Significance
Bruton’s tyrosine kinase (BTK) is targeted in treatment of immune cancers. As patients experience drug resistance, there is a need for alternative approaches to inhibit BTK. Other recently published findings clarify the role of the BTK pleckstrin homology (PH) domain in mediating activation via dimerization and sensing of ligand concentration at the membrane. Work presented here provides insight into the autoinhibitory BTK structure that has so far been elusive via crystallographic methods. In the resting state, the BTK PH domain binds to the activation loop face of the kinase domain and allosterically alters key sites within the kinase domain. The findings define a new regulatory site, the PH/kinase interface, that can be exploited in drug discovery efforts.
Keywords: kinase, regulation, PH domain
Abstract
The pleckstrin homology (PH) domain is well known for its phospholipid targeting function. The PH-TEC homology (PHTH) domain within the TEC family of tyrosine kinases is also a crucial component of the autoinhibitory apparatus. The autoinhibitory surface on the PHTH domain has been previously defined, and biochemical investigations have shown that PHTH-mediated inhibition is mutually exclusive with phosphatidylinositol binding. Here we use hydrogen/deuterium exchange mass spectrometry, nuclear magnetic resonance (NMR), and evolutionary sequence comparisons to map where and how the PHTH domain affects the Bruton’s tyrosine kinase (BTK) domain. The data map a PHTH-binding site on the activation loop face of the kinase C lobe, suggesting that the PHTH domain masks the activation loop and the substrate-docking site. Moreover, localized NMR spectral changes are observed for non–surface-exposed residues in the active site and on the distal side of the kinase domain. These data suggest that the association of PHTH induces allosteric conformational shifts in regions of the kinase domain that are critical for catalysis. Through statistical comparisons of diverse tyrosine kinase sequences, we identify residues unique to BTK that coincide with the experimentally determined PHTH-binding surface on the kinase domain. Our data provide a more complete picture of the autoinhibitory conformation adopted by full-length TEC kinases, creating opportunities to target the regulatory domains to control the function of these kinases in a biological setting.
Five nonreceptor TEC family tyrosine kinases are expressed in cells of hematopoietic lineage (1, 2). Two specific TEC kinases, Bruton’s tyrosine kinase (BTK) and IL-2–inducible T cell kinase (ITK), promote B cell and T cell signaling, respectively. Both kinases are cytoplasmic in resting cells and localize to the plasma membrane on receptor stimulation through binding of their pleckstrin homology (PH) domain to phosphatidylinositol-3,4,5-trisphosphate (PIP3), produced by the activity of PI3-kinase (3–7). The TEC family PH domain extends into a Tec homology (TH) motif, and so hereinafter we refer to this region as PHTH. The interaction of the BTK PHTH domain with PIP3 alters the conformation of the full-length kinase (8) and activates kinase function (8–10). In addition, the first crystal structures of the BTK PHTH domain revealed the “Saraste dimer” (11, 12), which likely forms at PIP3 membranes to promote trans-autophosphorylation (10, 13). The TEC family PHTH domain plays a clear role in activation, but the role of this domain in maintaining the inactive state of the kinase is less well understood.
Protein kinases are dynamic switches that shift between autoinhibited and activated ensembles in response to cellular signals (14, 15). For multidomain kinases, the conformational shift is accomplished through a variety of mechanisms. At the level of the kinase domain, ATP binding to the active site leads to activation loop phosphorylation (on Y551 in BTK) and triggers assembly of the regulatory spine, inward movement of the C-helix, formation of a salt bridge between a conserved lysine/glutamate pair, and conformational adjustments in the DFG motif (16, 17). In the multidomain TEC kinases, nucleotide preference and accessibility of the activation loop phosphorylation site are also controlled by noncatalytic domains (17). In addition to the N-terminal PHTH domain, the TEC kinases contain a proline-rich region (PRR) and Src homology 3 (SH3) and Src homology 2 (SH2) domains that impinge on the kinase domain to alter the conformational ensemble and thus the activation status of the enzyme. A crystal structure of the BTK SH3-SH2-kinase fragment has been solved (10) showing that the SH3 and SH2 domains of BTK assemble onto the distal side of the kinase domain (the surface opposite the activation loop), stabilizing the autoinhibited form of the kinase in a manner similar to that described for the SRC family (18–20) (Fig. 1A).
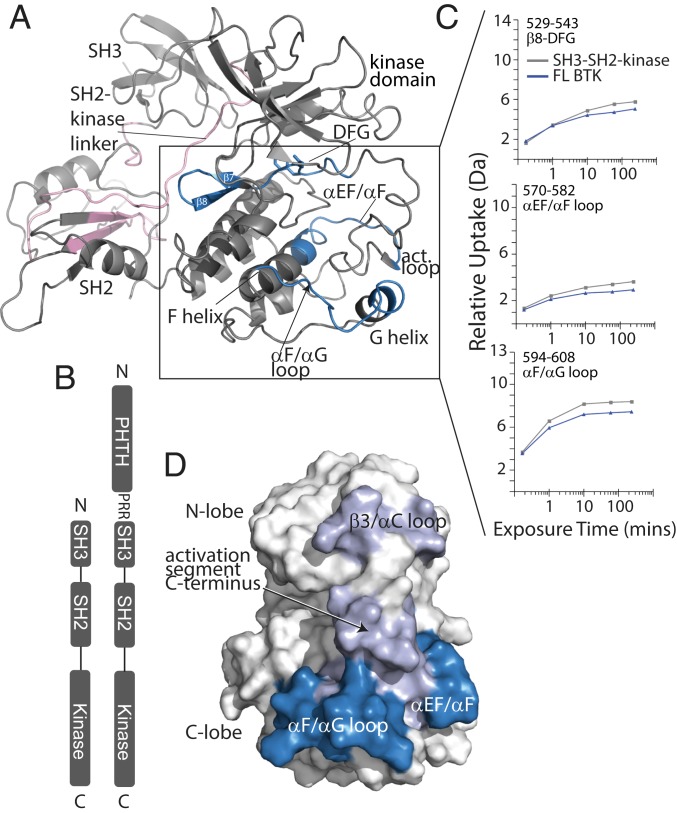
Fig. 1.
HDX-MS localizes PHTH interactions to the kinase domain C-lobe. (A) Structure of the autoinhibited BTK SH3-SH2-kinase fragment (Protein Data Bank [PDB] ID code 4XI2). Peptides showing protection (blue) or exposure (pink) across the time course of HDX are mapped onto the ribbon structure. The peptide showing the most protection (>1 Da) encompasses the αF/αG loop and extends into the G helix (peptides 594 to 608), and peptides that exhibit less protection (0.5 to 1.0 Da) are localized to the αEF/αF loop into the F helix and the β7/β8-DFG strand (peptides 570 to 582 and 529 to 543, respectively). (B) Domain structures for full-length BTK (FL BTK) and the SH3-SH2-kinase fragment. (C) Specific deuterium uptake curves showing HDX differences (DFL– DSH3-SH2-kinase, where D is relative deuterium incorporation) for BTK FL (dark blue) and SH3-SH2-kinase (gray). (D) Surface rendering of the BTK structure showing the location (blue) of the peptides that are protected in full-length BTK compared with the SH3-SH2-kinase fragment that lacks the PHTH domain. For the addition of PHTH to BTK kinase domain in trans, the αF/αG loop (blue), as well as the activation segment C terminus and β3/αC loop (lavender), show protection in the presence of PHTH. Experimental parameters and complete peptide deuterium uptake data are provided in Dataset S1.
The PHTH domain and PRR set the TEC family apart from the SRC family and, perhaps as a result of these additional domains, crystal structures of the full-length TEC kinases have not been solved. In addition to the BTK PHTH “Saraste dimer” (11), the structure of a construct that tethers the PHTH and kinase domains via a short linker (BTK PHTH-kinase) has been reported (10). In this PHTH-kinase structure, the PHTH domain contacts the N-lobe of the kinase domain (toward the distal side), but subsequent solution data (8) suggest that this binding mode may be a transient interaction rather than the primary mode of autoinhibition. The “Saraste dimer” is also observed in the PHTH-kinase structure, and since this PHTH dimer structure is compatible with PIP3 binding, the crystallographic insights might reveal a PHTH/kinase structure that accompanies membrane association and activation of BTK (8, 10). Recent work demonstrates a strict dependence of BTK activation (via dimerization) on PIP3 density at the membrane (21). Despite these advances in deciphering the BTK activation mechanism, a clear description of the autoinhibitory role of the PHTH domain in the full-length TEC kinases is lacking, however.
Biochemical evidence suggests that the PHTH domain negatively regulates kinase activity; deletion of PHTH leads to a 5-fold increase in kcat for ITK (22), and substrate phosphorylation of the BTK SH3-SH2-kinase fragment lacking the PHTH domain is faster than the same reaction catalyzed by the full-length BTK enzyme (10). Solution methods have mapped a regulatory surface centered on the β3-β4 loop of the PHTH domain; mutation of this region activates full-length BTK and ITK (8, 9). Thus, part of the regulatory picture is available, but our understanding of the TEC autoinhibitory mechanism remains incomplete without defining where and how the PHTH domain exerts its regulatory effect on the kinase domain. Data reported here localize the BTK PHTH regulatory interaction to the activation loop face of the kinase domain. This finding is consistent with previous biochemical data showing an increase in the accessibility of the ITK activation loop to an exogenous kinase either on deletion of the PHTH domain or on binding of the PHTH domain in the full-length enzyme to PIP3 (9). Evolutionary studies further suggest a modular evolution of TEC kinases in which BTK-specific residues define a unique PHTH-binding interface by building on the inactive SRC module.
Results
Hydrogen Deuterium Exchange Mapping.
To assess the interaction surface on BTK that mediates PHTH regulation, we used hydrogen/deuterium exchange mass spectrometry (HDX-MS) to probe the solvent-exchange behavior of backbone amides. Comparing HDX for the BTK SH3-SH2-kinase fragment that lacks the PHTH domain with full-length BTK (Fig. 1B) shows that exchange is reduced in the kinase domain and increased in the noncatalytic regions of full-length BTK (Fig. 1A). The peptides derived from the αEF/αF and αF/αG loops and the G-helix are protected from exchange in full-length BTK that includes the PHTH domain compared with the truncated SH3-SH2-kinase protein, which does not include PHTH (Fig. 1 A and C). These peptides cluster to a surface on the kinase domain C-lobe that surrounds the C terminus of the activation segment (Fig. 1D). Decreased deuterium uptake in these surface- exposed regions suggests that the PHTH domain interacts with and protects this part of the kinase domain in full-length BTK. The other region of reduced exchange in full-length BTK, β7/β8 through the DFG motif (Fig. 1 A and C), is not accessible in the autoinhibited BTK structure (10) and thus may be a secondary effect due to allosteric changes within the kinase domain on association of the PHTH domain. In contrast to protection in the C-lobe, increased deuterium exchange is measured in the SH2 and SH2-linker regions (Fig. 1A) indicating that this part of the autoinhibited structure is less protected from solvent in full-length BTK. The BTK SH3-SH2-kinase fragment used in the HDX-MS experiments lacks not only PHTH, but also the PRR. Therefore, the increased exchange observed in the SH2 and SH2-linker regions of full-length BTK compared with SH3-SH2-kinase could be due to the PRR competing with the SH2-kinase linker region for binding to SH3 (8), resulting in release of the SH3-SH2-linker region and the observed increase in exposure.
Based on the HDX-MS data indicating that the PHTH domain contacts the kinase domain in full-length BTK, we next made use of the separate kinase and PHTH domains to explore whether the interaction could be studied in trans. HDX was measured for the BTK kinase domain alone and in the presence of excess PHTH domain (Dataset S1). The results are consistent with the data acquired using full-length BTK; protection is observed on the activation loop face of the kinase domain, with the most reproducible and strongest region of protection at the C-terminal end of the activation segment (Fig. 1D). Protection is also observed in the αF/αG loop, coinciding with the peptide showing the greatest degree of protection in full-length BTK (Fig. 1 C and D). A small reduction in exchange is also observed for a peptide derived from the N-lobe β3/αC loop (Fig. 1D). Although the differences in deuterium exchange in this experiment are small, they are consistent with a weak regulatory interaction that is favored by the intramolecular nature of the contact in the full-length protein. To further probe this PHTH regulatory mechanism, we turned to an orthogonal solution-based biophysical approach.
Mapping the PHTH/Kinase Interface by NMR Spectroscopy.
NMR spectroscopy provides insight into protein-protein interactions at the single amino acid level. Provided that backbone amide resonance assignments are in hand, chemical shift mapping is a rapid approach to defining a protein-protein interaction interface. Sequential assignments were not available for the BTK kinase domain at the outset of this work, and so we proceeded to assign backbone amide resonance. The 1H-15N TROSY HSQC spectrum (SI Appendix, Fig. S1) is consistent with a folded domain, and 210 amide resonances (of 263 nonproline residues) are observed, of which 114 (54%) have been unequivocally assigned. The assigned residues are distributed across the entire BTK kinase domain structure (SI Appendix, Fig. S1), providing ample spectral probes for mapping the PHTH interaction.
With a subset of amide resonances assigned, we next titrated unlabeled BTK PHTH domain into the 15N-BTK kinase domain. Overall loss of peak intensity is observed throughout the titration due to the increased molecular weight of the PHTH/kinase complex. In addition, selected NMR resonances of the BTK kinase domain show more extensive loss of peak intensity (i.e., exchange broadening) on addition of PHTH, consistent with a protein-protein interaction in the intermediate exchange regime (Fig. 2A). Exchange-broadened resonances are identified by plotting the ratio of peak intensities for the BTK kinase domain in the presence of PHTH domain relative to the BTK kinase domain alone (Fig. 2B and Dataset S2). For each point of the titration, residues exhibiting intensity ratios less than the mean minus 1 SD were mapped onto the BTK kinase domain structure to determine regions that sense PHTH binding (Fig. 3 A and B). Exchange broadening is observed for residues on the activation loop face of the kinase domain (Fig. 3 A and B), as well as within the active site (Fig. 3C) and on the distal side of the N-lobe (Fig. 3 B and D). Residues on the distal surface of the kinase domain were not affected in HDX-MS. Moreover, this distal side of the kinase domain is occupied by the SH3/SH2-kinase linker in autoinhibited BTK (Fig. 3 B and D), and so NMR spectral changes on the activation loop face indicate the autoinhibitory site for PHTH domain binding.
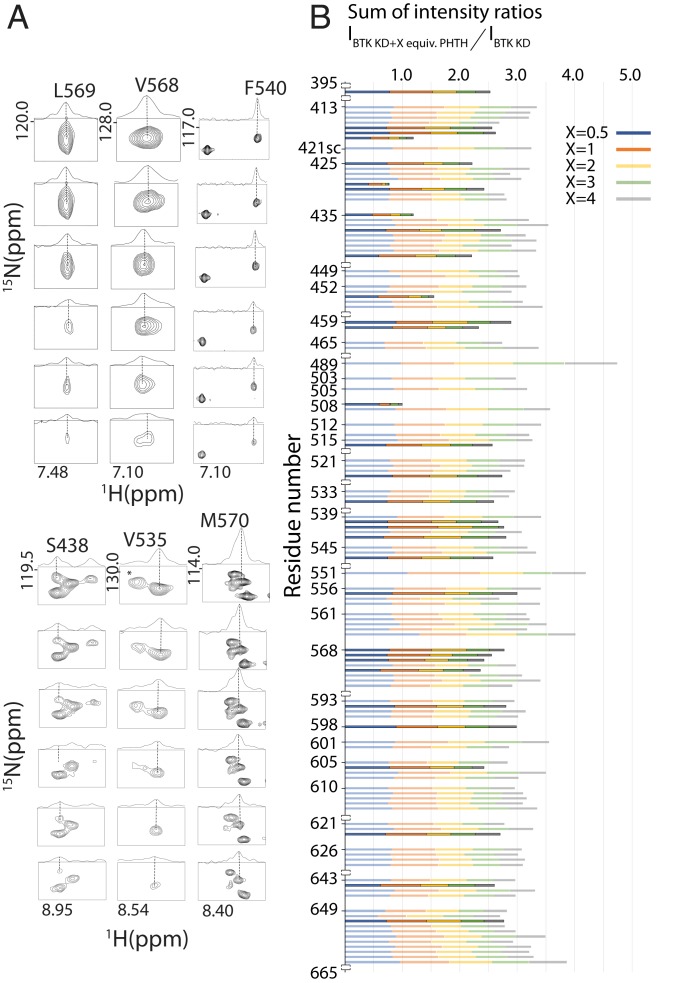
Fig. 2.
NMR spectral changes define selected BTK kinase domain residues that are affected by interaction with PHTH. (A) NMR data for selected kinase domain resonances showing line broadening throughout the titration of BTK PHTH. For each residue, the spectrum with no added PHTH is at the top, and each subsequent titration point (0.5, 1, 2, 3, and 4 equivalents of PHTH) is shown in order below. The asterisk indicates unassigned resonance that undergoes extensive broadening on addition of PHTH. (B) Histogram showing the sum of the intensity ratios (IBTK KD+PHTH/IBTK KD) for backbone amide resonances of the BTK kinase domain (KD at 200 μM) in the presence of 0.5, 1, 2, 3, or 4 molar equivalents of the PHTH domain. Columns are color-coded according to each point in the titration (Inset), and those that are bolded correspond to exchange-broadened residues as described in SI Appendix. All resonances are backbone amides except 421sc (tryptophan side chain).
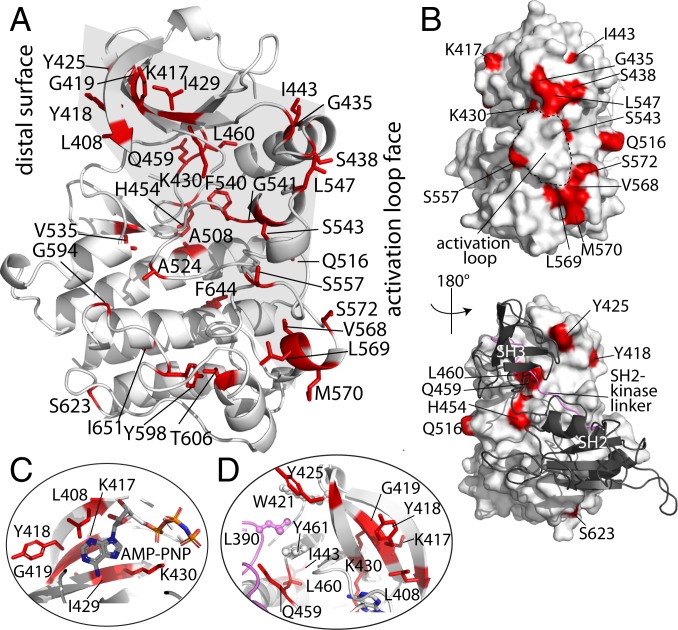
Fig. 3.
PHTH-induced NMR changes mapped onto the BTK kinase domain structure. (A) Residues corresponding to assigned BTK kinase domain resonances that show exchange broadening are shown in red (here and in all other panels) on the inactive structure of BTK kinase domain (PDB ID code 3GEN). The activation loop face and distal surface are labeled, and the gray-shaded area shows the concentration of residues between the activation loop face, the core of the active site, and the distal face. (B) Two views of the surface rendering of the autoinhibited BTK SH3-SH2-kinase structure (PDB ID code 4XI2). The N- and C-lobes of the kinase domain are indicated, and the activation loop is outlined by a dashed line (Top). The distal surface of the kinase domain is shown with the SH3-SH2 region in black ribbon (Bottom). (C) ATP-binding pocket showing kinase domain residues exhibiting spectral changes on the addition of PHTH. AMP-PNP is modeled into the structure based on PDB ID code 2DWB. (D) View of the residues on the distal face of the kinase domain (SH2-linker region in pink). The hydrophobic stack includes W421, L390 (from linker), and Y461 (labeled and shown in ball and stick).
Both HDX and NMR perturbations can be caused by direct binding and/or allosteric effects of the domain-domain interaction. Therefore, we used mutagenesis to probe the activation loop face of the BTK kinase domain to more directly define the recognition elements in this regulatory interaction. Five separate mutations, including 2 in the N-lobe above the activation loop (E434K and E439K) and 3 in the C-lobe below the activation loop (L569A, R562E, and R600S), were introduced into the BTK kinase domain. All residues targeted for mutation are located in peptides that show protection by HDX, show spectral perturbation by NMR, or are surface-exposed residues adjacent to those that exhibit NMR changes. Specific mutations either create a charge reversal or are based on mutations at these sites recorded in the COSMIC (Catalog of Somatic Mutations in Cancer) database (23). NMR spectra of BTK R562E, L569A, and E439K kinase domains were of poor quality due to protein precipitation during data acquisition, and so these mutants were not pursued further by either NMR or HDX, to avoid artifacts. The remaining 2 mutants, E434K and R600S, give rise to TROSY HSQC spectra that are nearly identical to the wild-type BTK kinase domain and thus suitable for further study.
Unlabeled BTK PHTH was titrated into 15N-labeled BTK E434K or R600S mutant kinase domains, followed by acquisition of 1H-15N TROSY HSQC spectra to assess whether and how the surface mutations influence the PHTH/kinase interaction (Dataset S2). Mapping the results of the NMR titrations onto the structure of the BTK kinase domain shows that mutation of R600 to serine results in fewer spectral changes on the activation loop face of the kinase domain compared with the wild-type PHTH/kinase titration. This suggests that mutation of R600 partially abrogates the PHTH/kinase interaction. In contrast, mutation of E434 has only a minor effect on the PHTH/kinase interaction, as indicated by spectral changes that more closely resemble that of the wild-type titration (SI Appendix, Fig. S2). These data support the HDX data (Fig. 1) indicating that the kinase domain C-lobe plays a key role in mediating the interaction between the BTK PHTH and kinase domains, while the N-lobe might be involved in the interaction but is not a primary binding determinant.
Evolutionary Constraints Associated with Functional Specialization of the Src Module in TEC Kinases.
The TEC kinases share a unique inactive conformation with SRC and ABL kinases and are collectively defined as the Src module kinases (20). The extent to which the Src module has diverged across specific lineages has not been examined, however. Using an unbiased, pattern-based classification of tyrosine kinase sequences (24), we identified co-conserved patterns that most distinguish Src module kinases from other tyrosine kinases, as well as those that uniquely distinguish TEC and BTK kinases (SI Appendix, Fig. S3). While many Src module-specific and TEC-specific residues appear to contribute to the unique inactive conformation of the kinase domain, a detailed analysis of these sequence features is beyond the scope of this work. We focus here on BTK-specific features of the BTK kinase domain.
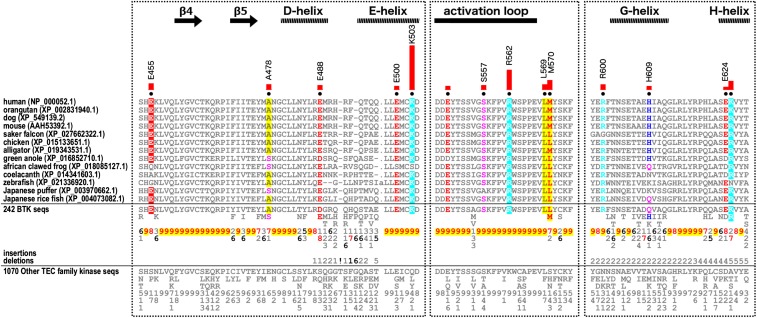
Comparisons of BTK sequences with other Tec kinase sequences (Fig. 4 and SI Appendix, Fig. S3) reveal BTK-specific evolutionary constraints imposed on residues in distinct regions of the kinase domain, including the experimentally determined PHTH interaction sites in the C-lobe (SI Appendix, Fig. S3). In particular, a patch of highly distinguishing BTK-specific residues locates to the front face of the kinase domain in the C-lobe—the same region identified through BTK PHTH/kinase experimental NMR and/or HDX-MS studies (Fig. 5 A and B). These include R600 and Y591 in the αFG loop; L569 and M570 in the αEF loop; E550, S557, and R562 in the activation segment; S436 in the β3/αC loop; and G535, L542, and S543 flanking the DFG motif (Fig. 5C). These residues are selective constraints in BTK, in that the nature of residues present at the corresponding position in other TEC kinases are biochemically different (Fig. 4). For example, BTK L569 and M570 are instead F or H/N in other TEC kinases. BTK-specific residues across the full alignment of the kinase domain are shown in SI Appendix, Fig. S3.
Fig. 4.
Contribution of specific BTK kinase domain residues to the PHTH interaction and unique sequence motifs defining BTK evolution. Sequence motifs unique to BTK kinases are shown. In the alignment, columns are highlighted where amino acids are highly conserved in BTK sequences but are nonconserved or biochemically dissimilar in other TEC family sequences. Histograms above the aligned sequences quantify the degree of divergence between BTK and other TEC sequences. Column-wise amino acid and insertion/deletion frequencies in BTK sequences and in other TEC family sequences are indicated in integer tenths, where a 5 indicates an occurrence of 50% to 60% in the given (weighted) sequence set. Kinase secondary structures are annotated above the histograms. Sequence numbering corresponds to the human BTK sequence.
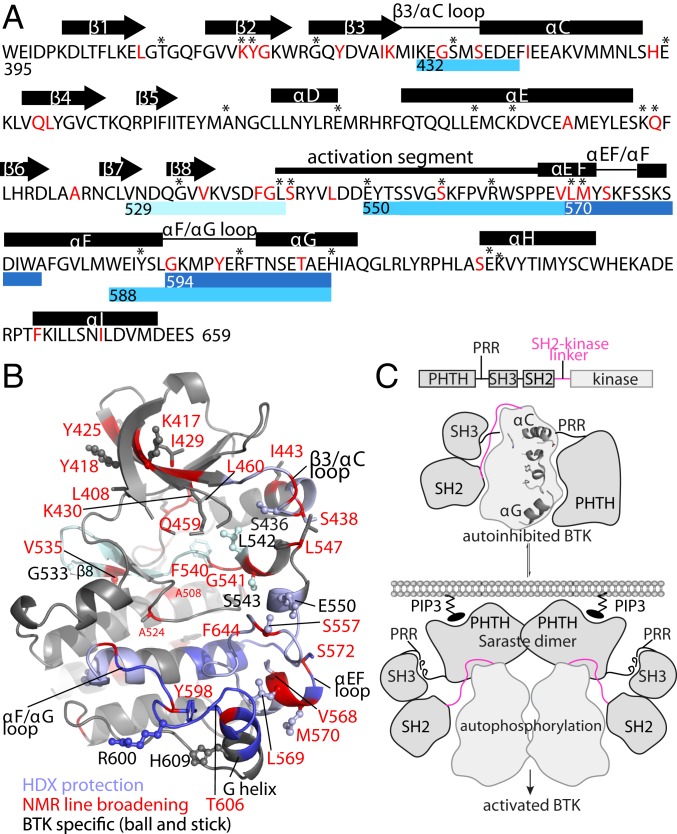
Fig. 5.
Combined data from HDX, NMR, and sequence conservation support a model for autoinhibited BTK. (A) Summary of HDX, NMR, and BTK-specific residues. Blue bars beneath the sequence of the BTK kinase domain indicate peptides that exhibit protection from exchange in full-length BTK compared with the SH3-SH2-kinase fragment (dark blue) or for the BTK kinase domain on the addition of excess PHTH domain (light blue). The peptide spanning the β8 strand into the DFG motif is indicated by a light cyan bar to indicate that this peptide shows no change in HDX before 10 min. Amino acids in red correspond to those residues for which NMR resonances undergo exchange broadening on the addition of PHTH to the BTK kinase domain. Asterisks above the sequence indicate BTK-specific residues. A secondary structure for the BTK kinase domain is shown above the primary sequence. (B) Data derived from NMR, HDX, and sequence comparison converge onto the β3/αC loop, activation loop, DFG region, αEF loop, G helix, and αF/αG loop. The blue ribbon represents peptides derived from HDX (dark blue, light blue, and light cyan as in A), red labels and red alpha carbons indicate residues identified by NMR, and BTK-specific residues are depicted in ball-and-stick format on the structure of inactive BTK (3GEN). (C) Model depicting stages of BTK regulation. The fully autoinhibited state, based on the crystal structure of the SH3-SH2-kinase fragment (10) and solution data presented here, consists of the SH3-SH2 region sandwiching the SH2-kinase linker (pink), with the distal side of the kinase domain and the PHTH domain on the opposite activation loop face. The domain structure of full-length BTK is provided above the model. Membrane-associated PIP3 competes with the kinase domain for binding of PHTH, releasing PHTH from its interaction with the kinase domain activation loop face. The crystal structure of the PHTH-kinase fragment (10) suggests that the PHTH domain contacts the N-lobe of the kinase domain in a manner compatible with PIP3 binding and formation of the “Saraste dimer.” The PRR may also release the SH3 domain from the distal side of the kinase domain (8) to promote a shift toward the active kinase. Autophosphorylation will result in activated BTK.
Discussion
The autoinhibitory conformation of the SRC module was determined 2 decades ago. Crystal structures of 2 members of the SRC family (18, 19) revealed the core regulatory unit shared across a number of eukaryotic protein tyrosine kinases (20). The SH3 domain sandwiches the SH2-linker region with the distal side of kinase domain N-lobe, and the SH2 domain mediates regulatory contacts with the C-lobe. The full-length TEC family kinases have been resistant to crystallization since they were first cloned in the early 1990s (25–31). Instead, solution methods provide significant insight into molecular-level regulation of these signaling molecules. We previously mapped inhibitory surfaces on the PHTH domains of both ITK and BTK and showed that PIP3 binding to the PHTH lipid-binding pocket competes with PHTH-mediated autoinhibition (8, 9). Moreover, PHTH association with the kinase domain reduces accessibility of the activation loop (9). These findings, along with the current findings based on HDX, NMR, and mutational analysis, support a model of fully autoinhibited BTK with the SH3 and SH2 domains on the distal side of the kinase domain (forming the Src module) and the PHTH domain on the activation loop face of the kinase domain in a manner that sterically hinders the lipid-binding pocket of PHTH (Fig. 5C). NMR spectral perturbations are also detected on the distal face of the kinase domain N-lobe on the addition of PHTH to the isolated kinase domain, suggesting the possibility of another interaction interface.
Even though such an arrangement has been captured crystallographically (10), the SH3-SH2-linker region in autoinhibited BTK occludes the distal surface mapped by NMR, suggesting that the putative PHTH/kinase interaction at this site may be accessible only as the full-length kinase opens on activation, possibly in the context of the PRR region displacing the SH3 domain from the SH2-kinase linker (Fig. 5C). Indeed, the PHTH domain in the structure of the tethered PHTH-kinase construct forms the well-characterized “Saraste dimer” that forms on localization of BTK to the membrane (10, 13, 21) and is likely mutually exclusive with the fully autoinhibited state. Thus, the molecular details of stepwise activation of BTK are coming into clearer view by virtue of findings generated by a range of experimental efforts.
The BTK-specific features have likely been accommodated through the selection of TEC-specific motifs during the evolution of the SRC module. For example, some of the most distinguishing BTK-specific residues locate to the αE helix (K503) and the αC-β4 loop (E455) on the back of the kinase domain, where they associate with the SH2 domain and SH2-kinase linker (SI Appendix, Fig. S3). These and other residues projecting from the distal side of the kinase domain that are not conserved among the TEC family suggest that the SH2-kinase linker- mediated autoinhibition may also rely on sequence features unique to BTK (32). Furthermore, the model for PHTH-mediated BTK regulation on the activation loop face of the kinase domain resembles a shared mechanism of autoinhibition across distinct kinase families. Three kinases outside of the TEC family—protein kinase C, protein kinase B (or AKT), and focal adhesion kinase (or PTK2)—contain regulatory subunits—C2, PH, and FERM domains, respectively—that dock onto their kinase domains in an autoinhibitory configuration that masks the activation loop face (SI Appendix, Fig. S4) (33–35). The regulatory domains in these kinases differ dramatically in size and sequence and, as we have shown for BTK, specialized regulatory surfaces on the activation loop face of numerous catalytic kinase domains have likely evolved in distinct contexts. In addition, the BTK-specific residues identified by pattern-based classification may play important roles in substrate recognition, given their location surrounding the G helix (36–38).
Solution NMR studies have previously demonstrated the malleability of the SRC module in response to inhibitor binding to the SRC active site (39). Similarly, the data presented here suggest that PHTH binding to the BTK kinase domain triggers allosteric changes within the kinase domain. Specifically, NMR evidence suggests that PHTH binding triggers structural and/or dynamic changes in the catalytic core of the kinase domain and in the ATP-binding pocket. It is also interesting to note that N-lobe residues that exhibit NMR spectral changes surround the previously identified “hydrophobic stack” present in the SRC module (40, 41). In BTK, the hydrophobic stack consists of L390 in the SH2-linker, which is sandwiched between W421 and Y461 in the kinase N-lobe (Fig. 3D). The stack is adjacent to W395, a key regulatory residue in the SH2-linker region (22, 42, 43). For the TEC kinase ITK, it has been previously shown that the L351A mutation (L390 in BTK) leads to release of the autoinhibitory SH3/linker-kinase interaction and increases the preference of the kinase domain for ATP over ADP (17). Thus, the NMR finding that the association of PHTH with the BTK kinase domain causes structural/dynamical perturbations around the hydrophobic stack suggests the possibility that the PHTH domain controls a key set of regulatory interactions among the C-lobe activation loop face, the active site, and the distal face of the kinase N-lobe.
Molecular details of the regulatory mechanisms are important as we strive to identify allosteric means to alter kinase function. The PHTH domain provides a unique means to target this hematopoietic TEC kinase family. Stabilizing the PHTH/kinase interaction is an appealing approach to allosterically inhibiting BTK function. Based on our data pointing to PHTH-dependent rearrangements within the active site of the kinase domain, a small molecule that stabilizes the PHTH/kinase interaction could enhance the efficacy of certain active site inhibitors. This strategy has the added benefit of PHTH autoinhibition simultaneously masking several critical features for activation: The PIP3-binding site, the PHTH dimerization interface, and the substrate-docking G helix. Like AKT (34), a small molecule that stabilizes the PHTH/kinase domains into the autoinhibited conformation may ultimately aid crystallization of a full-length TEC kinase, ending the long period between identification of the TEC gene family and a crystal clear view of the regulatory apparatus controlling TEC function in hematopoietic signaling.
Materials and Methods
All protein constructs and expression and purification methods used in this study, including uniform and selective 15N‐labeled proteins, have been described previously (8, 44). Procedures for HDX-MS have been described in detail previously (8) and in Dataset S1. NMR peak intensities were measured with SPARKY (45), and NMRviewJ (46) was used for all other data analysis. The intensity ratios were determined following the approach described in ref. 47 and as described in SI Appendix. For sequence alignment, tyrosine kinase sequences were extracted from the National Center for Biotechnology Information’s nonredundant sequence database (downloaded September 25, 2018) and aligned using curated sequence profiles and the Multiply-Aligned Profiles for Global Alignment of Protein Sequences (MAPGAPS) software suite (48–50). Sequences that did not span from at least the β3-lysine to the DFG-aspartate were deemed fragmentary and removed. A set of 13,639 representative diverse tyrosine kinase sequences (purged at 98% sequence identity) was used as input for the optimal multiple-category Bayesian Partitioning with Pattern Selection algorithm (omcBPPS) (51, 52), using a cluster size cutoff of 50 sequences to identify the major sequence families. Identification of motifs defining the Src module and BTK sequences (53) are described in SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants AI043957 (to A.H.A. and J.R.E.) and GM114409 (to N.K.). The HDX-MS experiments were partially supported by a research collaboration with the Waters Corporation.
Footnotes
Competing interest statement: A.H.A. has an equity interest in ImmVue Therapeutics, Inc., a company that may potentially benefit from the research results. The terms of this arrangement have been reviewed and approved by Iowa State University in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907566116/-/DCSupplemental.
References
- 1.Andreotti A. H., Joseph R. E., Conley J. M., Iwasa J., Berg L. J., Multidomain control over TEC kinase activation state tunes the T cell response. Annu. Rev. Immunol. 36, 549–578 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Berg L. J., Finkelstein L. D., Lucas J. A., Schwartzberg P. L., Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 23, 549–600 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Cantrell D., Signaling in lymphocyte activation. Cold Spring Harb. Perspect. Biol. 7, a018788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okoh M. P., Vihinen M., Pleckstrin homology domains of tec family protein kinases. Biochem. Biophys. Res. Commun. 265, 151–157 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Saito K., Scharenberg A. M., Kinet J. P., Interaction between the Btk PH domain and phosphatidylinositol-3,4,5-trisphosphate directly regulates Btk. J. Biol. Chem. 276, 16201–16206 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Hills L. B., Huang Y. H., Lipid and protein co-regulation of PI3K effectors Akt and itk in lymphocytes. Front. Immunol. 6, 117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fruman D. A., et al. , The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph R. E., Wales T. E., Fulton D. B., Engen J. R., Andreotti A. H., Achieving a graded immune response: BTK adopts a range of active/inactive conformations dictated by multiple interdomain contacts. Structure 25, 1481–1494.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devkota S., Joseph R. E., Boyken S. E., Fulton D. B., Andreotti A. H., An autoinhibitory role for the pleckstrin homology domain of interleukin-2-inducible tyrosine kinase and its interplay with canonical phospholipid recognition. Biochemistry 56, 2938–2949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., et al. , Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. eLife 4, e06074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baraldi E., et al. , Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure 7, 449–460 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Hyvönen M., Saraste M., Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: Molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16, 3396–3404 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Pechersky Y., Sagawa S., Pan A. C., Shaw D. E., Structural mechanism for Bruton’s tyrosine kinase activation at the cell membrane. Proc. Natl. Acad. Sci. U.S.A. 116, 9390–9399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornev A. P., Taylor S. S., Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 40, 628–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor S. S., Ilouz R., Zhang P., Kornev A. P., Assembly of allosteric macromolecular switches: Lessons from PKA. Nat. Rev. Mol. Cell Biol. 13, 646–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor S. S., Shaw A. S., Kannan N., Kornev A. P., Integration of signaling in the kinome: Architecture and regulation of the αC helix. Biochim. Biophys. Acta 1854, 1567–1574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Raußendorf F., de Ruiter A., Leonard T. A., A switch in nucleotide affinity governs activation of the Src and Tec family kinases. Sci. Rep. 7, 17405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W., Harrison S. C., Eck M. J., Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Sicheri F., Moarefi I., Kuriyan J., Crystal structure of the Src family tyrosine kinase Hck. Nature 385, 602–609 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Shah N. H., Amacher J. F., Nocka L. M., Kuriyan J., The src module: An ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Biol. 53, 535–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung J. K., et al. , Switch-like activation of Bruton’s tyrosine kinase by membrane-mediated dimerization. Proc. Natl. Acad. Sci. U.S.A. 116, 10798–10803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph R. E., Min L., Andreotti A. H., The linker between SH2 and kinase domains positively regulates catalysis of the Tec family kinases. Biochemistry 46, 5455–5462 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Forbes S. A., et al. , COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuwald A. F., Bayesian classification of residues associated with protein functional divergence: Arf and Arf-like GTPases. Biol. Direct 5, 66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyeck S. D., Berg L. J., Developmental regulation of a murine T-cell-specific tyrosine kinase gene, Tsk. Proc. Natl. Acad. Sci. U.S.A. 90, 669–673 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siliciano J. D., Morrow T. A., Desiderio S. V., itk, a T-cell-specific tyrosine kinase gene inducible by interleukin 2. Proc. Natl. Acad. Sci. U.S.A. 89, 11194–11198 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson S., et al. , Identification, cloning, and characterization of a novel human T-cell-specific tyrosine kinase located at the hematopoietin complex on chromosome 5q. Blood 82, 1561–1572 (1993). [PubMed] [Google Scholar]
- 28.Tanaka N., Asao H., Ohtani K., Nakamura M., Sugamura K., A novel human tyrosine kinase gene inducible in T cells by interleukin 2. FEBS Lett. 324, 1–5 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Vetrie D., et al. , The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 361, 226–233 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Yamada N., et al. , Structure and expression of novel protein-tyrosine kinases, Emb and Emt, in hematopoietic cells. Biochem. Biophys. Res. Commun. 192, 231–240 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Schaeffer E. M., Schwartzberg P. L., Tec family kinases in lymphocyte signaling and function. Curr. Opin. Immunol. 12, 282–288 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Register A. C., Leonard S. E., Maly D. J., SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family. Biochemistry 53, 6910–6923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antal C. E., Callender J. A., Kornev A. P., Taylor S. S., Newton A. C., Intramolecular C2 domain-mediated autoinhibition of protein kinase C βII. Cell Rep. 12, 1252–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu W. I., et al. , Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One 5, e12913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lietha D., et al. , Structural basis for the autoinhibition of focal adhesion kinase. Cell 129, 1177–1187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devkota S., Joseph R. E., Min L., Bruce Fulton D., Andreotti A. H., Scaffold protein SLP-76 primes PLCγ1 for activation by ITK-mediated phosphorylation. J. Mol. Biol. 427, 2734–2747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min L., Joseph R. E., Fulton D. B., Andreotti A. H., Itk tyrosine kinase substrate docking is mediated by a nonclassical SH2 domain surface of PLCgamma1. Proc. Natl. Acad. Sci. U.S.A. 106, 21143–21148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Q., Joseph R. E., Fulton D. B., Andreotti A. H., Substrate recognition of PLCγ1 via a specific docking surface on Itk. J. Mol. Biol. 425, 683–696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong M., et al. , Survey of solution dynamics in Src kinase reveals allosteric cross-talk between the ligand binding and regulatory sites. Nat. Commun. 8, 2160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonfloni S., et al. , The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J. 16, 7261–7271 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonfloni S., Frischknecht F., Way M., Superti-Furga G., Leucine 255 of Src couples intramolecular interactions to inhibition of catalysis. Nat. Struct. Biol. 6, 760–764 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Chopra N., et al. , Dynamic allostery mediated by a conserved tryptophan in the tec family kinases. PLoS Comput. Biol. 12, e1004826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joseph R. E., Xie Q., Andreotti A. H., Identification of an allosteric signaling network within Tec family kinases. J. Mol. Biol. 403, 231–242 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joseph R. E., et al. , Activation loop dynamics determine the different catalytic efficiencies of B cell- and T cell-specific tec kinases. Sci. Signal. 6, ra76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson B. A., Blevins R. A., NMR view: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Farrow N. A., et al. , Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S., The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Mohanty S., et al. , Hydrophobic core variations provide a structural framework for tyrosine kinase evolution and functional specialization. PLoS Genet. 12, e1005885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuwald A. F., Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics 25, 1869–1875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuwald A. F., A Bayesian sampler for optimization of protein domain hierarchies. J. Comput. Biol. 21, 269–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neuwald A. F., Evaluating, comparing, and interpreting protein domain hierarchies. J. Comput. Biol. 21, 287–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neuwald A. F., Surveying the manifold divergence of an entire protein class for statistical clues to underlying biochemical mechanisms. Stat. Appl. Genet. Mol. Biol. 10, 36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.