Significance
Mutations in inner kinetochore components induce centromere repositioning without alteration in the centromeric DNA sequence, revealing a feedback mechanism underlying the high epigenetic stability of the centromere. This also provides a desirable experimental system to explore the functional significance of centromere positioning in meiosis. We discovered that in a heterozygotic meiosis, a repositioned centromere generates a reproductive barrier, suggesting a functional role of evolutionary new centromeres in speciation; furthermore, in a homozygotic meiosis, chromosomes carrying repositioned centromeres frequently undergo the 2 stages of meiotic segregation in an inverted order, demonstrating high flexibility in the meiotic process.
Keywords: inner kinetochore, CENP-T-W-S-X complex, centromere repositioning, reproductive barrier, inverted meiosis
Abstract
The chromosomal position of each centromere is determined epigenetically and is highly stable, whereas incremental cases have supported the occurrence of centromere repositioning on an evolutionary time scale (evolutionary new centromeres, ENCs), which is thought to be important in speciation. The mechanisms underlying the high stability of centromeres and its functional significance largely remain an enigma. Here, in the fission yeast Schizosaccharomyces pombe, we identify a feedback mechanism: The kinetochore, whose assembly is guided by the centromere, in turn, enforces centromere stability. Upon going through meiosis, specific inner kinetochore mutations induce centromere repositioning—inactivation of the original centromere and formation of a new centromere elsewhere—in 1 of the 3 chromosomes at random. Repositioned centromeres reside asymmetrically in the pericentromeric regions and cells carrying them are competent in mitosis and homozygotic meiosis. However, when cells carrying a repositioned centromere are crossed with those carrying the original centromere, the progeny suffer severe lethality due to defects in meiotic chromosome segregation. Thus, repositioned centromeres constitute a reproductive barrier that could initiate genetic divergence between 2 populations with mismatched centromeres, documenting a functional role of ENCs in speciation. Surprisingly, homozygotic repositioned centromeres tend to undergo meiosis in an inverted order—that is, sister chromatids segregate first, and homologous chromosomes separate second—whereas the original centromeres on other chromosomes in the same cell undergo meiosis in the canonical order, revealing hidden flexibility in the perceived rigid process of meiosis.
Centromeres dictate the sites on chromosomes for kinetochore assembly and provide the foundation for spindle microtubule attachment to assure the faithful chromosome transmission during mitosis and meiosis (1, 2). For most eukaryotes, including humans and fission yeast, centromeres span a specific region—tens of kilobases in the fission yeast Schizosaccharomyces pombe, up to megabases in mammals and plants—of the chromatin, which are called monocentromeres. The position of the centromere on each chromosome is determined epigenetically by an evolutionarily conserved histone H3 variant, CENP-A (3). While the underlying centromeric DNA sequences are usually highly repetitive (called satellite or alphoid DNA), they diverge dramatically among species. Furthermore, the underlying DNA sequence does not determine the site of a centromere. The formation of a functional neocentromere, defined by a new centromere formed on an ectopic site other than that of the original centromere, is independent of the DNA sequence (4).
Centromere positioning on each chromosome is remarkably stable in all eukaryotes, although centromere repositioning due to neocentromere formation without alteration in the chromosomal marker order does occur sporadically in contemporary populations (e.g., 8 cases have been reported in humans) (5) and among related species (such as primates) on the evolutionary time scale (i.e., evolutionary new centromeres, ENCs) (6). It is postulated that a neocentromere may seed the formation of an ENC at a site devoid of satellite DNA, which is then matured through acquisition of repetitive DNA. ENCs and neocentromeres are considered as two sides of the same coin, manifestations of the same biological phenomenon at drastically different time scales and population sizes (7). Hence, understanding centromere repositioning may provide mechanistic insights into ENC emergence and progression.
CENP-A–containing chromatin directly recruits specific components of the kinetochore, called the constitutive centromere-associated network. The kinetochore is a proteinaceous machinery comprised of inner and outer parts, each compassing several subcomplexes. The inner kinetochore is proximal to or in direct contact with the CENP-A nucleosomes, linking the centromere to the outer kinetochore, which in turn physically binds the spindle microtubules (8). Mutations in the N-terminal tail of CENP-A reduce the centromeric localization of the inner kinetochore component CENP-T and cause centromere inactivation (9). Previous studies, including ours (10), have shown that kinetochore components may contribute to the stability of the centromeric chromatin organization pattern. Certain inner kinetochore components (e.g., Mis6 and Cnp3 in S. pombe; CENP-C and CENP-N in vertebrates) are required for maintaining proper levels of CENP-A nucleosomes in centromeres (11–14). Moreover, partial dysfunction of the kinetochore (e.g., mis6-302, mis12-537, and ams2∆) facilitates centromere inactivation and rescues the high rates of lethality caused by an engineered dicentric chromosome in fission yeast (15). In general, these data suggest functional cross-talk between the centromere and the kinetochore.
Centromere–kinetochore–microtubule attachment is crucial for accurate chromosome segregation in mitosis as well as meiosis. Meiosis occurs in eukaryotes that generate germ cells for sexual reproduction. Canonical meiosis involves 2 sequential nuclear divisions (meiosis I and meiosis II) following 1 round of DNA replication (16). The order of the segregation events is highly conserved. It is characterized by homologous chromosome pair separation during the first “reductional” division (meiosis I) followed by sister chromatids segregation during the second “equational” division (meiosis II). To accomplish faithful chromosome segregation in such a distinct manner, sister kinetochores must initially establish connection with only 1 pole of the spindle in meiosis I (called “mono-orientation”) so that they are cosegregated, whereas in meiosis II, they must establish biorientation with the 2 poles of the spindle in order to be segregated equally. In addition, crossover and sequential resolution of chromosome cohesion along chromosome arms in meiosis I and at centromeres in meiosis II are also necessary to fulfill the requirements of the specific order of meiotic nuclear divisions (17).
Interestingly, despite the common perception of the strict order of 2 nuclear divisions in canonical meiosis, inverted order of meiotic divisions (inverted meiosis) has been discovered in a few species with holocentromeres in which centromeres are distributed throughout the chromosomes, and is considered as one of several strategies specifically adopted by the holocentric organisms for meiosis (18–21). Surprisingly, a recent study in human female germline cells demonstrated that inverted meiosis also occurs in a monocentric organism (22). We here investigate in fission yeast how kinetochore mutations may affect centromere stability and report the functional consequences of repositioned centromeres in generating a reproductive barrier and inversion in the order of segregation events in meiosis.
Results
Inner Kinetochore Mutations Cause Various Levels of Pericentromeric Heterochromatin Spreading into Centromeric Core Regions.
Fission yeast haploid cells possess 3 chromosomes and exhibit a characteristic centromeric chromatin organization pattern (23). The central cores, consisting of mostly unique DNA sequences (cnt) and part of the innermost repeats (imr), are occupied by Cnp1/CENP-A nucleosomes interspersed with canonical H3 nucleosomes, whereas the flanking regions comprising repetitive DNA sequences (outermost repeats otr and part of imr) are packed into heterochromatin (24, 25), as marked by histone H3 lysine 9 methylation (H3K9me2) (26). The boundaries between the heterochromatin and the central cores are strictly delimited by tDNA elements (Fig. 1, diagrams) (27). We sought to investigate the mechanisms underlying the epigenetic stability of this centromeric chromatin organization.
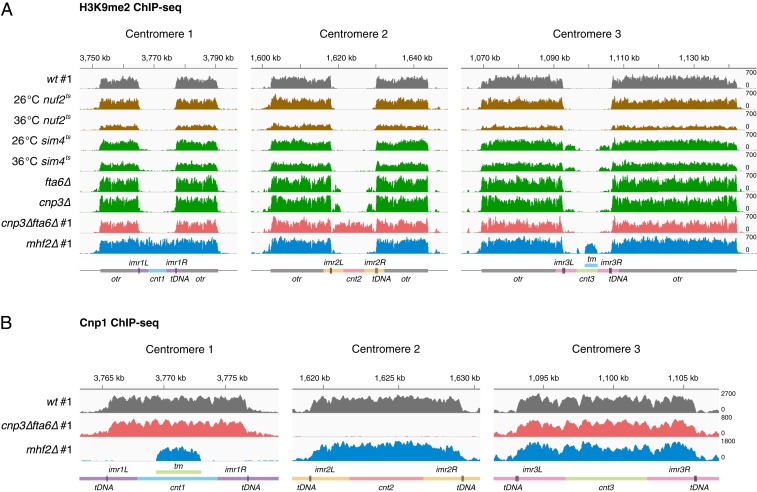
Fig. 1.
Inner kinetochore mutations cause various levels of pericentromeric heterochromatin spreading into centromeric core regions. (A) H3K9me2 ChIP-seq reads mapped to centromeric and pericentromeric regions of all 3 chromosomes in outer kinetochore mutants (brown), inner kinetochore mutants (green), mhf2∆ (blue), and cnp3∆fta6∆ (pink) compared to wild-type cells (gray). Strain names are as labeled. mhf2∆ (blue) and cnp3∆fta6∆ (pink) show complete occupancy of H3K9me2 on cnt1 and cnt2, respectively. Temperature sensitive (ts) strains were incubated at 26 °C (labeled as 26 °C), or incubated at 26 °C and shifted to 36 °C for 6 h (labeled as 36 °C). (B) Cnp1 ChIP-seq reads mapped to centromeric regions of all 3 chromosomes in mhf2∆ (blue) and cnp3∆fta6∆ (pink) compared to wild-type cells (gray). Tested strains were identical to that used in H3K9me2 ChIP-seq analysis as labeled in A. #1, Biological replicate 1. Diagrams illustrate the organization of centromeres 1, 2, and 3. tDNA, vertical lines; tm, segments with identical sequences in cnt1 and cnt3. The x axis, DNA coordinates on chromosomes 1, 2, and 3 according to reference genome (pombase.org); y axis, reads per million of ChIP-seq reads randomly assigned to the repetitive DNA sequences. The wild-type ChIP-seq raw data were previously published (10).
We hypothesized that the kinetochore may inversely affect the centromere. To test this, we systematically investigated the impact of kinetochore mutations on centromeric chromatin organization. Anti-H3K9me2 chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) were performed in mutants with either deletions of nonessential kinetochore component genes or conditional inactivation (temperature sensitive) mutations in the essential ones. These genes encode representative components of all inner and some outer kinetochore subcomplexes (8). Pericentromeric heterochromatin spreading into the core regions to various degrees was detected: Whereas the outer kinetochore mutants (nuf2-1 and mis12-537) showed no heterochromatin spreading, the inner kinetochore mutants (mis15-68, sim4-193, mal2-1, fta6∆, and cnp3∆) exhibited minor to major levels of heterochromatin encroaching into the core regions in 1 or 2 centromeres (Fig. 1A and SI Appendix, Fig. S1A). Noticeably, temperature-sensitive strains, for example sim4-193, displayed a similar heterochromatin spreading at the nonpermissive and permissive temperatures, indicating that centromeric chromatin organization is already impaired in cells under the viable condition. mhf2∆ (mammalian CENP-X homolog) exhibited complete heterochromatin occupancy in one centromere (cen1) but normal pericentromeric distribution in the other two (cen2 and cen3) (Fig. 1A and SI Appendix, Fig. S1A). The island-shape ChIP-seq signals detected on cnt3 in mhf2∆ are most likely caused by an artifact in informatics data processing as the border of the small island aligns precisely with the identical sequence in cnt1 and cnt3, which is biochemically highly unlikely (also see below). In a double-mutant cnp3∆fta6∆ strain, derived from genetic crossings, we found that cen2 was completely covered by heterochromatin (Fig. 1A and SI Appendix, Fig. S1A), suggesting that perturbations to centromeric chromatin by cnp3∆ and fta6∆ cumulatively led to centromere inactivation.
Anti-Cnp1 ChIP-seq detected no significant Cnp1 signal at centromeric cores occupied by heterochromatin but prominent levels of Cnp1 at the other two in both mhf2∆ and cnp3∆fta6∆ (Fig. 1B), confirming that only the original cen1 or cen2 was inactivated (designated as cen1inactive and cen2inactive hereafter) in these 2 strains, respectively. Due to the possible informatics artifact of the “island-shape” signals in the inactivated cen1 in mhf2∆, we cannot formally exclude the possibility that some Cnp1 persists there. However, the complete removal of Cnp1 occupancy and the fully coverage of H3K9me2 on the inactivated cen2 in cnp3∆fta6∆ are in favor of the scenario that the inactivated centromeres do not contain Cnp1. Taken together, these results demonstrate that the integrity of the inner kinetochore is required to maintain normal centromeric chromatin organization as well as distinct centromere identity. The effects appear specific to these mutants as we also examined mutants of genes encoding other centromere-interacting proteins known to affect centromeric Cnp1 incorporation, but detected no noticeable (mis16-53, mis18-262) or only minor (ams2∆ and sim3∆) heterochromatin spreading (SI Appendix, Fig. S1B) (28–30).
Single Depletion of CENP-T-W-S-X Components Induces Centromere Inactivation.
CENP-T-W-S-X is a conserved inner kinetochore complex in which each subunit contains a histone-fold domain that binds directly to DNA (31). To further explore the role of the CENP-T-W-S-X complex in maintaining centromere identity, we generated heterozygous deletion diploid strains de novo for the 3 nonessential components: wip1/CENP-W, mhf1/CENP-S, and mhf2/CENP-X (cnp20/CENP-T is essential for cell viability and is not included in this study) (12). Tetrad analysis of the meiotic progeny of each strain showed that most of the asci contained only two or fewer viable spores, demonstrating a significant reduction in meiotic progeny viability. However, the lethality was similar between the wild-type and mutant haploid progeny (SI Appendix, Table S1). Furthermore, among the surviving progeny, anti-Cnp1 ChIP-seq detected random inactivation in only 1 of the 3 centromeres in each of the 10 tested wip1∆, mhf1∆, or mhf2∆ haploid strains (Fig. 2 A and C and SI Appendix, Fig. S3A and Table S2), whereas the wild-type progeny from the same asci exhibited no heterochromatin occupancy in the centromeric cores by anti-H3K9me2 ChIP-seq but Cnp1 spreading into the pericentromeric regions by anti-Cnp1 ChIP-seq (Fig. 2B and SI Appendix, Fig. S3 B and C). Hence, centromere inactivation is tightly linked with the gene deletions. Centromere inactivation was not observed in the parental heterozygous deletion diploid cells (Fig. 2B), excluding the possibility that centromere inactivation occurred premeiotically. No haploid deletion strain was found carrying more than 1 inactivated centromere. We speculate that simultaneous inactivation of 2 or 3 centromeres may be incompatible with cell survival. In a few (6.1%) mhf2∆/+ asci containing 4 viable spores, the mhf2∆ progeny grew slower than the wild-type, formed minicolonies but frequently gained a growth advantage after restreaking several times (SI Appendix, Fig. S2). Among them, 2 mhf2∆ progeny carrying different inactivated centromeres (cen2inactive and cen3inactive, respectively) were recovered from the same ascus, further supporting the notion that centromere inactivation occurred postzygotically and independently in each progeny (Fig. 2C).
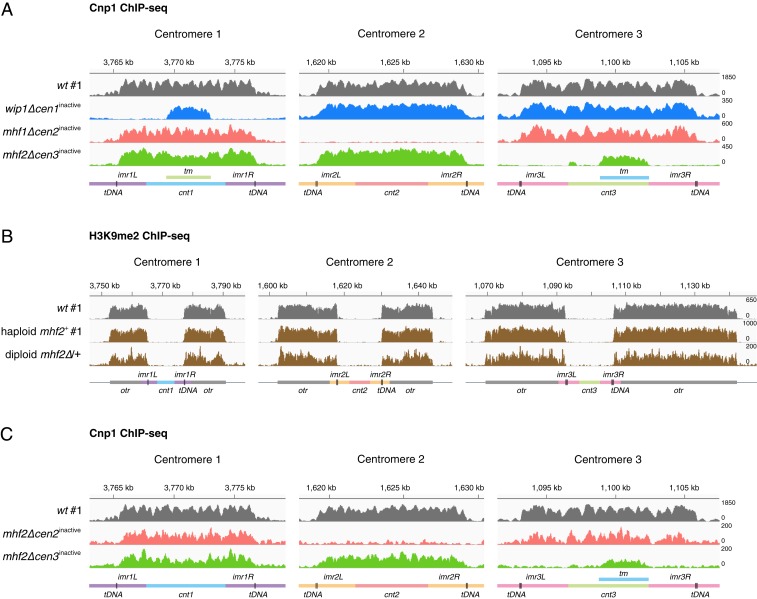
Fig. 2.
Single depletion of CENP-T-W-S-X components induces centromere inactivation. (A) Cnp1 ChIP-seq reads mapped to centromeric regions of all 3 chromosomes in randomly chosen wip1∆, mhf1∆, and mhf2∆ strains (cen1inactive blue, cen2inactive pink, cen3inactive green) compared to wild-type strain (cen1/2/3active gray). (B) H3K9me2 ChIP-seq reads mapped to centromeric and pericentromeric regions of all 3 chromosomes in meiotic haploid progeny mhf2+ and heterozygous deletion diploid mhf2∆/+ (brown) compared to wild-type cells (gray). (C) Cnp1 ChIP-seq reads mapped to centromeric regions of all 3 chromosomes in 2 mhf2∆ meiotic haploid progeny from the same tetrad (cen2inactive pink, cen3inactive green). #1, Biological replicate 1. Diagrams, x axis and y axis, same as in Fig. 1.
Neocentromeres Are Formed Preferentially in the Pericentromeric Regions.
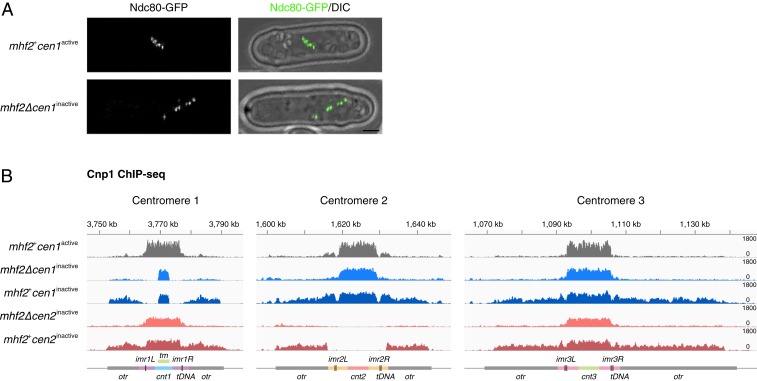
A previous study in fission yeast has shown that with complete excision of cen1 DNA by genome editing, a few cells (less than 0.1%) survive by either forming a neocentromere at a new location or fusing the acentric chromosome to another chromosome (32). To determine whether the surviving mhf2∆ cells acquired neocentromeres, we microscopically examined mhf2∆cen1inacitve cells expressing a green fluorescent protein-tagged outer kinetochore protein Ndc80-GFP for the presence of a complete set of kinetochores (3 pairs of sister kinetochores). Six discrete dots were resolved in a few M phase cells with sufficiently scattered kinetochores (Fig. 3A), suggesting that a functional kinetochore (and thereby, a neocentromere) was formed on chromosome 1 carrying cen1inacitve.
Fig. 3.
Neocentromeres are formed preferentially in the pericentromeric regions. (A) Six dots of outer kinetochore Ndc80-GFP observed in mhf2+cen1active (Upper) and mhf2∆cen1inactive (Lower) M phase cells treated with the thiabendazole (TBZ, 20 μg/mL). (Scale bar, 2 μm.) (B) Cnp1 ChIP-seq reads mapped to centromeric and pericentromeric regions of all 3 chromosomes in mhf2∆cen1inactive (blue) and mhf2+cen1inactive (dark blue); mhf2∆ cen2inactive (pink) and mhf2+cen2inactive (dark pink) compared to wild-type cells (gray). Diagrams, x axis and y axis, same as in Fig. 1.
To determine the locations of neocentromeres, anti-Cnp1 ChIP-seq was performed in mhf2∆cen1inactive and mhf2∆cen2inactive. While prominent levels of Cnp1 were present in the other 2 active and original centromeres, modest but clearly detectable Cnp1 appeared in the pericentromeric regions of the inactivated centromeres (Fig. 3B and SI Appendix, Fig. S4A). These neocentromeres are likely to be functional, considering that kinetochores were assembled successfully on all chromosomes (Fig. 3A). By crossing mhf2∆cen1inactive to wild-type (mhf2+cen1active), we recovered mhf2+cen1inactive exhibiting wild-type growth among the progeny (Table 1 and SI Appendix, Fig. S4B). This suggests that the inactivated state of the original centromere (and presumably the accompanying neocentromere) appear mitotically stable (at least within tens of cell growth generations) in the absence of the genetic lesion that induced it. In mhf2+cen1inactive and mhf2+cen2inactive, significant Cnp1 signals were detected in the pericentromeric regions of the inactivated centromeres by anti-Cnp1 ChIP-Seq (Fig. 3B and SI Appendix, Fig. S4C). Together with the low Cnp1 signal in mhf2∆cen1inactive and mhf2∆cen2inactive, these results are consistent with the possibility that either Cnp1 incorporation at the neocentromeres is low in mhf2∆ or the positions of Cnp1 nucleosomes might be divergent among individual mhf2∆ cells within a population. Three individual Ndc80-GFP dots (representing 3 pairs of sister kinetochores) in early mitosis were visualized in mhf2+cen1inactive after we introduced a conditional β-tubulin mutation (nda3-KM311) to allow the separation of the clustered centromeres at the restrictive temperature of nda3-KM311, further confirming the formation of a neocentromere (SI Appendix, Fig. S4D). Together, these results demonstrate that pericentromeric heterochromatin is the preferable site for neocentromere formation.
Table 1.
Incompatibility between neocentromeres and original centromeres causes a meiosis barrier
| Cross | Spore viability, % | Dissected asci (n) | Asci with 4 viable spores, % | Asci with 1 or no viable spores, % | Meiosis barrier |
| mhf2+cen1active × mhf2+cen1active | 93.1 | 130 | 80 | 1.54 | × |
| mhf2+cen1active × mhf2∆cen1inactive | 36.5 | 254 | 5.9 | 53.1 | √ |
| mhf2+cen1active × mhf2+cen1inactive | 25.2 | 272 | 4.04 | 69.5 | √ |
| mhf2+cen1inactive × mhf2+cen1inactive #1 | 89.8 | 108 | 68.5 | 1.9 | × |
| mhf2+cen1inactive × mhf2+cen1inactive #2 | 31.4 | 106 | 9.4 | 62.3 | √ |
| mhf2+cen1inactive × mhf2+cen2inactive | 19.8 | 377 | 0.8 | 81.2 | √ |
Cells with mismatched (mhf2+cen1active × mhf2∆cen1inactive, mhf2+cen1active × mhf2+cen1inactive, mhf2+cen1inactive × mhf2+cen1inactive #2, mhf2+cen1inactive × mhf2+cen2inactive) or matched (mhf2+cen1active × mhf2+cen1active, mhf2+cen1inactive × mhf2+cen1inactive #1) centromeres were crossed and subjected to tetrad dissection. Intact asci with 4 spores were dissected microscopically and scored for the number of viable spores. Spore viability is calculated as the ratio of the number of viable spores to the number of analyzed spores; >50% reduction in spore viability is defined as meiosis barrier and labeled as √, whereas no meiosis barrier is labeled as ×.
Neocentromeres Occupy the Pericentromeric Repetitive Sequences Asymmetrically.
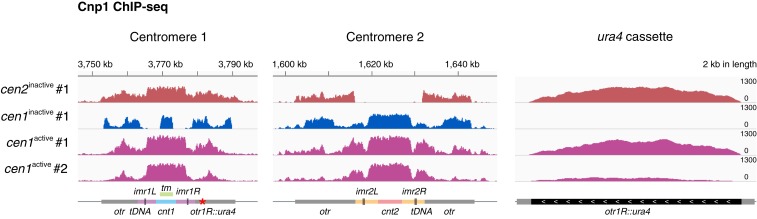
Cnp1 spreading into the pericentromeric regions, as annotated bioinformatically, appears to be ubiquitous for all centromeres (see, for example, Fig. 5A and SI Appendix, Figs. S3C, S7C, and S8C). Furthermore, Cnp1 occupancy in the neocentromeres appears symmetrical on the pericentromeric repeats at both left and right sides of cnt (Fig. 3B and SI Appendix, Figs. S7C and S8 B and C; see also Fig. 5). However, due to high DNA sequence similarity between the pericentromeric repeats in all 3 centromeres (33), it is unclear whether such Cnp1 spreading and occupancy represents the physical footprint of Cnp1. To this end, by genetic crossing, we generated new strains carrying a neocentromere and a reporter gene ura4 inserted into the right side of the repetitive regions of centromere 1 (otr1R::ura4) (see SI Appendix, Materials and Methods for details of genetic crosses). In strains carrying cen2inactive, Cnp1 signals were detected on otr1R::ura4 (Fig. 4 and SI Appendix, Fig. S5B), indicating Cnp1 occupancy at the otr repeats is not limited to the repositioned centromere, but rather is ubiquitous for all 3 centromeres. It is consistent with the observation that Cnp1 spreads onto all of the imr regions unique to each centromere. On the other hand, in 7 independent strains carrying cen1inactive, cells showed no significant Cnp1 incorporation into ura4 (Fig. 4 and SI Appendix, Fig. S5A). This excludes the possibility that Cnp1 occupancy on the repositioned centromere is symmetrical. Supporting this notion, in different strains exhibiting centromeric Cnp1 spreading, significant Cnp1 signals were detected on ura4 in some strains but not in others (Fig. 4 and SI Appendix, Fig. S5). Consistently, we also found that the levels of heterochromatin (H3K9me2) and Cnp1 occupancy are inversely correlated on the ura4 cassette (SI Appendix, Fig. S5B).
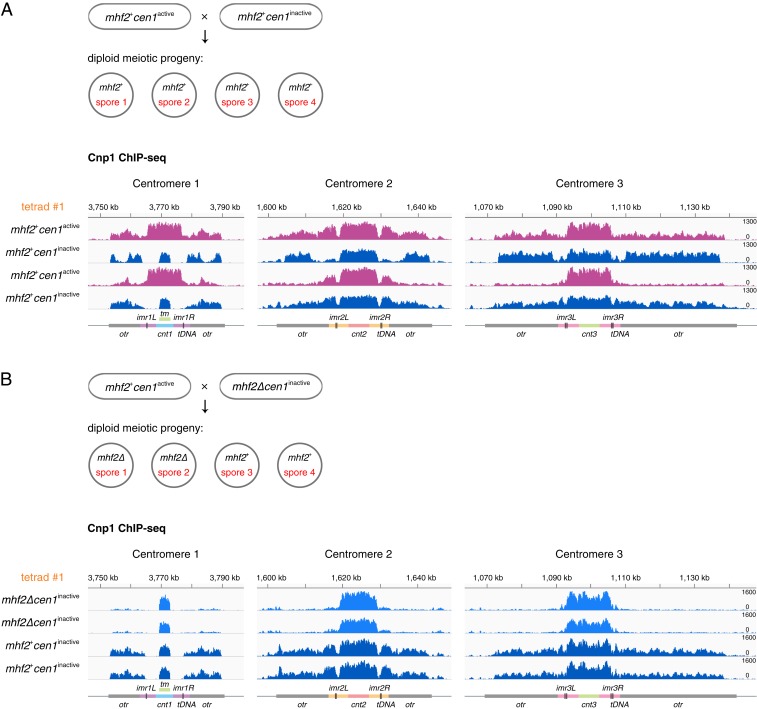
Fig. 5.
The endogenous centromere in mhf2+ cells tends to be converted to an inactivated centromere by mhf2∆ through meiosis. (A) Schematic illustrates the meiotic progeny of mhf2+cen1active × mhf2+cen1inactive in asci with 4 spores. Cnp1 ChIP-seq reads mapped to centromeric and pericentromeric regions of all 3 chromosomes in 4 viable meiotic progeny from the same ascus (tetrad #1) of mhf2+cen1active × mhf2+cen1inactive. mhf2+cen1inactive (dark blue) conformed to Mendelian inheritance (2: 2 segregation pattern). mhf2+cen1active (magenta) exhibits the spreading of Cnp1 into the pericentromeric regions. See SI Appendix, Fig. S7B for accompanying H3K9me2 ChIP-seq data. (B) Schematic illustrates the meiotic progeny of mhf2+cen1active × mhf2∆cen1inactive in asci with 4 spores. Same procedure as in A for analyzing mhf2+cen1active × mhf2∆cen1inactive. mhf2∆ conformed to Mendelian inheritance (2: 2 segregation pattern). cen1inactive was detected in mhf2+ (dark blue) and mhf2∆ (blue). See SI Appendix, Fig. S8B for accompanying H3K9me2 ChIP-seq data. Diagrams, x axis and y axis, same as in Fig. 1.
Fig. 4.
Neocentromeres occupy the pericentromeric repetitive sequences asymmetrically. Cnp1 ChIP-seq reads mapped to centromeric and pericentromeric regions of all 3 chromosomes in cen2inactive (dark pink), cen1inactive (dark blue), and cen1active with Cnp1 spreading (magenta). ura4 cassette was inserted into the right side of the otr1R (otr1R::ura4) and labeled by a red asterisk. Chromosomes 1 and 2 and ura4 cassette are shown. #1 and #2, Biological replicate 1 and 2. Diagrams, x axis and y axis, same as in Fig. 1.
Heterochromatin occupies the site of the inactivated centromere, and neocentromeres are formed at the pericentromeric regions that were originally occupied by heterochromatin. These findings prompted us to further investigate whether heterochromatin plays a role in centromere inactivation and neocentromere maintenance. To this end, we explored the possible impact of deletion of clr4, which encodes the only heterochromatin modification enzyme (H3K9 methyltransferase) for heterochromatin assembly in S. pombe (26). We deleted clr4 in mhf2∆cen2inactive (SI Appendix, Fig. S6B) or mhf2+cen1inactive haploid cells (SI Appendix, Fig. S6C) by DNA transformation and found that the inactivated centromeres and the neocentromeres were maintained. Together, these results suggest that mitotic maintenance of neocentromere does not require heterochromatin.
The Endogenous Centromere Tends to Be Converted Unilaterally to the Neocentromere in a Genetic Crossing Between Wild-Type and mhf2∆.
In genetic crosses between strains with mismatched centromeres (one carrying a neocentromere and the other an original centromere), a few asci produced 4 viable progeny, allowing reliable analysis of the inheritance of genetic lesions and epigenetic features (Table 1 and SI Appendix, Table S3). As expected, mhf2∆ conformed to Mendelian inheritance. Similarly, in mhf2+cen1inactive × mhf2+cen1active, cen1inactive and the associated neocentromere also conformed to Mendelian inheritance, suggesting that they are meiotically stable (Fig. 5A and SI Appendix, Fig. S7). Furthermore, the progeny without cen1inactive exhibited the spreading of centromeric Cnp1 into the pericentromeric regions in all centromeres, indicating a broad impact on centromeric chromatin due to cen1inactive through meiosis (Fig. 5A and SI Appendix, Fig. S7C). Cnp1 spreading in all original centromeres was also found in the wild-type meiotic progeny of wip1∆/+, mhf1∆/+, and mhf2∆/+ (SI Appendix, Fig. S3C). This broad alteration in centromeres might underscore the poor spore viability overall (SI Appendix, Table S1). In mhf2∆cen1inactive × mhf2+cen1active, 2 independent asci (each with 4 viable progeny) were examined and, surprisingly, all of the progeny carry the cen1inactive regardless of whether mhf2+ or mhf2∆ was in the haploid genome (Fig. 5B and SI Appendix, Fig. S8B). In addition, in 9 other mhf2+ progeny (derived from random spores) that were subjected to ChIP-seq analysis, 2 also exhibited cen1inactive, whereas 7 displayed Cnp1 spreading in all 3 centromeres (SI Appendix, Fig. S8C). Thus, cen1active had a propensity to be converted into cen1inactive in meiosis involving mhf2∆, most likely using cen1inactive on the homologous chromosome as the template. Although the mechanism remains unclear, this centromere conversion phenomenon highlights the pivotal role of mhf2 in maintaining centromere identity in wild-type cells and a plausible way of propagating the neocentromere in a cell population of mixed karyotypes.
Incompatibility Between Neocentromeres and Endogenous Centromeres Causes a Meiosis Barrier.
When crossed with wild-type (cen1/2/3active), mhf2∆cen1inactive showed high lethality of meiotic progeny. We also examined genetic crosses between wild-type (cen1/2/3active) and mhf2+cen1inactive, and found that mhf2+cen1inactive causes a more severe reduction in progeny viability than mhf2∆cen1inactive (Table 1). Strikingly, however, homozygotic meiosis (here, crossing between sister cells derived from the same ascus carrying the same neocentromere, mhf2+cen1inactive × mhf2+cen1inactive #1) exhibited near or at wild-type levels of spore viability (Table 1). These results demonstrate that cen1inactive is competent for meiosis but a mismatch in centromeres between a pair of homologous chromosomes generates a reproductive barrier.
To test this further, we performed a series of genetic crosses among strains with different (mhf2+cen1inactive × mhf2+cen2inactive, mhf2+cen1inactive × mhf2∆cen2inactive and mhf2∆cen1inactive × mhf2∆cen2inactive) or the same (mhf2+cen1inactive × mhf2∆cen1inactive) centromeres and determined spore viability (Table 1 and SI Appendix, Tables S3 and S4). Together, the results demonstrate that a mismatch between a neocentromere and the original centromere on any 1 chromosome alone causes poor spore viability, and that the spore viability is further reduced as the number of mismatched centromeres increased. We also found crosses between certain strains carrying the cen1inactive (each with a neocentromere somewhere in the pericentromeric regions of chromosome 1; i.e., mhf2+cen1inactive × mhf2+cen1inactive #2) showed significant loss of spore viability (Table 1). This is consistent with the notion that the locations of neocentromeres are asymmetric relative to the centromeric cores and may be different from each other in these 2 strains.
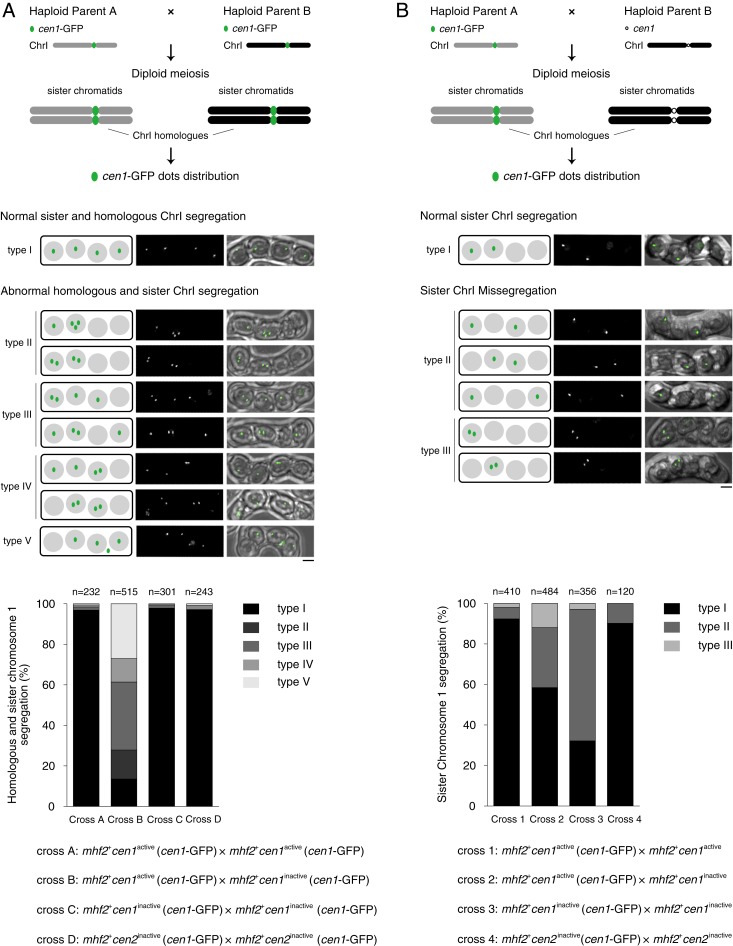
To obtain cytological evidence for the meiosis barrier caused by mismatched centromeres, we inserted a GFP tag at the lys1 locus close to cen1 using the lacOs/lacI-GFP system (designated as cen1-GFP hereafter) to microscopically track chromosome 1 segregation during meiosis. In this system, when chromosome 1 of both parental haploid cells are labeled with cen1-GFP, the 4-dot distribution pattern among the meiotic progeny indicates the segregation of sister and homologous chromosome 1 (Fig. 6A). When only 1 parental haploid cell carries the cen1-GFP, the 2-dot distribution pattern indicates the segregation of the tagged sister chromosome 1 (Fig. 6B). We identified and categorized the abnormal chromosome segregation patterns into different types. For example, type II and type III of 2-dot distribution suggest premature segregation of sister chromatids in meiosis I and missegregation of sister chromatids in meiosis II, respectively (Fig. 6B). As expected, we found severe sister and homologous chromosome segregation defects in both meiosis I and II in zygotic meiosis with mismatched centromere 1 (cross B in Fig. 6A and cross E in SI Appendix, Fig. S9A; cross 2 in Fig. 6B and cross 5 in SI Appendix, Fig. S9B). We also noticed that the meiotic defects were not confined to the mismatched centromeres; when mhf2+cen2active cells carrying the cen1-GFP were crossed to mhf2∆cen2inactive cells with a mismatched cen2, chromosome 1 also exhibited segregation defects (cross 6 in SI Appendix, Fig. S9B), but not as severe as that in mismatched centromeres (cross 2 in Fig. 6B and cross 5 in SI Appendix, Fig. S9B). Overall, these results demonstrate that mismatched centromeres between homologous chromosome pairs cause hybrid infertility due to severe meiotic defects, and thus constitute a meiosis barrier between the 2 strains.
Fig. 6.
Homozygotic meiosis with matched neocentromeres exhibits premature segregation of sister chromatids. (A) Schematic illustrates the meiotic segregation of chromosome 1 (ChrI) in the genetic cross between haploid parent A (gray) and haploid parent B (black), both cells carrying the chromosome 1 marker—lys1 locus decorated with GFP (lys1::lacOs/lacI-GFP)—designated as cen1-GFP (green dot). The distributions of cen1-GFP are categorized into 5 types (type I to type V). (Scale bar, 2 μm.) Cross A-B, mhf2+cen1active cells crossed to mhf2+cen1active and mhf2+cen1inactive cells, respectively; cross C, mhf2+cen1inactive cells crossed to mhf2+cen1inactive cells; cross D, mhf2+cen2inactive cells crossed to mhf2+cen2inactive cells. The cen1-GFP dots were scored in 4-spored asci to trace the meiotic segregation of homologous chromosome 1 and sister chromosome 1. n, the total 4-spored asci analyzed. (B) Schematic illustrates the meiotic segregation of chromosome 1 (ChrI) in the genetic cross between haploid parent A (gray) carrying the cen1-GFP and haploid parent B (black). Black circle, centromere 1 without GFP labeling (cen1). The distributions of cen1-GFP are categorized into 3 types (type I to type III). (Scale bar, 2 μm.) Crosses 1 and 2, mhf2+cen1active cells crossed to mhf2+cen1active and mhf2+cen1inactive cells, respectively; cross 3, mhf2+cen1inactive cells crossed to mhf2+cen1inactive cells; cross 4, mhf2+cen2inactive cells crossed to mhf2+cen2inactive cells. The cen1-GFP dots were scored in 4-spored asci to trace the meiotic segregation of sister chromosome 1. n, the total 4-spored asci analyzed.
Homozygotic Repositioned Centromeres Frequently Undergo Meiotic Segregation Events in an Inverted Order.
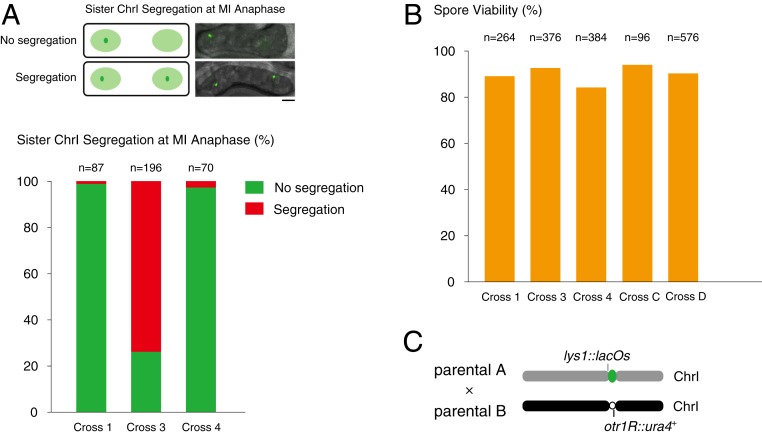
In homozygotic meiosis with the same neocentromere 1 or 2, 4 copies of chromosome 1 (visualized with cen1-GFP) were evenly segregated to the 4 spores in most asci (crosses C and D in Fig. 6A; crosses F and G in SI Appendix, Fig. S9A). Surprisingly, in homozygotic meiosis with the same neocentromere 1 but where only one was labeled with cen1-GFP, 73.5% of the zygotes at anaphase of meiosis I exhibited premature separation of sister cen1-GFP dots (cross 3 in Fig. 7A). Consistently, in 65 to 75% of the asci, 2 cen1-GFP dots no longer occupied sister-spore positions (cross 3 in Fig. 6B; crosses 7 and 8 in SI Appendix, Fig. S9B). Nonetheless, these crosses displayed wild-type levels of spore viability (Fig. 7B and SI Appendix, Fig. S9C), indicating eventual success in accurate meiotic chromosome segregation. This is in agreement with the results (Table 1) that spore viability of genetic crosses between cells carrying the same neocentromeres was comparable to the wild-type level (SI Appendix, Table S3). On the other hand, in a comparable genetic cross in which both strains carried cen2inactive but only 1 parental haploid was labeled with cen1-GFP, the majority of the labeled sister chromosome 1 segregation occurred in meiosis II and strictly followed the canonical order of meiosis I and meiosis II (cross 4 in Figs. 6B and 7A).
Fig. 7.
Homozygotic repositioned centromeres undergo meiotic segregation events in an inverted order and exhibit wild-type level spore viability. (A) Schematic illustrates the segregation pattern of sister chromosome 1 at anaphase during meiosis I (MI anaphase). (Scale bar, 2 μm.) The graph plots the percentage of MI anaphase cells displaying cosegregation (green) or presegregation (red) of sister chromosome 1 in homozygotic meiosis. n, The total counted MI anaphase cells. (B) Intact asci with 4 spores from the above genetic crosses were dissected microscopically and scored for the number of viable spores. Spore viability was calculated as the ratio of the number of viable spores to the number of analyzed spores. n, The total spores analyzed. (C) Schematic illustrates the assay calculating the crossover rates of regions nearby the centromere between parental A (lys1::lacOs/lacI-GFP) and parental B (otr1R::ura4).
To rule out the possibility that the observed early segregation of sister chromosome 1 may be due to highly frequent crossing-over between cen1 and the cen1-GFP tag (∼10 kb to the left edge of cen1), we tested the genetic linkage between otr1R::ura4 and cen1-GFP markers in these genetic crosses (Fig. 7C). We found 1.42% (n = 352) crossing over between these 2 loci in homozygotic repositioned centromere 1, comparable to that of the homozygotic original centromere 1 (1.47%, n = 272), ruling out the possibility of a recombination hotspot between the repositioned centromere 1 and the cen1-GFP tag.
Collectively, these results demonstrate that chromosomes carrying neocentromeres frequently invert the order of meiotic chromosome segregation events (i.e., sister chromatids segregate first and homologous chromosomes separate second) and that inverted meiosis is restricted to neocentromeres. Furthermore, the successful completion of meiosis and high progeny viability suggest that canonical and inverted meiosis on different chromosomes occur concomitantly in the same cell and thus, must be mechanistically compatible with each other.
Discussion
Centromere Repositioning Induced by the Inner Kinetochore Impairment.
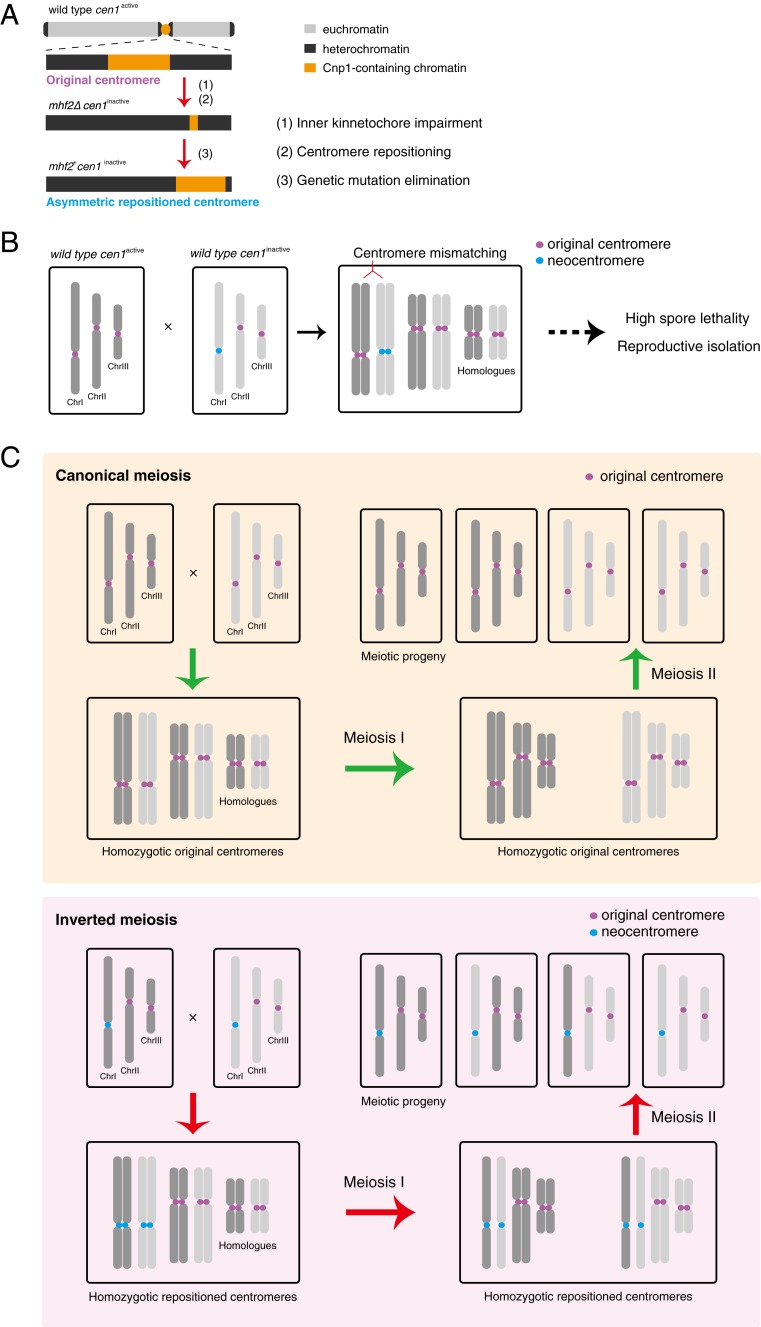
Our study shows that genetic abrogation of the inner kinetochore (such as subunits of the CENP-T-W-S-X complex) in fission yeast readily initiates centromere repositioning but is dispensable for the mitotic maintenance of neocentromeres (Fig. 8A). Together, these results reveal a fundamental and evolutionarily conserved role of the kinetochore in maintaining centromere epigenetic identity, and suggest that inducing neocentromere formation without incurring centromeric DNA changes or chromosomal rearrangements could be rapid and efficient in contrast to prior speculation that centromere repositioning might be a gradual, long evolutionary process (1, 34, 35).
Fig. 8.
Model diagram of centromere repositioning. (A) Centromere inactivation is induced by impairment of the inner kinetochore, with subsequent asymmetric neocentromere formation preferentially at the pericentromeric regions. (B) Centromere mismatching between an original centromere (cen1active) and a neocentromere (cen1inactive) generates a reproductive barrier by causing severe spore lethality. (C) Canonical meiosis with homozygotic original centromeres (Upper) and inverted meiosis with homozygotic neocentromeres (Lower) both generate four viable meiotic progeny. In canonical meiosis, sister chromatids are cosegregated in first meiotic division (meiosis I) and are separated in the second meiotic division (meiosis II). In inverted meiosis, sister chromatids are disassociated from each other during meiosis I and the homologs are segregated during meiosis II. For simplicity, other features of meiosis including crossover, recombination between homologs and random segregation of homologs are omitted in this diagram.
A recent study reported that in cultured chicken DT40 cells, knocking out nonessential constitutive kinetochore components, including CENP-S, enhances centromere drift within a defined small region on the chromosome upon prolonged cell proliferation, consistent with the role of the inner kinetochore CENP-T-W-S-X complex in stabilizing centromeres (36). However, in chicken cells, centromere drifting reflects enhanced dynamicity of centromeres between mitotic cell generations, whereas in fission yeast cells, once the centromere moves to a new location, it is stable mitotically and meiotically. Thus, in these 2 systems, CENP-T-W-S-X depletion may disrupt different aspects of centromere epigenetic stability.
In addition to the perturbation of inner kinetochore components, meiosis seems to actively facilitate the processes of centromere repositioning as its efficiency in the mhf2∆ progeny of heterozygous diploid (mhf2∆/+) meiosis or genetic crossings is 100% (15 independent strains tested), in contrast to that induced by the removal of whole cen1 DNA (about 0.06%) (32). We speculate that an unidentified step of meiosis may trigger centromeric chromatin reprogramming during neocentromere formation. We were unable to delete mhf2 directly in haploid cells and thus cannot determine whether it is possible to induce centromere repositioning in mitotic cells. However, its frequent occurrence through meiosis suggests an important role of meiotic processes in inducing the repositioned centromere. We also find that mhf2∆ tends to convert the original centromere in mhf2+ into the inactivated state, which should accelerate the propagation of neocentromeres in the population through meiosis.
Together, these results suggest that centromere repositioning should be a relatively prevalent phenomenon, yet few neocentromeres have been detected so far. One possible explanation is the experimental limitations on their detection: The karyotype of most organisms has traditionally been determined by cytogenetic analysis usually in only a few individuals except in clinical human samples; and small changes in centromere position may evade detection by classic cytogenetic techniques because of low resolution. In addition, neocentromeres and the mutations that induce their formation may cause detrimental effects on cell fitness resulting in their underrepresentation or elimination.
The Properties of Neocentromeres.
Neocentromeres have a propensity to locate on either side of the original inactivated centromere. This is in contrast to the scenario in which a neocentromere was formed near the subtelomeric heterochromatin regions upon complete removal of cen DNA, including the pericentromeric repeats in fission yeast (32), suggesting that pericentromeric regions are preferable to subtelomeric regions for neocentromere formation. In wild-type cells, a trace amount of Cnp1 was captured on the pericentromeric regions compared with the centromeric core regions (10, 37). Hence, it is likely that residual Cnp1 seeds the formation of neocentromeres and explains the preference for pericentromeric regions. Consistently, neocentromere formation at pericentromeric regions has been found in other organisms with complete removal of cen DNA (38–40).
Our data demonstrate that Cnp1 and H3K9me2 are mutually exclusive (SI Appendix, Fig. S5B), and that heterochromatin is not required for mitotic maintenance of the repositioned centromeres. Heterochromatin spreading is likely consequential to centromere perturbation and inactivation, although the possibility that heterochromatin to some extent contributes to neocentromere formation cannot be completely ruled out. It is also possible that other molecular features or processes in the pericentromeric region such as noncoding RNA transcription or small interfering RNA processing may favor neocentromere formation. Recently, 3D genomic architecture analysis suggested that neocentromeres physically interact with distant heterochromatin domains (41). Combining these observations, we propose that molecular processes or properties other than histone H3K9 methylation of the heterochromatin domain per se might play a pivotal role in neocentromere formation.
A Mechanism for Reproductive Barrier: Heterozygotic Repositioned Centromeres.
One important functional consequence of centromere repositioning in fission yeast is that mismatching between the original centromere and the neocentromere causes a reduction in the efficiency of meiosis (Fig. 8B). We notice that severe meiotic chromosome segregation defects are not limited to the mismatched centromeres, but are also detected (albeit to a lesser extent) in other centromere pairs, suggesting that the impact on meiosis is global (Fig. 6B and SI Appendix, Fig. S9B). The specific causes of these meiotic defects remains unclear and may be due to disruption of processes related to homologous centromeres, such as centromere pairing in early meiosis, or recombination repression at centromeres (42). We speculate that the potential conflict in executing different modes of meiosis (i.e., the canonical and the inverted meiosis) (Fig. 8C) for the same pair of homologous chromosomes could be a major cause for this reproductive barrier.
To our knowledge, no previous studies have described the specific functional impact or potential meiotic complications caused solely by centromere repositioning. On the other hand, ample evidence of ENCs strongly implies that neocentromeres may play an important role in speciation (5). However, direct experimental evidence is unavailable because established cases of heterogeneity in centromere positioning are lacking (a notable exception is orangutans, whose centromere of chromosome 12 exhibits alternative positions in about 20% of the population, but unfortunately, are not readily amendable to experimentation) (43, 44). By showing that heterozygous but not homozygous meiosis is defective, our results provide the experimental support that neocentromeres seen as ENCs represent an initiation step for genetic divergence during speciation (44–46). As a mechanism of establishing a meiosis barrier (Fig. 8B), the efficiency of mismatched centromeres is comparable to that of other known mechanisms in fission yeast including chromosomal rearrangements (47) and spore killer genes (48, 49).
Inverted Meiosis in Monocentric Organisms.
Unexpectedly, centromere repositioning frequently causes inversion of the order of 2 nuclear divisions in a homozygotic meiosis (Fig. 8C). Inverted meiosis has been observed in multiple species, all of which have holocentromeres. The canonical meiosis program requires sequential resolution of cohesion on chromosome arms and centromeric cohesion, which is incompatible with holocentromeres. Organisms with holocentromeres bypass this obstacle using various strategies, one of which being inverted meiosis (18–21). However, a recent study in human female germline, by tracking the distribution of homolog-specific genetic markers (SNPs) in mature oocytes and polar bodies during meiosis in vitro using single-cell whole-genome sequencing, has found that reversed segregation/inverted meiosis can also occur in a monocentric organism (22). Here, we present cytological evidence in fission yeast, a traditional model organism for studying canonical meiosis, that relocating a monocentromere to a nearby position frequently induces inverted meiosis, lending strong support to the notion that inverted meiosis might not be an exception but rather, may occur commonly in many organisms, and can be revealed by appropriate experimental approaches (for example, the human female meiosis study) or under suitable conditions, such as centromere repositioning in fission yeast.
Inversion in the order of meiotic chromosome segregation events imposes major mechanistic challenges. To segregate sister chromatids equally in meiosis I, sister kinetochores must now establish bipolar (biorientation) instead of monopolar attachment to the spindle (co-orientation) in canonical meiosis; and cohesion must be resolved completely along the whole chromosome, including the centromeres. We speculate that centromere repositioning somehow alters the local chromatin organization, rendering sufficient flexibility to sister kinetochore geometry that is compatible with both the biorientation required for sister chromatids separation and the co-orientation necessary for sister chromatids cosegregation. On the other hand, to ensure homologous chromosome disjunction in meiosis II in inverted meiosis, linkage between the homologs should be in place prior to their segregation. Further studies are needed to validate the existence and the molecular nature of such linkage, and to explore whether the established, recombination-dependent mechanisms or new, recombination-independent mechanisms are employed.
Materials and Methods
Details of the materials and methods, including strain construction, ChIP-seq and data analysis, microscopy, spore viability, and data availability are presented in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Shiv Grewal, Lilin Du, and Robin Allshire for providing strains and reagents; and Shelley Sazer, Zheng Zhou, Xing Guo, Jun Ma, and Mariano Rocchi for the valuable discussions. This work was supported by National 973 Plan for Basic Research Grant 2015CB910602 (to X.H.) and National Natural Science Foundation of China Grant 31628012 (to X.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE118016).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911745116/-/DCSupplemental.
References
- 1.Fukagawa T., Earnshaw W. C., The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cleveland D. W., Mao Y., Sullivan K. F., Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Westhorpe F. G., Straight A. F., The centromere: Epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7, a015818 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amor D. J., Choo K. H., Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71, 695–714 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall O. J., Chueh A. C., Wong L. H., Choo K. H., Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82, 261–282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocchi M., Archidiacono N., Schempp W., Capozzi O., Stanyon R., Centromere repositioning in mammals. Heredity 108, 59–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capozzi O., et al. , Evolutionary and clinical neocentromeres: Two faces of the same coin? Chromosoma 117, 339–344 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Cheeseman I. M., The kinetochore. Cold Spring Harb. Perspect. Biol. 6, a015826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folco H. D., et al. , The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 25, 348–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu M., He X., Ccp1 modulates epigenetic stability at centromeres and affects heterochromatin distribution in Schizosaccharomyces pombe. J. Biol. Chem. 293, 12068–12080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K., Chen E. S., Yanagida M., Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–2219 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K., Chang H. L., Kagami A., Watanabe Y., CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Falk S. J., et al. , Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L. Y., et al. , Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun. 8, 15775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato H., Masuda F., Takayama Y., Takahashi K., Saitoh S., Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 22, 658–667 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe Y., Geometry and force behind kinetochore orientation: Lessons from meiosis. Nat. Rev. Mol. Cell Biol. 13, 370–382 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Schvarzstein M., Wignall S. M., Villeneuve A. M., Coordinating cohesion, co-orientation, and congression during meiosis: Lessons from holocentric chromosomes. Genes Dev. 24, 219–228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabral G., Marques A., Schubert V., Pedrosa-Harand A., Schlögelhofer P., Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 5, 5070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heckmann S., et al. , Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 5, 4979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukhtanov V. A., et al. , Versatility of multivalent orientation, inverted meiosis, and rescued fitness in holocentric chromosomal hybrids. Proc. Natl. Acad. Sci. U.S.A. 115, E9610–E9619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ottolini C. S., et al. , Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47, 727–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allshire R. C., Ekwall K., Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7, a018770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami S., Matsumoto T., Niwa O., Yanagida M., Structure of the fission yeast centromere cen3: Direct analysis of the reiterated inverted region. Chromosoma 101, 214–221 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Steiner N. C., Hahnenberger K. M., Clarke L., Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol. Cell. Biol. 13, 4578–4587 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. S., Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Scott K. C., Merrett S. L., Willard H. F., A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16, 119–129 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Hayashi T., et al. , Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Chen E. S., Saitoh S., Yanagida M., Takahashi K., A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11, 175–187 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Dunleavy E. M., et al. , A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell 28, 1029–1044 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishino T., et al. , CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii K., et al. , Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321, 1088–1091 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Nakaseko Y., Adachi Y., Funahashi S., Niwa O., Yanagida M., Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 5, 1011–1021 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert I., Lysak M. A., Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27, 207–216 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Schubert I., What is behind “centromere repositioning”? Chromosoma 127, 229–234 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Hori T., et al. , Constitutive centromere-associated network controls centromere drift in vertebrate cells. J. Cell Biol. 216, 101–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi E. S., Cheon Y., Kang K., Lee D., The Ino80 complex mediates epigenetic centromere propagation via active removal of histone H3. Nat. Commun. 8, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang W. H., et al. , Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 24, 635–648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ketel C., et al. , Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5, e1000400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thakur J., Sanyal K., Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 23, 638–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura K., Komiya M., Hori T., Itoh T., Fukagawa T., 3D genomic architecture reveals that neocentromeres associate with heterochromatin regions. J. Cell Biol. 218, 134–149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellermeier C., et al. , RNAi and heterochromatin repress centromeric meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 107, 8701–8705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Locke D. P., et al. , Comparative and demographic analysis of orang-utan genomes. Nature 469, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolomeo D., et al. , Epigenetic origin of evolutionary novel centromeres. Sci. Rep. 7, 41980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura M., et al. , Evolutionary formation of new centromeres in macaque. Science 316, 243–246 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Wade C. M., et al. ; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team, Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zanders S. E., et al. , Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife 3, e02630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu W., et al. , A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. eLife 6, e26057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nuckolls N. L., et al. , wtf genes are prolific dual poison-antidote meiotic drivers. eLife 6, e26033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.