Significance
Ribosomes move along mRNAs in 3-nucleotide steps as they interpret codons that specify which amino acid is required at each position in the protein. There are multiple examples of genes with DNA sequences that do not match the produced proteins because ribosomes move to a new reading frame in the message before finishing translation (so-called frameshifting). This report shows that, when ribosomes stall at mRNA regions prone to cause frameshifting events, trailing ribosomes that collide with them can significantly change the outcome and potentially regulate protein production. This work highlights the principle that biological macromolecules do not function in isolation, and it provides an example of how physical interactions between neighboring complexes can be used to augment their performance.
Keywords: ribosome, translation, frameshift, bL9, dnaX
Abstract
Translational frameshifting involves the repositioning of ribosomes on their messages into decoding frames that differ from those dictated during initiation. Some messenger RNAs (mRNAs) contain motifs that promote deliberate frameshifting to regulate production of the encoded proteins. The mechanisms of frameshifting have been investigated in many systems, and the resulting models generally involve single ribosomes responding to stimulator sequences in their engaged mRNAs. We discovered that the abundance of ribosomes on messages containing the IS3, dnaX, and prfB frameshift motifs significantly influences the levels of frameshifting. We show that this phenomenon results from ribosome collisions that occur during translational stalling, which can alter frameshifting in both the stalled and trailing ribosomes. Bacteria missing ribosomal protein bL9 are known to exhibit a reduction in reading frame maintenance and to have a strong dependence on elongation factor P (EFP). We discovered that ribosomes lacking bL9 become compacted closer together during collisions and that the E-sites of the stalled ribosomes appear to become blocked, which suggests subsequent transpeptidation in transiently stalled ribosomes may become compromised in the absence of bL9. In addition, we determined that bL9 can suppress frameshifting of its host ribosome, likely by regulating E-site dynamics. These findings provide mechanistic insight into the behavior of colliding ribosomes during translation and suggest naturally occurring frameshift elements may be regulated by the abundance of ribosomes relative to an mRNA pool.
Naturally occurring translational frameshift motifs generally include a “slippery” messenger RNA (mRNA) sequence that contains an out-of-frame alternate transfer RNA (tRNA)−mRNA pairing option and adjacent stimulatory elements that interact with the ribosome to promote transient stalling or unseating (1). Although these features are clearly validated experimentally, much of the translation fidelity literature focuses on the behavior of ribosomes in isolation. Here, we show that ribosome collisions induced by translational stalling should also be considered as part of these frameshifting mechanisms and that ribosome collisions and overcompaction of polysomes may interfere with ribosome function.
The translation of codons within mRNA open reading frames (ORFs) is now understood in substantial detail (reviewed in refs. 2 and 3). Upstream of many bacterial ORFs, a short Shine−Dalgarno (SD) sequence is present that is complementary to a portion of the small ribosomal subunit RNA (4). During translation initiation, an interaction between these RNA segments helps to position the start codon in the peptidyl site (P-site) of the assembling ribosome, and the strength of complementarity can substantially alter the rate of translation initiation (5, 6). During translation elongation, codons in the adjacent A-site are evaluated for complementarity to the anticodon stems of aminoacylated tRNAs (aa-tRNA). When a match is found, the ribosome permits a chemical reaction between the amino acid on the A-site tRNA and the acyl bond that connects the nascent peptide to the P-site tRNA, thereby transferring the protein chain to the A-site tRNA. This reaction causes the 2 tRNAs to shift their orientations into the P/E A/P hybrid state, wherein the anticodon regions of the tRNAs remain in the P- and A-sites, but the molecules tilt such that the P-site tRNA’s acceptor end enters the exit site (E-site) and the A-site tRNA’s acceptor end enters the P-site. This rearrangement is accompanied by a movement of the uL1 stalk to partially close the tRNA E-site (7). At this stage, the ribosome binds to elongation factor G (EF-G), which couples the energy of guanosine triphosphate (GTP) hydrolysis to promote a transient rotation of the small subunit and to drive a 3-nucleotide ribosome translocation event. After translocation, the tRNA that was originally in the P-site temporarily resides in the E-site, the peptidyl-tRNA is fully positioned in the P-site, and the ribosome returns to a relaxed, nonrotated state awaiting a new aa-tRNA match in the A-site (3).
If there is a weak interaction between the tRNAs and the mRNA during translocation, a translational frameshift can occur if there is an alternative base pairing option in the vicinity (1). Likewise, if there is a delay in decoding or if there is mechanical stress on the ribosome from a nearby mRNA secondary structure, ribosomes can frameshift or hop over an mRNA segment (1). Aside from A- and P-site interactions, a cognate deacylated tRNA in the E-site can reduce frameshifting while it remains base-paired to the mRNA (8–12). In bacteria, allostery between the E-sites and A-sites has been observed during early translation cycles, in that E-site tRNAs are retained until A-site tRNAs are delivered (12). This synchrony is reduced once the nascent peptide reaches ∼4 amino acids long, and bacterial ribosomes switch to discarding E-site tRNAs irrespective of A-site occupancy (12, 13). In contrast to earlier models, it is now believed there is no connection between E-site occupancy and the fidelity of A-site tRNA selection (14). However, recent evidence suggests that an incoming mRNA secondary structure can alter the small subunit conformation and tRNA ejection rate from the E-site (15, 16).
Most naturally occurring frameshift motifs contain adjacent stimulator elements that encourage ribosomes to relocate. For example, the IS3 insertion element family contains a slippery sequence and a downstream mRNA pseudoknot to induce −1 frameshifting during production of their transposases (17–19). The dnaX gene in many bacteria contains a −1 frameshift element that is stimulated by both an upstream SD-like sequence and a downstream mRNA stem loop to control the ratio of Tau and Gamma proteins in the clamp loader complex of the DNA replisome (20–22). In another example, the frameshift element in the prfB gene uses an upstream SD-like stimulator and transient stalling at a UGA stop codon to enhance +1 frameshifting (23–25). In each example, ribosomes lose their grip on the mRNA and become repositioned in an alternate reading frame prior to resuming translation.
During a study of an extreme ribosome hopping event that occurs during translation of bacteriophage T4’s gene 60 (26), it was discovered that protein bL9 of the bacterial ribosome’s large subunit plays a role in maintaining translation fidelity (27). Since that discovery, several studies revealed that a lack of bL9 causes increased frameshifting, misincorporation, and ribosome hopping in a number of scenarios (1, 28, 29). Our group previously reported that Escherichia coli lacking bL9 become more sensitive to aminoglycosides and have a critical dependence on elongation factor P (EFP) (30). EFP enters stalled ribosomes through the E-site to enhance transpeptidation, most commonly during the synthesis of polyproline patches (31–35). Because bL9 is anchored at the base of the uL1 stalk near the E-site (36–38), we proposed a model in which bL9 functions at the interface between colliding ribosomes during translational stalling (30), an event that occurs more frequently in the absence of EFP. Biochemical and structural studies revealed that bL9 is dynamic and that its C domain can move between an extended conformation and a tucked-in conformation touching the small subunit (37, 39, 40). In the extended conformation found in crystals, bL9 reaches to a neighboring ribosome and blocks the GTPase factor binding site (39, 41). In addition to these structural insights, ribosome profiling studies of cells lacking EFP revealed extensive compaction of ribosomes preceding polyproline stalling motifs (42–44). Although there is no experimental evidence indicating bL9 acts on neighboring ribosomes (in trans), these observations inspired a mechanistic model that places bL9’s important role at collision interfaces.
In this report, we show that the load of ribosomes on mRNAs harboring the IS3, dnaX, and prfB regulatory elements significantly alters their programmed frameshift propensities. Using a series of engineered frameshift motifs, we discovered that trailing ribosomes exhibit increased −1 frameshifting when they collide with transiently stalled ones. We additionally discovered that ribosomes lacking bL9 are not only more sensitive to collisions, but also that they compact differently. By analyzing the nascent peptides produced from ribosomes stably stalled at a SecM motif, we established that bL9-deficient ribosomes can compact one codon closer to each other. As a consequence of this overcompaction, E-site tRNAs in the stalled ribosomes are more stably associated, which could affect allosteric small subunit behavior, transpeptidation efficiency, and access by translation factors such as EFP. Finally, we show that bL9 can function on its host ribosome (in cis) to suppress frameshifting at the beginning of an ORF in the absence of other ribosomes, supporting the idea that one of this protein’s major functions is to enhance its host ribosome’s grip on an mRNA. Collectively, these data provide a mechanistic model of bL9 function that explains its role in translation fidelity. Moreover, our observations raise the possibility that natural frameshift elements may be influenced by ribosome availability, which would tie translation efficiency and transient stalling to other important processes such as DNA replication (by altering the protein composition of clamp loaders) and transposition (by altering the production of transposases).
Results
A High Ribosome Load Reduces Frameshifting at the IS3 Pseudoknot.
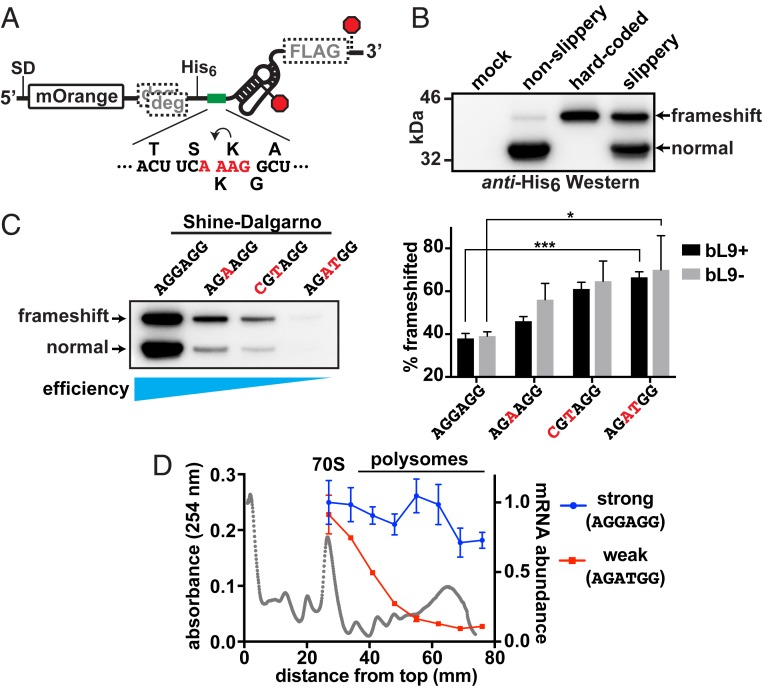
We evaluated the influence of ribosome abundance on −1 frameshifting using a well-characterized motif derived from insertion element IS3 that contains a slippery tetrad sequence (A_AAG) upstream of a pseudoknot stimulator (Fig. 1A). A prior study reported that this motif induced substantial −1 frameshifting, which was dependent on both the sequence of the slippery patch and the presence of the pseudoknot (19). We placed this motif in reporter plasmids that encoded a fluorescent protein (mOrange2) followed by the IS3 stimulator; upon −1 frameshifting, a larger fusion protein was generated that contained an appended FLAG epitope (Fig. 1A). Thus, the ratio of the 2 protein forms served as a readout of −1 frameshifting. These reporters also encoded ClpXP protease degrons in the −1 and +1 frames preceding the test region, so that any frameshifted products unrelated to the test region would not accumulate (45, 46). Control reporters included a version that lacked the slippery sequence and a version encoding the full-length fusion protein in frame.
Fig. 1.
Ribosome abundance on an mRNA influences frameshifting. A reporter construct based on the IS3 pseudoknot was used to evaluate the impact of ribosome load on −1 frameshifting. (A) Schematic of the reporter showing the location of the SD sequence that was altered to adjust translation initiation efficiency, the mOrange2 protein domain, out-of-frame ClpXP degrons (gray “deg”), a His6 tag, a test sequence (green bar), the IS3 pseudoknot stimulator, and a FLAG tag in the −1 frame. Stop codons for the 0 and −1 reading frames are marked with red octagons. The test sequence is displayed below, with the encoded amino acids in the 0 (top) and −1 (bottom) frames and slippery tetrad nucleotides in red. (B) Anti-His6 Western blot revealing the products produced from a mock culture, a nonslippery reporter version lacking the slippery tetrad (A_AAG to C_AAG), a hard-coded “frameshift” product, and a slippery reporter with a strong consensus SD sequence. The canonical normal product and −1 frameshifted products are indicated. (C) (Left) Western blot of reporters with SD sequences altered to reduce translation initiation. As the translation initiation efficiency waned, the percentage of frameshifted product increased. (Right) Bar graph of frameshifted product percentages from separate quantitative Westerns of 3 biological replicates. Error bars are 1 SD. Asterisks signify t test P values < 0.05 (*) and <0.001 (***). (D) Sucrose gradients of lysates prepared from cells that had expressed the reporters with the strongest and weakest SD were used to separate ribosome forms prior to extracting RNA for reverse-transcription RT-qPCR. An absorbance profile is shown in gray with the locations of the 70S monosomes and polysome peaks. Primers that amplified the 5′ ends of the reporter mRNAs and 16S rRNA were used to establish the message/ribosome ratios, which were normalized to the amount of strong mRNA observed in the 70S peak. Error bars represent 1 SD of 3 experimental replicates.
Cultures containing these plasmids were grown to early exponential phase and induced to express the reporters for 1 h. Samples were normalized by turbidity, harvested, and analyzed by Western blot (using an encoded His6 epitope) to determine the approximate abundance of each reporter form (Fig. 1B). Two major protein products were observed that corresponded to the canonically encoded, nonframeshifted “normal” product (∼32 kDa) and a larger, fusion product (∼36 kDa) arising from −1 frameshifting. These bands were not detected in cells expressing a mock plasmid. The construct containing a nonslippery (C_AAG) motif produced very low levels of frameshifted product, whereas the version with the slippery (A_AAG) sequence produced abundant frameshifted product that comigrated with a version that expressed an in-frame fusion (Fig. 1B). The upper band was also detected using anti-FLAG antibody in a separate Western blot, confirming it was a product of −1 frameshifting (SI Appendix, Fig. S1).
The frameshift reporter with the slippery sequence was modified to reduce the translation initiation frequency by mutating the SD sequence away from consensus. Several variants were compared to the parent reporter for overall expression efficiency and frameshifting (Fig. 1C). Remarkably, the level of frameshifting increased as translation initiation was reduced. In the most translationally attenuated case, frameshifting increased ∼2-fold such that ∼70% of the expressed reporter was frameshifted. We also compared the levels of frameshifting with these reporters in cells lacking bL9 (Fig. 1C). Although frameshifting increased as translation efficiency waned, there was no significant change to the levels compared to those found in bL9+ cells. These findings suggest that a higher abundance of ribosomes on the reporter mRNA suppressed frameshifting or that ribosomes acting alone were more prone to frameshifting on the IS3 element.
In a different experiment, cells that had expressed the reporters with the strongest and weakest SD sequences were separately fractionated on sucrose gradients, and total RNA was purified from the ribosome-containing fractions. Reverse transcription followed by real-time qPCR (RT-qPCR) was then used to establish the abundance of the reporter mRNAs relative to 16S ribosomal RNA (rRNA) in each fraction (Fig. 1D). The amount of each reporter mRNA in the 70S fractions was similar, but the message with the consensus (“strong”) SD sequence was significantly higher in the pool of polysomes, confirming that there was a higher ribosome load per message. We conclude that the ribosome load on these mRNAs was responsible for the observed changes in frameshifting levels, with higher ribosome loads suppressing frameshifting at the IS3 motif.
Ribosome Load also Alters Frameshifting Efficiency at the dnaX and prfB Motifs.
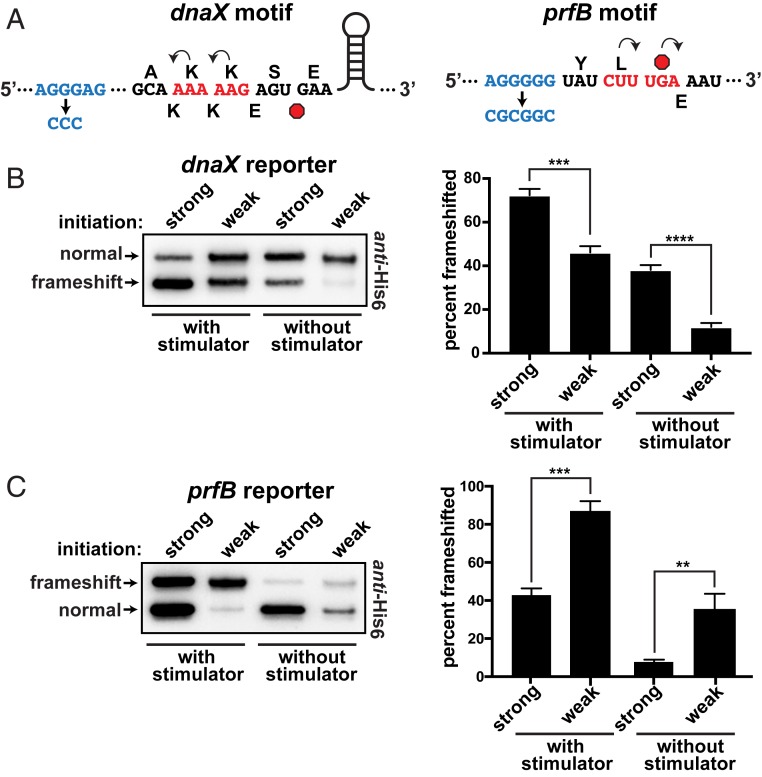
Many bacteria use programmed frameshifting to regulate expression of their dnaX and prfB genes. We placed the frameshift motifs from these genes in reporters with either a strong or a weak SD sequence to alter translation initiation and ribosome load (Fig. 2A). Both of these elements contain upstream SD-like stimulator sequences that help unseat ribosomes during frameshift events (25, 47). Because the presentation of these stimulators may be affected by trailing ribosomes, we also generated mutant versions of each motif with their SD-like stimulators inactivated to gauge their impact as a function of ribosome load.
Fig. 2.
Frameshifting at the dnaX and prfB motifs. (A) Schematics of the dnaX and prfB programmed frameshift motifs that were cloned into the reporter system. The upstream SD-like stimulators are shown in blue, with their deactivated versions below. The 0-frame amino acids are shown above and the frameshifted amino acids are shown below each sequence. Stop codons are red octagons. The dnaX motif also contains a downstream stem loop stimulator that was not altered. Note that, for the dnaX motif, −1 frameshifting produced the smaller translation product. (B) Frameshifting at the dnaX motif as a function of ribosome load. The construct with a strong SD sequence (AGGAGG) exhibited more frameshifting than a version with a weak SD sequence (AGATGG). Inactivation of the nearby SD-like stimulator reduced frameshifting and increased the influence of ribosome load. (Left) To even the abundances for presentation clarity, the anti-His6 Western had the strong samples diluted 35- and 25-fold for the with stimulator and without stimulator samples, respectively. (Right) Separate Westerns of 3 biological replicates were used to generate the data for the bar chart. (C) Frameshifting at the prfB motif. The construct with a strong SD sequence (AGGAGG) exhibited less frameshifting than the version with a weak SD sequence (AGAAGG); (Left) the strong versions were diluted 10-fold for the presented Western. Inactivation of the nearby SD-like stimulator reduced frameshifting and increased the influence of ribosome load. (Right) Separate Western blots of 3 biological replicates were used to generate the data for the bar chart. Error bars represent 1 SD. Asterisks indicate t test P values of <0.01 (**), <0.001 (***), and <0.0001 (****).
In contrast to the IS3 motif, −1 frameshifting at the dnaX motif was significantly lower when there were fewer ribosomes engaged with the mRNA. Quantification of reporter ratios indicated that frameshifting decreased from ∼70 to ∼45% when the SD sequence was mutated from strong to weak (Fig. 2B). Mutation of the dnaX SD-like stimulator element reduced frameshifting to ∼40%, and lowering ribosome load in this context reduced frameshifting even further to ∼10%. Thus, ribosome load significantly altered frameshifting propensity at the dnaX motif, and this impact was greater in the absence of the SD-like stimulator (2-fold vs. 4-fold).
Characterization of the reporters containing the prfB motif revealed that lowering ribosome load increased +1 frameshifting from ∼40 to ∼90%, similar to what was observed with the IS3 motif (Fig. 2C). When the SD-like stimulator was inactivated in this reporter set (while preserving the encoded amino acid sequence), +1 frameshifting dropped to ∼10%. Reducing the ribosome load in this context (i.e., without the stimulator element) caused a large increase in frameshifting to ∼50%. Taken together with the dnaX reporter data, the SD-like stimulator elements appear to dampen the influence of ribosome load.
Ribosome Load Alters Frameshifting Efficiency in a Cell-Free Translation System.
Our results showed that the abundance of ribosomes on a message affected frameshifting, for which we postulated the ribosomes collided with each other more frequently when they were more abundant on a message. Alternatively, some other influence on the translatability of the downstream reporter sequences may have affected the readouts from our assays. For example, differential degradation of mRNA downstream of the frameshift regions could have impacted the relative expression of the reporter proteins. Although these reporters contained a substantial length of untranslated mRNA 3′ of the test sites (an inactive lacZ ORF was transcribed beyond the FLAG sequence), differential ribosome occupancy may have indirectly altered the abundance of the region encoding the FLAG epitope. We evaluated this possibility by measuring the abundance of a reporter mRNA region located past the test site and compared that to the abundance of the beginning of the ORF for both the strong and weak SD reporters, using RT-qPCR. We did not observe significant differences in the relative abundances of RNA as a function of ribosome load (SI Appendix, Fig. S2), suggesting that differential mRNA levels cannot account for our results with the frameshift reporters.
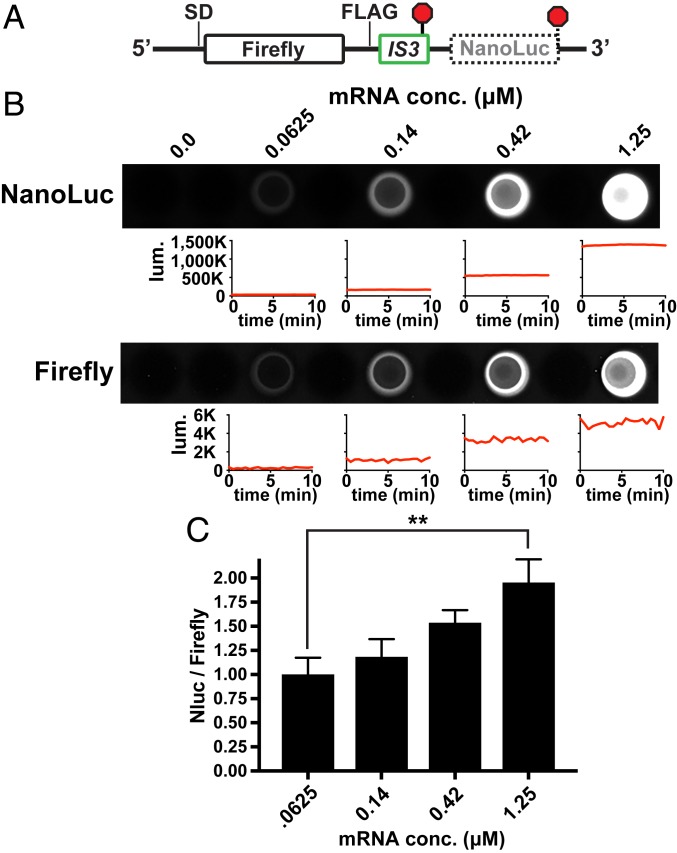
To further rule out the possibility of RNA turnover, and to limit the number of factors involved in altering frameshifting frequency, we employed an in vitro translation system comprising purified components. The frameshifted signals from our previously described reporters were too weak to detect using Western blots at this scale, so we constructed a dual luciferase reporter that allowed us to quantify frameshifted protein synthesis in vitro (Fig. 3). We inserted the IS3 element between 2 luciferase domains (Firefly and NanoLuc) that can be independently measured, with the downstream NanoLuc ORF situated in the −1 frame. The influence of translation initiation efficiency on downstream frameshifting events is difficult to evaluate in vitro because the overall translation activity in this system is poor compared to that found in vivo, so an experiment involving a direct titration of ribosomes would be complicated by the unknown ribosome reuse and reinitiation rates. As an alternative approach, we transcribed reporter mRNA with a strong SD sequence in vitro and then added varying concentrations of it as template into purified translation reactions. As the mRNA dose was increased, the production of NanoLuc increased relative to Firefly, revealing that having fewer ribosomes per message increased frameshifting in this system as well (similar to the IS3 element in vivo) (Fig. 3 B and C). Therefore, frameshifting efficiencies were altered as a function of ribosome load in the absence of RNases and many other cellular factors.
Fig. 3.
Ribosome load alters frameshifting in a cell-free translation system. An mRNA was designed to test the effect of ribosome load on frameshifting in a purified translation system. (A) Schematic of the reporter mRNA showing the IS3 element (detailed in Fig. 1) located between the coding regions for Firefly luciferase and an out-of-frame NanoLuc luciferase. (B) Reporter mRNA that was synthesized in vitro using T7 RNA polymerase was added at different final concentrations to translation reactions containing purified components (∼2 μM ribosomes). After 2 h of translation, the luminescence of each luciferase was independently measured in a 96-well plate. The plate was imaged, the 10-min-averaged luminescence values were subsequently recorded in a luminometer, and those values from experimental replicates were then averaged. Representative plots of luminescence signals vs. time are shown below wells of a plate after subtracting background signals from the no-RNA controls to illustrate signal stabilities. The NanoLuc signal was substantially brighter than the Firefly signal. (C) Plot of averaged data from 3 experimental replicates. The NanoLuc (Nluc)/Firefly ratio increased significantly as the mRNA abundance increased, which reduced ribosome load per message. Asterisks indicate a t test P value of <0.01 (**).
Ribosome Collisions Promote −1 Frameshifting in Trailing Ribosomes.
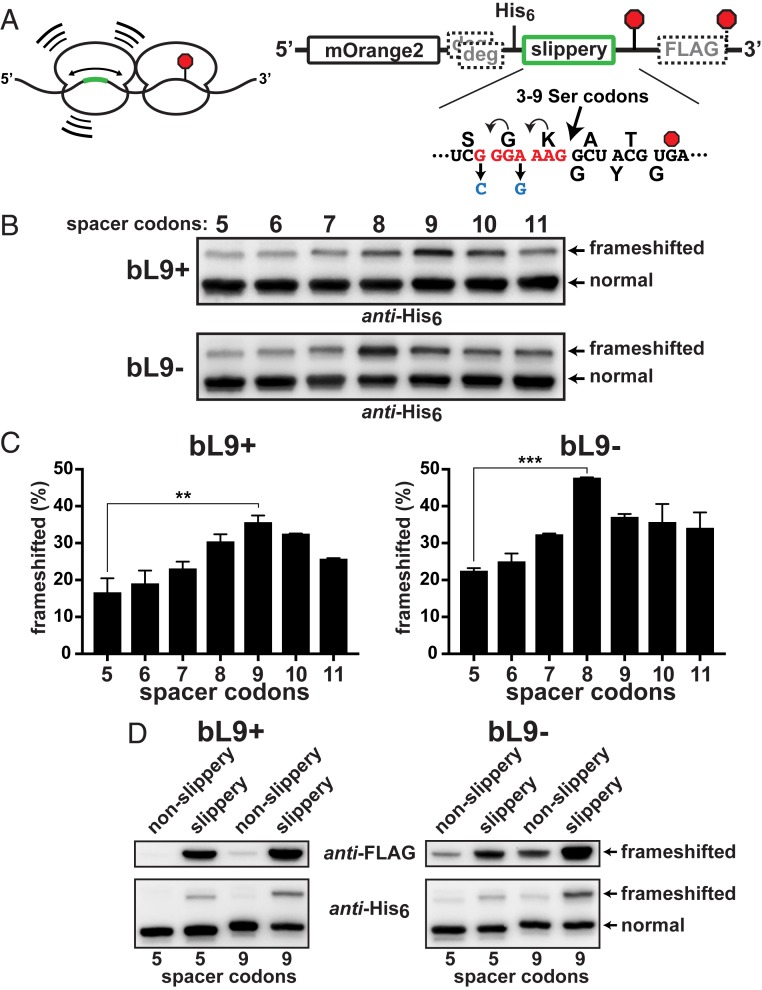
We considered the possibility that stalled ribosomes may be able to function as frameshift stimulators for trailing ribosomes during collisions, so we created a series of reporters designed to test the influence of transiently stalled leading ribosomes on the fidelity of trailing ribosomes that collide with them. Our strategy was to alter the position of a slippery sequence upstream of a stall site such that it would be located in the P/A-sites of trailing ribosomes as they collided with stalled ones (Fig. 4A). We chose to transiently stall the leading ribosomes with the slowly decoded UGA stop codon so that any trailing ribosomes that frameshifted would have an opportunity to translate into the downstream FLAG-encoding region. The simple UGA motif represents a common, naturally occurring element in E. coli, and it allowed us to evaluate frameshifting without the use of stimulatory elements and to eliminate RNA folding events that are likely to be affected by ribosome load (Fig. 4A). For this reporter collection, we employed a slippery heptad sequence (G_GGA_AAG) that is more prone to frameshifting than a slippery tetrad (19). The heptad’s position relative to the stall site was altered by inserting 3 to 9 Ser codons between the slippery sequence and the E-site codon of the stalled ribosome. This arrangement presented the slippery sequence to trailing ribosomes with 5- to 11-codon spacers between A-sites.
Fig. 4.
Stalled ribosomes are frameshift stimulators. A series of reporters was constructed to evaluate the influence of ribosome collisions on −1 frameshifting. (A, Left) Schematic of a collision between a ribosome transiently stalled at a stop codon (red octagon) and a trailing ribosome engaged with a slippery sequence located at different upstream positions (green). (Right) A detailed schematic. A slippery heptad sequence was positioned at varying distances 5′ to a slowly decoded UGA stop codon (red octagon). Nonslippery versions contained changes (blue) that hampered repositioning of the tRNAs. A downstream FLAG-tag appendage was encoded in the −1 frame. Out-of-frame ClpXP degrons preceded the test regions. The distance between the slippery sequence and the stalling stop codon was varied by inserting additional Ser codons such that the final A-site to A-site spacing ranged from 5 to 11 codons. (B) Anti-His6 Western blots of the reporter series that was expressed in bL9+ and bL9− cells. Frameshifting was evident in each case, but peaked with a 9-codon spacer in bL9+ and with an 8-codon spacer in bL9− cells. (C) Quantifications from 2 biological replicates were averaged and plotted. The peak frameshifting levels were significantly higher than the levels with the 5-codon spacer, and the bL9− cells exhibited more frameshifting overall. P values < 0.01(**) and <0.001 (***) are indicated. (D) (Top) Anti-FLAG and (Bottom) anti-His6 Westerns showing comparisons between reporters with 5 or 9 spacer codons with and without the slippery heptad.
After expression of each reporter (containing variable spacers) in parallel cultures, Western blot analyses indicated that there was a notable increase in frameshifting when the slippery motif was positioned from 8 to 10 codons upstream of the stop codon, with the majority occurring with a 9-codon spacer (Fig. 4B). Because this distance (27 nucleotides) is in agreement with the length of mRNA that ribosomes protect from RNases, we conclude that the trailing ribosomes exhibited increased −1 frameshifting when they were positioned on the slippery sequence while colliding with ribosomes transiently stalled at the UGA stop codon.
When this reporter series was expressed in cells lacking bL9, we again observed an increase in −1 frameshifting that was position-dependent (Fig. 4B). Interestingly, not only did the levels of frameshifting increase overall in the absence of bL9, the frameshifting peak became maximal when there was an 8-codon spacer between the slippery sequence and the stop codon. Quantification of experimental replicates confirmed that frameshifting had increased ∼2-fold when the trailing ribosomes were positioned on slippery sequences as they collided with the stalled ribosomes (Fig. 4C). Also, this analysis confirmed that the peak of frameshifting was one codon shorter in bL9− cells, which reinforces the conclusion that physical contact between ribosomes was required to enhance frameshifting on the slippery sequence.
E. coli ribosomes stalled at UGA stop codons are known to undergo a variety of recoding events, including hopping, wherein ribosomes slide to alternate codons that match their P-site anticodon (29). To establish the contribution of these alternatives to the frameshift readouts in our collision assays, we eliminated the slippery sequences from the 5- and 9-codon reporters and measured frameshifting. Inactivation of the slippery sequence substantially reduced −1 frameshifting with both reporters, indicating that the majority of the frameshift signals in the preceding experiment resulted from the trailing ribosomes (Fig. 4D). An alternate His6-tagged translation product was detectable in the absence of the slippery sequence that migrated slightly faster than the product of −1 frameshifting at the slippery sequence, and its abundance was higher in the absence of bL9. That product was also reactive to anti-FLAG antibody, indicating it was read from the −1 frame and may have arisen from hopping at the stop codon (the P-site tRNA had an alternative ACG landing codon in the −1 frame 11 nucleotides past the stop codon). Nonetheless, the majority of the frameshift signals from constructs containing the slippery sequence were generated by trailing ribosomes.
Ribosomes Compact One Codon Closer in the Absence of bL9.
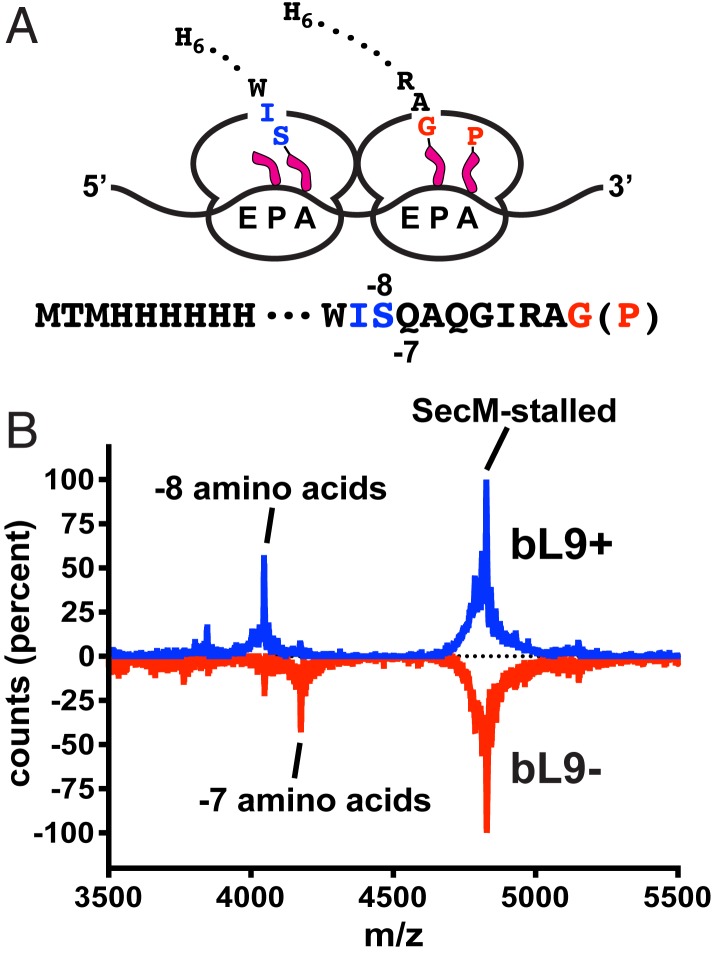
The preceding results indicated that an appreciable fraction of trailing bL9− ribosomes were able to advance one codon closer to the stalled ribosomes before the collisions promoted frameshifting. To directly test this idea, we designed an expression construct that formed very stable ribosome pile-ups so we could evaluate the spacing of ribosomes based on the emerging nascent peptides. The pileup ORF was designed to allow for the loading of up to 4 ribosomes and encoded an N-terminal His6 tag, a spacer sequence, and a robust SecM translational stalling motif (Fig. 5A) (48–50). When expression of this construct was induced for 20 min, cell growth was inhibited, and polysome profiles indicated that the ribosomes primarily accumulated as members of 2- to 3- and 4-somes (shown below).
Fig. 5.
Ribosomal protein bL9 alters interribosomal spacing. A construct was designed to stably stall ribosomes so that the positions of trailing ribosomes could be established by characterizing their nascent peptides. (A) Schematic of 2 colliding ribosomes and the nascent peptide sequences. The lead ribosome is stably engaged with a SecM stalling motif with peptide-tRNAGly in the P-site and tRNAPro in the A-site (red). The colliding ribosome contains a shorter nascent peptide, with deacylated tRNAIle in the P/E-site and peptide-tRNASer in the A/P-site (blue). The peptides contained N-terminal His6 tags that allowed for purification under denaturing conditions prior to mass spectroscopy. (B) MALDI-TOF mass spectra of peptides recovered from sucrose gradient polysome fractions of bL9+ (blue) and bL9− cells (red, inverted). The dominant mass for the SecM-stalled polysomes corresponded to a peptide ending at the Gly residue of the SecM sequence shown in A (theoretical m/z = 4,801.40 Da, observed = 4,801.14 Da). Peptides recovered from bL9+ polysomes also contained a prominent mass corresponding to a peptide that was 8 residues shorter than the SecM-stalled peptide (theoretical = 4,019.53 Da, observed = 4,019.42). The peptides from bL9− polysomes contained an additional peptide 7 residues shorter (theoretical = 4,147.66 Da, observed = 4,146.90 Da).
To establish the positions of ribosomes on the mRNA near the SecM motif, polysomes were collected from bL9+ and bL9− cells that had expressed the stall construct, and His6-containing peptides were recovered under denaturing conditions. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectroscopy was then used to characterize the masses of the recovered peptides (Fig. 5B). The dominant peaks corresponded to nascent peptides ending with the Gly residue of the SecM peptide. In the presence of bL9, an additional mass was observed that matched that of a peptide that was 8 amino acids shorter (ending with the Ser residue depicted in blue in Fig. 5A), which is consistent with ribosomes having 9-codon, P-site to P-site spacing and having accomplished transpeptidation to the A-site tRNA. In the absence of bL9, there was an additional abundant mass that corresponded to a peptide 7 amino acids shorter than the SecM-stalled peptide (ending with the adjacent Gln residue). Taken together with the collision frameshifting data above, it appears that, when ribosomes collided, attempts to translocate after transpeptidation provided an impetus for frameshifting. In the absence of bL9, these events occurred with the ribosomes one codon closer together.
E-Site tRNAs Are More Stably Associated with Condensed Polysomes Lacking bL9.
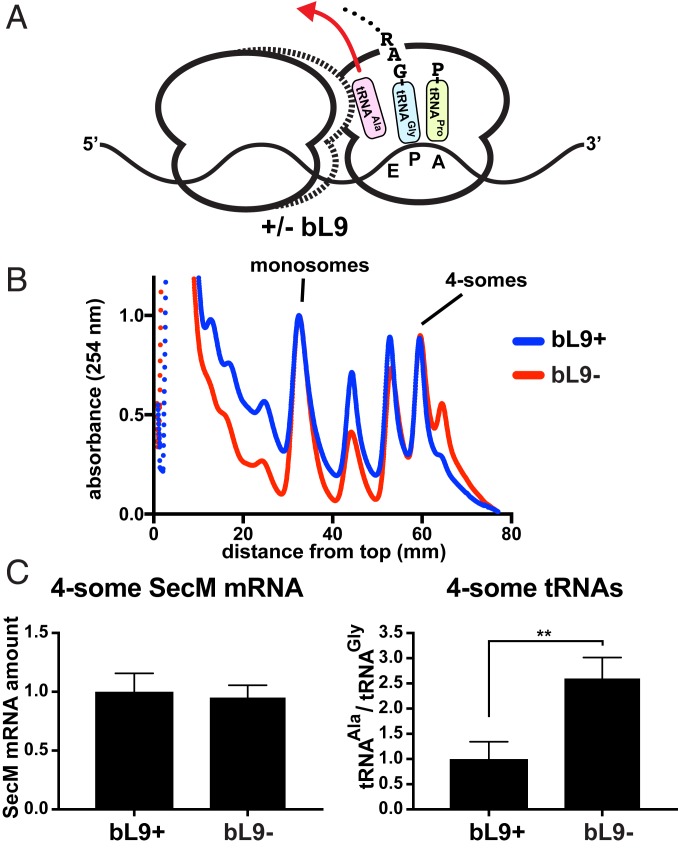
Led by our prior discovery that Δefp E. coli requires bL9 for appreciable growth, we considered the possibility that the overcompaction observed in stalled bL9− polysomes may have been responsible for poisoning the translation pool in this background. Based on the SecM-stall ORF sequence and confirmed by our mass analyses, most of the P-sites of the stalled ribosomes should have contained peptide-tRNAGly3, and the E-sites may have contained tRNAAla1B that survived the voyage from lysis through gradient centrifugation (51). Tight associations between overcompacted ribosomes in bL9− cells may have altered the retention of the E-site tRNAs (Fig. 6A), so we elected to determine their abundance relative to the P-site tRNAGly3.
Fig. 6.
Stalled ribosomes have higher E-site occupancy in the absence of bL9. The SecM stalling construct was used to generate compacted polysomes that were recovered to quantify tRNA abundances in compacted polysomes. (A) Schematic of a SecM collision complex with an E-site deacylated tRNAAla1B (pink), a P-site tRNAGly3 connected to the nascent peptide (celeste), and an A-site tRNAPro2/3 waiting to engage the peptidyltransferase center (chartreuse). In the absence of bL9, trailing ribosomes advance one codon closer to stalled ribosomes (dashed outline) and potentially influence the egress of E-site tRNAAla1B (red arrow). (B) A plasmid encoding a SecM-stall peptide (41 residues including the stalling Gly) was expressed for 20 min in bL9+ and bL9− cells, and lysates were resolved on sucrose gradients and fractionated. The absorbance profiles of representative gradients are shown with the bL9+ material (blue) and bL9− material (red) normalized to the abundance of monosomes. The positions of the monosomes and compacted 4-somes are indicated. An overcompacted 5-some peak was reproducibly more abundant in bL9− lysates. (C) RNA was purified from 4-some gradient peaks, converted to cDNA with dedicated primers, normalized, and used as templates in RT-qPCR reactions specific for the SecM-stall mRNA, as well as the tRNAAla1B and tRNAGly3 isoforms that had engaged the stalled ribosomes’ E- and P-site codons. For comparison, the level of SecM-stall mRNA and the ratio of tRNAAla1B to tRNAGly3 was normalized to the average amount found in the fractions recovered from bL9+ cells. Error bars represent SDs from 3 biological replicates and 5 qPCR measurements for each target RNA. A P value < 0.01 is indicated (**).
We developed a sensitive RT-qPCR protocol that allowed us to measure tRNA abundances in recovered polysome peaks so they could be compared to each other. In preliminary experiments, we observed that the qPCR values for tRNAAla1B were ∼3× overrepresented in 4-somes recovered from bL9− cells that had expressed the SecM-stall construct used for the mass analyses presented above. However, we noted that, in addition to the E-site SecM codon, there were 2 other Ala codons in our original design that may have been differentially engaged by trailing ribosomes if they shifted register. Therefore, we reengineered the SecM-stall mRNA such that the only Ala codon was at the E-site of the SecM motif. This redesigned version stalled ribosomes as well as the original and also led to the formation of primarily 2-, 3-, and 4-somes (Fig. 6B; for comparison to a normal polysome profile, see Fig. 1D). In addition, some 5-somes were present and reproducibly more abundant in lysates derived from bL9− cells. We suspect these polysomes resulted from overcompaction that cleared room for an additional ribosome on each mRNA.
RT-qPCR analyses indicated that the SecM-stall mRNA was detected at similar levels in the recovered 4-some peak RNA from bL9+ and bL9− cells, which was consistent with the observation that the amount of SecM-stalled 4-somes was comparable between these lysates (Fig. 6C). However, the amount of tRNAAla1B was again higher in the 4-some peaks recovered from bL9− cells. As an index of E-site tRNA retention, we calculated the ratio of tRNAAla1B to tRNAGly3 qPCR values for each biological replicate and found that tRNAAla1B was overrepresented by ∼2.5-fold (Fig. 6C). Because the stalled bL9− ribosomes had trailing ribosomes positioned one codon closer to their E-sites, we tentatively conclude that this overcompaction impeded the egress of the E-site tRNAs of the stalled ribosomes.
L9 Acts in cis to Reduce Frameshifting.
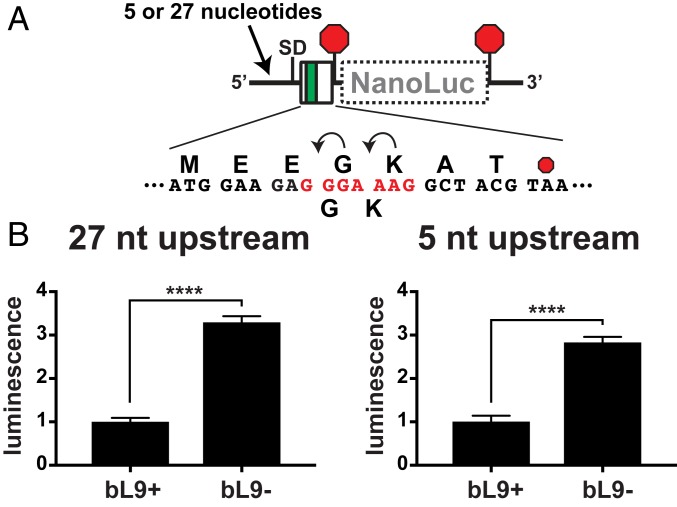
We entertained 2 mechanisms for how bL9 could increase fidelity during ribosome collisions. In one, bL9 acts in cis to stabilize its own ribosome during stalling or translocation attempts; in the other, bL9 interacts in trans with a trailing ribosome to somehow reduce interference. Support for a trans mechanism comes from a crystal packing form in which bL9 reaches between particles and blocks the docking site for translation GTPases (39, 52, 53). To evaluate a potential cis mechanism, we created a reporter that positioned a slippery motif near the beginning of a miniature ORF such that only one ribosome could be present at the slippery site (Fig. 7A). When expressed in bL9+ cells, −1 frameshifting allowed a low level of expression of the out-of-frame luciferase reporter (Fig. 7B). However, when this construct was expressed in bL9− cells, frameshifting increased substantially, suggesting that L9 had functioned in the context of isolated ribosomes (Fig. 7B). We did not detect significant differences in the abundance of reporter mRNAs between bL9+ and bL9− cells that could have accounted for this observation (SI Appendix, Fig. S3). We also generated a variant of the reporter that lacked the majority of the untranslated leader, leaving only 5 nucleotides upstream of the SD sequence (SI Appendix). Using this version, we again observed that frameshifting was significantly increased in bL9− cells (Fig. 7B), consistent with the model that bL9 can function to improve the grip of its host ribosome. In conclusion, bL9 reduced −1 frameshifting in a reporter system that limited mRNA occupancy to a single ribosome, indicating that bL9 can function in cis to improve translation fidelity. Although these experiments did not rule out a trans mechanism, the reduction in fidelity observed in these experiments is sufficient to account for the reported influence of bL9, at least in the context of frameshifting.
Fig. 7.
Ribosomal protein bL9 functions in cis to reduce frameshifting. A frameshift reporter was designed to evaluate the performance of single ribosomes as they engage a slippery sequence. (A) Schematic of reporters with a slippery heptad (red) located at the 5′ end of a mini ORF with a downstream NanoLuc ORF in the −1 frame. Transcripts containing either 27 or 5 nucleotides 5′ to the SD were compared. (B) After expression in bL9+ and bL9− cells, NanoLuc luminescence was measured and normalized to the level present in bL9+ cells. The level of frameshifting was significantly higher in cells lacking bL9. Error bars are SDs of 3 biological replicates. P values < 0.0001 (****) are indicated.
Discussion
This study revealed a connection between the abundance of ribosomes on a message and frameshifting propensity. We first observed this phenomenon using the IS3 motif, so one mechanism we initially entertained was that the higher ribosome traffic simply reduced the refolding rate of the stimulator pseudoknot. However, the influence of ribosome load was the opposite for the dnaX motif (which also uses a folded stimulator), and the prfB motif does not rely on a folded mRNA. Direct evidence for ribosome collisions being a cause of the observed changes in frameshifting came from the use of reporters that monitored the performance of trailing ribosomes as they collided with ribosomes that were transiently stalled. We also determined that protein bL9 can function to reduce frameshifting on isolated ribosomes and that it reduces the propensity of trailing ribosomes to block the E-sites of stalled ribosomes. Together, these observations provide a molecular mechanism that explains bL9’s influence on translation fidelity and also why cells lacking EFP (experiencing rampant stalling) are dependent on bL9.
Although we have no comprehensive model that can predict the influence of ribosome collisions on a given frameshifting motif, it is important to consider where and when the motivational stresses are encountered within each system. One model that explains reduced frameshifting with higher ribosome load at the IS3 element is that the trailing ribosomes may not encounter the pseudoknot stimulator once they have been queued. This scenario would occur because any ribosomes that successfully unfold the pseudoknot without frameshifting are expected to transiently stall at the downstream stop codon, which would then prevent the pseudoknot from reforming. In a similar model, if there is a mechanical coupling that allows the thrust of trailing ribosomes to be harnessed, then pseudoknot unfolding may have occurred more rapidly with higher ribosome load. In bacteria, translational stalling can lead to ribosome rescue if the A-site does not contain mRNA (54); however, this scenario is not likely to have occurred during these experiments (SI Appendix).
The influence of mRNA tension during frameshifting is another angle to consider when ribosomes encounter obstructions. In the cases of the dnaX and prfB motifs, attention should be drawn to the influence of the upstream SD-like stimulators. The conformation of the anti-SD in the 16S rRNA is dynamic, and its positioning near the mRNA exit site is variable, depending on interactions with the mRNA (SI Appendix, Fig. S4) (9, 25, 55, 56). In the case of dnaX, the SD-like stimulator is predicted to interact with the anti-SD element either prior to or soon after a ribosome stalls at the stem loop. A modeling of ribosome collisions revealed that such an SD:anti-SD interaction in dnaX can be preserved in the interribosomal space during collisions (discussed below). In a recently reported structure of a ribosome engaged with the dnaX motif, the bottom of the stimulator stem loop (2 codons ahead of the A-site) is partially unfolded and drawn toward the mRNA entrance channel (16). In addition, that particular translation complex was formed using an engineered mRNA containing a rather short spacing of the SD sequence of 7 nucleotides ahead of the P-site, whereas the natural SD-like stimulator of dnaX is positioned 5 nucleotides farther upstream (with the A in AGGA as a reference position in the SD sequence) (22, 57). Incidentally, the IS911 −1 frameshift element contains the same spatial arrangement of an SD-like patch and stem loop motif as in dnaX (58).
When SD:anti-SD interactions are allowed to form in the absence of mRNA tension, the mRNA relaxes to a position that places the P-site codon 10 nucleotides downstream of the SD sequence, which coincides with the predominant spacing of the SD sequence at initiation sites in E. coli (56, 57). Tension in the mRNA during translation of a dnaX-like motif due to a collision with a stem loop has been reported to reduce successful translocation after entry into the hybrid state (59). Therefore, we suspect that the choice of using a short nucleotide spacer after the SD stimulator element to form dnaX-like complexes for biochemical and structural studies serendipitously captured a translation state with “mRNA stress” resulting from a retracted SD:anti-SD helix and the downstream stimulator stem loop, which may explain why A-site tRNA was not bound as expected in one study (16, 60). Our observation that a high ribosome load promoted frameshifting at the dnaX motif suggests that trailing ribosomes either enhanced the activity of the SD-like stimulator or promoted the positioning of the lead ribosome with higher stress on the mRNA, or a combination of both (Fig. 8A).
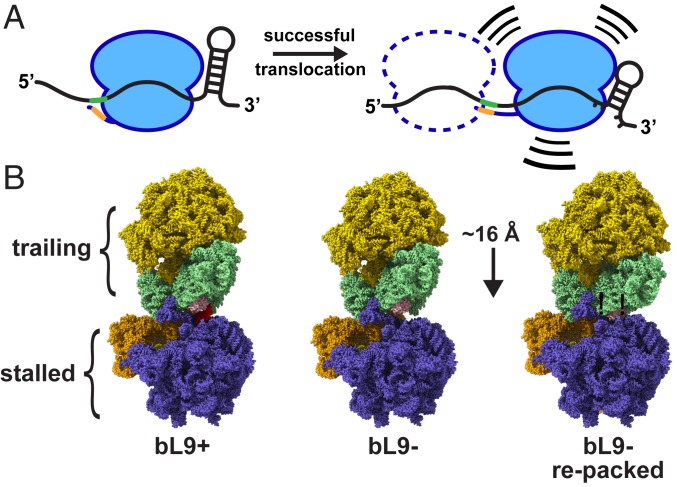
Fig. 8.
Models of mRNA tension and collisions. (A) Schematic of a ribosome in 2 locations relative to a frameshift element containing flanking stimulators. (Left) The SD sequence in the mRNA (green) interacts with the anti-SD (orange) in a relaxed manner, and the downstream mRNA stem loop is relaxed. (Right) A ribosome that has translocated farther into the motif, with a tensioned SD:anti-SD structure and a partially unfolded stem loop. The combination of the 2 flanking forces provides the energy to increase −1 frameshifting. A potential trailing ribosome is depicted as a dashed line. The trailing ribosome may promote entry of the lead ribosome into a compromised location while still allowing for a −1 frameshift. (B) Ribosome collision models with a stalled ribosome and a trailing ribosome. (Left) A cryo-EM structure of a ribosome awaiting translation termination (stalled, PDB ID 6ore; slate and orange) that is docked to a structure of a ribosome in the resting state (trailing, PDB 6osq; gold and light green) using a collision orientation similar to that observed in many crystal structures. Protein bL9 of the stalled ribosome is colored red, and protein uS4 of the trailing ribosome is colored pink. (Center) Protein bL9 was removed while maintaining the orientations of both particles. (Right) The trailing ribosome was reoriented and moved closer such that uS4 of the trailing ribosome accommodated the space previously occupied by bL9. This relocation reduced the intervening mRNA length by approximately one codon.
The influence of the upstream SD-like stimulator during prfB frameshifting has been well characterized, and base pairing between the SD and anti-SD sequences drives the +1 frameshift event (9, 25, 55). If colliding ribosomes simply pushed on the stalled ribosomes (or pulled on the mRNA), then frameshifting should have increased; yet, we observed a notable suppression of frameshifting with a high ribosome load. Base pairing between E-site tRNA and the mRNA is known to suppress frameshifting at the prfB motif (9, 25, 55). If collisions slowed the egress or positioning of E-site tRNAs, as we observed during SecM stalling, then this mechanism may explain the influence of ribosome load on this motif in our test system.
Discussions of ribosome collisions should account for the predicted spacing and orientations of the interacting particles. The positions of adjacent bacterial ribosomes found in crystal structures were used by another group to model condensed polysomes that were imaged by cryoelectron tomography (61). In the closest collision pairs, their modeling predicted 72 nucleotides between A-sites and a corresponding difference in nascent peptide length of 24 amino acids. Our data indicate that colliding ribosomes at a stall site have ∼9 codons of mRNA separating A-sites, and a nascent peptide difference of 8 amino acids. In an effort to explain these discrepancies, we computationally modeled potential ribosome collision complexes using several published structures, both from X-ray crystallography and cryogenic electron microscopy (cryo-EM). First, we established reference positions for the mRNA entry and exit sites using structures containing mRNAs so that we could also model particles without complete mRNA occupancy (SI Appendix, Fig. S5). Next, we calculated an average extended codon length of ∼16 Å by measuring codon lengths in a structure containing tensioned mRNA (SI Appendix, Fig. S5). Finally, the mRNA exit and entry sites were then linked in 2 adjacent ribosomes to monitor the intervening distance as the particles were repositioned in an effort to pack them closely together while minimizing the length of the intervening mRNA.
Using this approach, we found 3 collision orientations that satisfy our observation that an 8- to 9-codon spacer predominated between colliding particles (SI Appendix, Fig. S6). One of these orientations is the same as the one commonly observed in crystals containing bL9, with helix 33 of the trailing ribosome’s small subunit pointing toward the E-site of the stalled ribosome, adjacent to the uL1 stalk (Fig. 8B). The 2 other orientations required rotations from this position of ∼50° and then another ∼170° with a slight tilt (SI Appendix, Fig. S6). These orientations were possible using particles with bL9 extended from the stalled ribosome (observed in all crystals containing it) and also with bL9 folded against the side of the ribosome (observed in all cryo-EM structures). Thus, there is no structural restriction preventing adjacent ribosomes from having an 8- to 9-codon mRNA spacer between A-sites, and there are multiple options for the collision orientations. In addition, it appears there can be slack in the mRNA connecting adjacent in-frame ribosomes, so there can be room for a −1 nucleotide frameshift.
We also considered the influence of bL9 as we modeled collisions. The orientation in which an extended bL9 blocks the GTPase factor binding site on the trailing ribosome is only observed in crystals. Because bL9 is involved in regulating translational bypassing, others have proposed that bL9 docking near the A-site of a trailing ribosome may be used to influence the formation of secondary structures in the mRNA to promote takeoff of the ribosome during hopping (53). A cryo-EM structure is now available of a ribosome at the beginning of a gene60 bypassing event, and it shows the presence a small mRNA stem loop that blocks the A-site (15). However, this feature is more than 50 Å away from the tip of bL9 as it is positioned in the crystals, so it is unlikely that there is direct contact during bypassing.
We also interrogated the collision models in an attempt to explain the observation that ribosomes predominantly compacted one codon closer together when they lacked bL9. We found that bL9 of the stalled ribosome made contact with the trailing ribosome in most of the modeled collisions with short mRNA linkers. For example, using a structure of a ribosome awaiting a release factor and a trailing ribosome in the resting state (akin to our experimental system that evaluated spacing), bL9 contacted uS4 of the trailing ribosome (Fig. 8B). By computationally removing bL9, a clearing was made that allowed the trailing ribosome to pack closer. Interestingly, this repositioning shortened the mRNA linker by ∼15 Å to 17 Å, depending on the model, which is sufficient to explain the observation that bL9− ribosomes tended to pack closer together. However, we would like to point out that polysomes containing bL9 can also compact one codon closer together than normal, just not as often. It is also important to note that these models do not include bS1, which is commonly removed from ribosomes for structural and biochemical studies but may be involved in mRNA positioning near the exit site or alter collision orientations. Taken together, our modeling and experimental data do not rule out a function of bL9 that shuts down a trailing ribosome, but such a mechanism would require an extended bL9 conformation and only one of several possible collision orientations.
Our previous genetic discovery that cells lacking bL9 require EFP led us to consider the mechanistic linkage between a protein with a role in improving fidelity and a protein that accelerates transpeptidation, which seem to be at odds because speeding up transpeptidation should make fidelity defects worse. Recent reports suggest EFP surveils most ribosomes, not just those that are stalled (62); however, whatever interaction EFP has with the majority of ribosomes is relatively weak because it is not detectable as a ribosomal cofactor in sucrose gradients (30). Nonetheless, a lingering empty E-site is a sign of stalling, and EFP requires E-site access to facilitate transpeptidation.
Because EFP functionality is not compromised in bL9− cells, the majority of E-sites can still be accessed. In addition, full-length proteins are made in the absence of EFP, so the transpeptidation stalls must eventually resolve. In light of our data on the behavior of SecM-stalled polysomes, one model that explains why a bL9− EFP−combination is so toxic is that catastrophic train wrecks may form with overly compacted polysomes and jammed E-sites. In this situation, an attempt by the P-site tRNA to enter the P/E hybrid state may fail if the ribosome is unable to clear its E-site tRNA or if there is an obstruction blocking E-site closure. Another model is that, in the absence of bL9, the trailing ribosomes unseat stalled ribosomes because of unregulated translocation efforts.
The eukaryotic EFP ortholog eIF5A is essential, and ribosomal E-sites are empty less often, which may explain why there appears to be no functional ortholog of bL9 outside of bacteria (12, 63, 64). Because translation initiation is slower than elongation, compacted ribosomes are a hallmark of slow or stalled translation. This feature has been identified as a trigger for ribosome rescue from 3′ untranslated regions in yeast (65), mRNA cleavage during stalling in yeast (66), and translation quality control in mammals (67). Nonetheless, in all translation systems, any events that alter the rate of elongation, including mutations within the ribosome, are expected to also influence the rates of ribosome collisions.
An absence of bL9 caused an increase in −1 frameshifting by individual ribosomes at the 5′ end of an ORF when the nascent peptide was only 5 amino acids long. A conclusion from this observation is that bL9 primarily influences the grip of its host ribosome, and may not regulate a trailing ribosome’s translocation efforts. An alternative conclusion is that ribosomes assembled in bL9− cells are inherently more prone to miscoding for some indirect reason, perhaps because of the incomplete 16S rRNA processing we previously reported (30). Arguing against this idea is the observation that bL9− ribosomes performed the same as wild-type ones in our assays that titrated the dose of ribosomes on reporter mRNAs.
This study highlights an important consideration in the interpretation and design of translation reporter systems: Changes in initiation or elongation rates can alter collision rates, which may unpredictably affect programmed recoding or E-site behavior. The long history of experimentation on bL9 stemmed from investigations of the remarkable translation bypass that occurs in T4’s gene60 (1, 26, 27). Although we did not characterize the gene60 motif in this study, we predict that ribosome loading influences that system as well. Likewise, it seems reasonable that ribosome load should influence the ability of repetitive extragenic palindromic (REP) sequences to regulate translation termination (68). Finally, because we observed that ribosome loading changes the translation outcomes for important regulatory elements, an exciting possibility is that these and other reprogramming motifs might be used to gauge the occupancy or health of the ribosome pool as transcriptomes are altered during adaptive responses.
Materials and Methods
Additional details of experimental procedures can be found in SI Appendix.
Bacterial Strains.
Cloning was performed using DH5α (Invitrogen), and plasmids were transferred to SM1344 for experimentation (a Δrna derivative of BW30270, which is a prototrophic, rph+ MG1655 strain from the Yale Coli Genetic Stock Center, #7925). This cell line was authenticated by our group using whole genome sequencing. The ΔrplI strain was generated using P1 transduction from a donor having the bL9 ORF replaced with a kanamycin resistance marker. Strains with reporter plasmids were maintained with antibiotic selection in lysogeny broth medium (0.5% NaCl) supplemented with 0.2% glucose. Quantitative experiments were performed using a defined Mops-buffered medium (SI Appendix).
In Vitro Translation Assays.
RNA transcripts were prepared from a plasmid template (SI Appendix) and added to purified in vitro translation reactions (PURExpress; New England Biolabs). Reactions were incubated at 37 °C for 2 h, and 2 μL of reaction mix (out of 10 μL total) were diluted into 48 μL of a phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.5 mM MgCL2, pH 7.4) in white 96-well plates for serial detection of Firefly and NanoLuc luminescence (NanoDLR; Promega).
Mass Spectroscopy of Nascent Peptides.
Fractions from sucrose gradients were incubated for 1 h under mildly alkaline conditions with nickel− nitrilotriacetic acid agarose. Unbound material was removed by washing, and the bound His6-containing peptides were eluted, desalted, and concentrated prior to mixing with crystallization matrix for MALDI mass spectroscopy (SI Appendix).
Measurement of Relative tRNA Abundances.
RNA was purified from sucrose gradient fractions and converted to complementary DNA (cDNA) using dedicated primers (SI Appendix). RT-qPCR was carried out using SsoFast EvaGreen Supermix (Bio-Rad) according to the manufacturer’s instructions in a CFX96 Real-Time PCR Detection System (Bio-Rad). Dedicated target primer pairs were designed to generate ∼110- to 120-base pair amplicons using MUSCLE software sequence alignments of E. coli tRNAs as a guide (69). Target specificity was confirmed for each primer pair by Sanger sequencing the qPCR amplicons.
Quantitative PCR Analysis.
Raw amplification data were exported from the thermal cycler and converted to comma-separated data tables, and template abundances were quantified by global fitting using the qPyCR software implementation of a global fitting algorithm (SI Appendix) (70). This method allowed for template abundances to be established without propagation of errors as exponentials. The resulting variances from this approach were generally less than 1% compared to the ∼50% error associated with the 2-ΔΔCT method (71) with the same RT data.
Collision Modeling.
Molecular modeling, superimpositions, distance measurements, and image generation were all performed using ChimeraX (72). Structure coordinate files were obtained from the RCSB Protein Data Bank (PDB): E. coli empty 70S, X-ray/crystal, PDB ID 4v4q (52); E. coli with mRNA, X-ray/crystal, PDB ID 4v50 (73); E. coli 70S empty A-site, cryoEM, PDB ID 5mdz (74); E. coli 70S release complex, cryoEM, PDB ID 6ore (75); E. coli 70S after termination, cryoEM, PDB ID 6osq (75); E. coli SecM complex, cryoEM, PDB ID 3jbu (50); Thermus thermophilus 70S rRNA modifications, X-ray/crystal, PDB ID 4y4o (76); T. thermophilus elongation complex, X-ray/crystal, PDB ID 4v6f (56); T. thermophilus with dnaX mRNA, cryoEM, PDB ID 5uq7 (16); and T. thermophilus lacking bL9 with EF-G bound, PDB ID 4v9h (77).
Supplementary Material
Acknowledgments
We thank Nikhil Bose and Kelly Rosch for their editorial comments. We also thank Yury Polikanov, Christine Dunham, and Eric Hoffer for their assistance in model building. This work was supported by NIH Grant 1R01GM118896.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910613116/-/DCSupplemental.
References
- 1.Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., Baranov P. V., Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G., New structural insights into translational miscoding. Trends Biochem. Sci. 41, 798–814 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Rodnina M. V., Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10, a032664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang B., Halgamuge S., Tang S. L., Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373, 90–99 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Studer S. M., Joseph S., Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 22, 105–115 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Salis H. M., Mirsky E. A., Voigt C. A., Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J., Gonzalez R. L. Jr, Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu. Rev. Biochem. 79, 381–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders C. L., Curran J. F., Genetic analysis of the E site during RF2 programmed frameshifting. RNA 13, 1483–1491 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaraj A., Shoji S., Holbrook E. D., Fredrick K., A role for the 30S subunit E site in maintenance of the translational reading frame. RNA 15, 255–265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tinoco I. Jr, Kim H.-K., Yan S., Frameshifting dynamics. Biopolymers 99, 1147–1166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q., Fredrick K., Intersubunit bridges of the bacterial ribosome. J. Mol. Biol. 428, 2146–2164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C., et al. , Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 16980–16985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C., et al. , Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 20, 582–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petropoulos A. D., Green R., Further in vitro exploration fails to support the allosteric three-site model. J. Biol. Chem. 287, 11642–11648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agirrezabala X., et al. , Ribosome rearrangements at the onset of translational bypassing. Sci. Adv. 3, e1700147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Hong S., Ruangprasert A., Skiniotis G., Dunham C. M., Alternative mode of E-site tRNA binding in the presence of a downstream mRNA stem loop at the entrance channel. Structure 26, 437–445.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekine Y., Eisaki N., Ohtsubo E., Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235, 1406–1420 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Licznar P., et al. , Programmed translational -1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 22, 4770–4778 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma V., et al. , Analysis of tetra- and hepta-nucleotides motifs promoting -1 ribosomal frameshifting in Escherichia coli. Nucleic Acids Res. 42, 7210–7225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchihashi Z., Kornberg A., Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 87, 2516–2520 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen B., Gesteland R. F., Atkins J. F., Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: Programmed efficiency of 50%. J. Mol. Biol. 271, 47–60 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen B., Wills N. M., Gesteland R. F., Atkins J. F., rRNA-mRNA base pairing stimulates a programmed -1 ribosomal frameshift. J. Bacteriol. 176, 6842–6851 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craigen W. J., Caskey C. T., Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322, 273–275 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Kawakami K., Nakamura Y., Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor 2. Proc. Natl. Acad. Sci. U.S.A. 87, 8432–8436 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devaraj A., Fredrick K., Short spacing between the Shine-Dalgarno sequence and P codon destabilizes codon-anticodon pairing in the P site to promote +1 programmed frameshifting. Mol. Microbiol. 78, 1500–1509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W. M., et al. , A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science 239, 1005–1012 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Herbst K. L., Nichols L. M., Gesteland R. F., Weiss R. B., A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 91, 12525–12529 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herr A. J., Nelson C. C., Wills N. M., Gesteland R. F., Atkins J. F., Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol. 309, 1029–1048 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Seidman J. S., Janssen B. D., Hayes C. S., Alternative fates of paused ribosomes during translation termination. J. Biol. Chem. 286, 31105–31112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naganathan A., Wood M. P., Moore S. D., The large ribosomal subunit protein L9 enables the growth of EF-P deficient cells and enhances small subunit maturation. PLoS One 10, e0120060 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaha G., Stanley R. E., Steitz T. A., Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science 325, 966–970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ude S., et al. , Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Doerfel L. K., et al. , EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Woolstenhulme C. J., et al. , Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, E878–E887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajkovic A., Ibba M., Elongation factor P and the control of translation elongation. Annu. Rev. Microbiol. 71, 117–131 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Nag B., et al. , Monoclonal antibodies to Escherichia coli ribosomal proteins L9 and L10. Effects on ribosome function and localization of L9 on the surface of the 50 S ribosomal subunit. J. Biol. Chem. 266, 22129–22135 (1991). [PubMed] [Google Scholar]
- 37.Lieberman K. R., et al. , The 23 S rRNA environment of ribosomal protein L9 in the 50 S ribosomal subunit. J. Mol. Biol. 297, 1129–1143 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Yusupov M. M., et al. , Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Selmer M., Gao Y.-G., Weixlbaumer A., Ramakrishnan V., Ribosome engineering to promote new crystal forms. Acta Crystallogr. D Biol. Crystallogr. 68, 578–583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer N., et al. , Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567–570 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Gao Y.-G., et al. , The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullwinkle T. J., et al. , (R)-β-lysine-modified elongation factor P functions in translation elongation. J. Biol. Chem. 288, 4416–4423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hersch S. J., Elgamal S., Katz A., Ibba M., Navarre W. W., Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J. Biol. Chem. 289, 28160–28171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woolstenhulme C. J., Guydosh N. R., Green R., Buskirk A. R., High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottesman S., Roche E., Zhou Y., Sauer R. T., The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flynn J. M., et al. , Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 10584–10589 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen B., et al. , Upstream stimulators for recoding. Biochem. Cell Biol. 73, 1123–1129 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Nakatogawa H., Ito K., The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Tsai A., Kornberg G., Johansson M., Chen J., Puglisi J. D., The dynamics of SecM-induced translational stalling. Cell Rep. 7, 1521–1533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., et al. , Mechanisms of ribosome stalling by SecM at multiple elongation steps. eLife 4, e09684 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong H., Nilsson L., Kurland C. G., Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Schuwirth B. S., et al. , Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Dunkle J. A., Cate J. H. D., “The packing of ribosomes in crystals and polysomes” in Ribosomes: Structure, Function, and Dynamics, Rodnina M. V., Wintermeyer W., Green R., Eds. (SpringerWien, NewYork, 2011), vol. 1, pp. 65–73. [Google Scholar]
- 54.Huter P., Müller C., Arenz S., Beckert B., Wilson D. N., Structural basis for ribosome rescue in bacteria. Trends Biochem. Sci. 42, 669–680 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Márquez V., Wilson D. N., Tate W. P., Triana-Alonso F., Nierhaus K. H., Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell 118, 45–55 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Jenner L. B., Demeshkina N., Yusupova G., Yusupov M., Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Shultzaberger R. K., Bucheimer R. E., Rudd K. E., Schneider T. D., Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 313, 215–228 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Rettberg C. C., Prère M. F., Gesteland R. F., Atkins J. F., Fayet O., A three-way junction and constituent stem-loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 286, 1365–1378 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Kim H. K., et al. , A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 111, 5538–5543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin P., Yu D., Zuo X., Cornish P. V., Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 15, 185–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandt F., et al. , The native 3D organization of bacterial polysomes. Cell 136, 261–271 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Mohapatra S., Choi H., Ge X., Sanyal S., Weisshaar J. C., Spatial distribution and ribosome-binding dynamics of EF-P in live Escherichia coli. MBio 8, e00300-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., Hershey J. W., Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3105–3114 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park M. H., Wolff E. C., Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 293, 18710–18718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guydosh N. R., Green R., Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156, 950–962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simms C. L., Yan L. L., Zaher H. S., Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juszkiewicz S., et al. , ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang W., Rudd K. E., Deutscher M. P., A role for REP sequences in regulating translation. Mol. Cell 58, 431–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carr A. C., Moore S. D., Robust quantification of polymerase chain reactions using global fitting. PLoS One 7, e37640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berk V., Zhang W., Pai R. D., Cate J. H., Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. U.S.A. 103, 15830–15834 (2006). Erratum in: Proc. Natl. Acad. Sci. U.S.A.103, 19931 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.James N. R., Brown A., Gordiyenko Y., Ramakrishnan V., Translational termination without a stop codon. Science 354, 1437–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu Z., et al. , The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nat. Commun. 10, 2579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polikanov Y. S., Melnikov S. V., Söll D., Steitz T. A., Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tourigny D. S., Fernández I. S., Kelley A. C., Ramakrishnan V., Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.