Significance
What factors maintain genetic variation in natural populations? Opposing selection pressures on protein stability and catalytic activity are thought to maintain variation along thermal gradients in many enzymes. We examined a classic hypothesis of temperature-mediated balancing selection, the alcohol dehydrogenase enzyme of Drosophila melanogaster, in which 2 latitudinally distributed variants are thought to be maintained by an activity/stability trade-off. Using in vitro and in vivo assays and population genetic analyses, we found no evidence of the predicted biochemical or fitness trade-offs and no signature of balancing selection. Rather, one variant confers greater activity and survival in the presence of ethanol, irrespective of temperature. Variation in Adh, and possibly other enzymes, must therefore be caused by other factors correlated with temperature.
Keywords: evolutionary genetics, evolutionary biochemistry, adaptation, activity/stability trade-off, alcohol physiology
Abstract
Polymorphism in the alcohol dehydrogenase (ADH) protein of Drosophila melanogaster, like genetic variation in many other enzymes, has long been hypothesized to be maintained by a selective trade-off between thermostability and enzyme activity. Two major Adh variants, named Fast and Slow, are distributed along latitudinal clines on several continents. The balancing selection trade-off hypothesis posits that Fast is favored at high latitudes because it metabolizes alcohol faster, whereas Slow is favored at low latitudes because it is more stable at high temperatures. Here we use biochemical and physiological assays of precisely engineered genetic variants to directly test this hypothesis. As predicted, the Fast protein has higher catalytic activity than Slow, and both the Fast protein and regulatory variants linked to it confer greater ethanol tolerance on transgenic animals. But we found no evidence of a temperature-mediated trade-off: The Fast protein is not less stable or active at high temperatures, and Fast alleles increase ethanol tolerance and survivorship at all temperatures tested. Further, analysis of a population genomic dataset reveals no signature of balancing selection in the Adh gene. These results provide strong evidence against balancing selection driven by a stability/activity trade-off in Adh, and they justify caution about this hypothesis for other enzymes except those for which it has been directly tested. Our findings tentatively suggest that environment-specific selection for the Fast allele, coupled with demographic history, may have produced the observed pattern of Adh variation.
A major goal in evolutionary biology is to understand the evolutionary forces and biological factors that maintain genetic variation within and across populations. Multiple variants may be stably maintained by balancing selection—such as heterozygote advantage, frequency-dependent selection, or selection that is spatially or temporally heterogeneous—or by other forces, such as mutation and migration; variation may also exist transiently because of drift or directional selection on an allele that has not yet reached fixation. Distinguishing among these possibilities is challenging, because various evolutionary scenarios can produce similar sequence patterns (1–4). Moreover, analyses of genetic patterns do not illuminate the biological and environmental factors upon which selection acts. An effective strategy is to experimentally test specific hypotheses concerning the functional and fitness effects of genetic variation under specific environmental conditions (5–11).
The alcohol dehydrogenase (Adh) gene of Drosophila melanogaster is a classic case in which variation is thought to be maintained by balancing selection with a specific biochemical and physiological basis (1, 12–17). D. melanogaster larvae and adults use ethanol-rich rotting fruit as a dietary resource and have greater ethanol tolerance than closely related species. Numerous loci contribute to ethanol tolerance (18), but Adh’s role has been of particular interest. The ADH enzyme catalyzes the first step of the ethanol detoxification pathway and is a major determinant of tolerance (19, 20). Two classes of Adh alleles, encoding enzymes named Fast and Slow because of their migration speeds on electrophoretic gels, segregate in natural populations (21). The 2 classes differ by a single amino acid (SI Appendix, Fig. S1; lys [K] in Slow or thr [T] in Fast at site 192) and by many synonymous nucleotide differences in the coding sequence, introns, and flanking regions. This variation has been hypothesized to be maintained by balancing selection because of a trade-off between catalytic activity and thermostability caused by the K192T polymorphism (22–27).
This hypothesis was formulated decades ago based on several observations. First, Fast and Slow alleles are distributed along stable latitudinal clines in Australia and North America: Fast is more frequent at higher latitudes, where temperatures tend to be cooler, and nearly absent at lower, hotter latitudes (28–30). This distribution mirrors the phenotypic pattern: High-latitude populations are generally more ethanol-tolerant than those from low latitudes (31, 32), and fly strains containing Fast and Slow haplotypes differ in ethanol tolerance under some conditions (14, 23, 33, 34). Second, lysate from flies with the Fast genotype has greater catalytic activity against ethanol than lysate from Slow-genotype flies, but this order is reversed at higher assay temperatures (23–27, 35, 36). Third, in some laboratory selection experiments, the Fast allele increased in frequency over generations in the presence of high ethanol concentrations, indicating that Adh variants can be targets of alcohol-mediated selection (14, 34). Fourth, the first studies to apply widely used population genetics tests for balancing selection found that patterns of sequence diversity near K192T were consistent with predictions of balancing selection (16, 37–39). Finally, studies of other enzymes found that thermostable variants are often more frequent in hotter environments (40–42), and an intrinsic trade-off was proposed between stability and activity, putatively because excess stability could interfere with the structural flexibility required for fast catalysis (refs. 40–44, but see refs. 45–47).
These findings are consistent with the trade-off hypothesis, but the precise relationships between the amino polymorphism, temperature-sensitive effects on biochemical activity, and fitness in the presence of ethanol remain unknown. The relevant phenotypes—ethanol metabolism, overall ethanol tolerance, temperature sensitivity, and the heat stress response—are affected by not only Adh but a host of other genes. Various strains carrying Fast and Slow protein electromorphs may have incorporated different forms of this background variation, confounding causal inference about the phenotypic effects of Adh variation. Indeed, some of these other genetic factors are known to vary geographically (48–54). This background variation may also explain why some but not all studies have produced results consistent with the trade-off hypothesis (32, 55–64). Efforts to better control for genetic background (49, 65) made the experiments somewhat more precise, but the effects of K192T and other Adh polymorphisms on stability, function, and fitness have not been isolated, and the specific predictions of the trade-off hypothesis have not been tested. Here we precisely engineer genetic variants and characterize their biochemical and fitness-related properties to test the predictions of the hypothesis that balancing selection, mediated by a trade-off between enzyme activity and stability caused by the K192T polymorphism, explains the maintenance and distribution of the Fast and Slow alleles.
Results
Effects In Vitro of Polymorphism K192T.
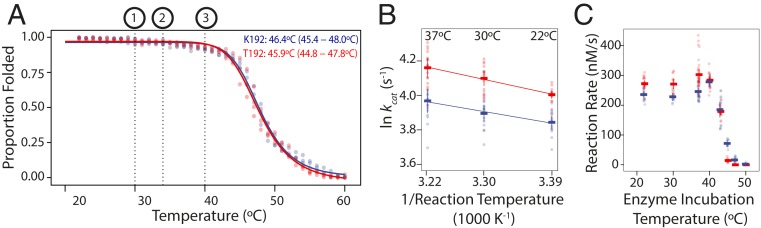
The hypothesis of an activity/stability trade-off makes testable predictions about the ADH protein’s in vitro properties. First, it predicts that T192 should reduce stability compared to K192. We synthesized ADH coding sequences that varied only at the polymorphic amino acid site and used a bacterial protein expression system to express and purify the 2 enzyme variants. We used circular dichroism (CD) spectrometry to monitor the loss of secondary structure as temperature increased. Contrary to the prediction, we found that the melting temperatures (Tm, the point at which 50% of secondary structure is lost) of the K192 and T192 enzymes are statistically indistinguishable (Fig. 1A). Both proteins are almost entirely folded up to 42 °C, well above the expected physiologically relevant range, including temperatures at which D. melanogaster heat shock response is induced (66), males become sterile (67), lethality occurs (SI Appendix, Fig. S2A), and short-term exposure incapacitates flies (SI Appendix, Fig. S2B).
Fig. 1.
Effect of K192T on ADH thermal stability and catalytic activity in vitro. (A) CD measurements of the proportion of folded protein across a temperature gradient for heterologously expressed ADH proteins containing K192 (blue) or T192 (red). Each circle shows one of 3 replicate measurements; solid lines, best-fit curves. Temperature at which 50% of secondary structure is lost (Tm) is listed, with 95% confidence interval. Dotted lines, temperatures at which the following occur: 1) induction of organismal heat shock response and male sterility, 29 °C to 30 °C; 2) death following chronic exposure, 34 °C to 35 °C; and 3) rapid death/incapacitance, 40 °C. (B) Effect of reaction temperature on maximum catalytic rate per unit enzyme (kcat) at saturating ethanol and cofactor concentrations. Circles show replicate measurements; horizontal bars show mean with 95% confidence interval. The reaction mixture including enzyme was incubated and reaction rates were measured at the temperature listed. Solid line, best-fit linear regression using expected Arrhenius relationship between reaction rate and temperature. There is a significant effect of temperature (F test, P < 0.01) and genotype (P < 0.01), but no genotype−temperature interaction (P = 0.46). (C) Effect of enzyme heat stress on maximal catalytic rate at saturating concentrations; 500 nM enzyme was incubated for 1 h at the temperature plotted; reaction mixture was then added and rate was measured at 22 °C.
Alternatively, T192 might impair catalytic activity at high temperature, without affecting overall structural stability. We used a spectrophotometric assay to measure ADH activity of controlled quantities of purified enzyme at reaction temperatures from 22 °C to 37 °C. Contrary to prediction, T192 has higher maximal activity per unit enzyme (kcat) at all temperatures tested (Fig. 1B). We also examined whether the 2 variants differ in their resistance to short-term increases in temperature, by incubating each protein at temperatures ranging from 22 °C to 50 °C for 1 h and then measuring catalytic activity at 22 °C. T192 again displayed higher activity at all temperatures below 40 °C; above this temperature, the activity of both variants rapidly declined (Fig. 1C). We observed the same patterns using different alcohols (SI Appendix, Fig. S3). These experiments show that T192 unconditionally confers greater activity than K192 and are inconsistent with a proposed deficit in stability or activity at high temperatures.
Effects In Vivo of Polymorphism K192T.
It is possible that the T192 variant may impose temperature-dependent costs in vivo that are not apparent in vitro. For example, the cellular environment in vivo may modify enzyme activity, or physiological/fitness effects may be mediated by mechanisms other than a difference in maximal ethanol turnover rate. To isolate the effect of K192T on survival in ethanol-rich environments, we engineered transgenic flies that differ solely by this one amino acid in the ADH protein. We prepared 2 versions of the ADH coding sequence, one coding for lys and the other for thr at site 192, both in the context of a naturally occurring Slow allele of the Adh locus, including cis-regulatory regions. These were then integrated into the same genomic location in an Adh-null strain.
The trade-off hypothesis predicts that the T192 strain’s resistance to ethanol should be greater than that of K192 strains at colder temperatures, but lower at higher temperatures, at which the T192 enzyme would denature, slowing ethanol detoxification. We first tested this hypothesis in larvae at temperatures within the operable temperature range of D. melanogaster. We measured mortality of 4,000 larvae of each genotype at variable ethanol concentrations and temperatures and calculated the ethanol dose at which half of the organisms die (LD50). At 22 °C, LD50 of K192 and T192 strains were not significantly different (Fig. 2A), and there was no statistical effect of genotype on mortality (SI Appendix, Table S1). At 27 °C, mortality of both strains was higher, but T192 larvae were significantly more ethanol-resistant than K192 larvae (Fig. 2B and SI Appendix, Table S1). The K192T polymorphism therefore affects larval ethanol tolerance, but the direction of temperature dependence is opposite to that predicted by the trade-off hypothesis.
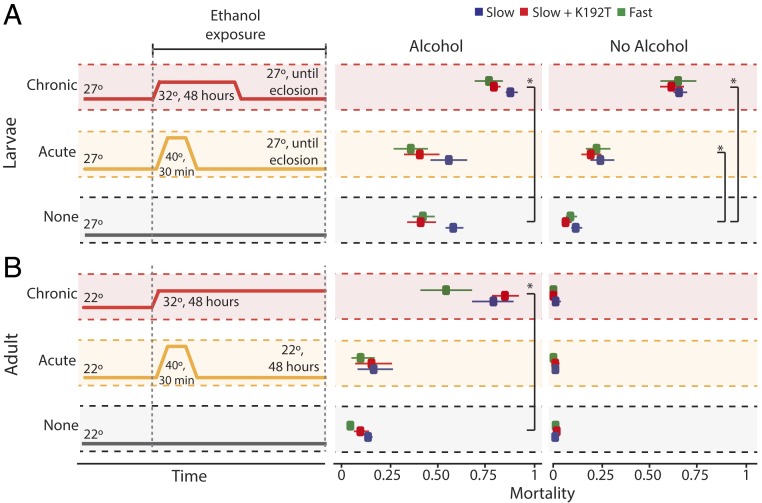
Fig. 2.
Effect of temperature and Adh genotype on ethanol tolerance in transgenic flies. Adh-null strains were transformed with a naturally occurring Slow allele (blue), the Slow allele modified with mutation K192T (red), or a naturally occurring Fast allele containing T192 and linked polymorphisms (green). Larvae (A and B) and adult females (C and D) were assayed for survival in the presence of increasing ethanol concentrations at different temperatures. Circles and error bars show the mean and SEM of the fraction of individuals dead in each treatment. For larvae, survival until eclosion was measured in 8 to 10 replicates of 30 larvae each per treatment; for adults, survival after 48 h of exposure was measured in 4 to 8 replicates of 25 to 30 individuals each per treatment. For each genotype, the LD50 is shown, with SE. Asterisks, significant differences between genotypes (P < 0.05, Wald’s test).
We next tested whether K192T affects ethanol resistance in a temperature-dependent fashion when flies are exposed to ethanol as adults. We measured mortality of ∼2,000 flies of each sex and strain at variable ethanol concentrations and temperatures. We found no statistically significant difference between the LD50 values of K192 and T192 strains at either 22 °C or 27 °C (Fig. 2 C and D and SI Appendix, Table S2). Males were more ethanol-tolerant than females, and both genotypes tolerated more ethanol at lower temperatures, but under no conditions tested did K192 flies show higher survivorship than T192 flies.
Another possibility is that T192 might have deleterious effects at temperatures beyond the normal physiological range. We assayed larval and adult mortality after chronic heat stress (48 h at 32 °C) or acute heat treatment (30 min at 40 °C) in the presence or absence of an intermediate ethanol concentration (Fig. 3). Both types of heat stress increased mortality in larvae (P < 0.01; SI Appendix, Table S3), and chronic heat stress reduced ethanol tolerance in adults (SI Appendix, Table S4). But the mortality of T192 flies was no greater than that of the K192 flies under any conditions tested (Fig. 3).
Fig. 3.
Effect of heat stress and Adh genotype on ethanol tolerance in transgenic flies. Transgenic (A) larvae and (B) adults were subject to chronic or acute heat stress in the presence or absence of alcohol. (Left) Schematic of treatments. For larvae, 8.5% ethanol exposure was initiated at late second or early third instar and continued through heat treatment until eclosion, when survivorship was measured. For adults, 5% ethanol exposure was continued through 48 h of heat treatment. (Right) Fraction dead at measurement in each group. Points and error bars, mean and 95% CI of 8 to 10 replicates of 30 larvae or 25 to 30 adults each. Asterisks, increased mortality compared to no-heat-stress control (P < 0.01, Wald’s test).
These results are all inconsistent with the predictions of the hypothesis that balancing selection maintains the Fast and Slow alleles in different environments because amino acid T192 increases ethanol resistance at low temperatures but reduces it at high temperatures. High ethanol concentrations are toxic, and toxicity increases at high temperatures. At all temperatures tested, however, T192 either increases ethanol tolerance or has no effect. Under no conditions did T192 confer greater mortality than K192, providing no evidence of a fitness trade-off.
Effects In Vivo of Linked Regulatory Polymorphisms.
An alternative possibility to explain the maintenance of Fast and Slow alleles and their latitudinal pattern is that polymorphisms at sites other than amino acid 192, such as nucleotide differences at regulatory sites (48), may increase the fitness of Fast-carrying flies at low temperature but decrease it at high temperature. We tested this possibility by engineering transgenic flies containing a naturally occurring Fast Adh allele, which corresponds precisely in its boundaries to that of our Slow and Slow+T192 alleles. The Fast allele construct comprises the entire coding sequence, all introns, and flanking regions that include all previously characterized cis-regulatory regions known to affect Adh expression, including the Δ1 indel polymorphism that cosegregates with T192 (49, 50, 68). We compared ethanol resistance of these transgenic Fast-allele flies at varying temperature to those carrying the Slow and Slow+T192 alleles described above.
We found that the Fast allele had no significant effect on larval ethanol tolerance compared to Slow+T192, whether they were raised at 22 °C or 27 °C or exposed to chronic or acute heat shock (Figs. 2 A and B and 3A); the included regulatory polymorphisms therefore did not affect larval ethanol tolerance. In adults, however, the Fast haplotype significantly increased ethanol tolerance compared to Slow+T192 in all temperature treatments (Figs. 2 B and D and 3B). Linked nucleotide polymorphisms in the Fast allele therefore increase ethanol tolerance only in adults, in contrast to amino acid polymorphism T192, which increases ethanol tolerance only in larvae. The transgenic Fast allele we used has been shown to increase expression compared to the Slow allele, and the alcohol dehydrogenase activity in lysate from these flies is similar to that of natural Fast and Slow strains, respectively (68). Both coding and regulatory aspects of the Fast allele therefore contribute to increased ethanol resistance, at different life stages. No temperature-dependent trade-off between genotypes was present at any stage or treatment.
No Sequence Signature of Balancing Selection on K192T.
The lack of in vivo or in vitro evidence for a fitness trade-off predicted by the balancing selection hypothesis led us to revisit the previously observed signature of balancing selection in Adh sequences (16, 37, 38). Previous analyses found that, when allele classes are defined by the amino acid at site 192, there is more between-class nucleotide diversity in the surrounding region than predicted by a specific parametric model given a neutral scenario at demographic equilibrium (37). Those analyses were based on relatively small samples from the geographically distinct populations available at the time; it is possible that sampling artifacts, demographic differences, or other evolutionary processes might have counterfeited or diluted a signature of balancing selection (3, 69). Further, as the authors of the original study noted, the neutral model used may not have been representative of historical evolutionary conditions at Adh (37).
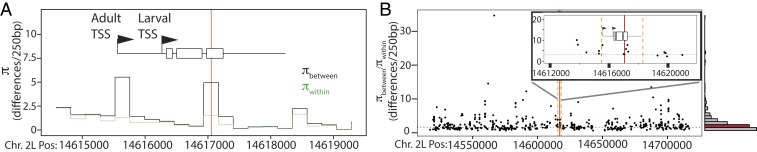
We therefore tested whether a signature of balancing selection is associated with amino acid site 192, using Adh gene sequences from 163 individuals in a single panmictic population (Raleigh, NC). Both Fast and Slow segregate at intermediate frequencies in this population (70), which is located within a latitudinal cline (28, 29). When haplotypes were classified based on the presence of K or T at site 192, nucleotide diversity between classes (πbetween) was higher than diversity within classes (πwithin) in the 250-base pair (bp) window centered on that site (Fig. 4A), a pattern similar to previous observations (37). πwithin is greater in the K192 class (SI Appendix, Fig. S3), consistent with the fact that K192 is the ancestral state (55, 71).
Fig. 4.
Genetic variation near Adh. (A) Within- and between-class nucleotide diversity when D. melanogaster haplotypes from Raleigh, NC, are classified by the amino acid at site 192. For each 250-bp window across the locus, the mean nucleotide difference over all pairs of sequences within each class (πwithin, green) and between classes (πbetween, black) is shown. Red line, K192T; rectangles, Adh exons; arrows, transcription start sites (TSS); black line, flanking regions encompassing all known natural cis-regulatory variants (68); SI Appendix, Fig. S4. (B) Distribution of excess πbetween/πwithin in the 200-kb region surrounding Adh. Each dot plots one SNP (frequency 0.25 to 0.35); πbetween/πwithin in a 250-bp window around it was calculated when haplotypes were classified based on the nucleotide at that site. Red line, K192T; orange, boundaries of Adh locus as in A; histogram, distribution of πbetween/πwithin across all SNPs; red bin, K192T. (Inset) Close view of plot across Adh locus.
But an excess of between-class diversity is, by definition, always expected around whatever site is used as the focal polymorphism to define classes. Diagnosing balancing selection requires assessing whether this excess is greater than that otherwise expected, given the classification effect and the many other evolutionary and demographic conditions that affect diversity in the same genomic region. If a site has evolved under balancing selection, then the excess of between-class diversity around that site is expected to be greater than around other putatively neutral polymorphic sites in the same region when those sites are used to define allele classes. We therefore compared the excess between-class diversity (πbetween/πwithin) around focal site 192 to the background distribution of the same statistic across a >200-kb region surrounding the Adh open reading frame (ORF), which extends far beyond the known Adh regulatory region and the expected scale of genetic linkage (70). For every single-nucleotide polymorphism (SNP) in this region with frequency similar to that of site 192, we classified alleles based on the state at that site and then calculated πbetween/πwithin in the 250-bp window around that site.
We found no signature of balancing selection associated with K192T. More than 60% of SNPs of similar frequency across the region have πbetween/πwithin higher than that of K192T (Fig. 4B). No other variants in the Adh coding or known regulatory regions are associated with an elevated πbetween/πwithin, either.
Discussion
The D. melanogaster Adh gene was once called the most convincing case of selection acting on natural genetic variation (72). Despite scores of studies investigating Adh variation, the specific genetic targets and biological mechanisms driving its evolution have remained elusive (19, 73). Historically, several technical factors made Adh an attractive system for probing the evolutionary factors that shape natural genetic variation. The ADH protein is abundantly expressed, and it exhibits genetic variation that can be readily observed using protein electrophoresis (19). ADH enzyme activity can be easily assayed in vitro, which raised the possibility of experimentally linking genetic variation in the protein to differences in its biochemical function. And ADH’s role in ethanol metabolism, together with the physiological and ecological importance of ethanol tolerance in D. melanogaster, meant that biochemistry could be plausibly linked to both physiology and environment-specific fitness. Adh therefore became one of the first systems in which an integrated experimental account connecting genotype, molecular function, phenotype, and fitness could be attempted (1, 72).
The hypothesis we tested—that the latitudinal clines of Adh variants observed in nature (25, 28) are maintained by balancing selection acting on a temperature-driven trade-off between protein stability and enzyme activity—was formulated long ago based on numerous lines of suggestive evidence. Technical limitations prevented decisive tests, because neither the amino acid polymorphism nor linked variants in the Fast allele class were fully isolated from other forms of genetic variation, which may plausibly contribute to ethanol- and heat-related fitness components and may be geographically structured in natural populations. We therefore used precise manipulative genetic approaches to specifically test the predicted effects of the amino acid polymorphism on protein activity and thermostability in vitro and on organismal heat sensitivity and ethanol tolerance in vivo. Using proteins that differ only at amino acid 192, expressed heterologously at matched concentrations and assayed under identical conditions, we found that the polymorphism does not cause the predicted biochemical trade-off; rather, T192 increases ADH activity in a temperature-independent fashion and has no measurable effect on in vitro thermal stability relative to K192. In vivo, the predicted fitness and physiological trade-off is not evident, either: Transgenic flies carrying T192 are more ethanol-tolerant than otherwise identical flies bearing K192, and introducing the cis-regulatory variants that are tightly linked to T192 further increases ethanol tolerance, but these effects are all independent of temperature. Even the sequence signature of balancing selection at the K192T polymorphism disappears when a similar test is applied to contemporary sequence samples and the background distribution in nucleotide diversity across Adh’s region of the genome is accounted for. These results cannot be reconciled with the hypothesis that balancing selection mediated by a stability/activity trade-off explains the Fast/Slow genetic variation in natural populations.
Why do our results differ from some previous findings and agree with others? First, our in vitro experiments, like much earlier work using cell lysates, show that the Fast protein has higher catalytic activity than the Slow protein (24, 26, 32, 35, 36, 56, 62, 64, 74, 75). Some previous studies found that this order is reversed at higher temperatures, but, unlike our assays—which used heterologously expressed, purified proteins at matched concentrations—those experiments did not isolate the ADH protein from other cellular factors (24, 26, 33, 35, 36, 74); for example, factors involved with heat stress also vary latitudinally (51, 76) and may explain a difference in temperature sensitivity between lysates. Second, we found that the Fast protein does not differ in thermostability from the Slow protein, using purified ADH protein and a direct assay of structural denaturation with increasing temperature; no previous study directly measured the structural stability of ADH proteins. Third, our in vivo results are consistent with many previous studies showing that Fast-containing natural strains have greater ethanol tolerance (14, 26, 34, 57, 58, 74). They contrast with earlier findings that Slow-containing strains have higher survivorship at higher temperatures (25, 26, 36, 74), but those studies, unlike our experiments using precisely engineered transgenic variants, did not isolate the causal role of the Adh allele from that of other cosegregating loci that may be enriched at lower latitudes (29, 52, 53). Finally, earlier work used recombinant lines to demonstrate and isolate the effects of Adh coding variants on Adh activity and noncoding variants on Adh expression level (49, 68, 75); we found that both coding and noncoding portions of the Fast allele increase ethanol tolerance, consistent with those results. Neither effect was temperature-dependent in our experiments; the earlier studies did not investigate heat sensitivity.
Our work does not rule out the possibility that other kinds of balancing selection might maintain Adh genetic variation along latitudinal gradients (77). Indeed, innumerable hypotheses could be articulated concerning selective factors that might act on genetic variation in Adh via biochemical and physiological properties other than an activity/stability trade-off. Our experiments were conducted in a specific strain of flies, and it is possible that this strain might buffer a temperature-dependent fitness effect of the Fast allele that would manifest in some other genetic backgrounds; however, our in vitro experiments show that the protein’s intrinsic thermal stability does not differ between Fast and Slow enzymes, and there is no temperature at which the T192 enzyme’s intrinsic catalytic activity becomes slower than that of K192. Thus, any fitness trade-off that might be revealed in a different genetic background would support balancing selection only through a mechanism different from the classical stability/activity trade-off. We did not detect evidence of balancing selection in Adh sequences using a classic nucleotide diversity-based test, but large population sizes and high recombination rates in D. melanogaster can rapidly erode the sequence patterns that this method is designed to detect (3, 6).
Our findings are inconsistent with the idea that an intrinsic trade-off between enzyme activity and thermal stability shapes the evolution of many proteins across thermal and genetic environments (40–44, 78). In ADH, T192 increases activity compared to K192 without reducing the protein’s thermal stability. Although mutations that increase activity often do decrease stability (42, 43), this observation may simply reflect the fact that the majority of all mutations reduce stability rather than a necessary trade-off (47). Indeed, numerous studies have identified mutations that increase both activity and stability (46, 79) or increase stability without reducing activity or function (11, 80, 81). Proteins from organisms that inhabit lower-temperature environments are often less stable than those from high-temperature environments (46), but this does not imply a trade-off: The same relationship would arise if proteins need only be stable enough to predominantly occupy folded conformations at the temperatures to which they are exposed, with excess stability having no effect on fitness (45).
Our results suggest a different hypothesis that may partially explain the maintenance of the Fast and Slow alleles. The Fast allele increases ethanol tolerance, irrespective of temperature, suggesting that it may be selected for in environments with higher levels of ethanol, while Fast and Slow may segregate with nearly equal fitness in lower-ethanol environments. If ethanol is more abundant in the environments of D. melanogaster at higher latitudes and sufficient gene flow occurs across this gradient, this scenario could also explain the clinal distribution of the alleles. This hypothesis is consistent with the observation that flies from higher latitudes have greater ethanol tolerance (31, 32) and with the observation that nucleotide diversity is very low in the Fast haplotype class (37), as expected if the haplotype originated recently and swept to increased frequency in some populations (39, 55). Our experiments provide evidence for several links predicted by this hypothesis among genotype, function, and fitness components in the presence of ethanol. But we know of no evidence to support the central premise that concentrations of ethanol in D. melanogaster’s environment are related to latitude. An alternative is that the clinal distribution of Fast and Slow could be caused by linkage to other alleles that mediate adaptation to latitude-correlated environments or by residual effects of geographically structured colonization patterns by European and African founder populations (82). Targeted experiments and population genetic analyses are therefore necessary to test and modify the hypothesis we propose to explain the observed patterns of genetic variation in Adh.
Methods
Protein Expression and Purification.
The ADH Slow allele coding sequence, which contains a Lys192 codon (K192), was synthesized with codon bias optimized for bacterial expression (GenScript). An identical sequence but containing Thr192 codon (T192) was generated from the Slow template by Phusion Site-Directed Mutagenesis (ThermoFisher). Sequences were cloned into pLIC-(His6)maltose binding protein fusion plasmids, verified by Sanger sequencing, and transformed into Escherichia coli BL21 (DE3) Rosetta. Protein was expressed and purified from transformed cells using nickel-affinity and column chromatography (detailed in SI Appendix).
Stability and Activity Assays.
Protein was concentrated to 10 μM in 50 mM sodium phosphate buffer. Absorbance was measured at 222 nM using a Jasco J-1500 CD Spectrometer at temperature increments of 2 °C from 22 °C to 60 °C, with 2 min of equilibration per temperature. Absorbance was measured twice per temperature, and this procedure was repeated 3 times on separate days. Proportion of folded protein and Tm were estimated using a 2-state 5-parameter logistic regression curve. Initial reaction rates were measured as reduced nicotinamide-adenine dinucleotide (NADH) coproduct formation (340 nm) for the first 90 s of reaction after enzyme addition at saturating conditions (500 nM NADH, 1 mM NAD, 200 mM alcohol, 50 mM KaPO4 buffer, pH 7.6). Effect of reaction temperature was assessed by incubating the reaction mixture for 1 h at 22 °C, 30 °C, or 37 °C and then adding the enzyme, which had been incubated at 22 °C. Effect of heat treatment on activity was measured by first incubating enzyme at temperatures ranging from 22 °C to 50 °C for 1 h and then adding the enzyme to reaction mixtures that had been incubated at 22 °C; 10 to 15 replicates were assessed for each enzyme−temperature combination. Effects of genotype, temperature, and genotype−temperature interaction on reaction rate were estimated by linear regression (lm() function) in R.
Transgenic Animals.
D. melanogaster that differ only at their Adh locus were made using the PhiC31-attp transgenesis system (SI Appendix and refs. 65 and 68). The Slow allele was derived from a naturally occurring Canton-S fly strain. The Slow+K192T is identical to the Slow allele except for a Lys−Thr coding change at amino acid 192. The Fast allele is from Florida strain FL9 (68). All constructs have identical boundaries and are the same ones used in refs. 65 and 68; they include the entire ADH coding sequence, the cotranscribed ADHR coding sequence, and 2.9 kb upstream and 1.6 kb downstream. Constructs were transformed into Canton-S Adh-null flies at the same landing site and bred to homozygosity.
Ethanol Survivorship.
Developmentally staged larvae and adults were placed in media with various concentrations of ethanol at 22 °C or 27 °C (4–8 replicates of 25 to 30 adults of each sex per treatment and 8 to 10 replicates of 30 larvae per treatment; ethanol concentration ranged from 0 to 10% for adults and 0 to 15% for larvae). Survivorship was measured as proportion eclosed (larvae) or alive after 48 h of exposure (adults). For heat stress, adult flies raised at 22 °C were placed into vials with 0% or 5% ethanol at 32 °C for 48 h or 40 °C for 30 min and then returned to 22 °C for 48 h. For each treatment, 4 to 8 replicates of 25 to 30 adults were assayed under each condition. For larvae, developmentally staged larvae were raised at 27 °C, placed in vials with 0% or 8.5% ethanol and then subjected to heat treatments (no treatment: 27 °C for 48 h; chronic: 32 °C for 48 h; acute: 40 °C for 30 min), and then returned to 27 °C; 8 to 10 replicates of 30 larvae per genotype and treatment were assayed, and the proportion of larvae surviving to eclosion was recorded. LD50 was estimated and the effects of ethanol concentration, genotype, temperature and sex on mortality were assessed using logistic regression and the quasibinomial link function, as implemented in glm() in R. Only interaction terms that significantly improved fit (F test) were included in the final fitted model. Additional details are provided in SI Appendix.
Sequence Analysis.
The Drosophila Genome Research Panel dataset was acquired and processed with scripts from Drosophila Genome Nexus (https://www.johnpool.net/genomes.html). Samples missing data information at >20% of sites in the Adh region were excluded. Sampled haplotypes were classified as Fast or Slow based on amino acid state at site 192 (coordinate 14617051, Flybase 5.0, Drosophila Genome Nexus 1.0). The 5-kb region centered on this site was divided into 250-bp windows; in each window, πwithin was calculated as the mean proportion of sequence differences across all pairwise comparisons within both classes; πbetween is the mean for all pairs in different classes. For the background distribution, we used a 200-kb region approximately centered on the Adh ORF (14517051–14717051), which contains 919 SNPs with minor allele frequency 0.25 to 0.35 (compare 0.30 for K192). For each such SNP, all haplotypes in the panel were classified based on the state at that SNP, and πbetween/πwithin was calculated for the 250-bp window centered on it.
Supplementary Material
Acknowledgments
We thank members of the J.W.T. laboratory and Evan Koch for advice and comments; Marty Kreitman for discussion and laboratory space; and David Loehlin for fly strains. The project was supported by an NSF grant (DEB-1501877 to J.W.T. and M.A.S.), an NSF graduate research fellowship (M.A.S.), NIH training grant (T32-GM007197 to M.A.S.), and NIH grants (R01-GM104397 and R01GM121931 to J.W.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909216116/-/DCSupplemental.
References
- 1.Lewontin R. C., The Genetic Basis of Evolutionary Change (Columbia University Press, New York, NY, 1974). [Google Scholar]
- 2.Gillespie J. H., The Causes of Molecular Evolution (Oxford University Press, 1994). [Google Scholar]
- 3.Charlesworth D., Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2, e64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vitti J. J., Grossman S. R., Sabeti P. C., Detecting natural selection in genomic data. Annu. Rev. Genet. 47, 97–120 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Karasov T. L., et al. , The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512, 436–440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unckless R. L., Howick V. M., Lazzaro B. P., Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr. Biol. 26, 257–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad K. V. S. K., et al. , A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337, 1081–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnen C. R., et al. , Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339, 1312–1316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett R. D. H., Hoekstra H. E., Molecular spandrels: Tests of adaptation at the genetic level. Nat. Rev. Genet. 12, 767–780 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Jones M. R., et al. , Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360, 1355–1358 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Jones F. C., et al. ; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team , The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Futuyma D., Evolution (Sinauer Associates, Sunderland, MA, ed. 3, 2013). [Google Scholar]
- 13.Li W.-H., Molecular Evolution (Sinauer Associates, Sunderland, MA, 1997). [Google Scholar]
- 14.van Delden W., Boerema A. C., Kamping A., The alcohol dehydrogenase polymorphism in populations of Drosophila melanogaster. I. Selection in different environments. Genetics 90, 161–191 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Delden W., “The alcohol dehydrogenase polymorphism in Drosophila melanogaster” in Evolutionary Biology, Hecht M. K., Wallace B., Prance G. T., Eds. (Springer, Boston, MA, 1982), vol. 15, pp. 187–222. [Google Scholar]
- 16.Hudson R. R., Kreitman M., Aguadé M., A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barton N. H., Richard Hudson and Norman Kaplan on the coalescent process. Genetics 202, 865–866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry J. D., Mechanisms of naturally evolved ethanol resistance in Drosophila melanogaster. J. Exp. Biol. 217, 3996–4003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers G. K., “The Drosophila alcohol dehydrogenase gene–enzyme system” in Advances in Genetics, Caspari E. W., Scandalios J. G., Eds. (Academic, 1988), pp. 39–107. [Google Scholar]
- 20.Ashburner M., Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. Bioessays 20, 949–954 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Johnson F. M., Denniston C., Genetic variation of alcohol dehydrogenase in Drosophila melanogaster. Nature 204, 906–907 (1964). [DOI] [PubMed] [Google Scholar]
- 22.Milkman R., Further evidence of thermostability variation within electrophoretic mobility classes of enzymes. Biochem. Genet. 14, 383–387 (1976). [DOI] [PubMed] [Google Scholar]
- 23.Sampsell B., Sims S., Effect of adh genotype and heat stress on alcohol tolerance in Drosophila melanogaster. Nature 296, 853–855 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Day T. H., Hillier P. C., Clarke B., Properties of genetically polymorphic isozymes of alcohol dehydrogenase in Drosophila melanogaster. Biochem. Genet. 11, 141–153 (1974). [DOI] [PubMed] [Google Scholar]
- 25.Johnson F. M., Powell A., The alcohol dehydrogenases of Drosophila melanogaster: Frequency changes associated with heat and cold shock. Proc. Natl. Acad. Sci. U.S.A. 71, 1783–1784 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alahiotis S. N., Adaptation of Drosophila enzymes to temperature IV. Natural selection at the alcohol-dehydrogenase locus. Genetica 59, 81–87 (1982). [Google Scholar]
- 27.Adrion J. R., Hahn M. W., Cooper B. S., Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 31, 434–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakeshott J. G., et al. , Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36, 86–96 (1982). [DOI] [PubMed] [Google Scholar]
- 29.Cogni R., et al. , On the long-term stability of clines in some metabolic genes in Drosophila melanogaster. Sci. Rep. 7, 42766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umina P. A., Weeks A. R., Kearney M. R., McKechnie S. W., Hoffmann A. A., A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Merçot H., Defaye D., Capy P., Pla E., David J. R., Alcohol tolerance, ADH activity, and ecological niche of Drosophila species. Evolution 48, 746–757 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montooth K. L., Siebenthall K. T., Clark A. G., Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J. Exp. Biol. 209, 3837–3850 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Briscoe D. A., Robertson A., Malpica J.-M., Dominance at Adh locus in response of adult Drosophila melanogaster to environmental alcohol. Nature 255, 148–149 (1975). [DOI] [PubMed] [Google Scholar]
- 34.Cavener D. R., Clegg M. T., Multigene response to ethanol in Drosophila melanogaster. Evolution 35, 1–10 (1981). [DOI] [PubMed] [Google Scholar]
- 35.McKay J., Variation in activity and thermostability of alcohol dehydrogenase in Drosophila melanogaster. Genet. Res. 37, 227–237 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Stephanou G., Alahiotis N., Adaptive significance of the action of the Drosophila melanogaster alcohol dehydrogenase locus through the heat shock protein system. Genetica 69, 59–68 (1986). [Google Scholar]
- 37.Hudson R. R., Kaplan N. L., The coalescent process in models with selection and recombination. Genetics 120, 831–840 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreitman M., Hudson R. R., Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics 127, 565–582 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth B., Charlesworth D., Population genetics from 1966 to 2016. Heredity 118, 2–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somero G., Temperature adaptation of enzymes: Biological optimization through structure-function compromises. Annu. Rev. Ecol. Evol. Syst. 9, 1–29 (1978). [Google Scholar]
- 41.Somero G. N., Proteins and temperature. Annu. Rev. Physiol. 57, 43–68 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Fields P. A., Review: Protein function at thermal extremes: Balancing stability and flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129, 417–431 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Minasov G., Shoichet B. K., Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320, 85–95 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Shoichet B. K., Baase W. A., Kurokit R., Matthews B. W., A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. U.S.A. 92, 452–456 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taverna D. M., Goldstein R. A., Why are proteins marginally stable? Proteins 46, 105–109 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Arnold F. H., Wintrode P. L., Miyazaki K., Gershenson A., How enzymes adapt: Lessons from directed evolution. Trends Biochem. Sci. 26, 100–106 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Romero P. A., Arnold F. H., Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry A., Kreitman M., Molecular analysis of an allozyme cline: Alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134, 869–893 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stam L. F., Laurie C. C., Molecular dissection of a major gene effect on a quantitative trait: The level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 144, 1559–1564 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King E. G., et al. , Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22, 1558–1566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frydenberg J., Hoffmann A. A., Loeschcke V., DNA sequence variation and latitudinal associations in hsp23, hsp26 and hsp27 from natural populations of Drosophila melanogaster. Mol. Ecol. 12, 2025–2032 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann A. A., Willi Y., Detecting genetic responses to environmental change. Nat. Rev. Genet. 9, 421–432 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Fry J. D., Donlon K., Saweikis M., A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: Evidence for selection mediated by dietary ethanol. Evolution 62, 66–75 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Chakraborty M., Fry J. D., Evidence that environmental heterogeneity maintains a detoxifying enzyme polymorphism in Drosophila melanogaster. Curr. Biol. 26, 219–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begun D. J., Betancourt A. J., Langley C. H., Stephan W., Is the Fast/Slow allozyme variation at the Adh locus of Drosophila melanogaster an ancient balanced polymorphism? Mol. Biol. Evol. 16, 1816–1819 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Chambers G. K., Wilks A. V., Gibson J. B., Variation in the biochemical properties of the Drosophila alcohol dehydrogenase allozymes. Biochem. Genet. 22, 153–168 (1984). [DOI] [PubMed] [Google Scholar]
- 57.Oakeshott J. G., Selection at the alcohol dehydrogenase locus in Drosophila melanogaster imposed by environmental ethanol. Genet. Res. 26, 265–274 (1975). [DOI] [PubMed] [Google Scholar]
- 58.Vigue C. L., Weisgram P. A., Rosenthal E., Selection at the alcohol dehydrogenase locus of Drosophila melanogaster: Effects of ethanol and temperature. Biochem. Genet. 20, 681–688 (1982). [DOI] [PubMed] [Google Scholar]
- 59.Heinstra P. W., Thörig G. E., Scharloo W., Drenth W., Nolte R. J., Kinetics and thermodynamics of ethanol oxidation catalyzed by genetic variants of the alcohol dehydrogenase from Drosophila melanogaster and D. simulans. Biochim. Biophys. Acta 967, 224–233 (1988). [DOI] [PubMed] [Google Scholar]
- 60.McKenzie J. A., Parsons P. A., Microdifferentiation in a natural population of Drosophila melanogaster to alcohol in the environment. Genetics 77, 385–394 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenzie J. A., McKechnie S. W., Ethanol tolerance and the Adh polymorphism in a natural population of Drosophila melanogaster. Nature 272, 75–76 (1978). [DOI] [PubMed] [Google Scholar]
- 62.Van Delden W., Kamping A., The alcohol dehydrogenase polymorphism in populations of Drosophila melanogaster IV. Survival at high temperature. Genetica 51, 179–185 (1980). [Google Scholar]
- 63.Cohan F. M., Graf J.-D., Latitudinal cline in Drosophila melanogaster for knockdown resistance to ethanol fumes and for rates of response to selection for further resistance. Evolution 39, 278–293 (1985). [DOI] [PubMed] [Google Scholar]
- 64.Gibson J. B., Wilks A. V., The alcohol dehydrogenase polymorphism of Drosophila melanogaster in relation to environmental ethanol, ethanol tolerance and alcohol dehydrogenase activity. Heredity 60, 403–414 (1988). [DOI] [PubMed] [Google Scholar]
- 65.Laurie C. C., Stam L. F., The effect of an intronic polymorphism on alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 138, 379–385 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindquist S., Regulation of protein synthesis during heat shock. Nature 293, 311–314 (1981). [DOI] [PubMed] [Google Scholar]
- 67.Rohmer C., David J. R., Moreteau B., Joly D., Heat induced male sterility in Drosophila melanogaster: Adaptive genetic variations among geographic populations and role of the Y chromosome. J. Exp. Biol. 207, 2735–2743 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Loehlin D. W., Ames J. R., Vaccaro K., Carroll S. B., A major role for noncoding regulatory mutations in the evolution of enzyme activity. Proc. Natl. Acad. Sci. U.S.A. 116, 12383–12389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hahn M., Molecular Population Genetics (Sinauer Associates, Sunderland, MA, 2018). [Google Scholar]
- 70.Mackay T. F. C., et al. , The Drosophila melanogaster Genetic Reference Panel. Nature 482, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siddiq M. A., Loehlin D. W., Montooth K. L., Thornton J. W., Experimental test and refutation of a classic case of molecular adaptation in Drosophila melanogaster. Nat. Ecol. Evol. 1, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewontin R. C., Twenty-five years ago in genetics: Electrophoresis in the development of evolutionary genetics: Milestone or millstone? Genetics 128, 657–662 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heinstra P. W., Evolutionary genetics of the Drosophila alcohol dehydrogenase gene-enzyme system. Genetica 92, 1–22 (1993). [DOI] [PubMed] [Google Scholar]
- 74.Sampsell B., Sims S., Effect of adh genotype and heat stress on alcohol tolerance in Drosophila melanogaster. Nature 296, 853–855 (1982). [DOI] [PubMed] [Google Scholar]
- 75.Laurie C. C., Bridgham J. T., Choudhary M., Associations between DNA sequence variation and variation in expression of the Adh gene in natural populations of Drosophila melanogaster. Genetics 129, 489–499 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McColl G., McKechnie S. W., The Drosophila heat shock hsr-omega gene: An allele frequency cline detected by quantitative PCR. Mol. Biol. Evol. 16, 1568–1574 (1999). [DOI] [PubMed] [Google Scholar]
- 77.Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10, e1004775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DePristo M. A., Weinreich D. M., Hartl D. L., Missense meanderings in sequence space: A biophysical view of protein evolution. Nat. Rev. Genet. 6, 678–687 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Knies J. L., Cai F., Weinreich D. M., Enzyme efficiency but not thermostability drives cefotaxime resistance evolution in TEM-1 β-lactamase. Mol. Biol. Evol. 34, 1040–1054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serrano L., Day A. G., Fersht A. R., Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J. Mol. Biol. 233, 305–312 (1993). [DOI] [PubMed] [Google Scholar]
- 81.Gong L. I., Suchard M. A., Bloom J. D., Stability-mediated epistasis constrains the evolution of an influenza protein. Elife 2, e00631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergland A. O., Tobler R., González J., Schmidt P., Petrov D., Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol. Ecol. 25, 1157–1174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.