demonstrated that the Hanoi variant k-mers were derived from PPE18 and KwaZulu-Natal’s k-mers initially annotated with PPE18 were derived from PPE19. Such a high degree of sequence similarity was also observed in significant unitigs identified in both study sets with the DBGWAS approach. Therefore, these k-mers and unitigs were designated as PPE18/19 in our study (Table 3).
Individual variant analysis using a genome-wide approach
When mapping the aforementioned significant k-mers of Hanoi samples on the H37Rv genome, we identified variants corresponding to the significant k-mers. Next, we attempted to confirm the variants by SNV- and small indel-based GWAS or BLAST search, and finally identified mutations encoding E99A and A101T, in PPE18, E173* (Glu173Stop) in the gid, F508S in emrB, P34P in Rv1588c, and Q141P in pncA. They were significantly associated with cluster[+]/S315T[+] (P values were 1.941E-09 for both PPE18 SNVs, and the P values were 2.991E-06, 1.507E-08, 1.511E-08, and 4.919E-07 for gid, emrB, Rv1588c, and pncA SNV, respectively) (Table 3). The PROVEAN web server24 and SIFT_4G tool25 predicted gid-E173* and emrB-F508S as deleterious or that affecting the protein function.
For the KwaZulu-Natal samples, mapping results showed SNVs in emrB (I461I) and pncA (T153fs) (variant-based GWAS P values = 1.077E-06 and 6.596E-11, respectively). Initially, the SNV in the PPE18/19 was not clearly mapped to the reference for the aforementioned reason. After the in-depth search for de novo assembled contigs, Q286R in PPE19 was identified. Large deletions were found in gid in 101 of the 337 samples (from 120 to 675 bp). Rv1588c did not show any SNV in the corresponding area of the k-mers (Table 3).
SNVs and small indels from VCF files, significantly associated with the cluster[+]/S315T[+] of the Hanoi cohort obtained by GWAS, included 329 SNVs and indels other than the six shown above (Supplementary Table S3) (the best P = 1.754E-13, Supplementary Table S4). None of them were lineage-specific SNVs that have been reported elsewhere22. After further excluding variants in ambiguous PE and PPE genes, synonymous SNVs, a mutation causing S315T, and those with frequency no more than two in case or control samples, 144 SNVs and small indels were extracted in the samples from Hanoi. Of these, 77 SNVs correlated well with the principal component (PC)-7 (Supplementary Table S4), which corresponded to a phylogenetic branch of the ancient Beijing strains (Supplementary Fig. S5), with a high percentage (92.9%) of INH resistance.
We further analyzed the structural variants by detecting zero or low-depth areas when H37Rv and four complete genomes in Hanoi (AL123456, and AP018033 to AP018036) were used as references to be mapped. We found two groups of gene deletions associated with this phenotype. The first group included 1-bp deletion in Rv1043c, 52-bp deletion in Rv2286c, 3-bp insertion in Rv0790c, 1-bp deletion in Rv3230c, and a big deletion in Rv2025c, and these correlated well with PC-9 (Table 4), which corresponded to the same branch of ancient Beijing strains shown in SNV analysis. The second group included a 238-bp deletion in accD2, 2-bp deletion in eis, a 459-bp insertion in Rv3077, a 12-bp deletion in Rv2690c, and correlated with PC-12 (Table 4) corresponding to another branch of modern Beijing strains.
Table 4.
Genes with deletions/insertions significantly associated with clustered strains harboring katG-S315T mutation among Hanoi samples obtained by GWAS, and corresponding k-mers.
| Reference gene* | Locus in H37Rv (AL123456) | Deletion/insertion | P value | Principal component | K-mers | Strongest P value |
|---|---|---|---|---|---|---|
| Details | No of significant k-mers/No of k-mers | |||||
| AL123456.3H37Rv|00828|Rv0790c | Rv0790c | 3-bp insertion at 884090 to 884092 | 1.507E-08 | 9 | 42/46 | 1.43E-08 |
| AL123456.3H37Rv|01092|Rv1043c | Rv1043c | 1-bp deletion at 1166499 | 1.941E-09 | 9 | 45/61 | 1.43E-08 |
| AL123456.3H37Rv|02117|Rv2025c | Rv2025c | Deletion of almost all Rv2025c gene, also Rv2024c (789-bp) | 9.053E-08 | 9 | 819/833 | 1.43E-08 |
| AL123456.3H37Rv|02393|Rv2286c | Rv2286c | 52-bp deletion from 2559452 to 2559503 | 1.507E-08 | 9 | 69/123 | 1.43E-08 |
| AL123456.3H37Rv|03394|Rv3230c | Rv3230c | 1-bp deletion at 3607421 | 9.053E-08 | 9 | 59/63 | 1.84E-09 |
| AL123456.3H37Rv|02535|eis | Rv2416c | 2-bp deletion at 2714527–2714528 | 3.219E-06 | 12 | 0/60 | 1.32E-05 |
| AP018033.1HN024|01030|accD2 | Best hit with Rv0974c | 238-bp deletion from 1086195 to 1086432 | 2.991E-06 | 12 | — | — |
| AP018033.1HN024|02797|HN024_02796 | Best hit with Rv2690c | 12-bp deletion | 1.720E-05 | 12 | — | — |
| AP018036.1HN506|03217|HN506_03216 | Best hit with Rv3077 | 459-bp insertion | 1.720E-05 | 12 | — | — |
PC-9 correlated gene variants were observed in 14 samples (HN-042, HN-065, HN-120, HN-127, HN-152, HN-154, HN-170, HN-179, HN-186, HN-222, HN-253, HN-391, HN-441, HN-456) that belong to ancient Beijing sublineages.
PC-12 correlated gene variants were observed in 10 samples (HN-019, HN-075, HN-109, HN-137, HN-169, HN-324, HN-330, HN-435, HN-462, HN-506) that belong to modern Beijing sublineages.
*Genome sequences of clinical isolates in our Hanoi cohort, AP018033 to AP018036 as well as AL123456 (H37Rv genome) were used for reference.
Convergence-based phyOverlap analysis
Seeking variants caused by convergent evolution is another alternative for detecting mutations supporting drug resistance, such as compensatory mutations26. We also tried to identify the phenotype-associated variants caused by convergence evolution. As expected, rpoB and pncA were also associated with katG-S315T mutation alone. However, no genes and inter-genic regions were significantly associated with the presence of cluster[+]/S315T[+] in both KwaZulu-Natal and our Hanoi panels (Table 5, Supplementary Table S5, Fig. 2b). One PPE gene, two PE genes, and one intergenic region, PPE47, PE_PGRS55, PE_PGRS20 and PE_PGRS3-PE_PGRS4 were extracted in the Hanoi samples only (Table 5).
Table 5.
Genes detected by the phyOverlap method and their significant associations with the Hanoi strains harboring the katG-S315T mutation, with and without clustering, and loci shared with the KwaZulu-Natal study population.
| Locus | Gene name | No of SNV | Parsimony score | P value | FDR (q value) | Remark |
|---|---|---|---|---|---|---|
| Associated with cluster[+]/S315T[+] | ||||||
| Rv3021c | PPE47 | 12 | 73 | 0.00000 | 0.00000 | PE/PPE |
| Rv3511 | PE_PGRS55 | 13 | 18 | 0.00000 | 0.00000 | PE-PGRS family protein |
| Rv0278c-Rv0279c | PE_PGRS3-PE_PGRS4 | 2 | 37 | 0.00002 | 0.02434 | PE-PGRS family protein |
| Rv1068c | PE_PGRS20 | 13 | 27 | 0.00002 | 0.02434 | PE-PGRS family protein |
| Associated with S315T[+] | ||||||
| Rv0667 | rpoB | 14 | 27 | 0.00000 | 0.00000 | Rifampicin-resistance related gene |
| Rv0682 | rpsL | 2 | 21 | 0.00000 | 0.00000 | Streptomycin-resistance related gene |
| Rv0758 | phoR | 8 | 8 | 0.00002 | 0.01217 | Sensor part of a two component regulatory system |
| Rv2043c | pncA | 10 | 11 | 0.00000 | 0.00000 | Pyrazinamide-resistance related gene |
| Rv3021c | PPE47 | 12 | 73 | 0.00000 | 0.00000 | PE/PPE |
| Rv3169 | Rv3169 | 5 | 5 | 0.00002 | 0.01217 | Unknown |
| Rv3428c-Rv3429 | Rv3428c-PPE59 | 28 | 247 | 0.00000 | 0.00000 | |
| Rv3647c-Rv3648c | Rv3647c-cspA | 4 | 4 | 0.00006 | 0.03245 | |
| Rv3680-Rv3681c | Rv3680-whiB4 | 1 | 78 | 0.00000 | 0.00000 | |
The genes in bold are shared between Hanoi and KwaZulu-Natal study population.
PPE46 and PPE47 share large portions of identical nucleotide sequences. By BLAST search for variants in the de novo assembled contigs in addition to genome-wide screening of zero or low-depth areas (<15% of average depth) in the reference genome, we found that 37 (4 in L1, 19 in L2, and 14 in L4) strains had large deletions in PPE47 (Supplementary Fig. S6) and all had fusion with PPE46, resulting in PPE46-like chimeric genes in 35 isolates and PPE47-like chimeric genes in 2 isolates (Supplementary Fig. S7). SNVs identified from 3379708 to 3379763 of AL123456.3 (H37Rv) in the PPE47 region were the main reason for the significant association in the phylogenetic convergence test. In other candidates from phyOverlap, specific variants were not validated, presumably owing to the difficulties in mapping short reads followed by ambiguous base calling within PE_PGRS genes.
Analyses using logistic regression models adjusted for host confounders in Hanoi samples
By multivariate analyses using conventional logistic regression models after adjustment for patients’ gender, age, living area, as well as Mtb lineages, all k-mers from PPE18/19, emrB, and a part of k-mers from gid showed positive associations with clustered strains carrying katG-S315T mutations in the Hanoi study population (adjusted odds ratio [aOR] with 95% confidence interval [CI] = 13.20 [3.49–49.96], 11.98 [3.24–44.29], and 12.42 [2.81–54.90], respectively), whereas Rv1588c and pncA k-mers showed negative associations (aOR with 95% CI = 0.08 [0.02–0.31], and 0.01 [0.00–0.25], respectively). All variants corresponding to significant k-mers showed positive associations with cluster[+]/S315T[+]. The PPE46/47-like chimeric gene also showed positive association with cluster[+]/S315T[+] (aOR with 95% CI = 6.81 [2.13–21.72]) (Supplementary Table S6).
Discussion
We identified a variety of drug resistance-conferring mutations prevailing in Hanoi, northern Vietnam, which appeared most frequently in the East Asian Mtb lineage L2 particularly in ancient Beijing sublineage, and less in L1, and then least in L4. Large deletions that were not detected by conventional variant calling from mapped short reads were also found in pncA. Using the bacterial GWAS approach, we extracted candidate genes that were significantly associated with the clustered strains harboring the katG-S315T mutation and that were common to the two independent data sets—our cohort panel and a previous South African study, the KwaZulu-Natal cohort by Cohen et al.19—suggesting that expansions of INH-resistance TB can be facilitated by pathogen factors, at least in part.
A major drug-resistance conferring mutation, katG-S315T, accounted for 85.3% of the INH-resistance strains in our study population, which was similar to that reported in other studies in Vietnam, e.g., 73.2% was reported by Minh et al.13, 73.6% was reported by Huyen et al.27, 81.3% by Nguyen et al.15 and 78.4% was reported in Southeast Asian countries28. This katG-S315T is known as a mutation with low-fitness cost, spreads to Beijing strains and others, and is more likely to be clustered23,29,30. The acquisition of katG-S315T mutation precedes other resistance mutations, including RMP4,5,19. Indeed, this mutation occurs more frequently in MDR-TB strains than other mutations30,31, and it has an important implication in the transmission and control of MDR-TB.
In this setting, we performed bacterial GWAS with phenotypic combination of clusters defined by <6 SNVs32 and katG-S315T mutation as a surrogate, and searched for pathogenic variants contributing to the spread of INH-resistance. We identified five genes, namely, PPE18/19, gid, emrB, Rv1588c, and pncA, which were shared by two different sample panels in Asia and Africa with different major Mtb lineages, L2 and L4, respectively.
Of the five genes extracted from the k-mer GWAS, except for drug-resistance-conferring genes, the best P values were obtained from PPE18/19 both in the Hanoi population and in the KwaZulu-Natal cohort. The association was mainly observed in L2 of Hanoi’s samples and L4 of KwaZulu-Natal’s samples. PPE18/19 genes are members of a multigene family and share high sequence similarity with another PPE gene, PPE6033. Among these PPE genes, homologous recombination events frequently occur and contribute to the sequence diversity34,35. Among nonsynonymous SNVs and small indels detected in the PPE18 gene of our study population leading to amino acid change in Mtb39A, two SNVs were significantly associated with cluster[+]/S315T[+], suggesting the spread of INH-resistant TB. Although their role in virulence is not fully understood, the PPE18 protein, also known as Mtb39A, was shown to downregulate the proinflammatory response and Th1-type immunity, interacting with host TLR236, and facilitated survival and multiplication of Mtb bacilli in a mouse model37.
The T-cell epitopes in Mtb39A38–40 lead to strong T-cell proliferation and IFN-gamma production38. Therefore it is used as a subunit for the human TB vaccine candidate Mtb72F and its successor M72. It has been proved to be immunogenic and can stimulate both cellular and humoral immune response41,42. By comparing with the lists of the T-cell epitopes published elsewhere38,40,43,44, nonsynonymous mutations E99A and A101T in PPE18 identified in the Hanoi population and Q286R in PPE19 in the KwaZulu-Natal’s panel were both found to be included in the sequences acting as T-cell epitopes for cellular immunity. Despite the presence of PPE18 variants in the Hanoi strains, the original T-cell epitope sequences were mostly conserved in either PPE19 or PPE60, when the BLAST search was applied. Variants of these PPE genes associated with spread of INH-resistance strains may thus increase the antigenic diversity of the bacilli, which may help evade or exploit human immunity by unidentified mechanisms through the process of human-Mtb coevolution33,35,45.
Mutations in the gid gene have been reported to be associated with low-level resistance to SM46, which has been used as the first-line drug since a long time until recently. Because gid encodes methyltransferase that is responsible for the methylation of 16 S rRNA involved in the translational fidelity47, it is thus conceivable that the gid mutation may modulate the fitness of INH-resistance conferring mutations through the change in mRNA translational fidelity. Our study revealed that gid k-mers with E173* mutant were significantly associated with cluster[+]/S315T[+] and S315T[+], but not with cluster[+] alone, indicating that a concurrence of gid-E173* and katG-S315T may facilitate transmission even after controlling for population structure. The concurrence of gid 130-bp deletion and katG-S315T is the first step toward XDR-level drug resistance in Africa19. In our study population, all strains with gid-E173* mutation had at least one genetic mutation conferring resistance to first-line drugs. This non-sense mutation gid-E173* may facilitate the expansion of katG-S315T mutant strains. Indeed, other groups have reported that SM-resistant strains seem to be more clustered in Vietnam48.
Efflux pumps, including emrB, have been reported to be associated with pathogenicity since the up-regulation of efflux gene expression is involved in the development of resistance to anti-TB drugs49 and a wide array of physiologic processes such as the growth kinetics or transportation of a variety of compounds50. The combination of the katG-S315T mutant with emrB variant F508S in the Hanoi study may thus increase the drug efflux activity and facilitate Mtb survival and spread, by mitigating drug pressure. emrB (Rv0783c) belongs to the major facilitator superfamily (MFS) characteristically energized by the proton motive force (H+ or Na+)50,51, and may confer low-level resistance to RMP52. Although the role of I461I in emrB in KwaZulu-Natal’s population is unknown at present, this synonymous mutation (c.1383 C > A, ATC > ATA) is very rare in terms of codon usage of Mtb53, and the significance of codon usage bias and t-RNA modification should be taken into account, because rare codons are sometimes advantageous to the survival of Mtb under stress conditions54. When exposed to INH, various MFS efflux pump genes were reported to be overexpressed50, and these may induce sustained increased efflux activity with selection and stabilization of drug-resistant mutations55. This may also be relevant to the acquisition of additional drug resistance. Further studies are required for elucidating the function of the mutations in efflux pump genes. Indeed, SNVs in efflux pump genes are often found in XDR strains but not in drug-susceptible strains51, although it is often difficult to identify the extrusion of a drug to a specific gene50,56. The association pattern characterized by the presence or absence of the variant of emrB with the phenotype was quite similar to that of PPE18/19, whose phenomenon was shown beyond lineages, L2 in Hanoi, and L4 in KwaZulu-Natal. Although membrane localization of the PPE genes may be functionally linked to efflux pump activities, it is currently unknown.
Rv1588c is a partial REP13E12 repeat protein57. Although k-mers carrying the reference sequences in Rv1588c showed negative association with the clustered strains harboring katG-S315T in the two panels, their functional significance remains unclear. As a variant, only a synonymous variant P34P was found in Hanoi, which was associated with ancient Beijing sublineage.
The reference sequence (=wild type) k-mers in pncA were also associated negatively with the phenotype in the Hanoi cohort, and variant-carrying k-mers showed a positive association in the KwaZulu-Natal cohort. As a variant found in Hanoi, Q141P in pncA has been reported as a high-confidence mutation leading to PZA resistance58,59. PZA resistance is often observed among MDR-TB isolates60,61. Thus, the possibility of pncA mutations facilitating the transmission of katG-S315T mutant Mtb isolates may make TB management more challenging.
Convergence-based phyOverlap analysis, which is a different approach, revealed that four different gene/intergenic regions only in the Hanoi study population that may have been caused by convergence evolution were significantly associated with the clustered strains carrying the katG-S315T mutation; and these four were present in the PE or PPE region. The impact of genetic variation on the function of PE or PPE proteins remains largely unknown35. However, at least large deletions between PPE46 and PPE47 genes were observed in all three lineages (L1, L2, and L4) in Hanoi, and these were positively associated with the spread of INH-resistant TB. The deletion between the identical sequences of the two PPE genes leads to in-frame gene fusions through homologous recombination62, and a relatively high prevalence, indicating a clonal expansion of Haarlem strains (L4) in Tunisia34, suggested that the generation of the new chimeric genes may facilitate antigenic diversity and provide new determinants for pathogen’s virulence across the lineages.
Further analyses using logistic regression models confirmed that all variants corresponding to significant k-mers of the five genes and even variants detected by the phylogenetic convergence test in Hanoi samples were positively associated with the spread of INH-resistant TB, even after adjustment for other possible confounders.
Our study has some limitations. First, we were not able to trace the epidemiological link among the patients to corroborate the transmission chain. Hanoi is the capital city of Vietnam with on-going urbanization and this city consists of a floating population coming from many other provinces; thus, pursuing an epidemiological link is rather difficult. However, we have detailed information on the patients’ residential districts and we have added this information to the logistic regression analyses. Second, our samples were obtained in a population-based setting in an Asian city; but to generalize our finding, we analyzed another African cohort set available in the public by using the same methodology. Third, performing in vitro experiments to elucidate the functional significance of each genetic variant was beyond our scope owing to resource limitation. Nevertheless, the extracted genes were associated with the spread of INH-resistant strains carrying katG-S315T mutation and these reached statistically significant levels by using the bacterial GWAS approach based on LMM.
Previous studies suggest that the katG gene’s physiological function is not largely reduced by S315T substitution63. Its catalase-peroxidase-peroxynitritase activities may play a part to protect Mtb against reactive oxygen and nitrogen species derived from the phagocyte oxidative burst in human macrophages63,64. This may link KatG with other pathogen factors relevant to immune evasion or virulence such as PE/PPE65,66, although possible additive or synergistic effects on fitness should be further investigated. It is desirable to conduct validation studies in different populations. These Mtb genes are attractive candidates, presumably because of their relevance to the pathogen’s virulence, and they could be important sources to consider in in vitro and in animal models.
In conclusion, WGS data demonstrated the status of primary drug resistance at gene levels in the Hanoi city, and bacterial GWAS was performed to identify candidate genes that may facilitate the spreading of INH-resistant strains. Our findings provide new insights into the pathogenic mechanisms possibly mediated by the candidate genes including PE/PPE, by which drug-resistant Mtb can maintain epidemiological fitness and spread in high-burden countries such as in Asia and Africa.
Methods
Study sites, patient recruitment, and sample collection
This was a part of our cohort study of patients who were over 16 years of age and who were newly diagnosed with smear-positive pulmonary TB without any treatment history in Hanoi, Vietnam during 2007–2009, in which basic data with clinical interpretation were published in a previous report12,67–69. In brief, we included 7 of the 14 districts in Hanoi as the catchment area, where more than half of new smear-positive TB patients in the city were diagnosed and treated in the area during the study period12. Sputum specimens were collected before starting the treatment, 92.7% of which revealed culture-positive, and drug susceptibility testing for first-line drugs was performed using the WHO standard proportional method12. The patients’ clinico-epidemiological information was also collected. Written informed consent was obtained from all the patients.
Ethics statement
This cohort study was approved by the Ethical Committees of the Ministry of Health, Vietnam, National Center for Global Health and Medicine, and the Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association, Japan. All experiments were performed in accordance with relevant guidelines and regulations. In the case of minors, their parents provided written informed consent.
WGS
Mycobacterial DNA samples from Löwenstein-Jensen culture media were extracted using the Isoplant kit (Nippon Gene, Tokyo, Japan) and analyzed using Illumina HiSeq and MiSeq systems (Illumina, San Diego, CA, USA). These experiments were performed using a class II safety cabinet in a biosafety level 3 laboratory to prevent contamination. For Hiseq. 2500, a library of WGS was prepared using an automated sample preparation system (Agilent Technologies Inc.) with the TruSeq DNA PCR free sample prep kit (Illumina). For Miseq, a library was prepared from 200 ng of genomic DNA with the TruSeq Nano DNA LT Sample Preparation Kit (Illumina), following the manufacturer’s instructions. Paired-end (2 × 150 bp) sequencing was performed using Hiseq. 2500. For Miseq, the paired-end (2 × 250 bp or 2 × 300 bp) sequencing system was used. The sequence data are available in the DDBJ/EMBL/GenBank databases under the accession numbers DRA008666-7 and DRA008677.
Extracting single nucleotide variants (SNVs) for Mtb lineages/sublineages and genetic clustering
Briefly, after trimming and excluding severely contaminated samples, sequence reads were mapped to the H37Rv genome (AL123456.3) by using BWA-MEM 0.7.15 (https://github.com/lh3/bwa), followed by variant calling with the Genome analysis toolkit (GATK version 3.7)70. Only paired-end fastq files with average depth more than 25X were accepted for the analysis. The criteria set for identifying SNVs and small indels included Q30 minimum base call quality score and a minimum coverage depth of 10X. Drug resistance-conferring mutations, including small indels and lineage-specific variations, were extracted using the TB-Profiler version 0.3.79,22. The Beijing genotype of lineage-2 (L2.2) was further classified into ancient and modern Beijing sublineages by detecting the SNVs at the nucleotide position 649,345, which is equivalent to the presence of IS6110 in the NTF region71.
Large deletions were screened throughout the mapped reads by seeking zero or low-depth areas (<15% of the average depth) using an in-house python script and then visualized for confirmation with the Integrative Genomics Viewer (IGV) version 2.3.91. For this deletion screening, complete genome sequences of the clinical isolates in our Hanoi cohort, AP018033 to AP01803672,73 as well as H37Rv genome, were used as reference sequences. After excluding ambiguous variants in categories of repetitive and insertion sequences and phages, genetic clusters were defined by the pairwise differences of no more than five SNVs74. A phylogenetic tree was constructed by the maximum likelihood method using RAxML version 8.2.875 and then visualized with plotTree (https://github.com/katholt/plotTree) by using a lineage-7 strain ERR181435 as an out-group.
Analyses of bacterial GWAS
The associations between the phenotypes and the presence or absence of 31-bp short sequences, k-mers, in the genome were investigated using a genome-wide efficient mixed model association algorithm, the GEMMA software (https://github.com/genetics-statistics/GEMMA). At first, the DSK software (https://github.com/GATB/dsk) was used for listing all the unique 31-bp DNA k-mers, and then their presence or absence in all the samples was analyzed as mentioned above20.
DBGWAS, an extended k-mer-based GWAS tool with compacted De Bruijn graph76, was further used to confirm the genetic variants associated with the phenotypes of interest. Sequence reads were assembled using SPAdes v3.13.077 and Platanus 1.2.478 when appropriate, and the generated contigs were used for BLAST search (ncbi-blast-2.8.1+) to identify the location of the phenotype-associated k-mers. Bonferroni correction was applied for multiple testing; the threshold of the significance after correction was calculated as 0.05 divided by the number of variants identified in the study samples.
To investigate whether any variant, including SNVs or indels in the whole genome, has any possible association with the phenotypes of interest, we used bugwas R package20 with built-in GEMMA. The bi-allelic SNVs were used to calculate the relatedness matrix of the samples for LMMs to control for the population structure. Likelihood ratio tests were used for assessing the significant associations.
Phylogenetic convergence tests (phyOverlap)26 were also performed to identify the convergent variants associated with the phenotypes. Herein, Benjamini-Hochberg adjustment at 0.05 false discovery rate level was applied as the q value for phyOverlap.
To compare the findings obtained from the GWAS analysis of our 332 samples in Vietnam, another set of WGS data from 337 clinical isolates, which were collected in the KwaZulu-Natal province of South Africa from 2008 to 2013 in a study conducted by Cohen et al.19 to investigate the emergence of drug-resistant TB (hereafter referred to as KwaZulu-Natal study), were retrieved from the public database, and analyzed in a similar way.
Other statistical analyses
Chi square and Fisher’s exact tests were performed to compare the differences in the proportions among the groups. Venn diagram (R version 3.4.4 VennDiagram package) was used to demonstrate the common gene(s) harboring variants, including k-mers, associated with different phenotypes. Possible associations between the given genetic variations and INH-resistant clusters, adjusted for Mtb lineages and patients’ age, gender, and living area were further studied using logistic regression models. These analyses were performed using STATA version 12 (StataCorp, College Station, TX, USA), and P values less than 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
The authors would like to thank Dr. Luu Thi Lien for her leadership and Dr. Shinsaku Sakurada for his monitoring of the on-site implementation of the original cohort study, and Ms. Ikumi Matsushita for her technical support and resource contribution. The authors would like to thank Enago (www.enago.jp) for the English language review. This research was supported by AMED under Grant Number JP19fm0108001 (Japan Initiative for Global Research Network on Infectious Diseases; J-GRID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
N.T.L.H. supervised the on-site implementation, performed the analyses, and wrote and finalized the manuscript. M.H. performed whole genome sequencing and analyzed the sequencing results. S.M. provided technical support and performed genotyping. J.O. provided statistical advice and support. A.M. and S.S. provided technical support for the microbiological experiments. P.H.T., H.V.H. and V.C.C. supervised on-site sample and data collection. N.P.H. and N.V.H. supervised on-site bacterial sample management. N.K. conceptualized the project, wrote the in-house scripts for data analyses, performed the analyses, and corrected and finalized the manuscript.
Data availability
All data pertaining to the manuscript have been provided in the forms of tables and figures. Supporting Information is available as Supplementary Tables S1–S6 and Figs S1–S7. Datasets pertaining to the sequence searches described here are available from the corresponding author on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nguyen Thi Le Hang and Minako Hijikata.
Supplementary information
is available for this paper at 10.1038/s41598-019-51812-7.
References
- 1.WHO. World Health Organization. Global Tuberculosis Report 2018 (2018).
- 2.Bastos ML, et al. Treatment outcomes of patients with multidrug-resistant and extensively drug-resistant tuberculosis according to drug susceptibility testing to first- and second-line drugs: an individual patient data meta-analysis. Clin Infect Dis. 2014;59:1364–1374. doi: 10.1093/cid/ciu619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pooran A, Pieterson E, Davids M, Theron G, Dheda K. What is the cost of diagnosis and management of drug resistant tuberculosis in South Africa? PLoS One. 2013;8:e54587. doi: 10.1371/journal.pone.0054587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller B, Borrell S, Rose G, Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2013;29:160–169. doi: 10.1016/j.tig.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manson AL, et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet. 2017;49:395–402. doi: 10.1038/ng.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menzies Dick, Benedetti Andrea, Paydar Anita, Royce Sarah, Pai Madhukar, Burman William, Vernon Andrew, Lienhardt Christian. Standardized Treatment of Active Tuberculosis in Patients with Previous Treatment and/or with Mono-resistance to Isoniazid: A Systematic Review and Meta-analysis. PLoS Medicine. 2009;6(9):e1000150. doi: 10.1371/journal.pmed.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagg HR, Lipman MC, McHugh TD, Jenkins HE. Isoniazid-resistant tuberculosis: a cause for concern? Int J Tuberc Lung Dis. 2017;21:129–139. doi: 10.5588/ijtld.16.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nebenzahl-Guimaraes H, Jacobson KR, Farhat MR, Murray MB. Systematic review of allelic exchange experiments aimed at identifying mutations that confer drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2014;69:331–342. doi: 10.1093/jac/dkt358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coll F, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nhung NV, Hoa NB, Sy DN, Hennig CM, Dean AS. The fourth national anti-tuberculosis drug resistance survey in Viet Nam. Int J Tuberc Lung Dis. 2015;19:670–675. doi: 10.5588/ijtld.14.0785. [DOI] [PubMed] [Google Scholar]
- 12.Hang NT, et al. Primary drug-resistant tuberculosis in Hanoi, Viet Nam: present status and risk factors. PLoS One. 2013;8:e71867. doi: 10.1371/journal.pone.0071867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minh NN, et al. Molecular characteristics of rifampin- and isoniazid-resistant mycobacterium tuberculosis strains isolated in Vietnam. J Clin Microbiol. 2012;50:598–601. doi: 10.1128/JCM.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nhu NT, et al. Association of streptomycin resistance mutations with level of drug resistance and Mycobacterium tuberculosis genotypes. Int J Tuberc Lung Dis. 2012;16:527–531. doi: 10.5588/ijtld.11.0202. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen HQ, et al. Quadruple-first line drug resistance in Mycobacterium tuberculosis in Vietnam: What can we learn from genes? Infect Genet Evol. 2017;50:55–61. doi: 10.1016/j.meegid.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Papaventsis D, et al. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect. 2017;23:61–68. doi: 10.1016/j.cmi.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 18.Casali N, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen KA, et al. Evolution of Extensively Drug-Resistant Tuberculosis over Four Decades: Whole Genome Sequencing and Dating Analysis of Mycobacterium tuberculosis Isolates from KwaZulu-Natal. PLoS Med. 2015;12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earle SG, et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol. 2016;1:16041. doi: 10.1038/nmicrobiol.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50. doi: 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 22.Coll F, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagneux S, et al. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006;2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 26.Hicks ND, et al. Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat Microbiol. 2018;3:1032–1042. doi: 10.1038/s41564-018-0218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huyen MN, et al. Epidemiology of isoniazid resistance mutations and their effect on tuberculosis treatment outcomes. Antimicrob Agents Chemother. 2013;57:3620–3627. doi: 10.1128/AAC.00077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One. 2015;10:e0119628. doi: 10.1371/journal.pone.0119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ameeruddin NU, Luke Elizabeth H. Impact of isoniazid resistance on virulence of global and south Indian clinical isolates of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94:557–563. doi: 10.1016/j.tube.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Ano H, et al. Relationship between the isoniazid-resistant mutation katGS315T and the prevalence of MDR-/XDR-TB in Osaka, Japan. Int J Tuberc Lung Dis. 2008;12:1300–1305. [PubMed] [Google Scholar]
- 31.Unissa AN, Selvakumar N, Narayanan S, Suganthi C, Hanna LE. Investigation of Ser315 substitutions within katG gene in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from south India. Biomed Res Int. 2015;2015:257983. doi: 10.1155/2015/257983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. The Lancet Infectious Diseases. 2013;13:137–146. doi: 10.1016/s1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McEvoy CR, et al. Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints. PLoS One. 2012;7:e30593. doi: 10.1371/journal.pone.0030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karboul A, et al. Frequent homologous recombination events in Mycobacterium tuberculosis PE/PPE multigene families: potential role in antigenic variability. J Bacteriol. 2008;190:7838–7846. doi: 10.1128/JB.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96:901–916. doi: 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 36.Nair S, Pandey AD, Mukhopadhyay S. The PPE18 protein of Mycobacterium tuberculosis inhibits NF-kappaB/rel-mediated proinflammatory cytokine production by upregulating and phosphorylating suppressor of cytokine signaling 3 protein. J Immunol. 2011;186:5413–5424. doi: 10.4049/jimmunol.1000773. [DOI] [PubMed] [Google Scholar]
- 37.Bhat KH, Ahmed A, Kumar S, Sharma P, Mukhopadhyay S. Role of PPE18 protein in intracellular survival and pathogenicity of Mycobacterium tuberculosis in mice. PLoS One. 2012;7:e52601. doi: 10.1371/journal.pone.0052601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dillon DC, et al. Molecular characterization and human T-cell responses to a member of a novel Mycobacterium tuberculosis mtb39 gene family. Infect Immun. 1999;67:2941–2950. doi: 10.1128/iai.67.6.2941-2950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewinsohn DA, Lines RA, Lewinsohn DM. Human dendritic cells presenting adenovirally expressed antigen elicit Mycobacterium tuberculosis–specific CD8+ T cells. Am J Respir Crit Care Med. 2002;166:843–848. doi: 10.1164/rccm.2110094. [DOI] [PubMed] [Google Scholar]
- 40.Mortier MC, Jongert E, Mettens P, Ruelle JL. Sequence conservation analysis and in silico human leukocyte antigen-peptide binding predictions for the Mtb72F and M72 tuberculosis candidate vaccine antigens. BMC Immunol. 2015;16:63. doi: 10.1186/s12865-015-0119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed S, Lobet Y. Tuberculosis vaccine development; from mouse to man. Microbes Infect. 2005;7:922–931. doi: 10.1016/j.micinf.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Rodo MJ, et al. A comparison of antigen-specific T cell responses induced by six novel tuberculosis vaccine candidates. PLoS Pathog. 2019;15:e1007643. doi: 10.1371/journal.ppat.1007643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaseitsiwe S, et al. Pattern recognition in pulmonary tuberculosis defined by high content peptide microarray chip analysis representing 61 proteins from M. tuberculosis. PLoS One. 2008;3:e3840. doi: 10.1371/journal.pone.0003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaseitsiwe S, et al. Peptide microarray-based identification of Mycobacterium tuberculosis epitope binding to HLA-DRB1*0101, DRB1*1501, and DRB1*0401. Clin Vaccine Immunol. 2010;17:168–175. doi: 10.1128/CVI.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebert AM, et al. DNA polymorphisms in the pepA and PPE18 genes among clinical strains of Mycobacterium tuberculosis: implications for vaccine efficacy. Infect Immun. 2007;75:5798–5805. doi: 10.1128/IAI.00335-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto S, et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 47.Wong SY, et al. Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:6311–6318. doi: 10.1128/AAC.00905-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buu TN, et al. Increased transmission of Mycobacterium tuberculosis Beijing genotype strains associated with resistance to streptomycin: a population-based study. PLoS One. 2012;7:e42323. doi: 10.1371/journal.pone.0042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigues L, Parish T, Balganesh M, Ainsa JA. Antituberculosis drugs: reducing efflux=increasing activity. Drug Discov Today. 2017;22:592–599. doi: 10.1016/j.drudis.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Louw GE, et al. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother. 2009;53:3181–3189. doi: 10.1128/AAC.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanji A, et al. Single nucleotide polymorphisms in efflux pumps genes in extensively drug resistant Mycobacterium tuberculosis isolates from Pakistan. Tuberculosis (Edinb) 2017;107:20–30. doi: 10.1016/j.tube.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Pang Y, et al. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:893–900. doi: 10.1128/AAC.01024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gun L, Yumiao R, Haixian P, Liang Z. Comprehensive Analysis and Comparison on the Codon Usage Pattern of Whole Mycobacterium tuberculosis Coding Genome from Different Area. Biomed Res Int. 2018;2018:3574976. doi: 10.1155/2018/3574976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chionh YH, et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat Commun. 2016;7:13302. doi: 10.1038/ncomms13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machado D, et al. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One. 2012;7:e34538. doi: 10.1371/journal.pone.0034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garima K, et al. Differential expression of efflux pump genes of Mycobacterium tuberculosis in response to varied subinhibitory concentrations of antituberculosis agents. Tuberculosis (Edinb) 2015;95:155–161. doi: 10.1016/j.tube.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology. 1999;145(Pt 4):881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 58.Chang KC, Yew WW, Zhang Y. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis: a systematic review with meta-analyses. Antimicrob Agents Chemother. 2011;55:4499–4505. doi: 10.1128/AAC.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miotto P, et al. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. MBio. 2014;5:e01819–01814. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia Q, et al. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob Agents Chemother. 2015;59:1690–1695. doi: 10.1128/AAC.04541-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pang Y, et al. Prevalence and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. BMC Infect Dis. 2017;17:711. doi: 10.1186/s12879-017-2761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marmiesse M, et al. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology. 2004;150:483–496. doi: 10.1099/mic.0.26662-0. [DOI] [PubMed] [Google Scholar]
- 63.Unissa AN, Subbian S, Hanna LE, Selvakumar N. Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol. 2016;45:474–492. doi: 10.1016/j.meegid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 65.Cui Ze-Jia, Yang Qing-Yong, Zhang Hong-Yu, Zhu Qiang, Zhang Qing-Ye. Bioinformatics Identification of Drug Resistance-Associated Gene Pairs in Mycobacterium tuberculosis. International Journal of Molecular Sciences. 2016;17(9):1417. doi: 10.3390/ijms17091417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bainomugisa, A. et al. A complete high-quality MinION nanopore assembly of an extensively drug-resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microb Genom4, 10.1099/mgen.0.000188 (2018). [DOI] [PMC free article] [PubMed]
- 67.Hung NV, et al. Clonal expansion of Mycobacterium tuberculosis isolates and coexisting drug resistance in patients newly diagnosed with pulmonary tuberculosis in Hanoi, Vietnam. BMC research notes. 2013;6:444. doi: 10.1186/1756-0500-6-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maeda S, et al. Mycobacterium tuberculosis strains spreading in Hanoi, Vietnam: Beijing sublineages, genotypes, drug susceptibility patterns, and host factors. Tuberculosis (Edinb) 2014;94:649–656. doi: 10.1016/j.tube.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 69.Hang NT, Maeda S, Keicho N, Thuong PH, Endo H. Sublineages of Mycobacterium tuberculosis Beijing genotype strains and unfavorable outcomes of anti-tuberculosis treatment. Tuberculosis (Edinb) 2015;95:336–342. doi: 10.1016/j.tube.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 70.Hijikata M, et al. Spoligotyping and whole-genome sequencing analysis of lineage 1 strains of Mycobacterium tuberculosis in Da Nang, Vietnam. PLoS One. 2017;12:e0186800. doi: 10.1371/journal.pone.0186800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chuang PC, Chen HY, Jou R. Single-nucleotide polymorphism in the fadD28 gene as a genetic marker for East Asia Lineage Mycobacterium tuberculosis. J Clin Microbiol. 2010;48:4245–4247. doi: 10.1128/JCM.00970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wada, T. et al. Complete Genome Sequence of a Mycobacterium tuberculosis Strain Belonging to the East African-Indian Family in the Indo-Oceanic Lineage, Isolated in Hanoi, Vietnam. Genome Announc5, 10.1128/genomeA.00509-17 (2017). [DOI] [PMC free article] [PubMed]
- 73.Wada, T. et al. Complete Genome Sequences of Three Representative Mycobacterium tuberculosis Beijing Family Strains Belonging to Distinct Genotype Clusters in Hanoi, Vietnam, during 2007 to 2009. Genome Announc5, 10.1128/genomeA.00510-17 (2017). [DOI] [PMC free article] [PubMed]
- 74.Nikolayevskyy, V. et al. Role and value of whole genome sequencing in studying tuberculosis transmission. Clin Microbiol Infect, 10.1016/j.cmi.2019.03.022 (2019). [DOI] [PubMed]
- 75.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaillard M, et al. A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events. PLoS Genet. 2018;14:e1007758. doi: 10.1371/journal.pgen.1007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data pertaining to the manuscript have been provided in the forms of tables and figures. Supporting Information is available as Supplementary Tables S1–S6 and Figs S1–S7. Datasets pertaining to the sequence searches described here are available from the corresponding author on request.