Significance
Human blood dendritic cells (DCs) are composed of different subsets endowed with specific and complementary functions in the course of an immune response against pathogens such as viruses. A new population of circulating precursors of DCs (pre-DCs) has been recently identified by high-dimensional approaches. Besides their role in DC ontogeny, pre-DCs remained uncharacterized upon pathogen encounter. Here we show that their constitutive expression of Siglec-1, a receptor known to bind HIV-1 particles, confers them with a unique set of properties with regards to HIV-1, including viral capture, infection, and transfer. Thus, circulating pre-DCs may contribute to the antiviral immune response by providing a source of viral antigens to other DC populations.
Keywords: blood dendritic cells, pre-DC, HIV-1, Siglec-1, virus-containing compartments
Abstract
The human dendritic cell (DC) lineage has recently been unraveled by high-dimensional mapping, revealing the existence of a discrete new population of blood circulating DC precursors (pre-DCs). Whether this new DC population possesses specific functional features as compared to the other blood DC subset upon pathogen encounter remained to be evaluated. A unique feature of pre-DCs among blood DCs is their constitutive expression of the viral adhesion receptor Siglec-1. Here, we show that pre-DCs, but not other blood DC subsets, are susceptible to infection by HIV-1 in a Siglec-1–dependent manner. Siglec-1 mediates pre-DC infection of CCR5- and CXCR4-tropic strains. Infection of pre-DCs is further enhanced in the presence of HIV-2/SIVmac Vpx, indicating that Siglec-1 does not counteract restriction factors such as SAMHD1. Instead, Siglec-1 promotes attachment and fusion of viral particles. HIV-1–infected pre-DCs produce new infectious viral particles that accumulate in intracellular compartments reminiscent of the virus-containing compartment of macrophages. Pre-DC activation by toll-like receptor (TLR) ligands induces an antiviral state that inhibits HIV-1 fusion and infection, but Siglec-1 remains functional and mediates replication-independent transfer of HIV-1 to activated primary T lymphocytes. Altogether, Siglec-1–mediated susceptibility to HIV-1 infection of pre-DCs constitutes a unique functional feature that might represent a preferential relationship of this emerging cell type with viruses.
Dendritic cells (DCs) are professional pathogen-sensing and antigen-presenting cells that are central to the initiation and regulation of immune responses (1). The DC population is classified into 2 lineages: plasmacytoid DC (pDC) and conventional DC (cDC), the latter comprising cDC1 and cDC2 subpopulations (2). Recently, DC ontogeny has been the object of intensive work using new high-dimensional approaches, including single-cell level in mice (3) and in humans (4–6). Our study identified a population of precursors of DC (pre-DCs) that originate from bone marrow and circulate in the blood before reaching tissues where they can differentiate into conventional DCs (5). Although human pre-DCs share many markers with pDCs, they are clearly different morphologically, phenotypically, and functionally (5).
DCs are targets for HIV-1, although with lower efficiency than activated CD4+ T lymphocytes and macrophages (7, 8). DCs are thought to participate in the establishment of the infection due to the expression of lectin-like receptors, allowing them to capture and transfer HIV-1 to CD4+ T cells (9, 10). Since the seminal study of Steinman and coworkers, it is clear that DCs possess the capacity to capture and transfer HIV particles with a high efficiency to T cells (7). The process called trans-infection involves the lectin-like receptor Siglec-1 (CD169) present on the surface of DCs but still remains incompletely understood and essentially documented with in vitro-derived DCs from monocytes.
The capacity of the various DC subsets to initiate antiviral T cell responses through antigen presentation and to respond to infection is highly organized in space and time (11). pDCs detect viruses, rapidly respond by producing large amounts of type I IFN, and may be, at least partially, responsible for the high levels of IFNα measured during the acute phase of HIV-1 infection (12). cDC1s are resistant to HIV-1 infection, but can uptake HIV-1–infected cells to prime specific T cell responses (11). In contrast to pDCs and cDC1s, cDC2s are susceptible to HIV-1 infection in vitro and may thus provide, after dying and being phagocytosed, a source of viral antigens for cDC1 leading to cross-presentation (11).
Whether the recently discovered pre-DC subset is endowed with specific functions as compared to the other DC subsets upon pathogen encounter remained to be determined. Importantly, Siglec-1 is one of the very few specific and constitutive markers of pre-DCs (4, 13). Siglec-1 is an IFN-induced receptor that can bind gangliosides, including GM3 that is part of the HIV-1 membrane and mediate efficient viral capture and transmission to T cells in vitro (14–18). Marginal zone macrophages also express Siglec-1 and can mediate retroviral transfer to target cells in vivo (19). Hence pre-DCs appear as a potentially critical population of cells with regards to HIV-1, prompting us to study its relationship to this virus as compared to the other blood DC subsets. We found that pre-DCs are susceptible to infection by R5- and X4-tropic HIV-1. Notably, HIV-1 capture by pre-DCs and HIV-1 infection of pre-DCs are Siglec-1 dependent. Upon TLR-mediated activation, pre-DCs up-regulate RAB15 expression and become resistant to HIV-1 fusion and thus to infection. Importantly, activated pre-DCs switch to a replication-independent mechanism of virus transfer to activated primary T lymphocytes mediated by Siglec-1. These results show that constitutive expression of Siglec-1 by freshly isolated pre-DCs appears to be critical for the replication cycle of HIV-1 in these cells, from the capture of the viruses to the localization of the production of new viral progeny or to virus transfer.
Results
Pre-DC Subset Is Susceptible to HIV-1 Infection.
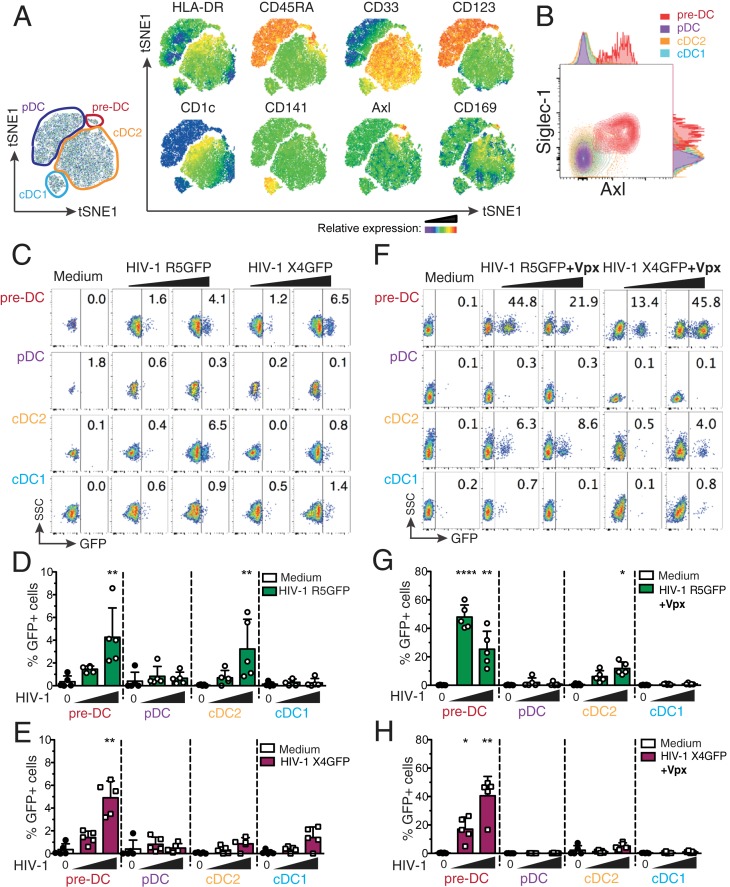
We first set up an isolation strategy of the 4 blood DC subsets: pre-DC, pDC, cDC1, and cDC2, by a combination of magnetic bead negative selection and flow cytometry sorting (SI Appendix, Fig. S1A). The sorted populations had the expected (4, 5) frequency and morphology at the ultrastructural level (SI Appendix, Fig. S1 B–D). Among the specific markers of pre-DCs, Axl and Siglec-1 stand out as the 2 most discriminative between DC subsets (Fig. 1 A and B and refs. 4 and 5).
Fig. 1.
Comparative analysis of human blood DC subsets for their susceptibility to HIV-1 strains. (A) Representative tSNE plots of flow cytometry data from HLA-DR+Lin(CD3/CD14/CD16/CD19/CD34)– PBMCs, showing gates defining the indicated DC subsets and relative expression of the indicated markers. (B) Representative dot plot of Siglec-1 and Axl expression on pre-DCs (HLA-DR+Lin–CD33intCD45RA+CD123+Axl+), pDCs (HLA-DR+Lin–CD33−CD45RA+CD123+Axl−), cDC2s (HLA-DR+Lin–CD33+CD45RA−CD123−CD1c+), and cDC1s (HLA-DR+Lin–CD33+CD45RA−CD123−CD141+). (C) GFP expression in blood DC subsets sorted and infected for 48 h with HIV-1 R5GFP or X4GFP. Freshly prepared viral preparations used for the experiments depicted from C to H were titrated on GHOST reporter cells in parallel and used at a MOI of 0.994 ± 0.112 (mean ± SEM). Representative dot plot analyses are presented with the percentages of GFP+ cells indicated in each dot plot. (D) Quantification with HIV-1 R5GFP infection or (E) with HIV-1 X4GFP as in C, n = 5 independent donors combined from 3 experiments. Individual donors are displayed with bars representing mean ± SD. (F) As in C using the indicated viruses complemented with Vpx. (G) Quantification of infection with HIV-1 R5GFP or (H) with HIV-1 X4GFP, both supplemented with Vpx, n = 5 independent donors combined from 2 to 4 experiments. Individual donors are displayed with bars representing mean ± SD. *P < 0.05, **P < 0.01, ****P < 0.0001.
Given that Siglec-1 can bind HIV particles, we evaluated pre-DC susceptibility to HIV-1 infection as compared to the other blood DC populations purified from healthy donor blood. When exposed to a CCR5-tropic HIV-1 encoding GFP (HIV-1 R5GFP), pre-DCs and cDC2s were infected to a similar extent (mean ± SD 4.2 ± 2.5% and 3.2 ± 2.6% of infected cells, respectively, after 48 h), while cCD1s and pDCs remained refractory to infection (Fig. 1 C and D). Interestingly, a CXCR4-tropic HIV-1 (HIV-1 X4GFP) yielded higher rates of infection with pre-DCs (4.9 ± 1.4%) compared with any of the other subsets, including cDC2, which all remained very low (Fig. 1 C and E).
The low susceptibility of DCs to HIV-1 infection has been linked to the presence of SAMHD1, a restriction factor that degrades cytosolic dNTPs, thus limiting the retrotranscription of the viral RNA and the infection rates (20–22). However, no difference in SAMHD1 mRNA level was detected among all DC subsets, including pre-DC (SI Appendix, Fig. S2A). Due to the very low numbers of pre-DCs isolated, we were unable to follow SAMHD1 phosphorylation that negatively regulates its activity (22, 23). Nevertheless, SAMHD1 activity can be efficiently counteracted by the accessory protein Vpx from HIV-2 (20–22, 24). Accordingly, complementation of our HIV-1 strains with Vpx (using a Vpr–Vpx fusion construct) highly increased the rates of infection that reached up to 60% for pre-DCs and ∼18% for cDC2s with HIV-1 R5GFP (Fig. 1 F and G), establishing that SAMHD1 remains in part active in these 2 populations, as observed with monocyte-derived macrophages (MDMs) (20). Interestingly, with HIV-1 X4GFP complemented with Vpx, pre-DC was the only population infected (up to 49.5%, Fig. 1 F and H). Of note, monocyte-derived DCs (MDDCs) remained resistant to both viruses, and only slightly susceptible to HIV-1 R5GFP when complemented with Vpx (SI Appendix, Fig. S2 B and C), as previously shown (7, 25, 26).
We noticed that even in the presence of Vpx when using a R5-tropic HIV-1, cDC2s remained much less susceptible to the infection than pre-DCs under the same conditions (up to 18% versus 60%, respectively) (Fig. 1G), suggesting that besides SAMHD1, other mechanisms might limit the infection of cDC2s.
Specific Siglec-1 Expression by Pre-DCs Confers an Advantage over Other DC Subsets for HIV-1 Capture and Infection.
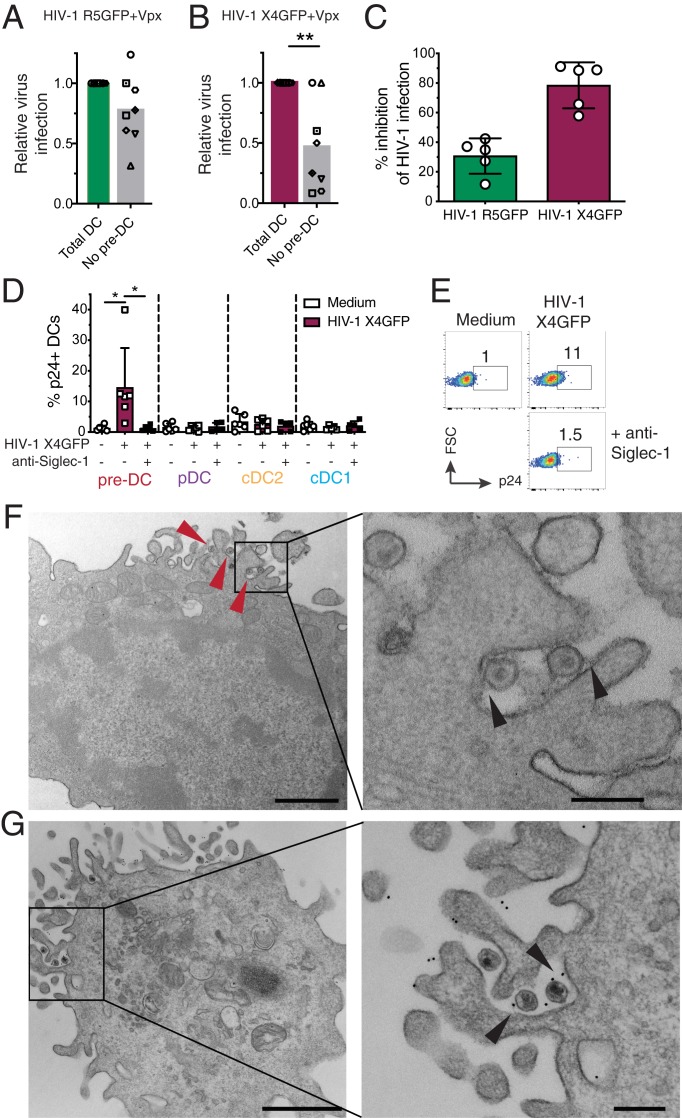
The constitutive expression of Siglec-1 by pre-DCs raised the question of their capacity to compete with the other blood DC subsets and to catch circulating viral particles. To approach this point in vitro, total DCs were isolated from peripheral blood mononuclear cells (PBMCs) by enrichment using the pan-DC magnetic sorting procedure followed by flow cytometry sorting of all Lin− HLA-DR+ cells that were used as “total DC,” or further sorted to obtain a DC population called “no pre-DC,” by excluding pre-DCs based on their expression of CD123 and Axl. Both populations were exposed either to HIV-1 R5GFP or to HIV-1 X4GFP complemented with Vpx in order to maximize the detection of infected cells. The rates of infection, as measured at 48 h postinfection by GFP expression, were reduced in the absence of pre-DCs by 22.5% with the R5-tropic virus (n = 8, Fig. 2A). The reduction was even more pronounced with the X4-tropic virus: an average decrease of 53.3% of DC infection in the absence of pre-DCs (Fig. 2B). These data indicated that despite pre-DC low representation among blood DCs, this cell population has a better capacity to capture HIV-1 and to get efficiently infected.
Fig. 2.
Pre-DC preferential infection is associated with Siglec-1–mediated capture of HIV-1. (A) Quantification of relative infection of total DCs (CD45+Lin(CD3/CD14/CD16/CD19/CD20/CD34)–HLA-DR+) or total DCs depleted of pre-DCs (CD33+CD45RAintCD123+Axl+) exposed to HIV-1 R5GFP + Vpx or (B) to HIV-1 X4GFP + Vpx. GFP expression was analyzed 48 h postinfection. (C) Relative quantification of the inhibition of the HIV-1 infection by preincubation with an anti–Siglec-1 mAb. Pre-DCs, preincubated with an anti–Siglec-1 mAb or not before, were infected for 48 h with HIV-1 X4GFP or R5GFP. Infection rates were quantified by following GFP expression. (D) Quantification of HIV-1 capture by the 4 DC subsets. Sorted DCs were preincubated for 30 min with an anti–Siglec-1 mAb or not, then exposed or not to HIV-1 X4GFP for 2 h at 37 °C, washed, stained for p24 to detect bound particles, and assayed by flow cytometry (HIV-1 X4GFP viral particles are not GFP+ by themselves), n = 5 or 6 independent donors combined in 5 experiments. Individual donors are displayed with bars representing mean ± SD. (E) Representative dot plot analysis of HIV-1 capture by pre-DCs performed as in D. (F) Ultrastrucutral analysis of pre-DCs exposed for 2 h to HIV-1 R5GFP, washed, and processed for EM. An epon section is presented, with a magnified view on the Right. Arrowheads point to virions captured by pre-DCs. On the magnified view, arrowheads point to protein linking the virions to the plasma membrane of the cells. (Scale bar, 2 µm [Left image] and 0.5 µm for the magnified view.) (G) Immuno-EM analysis of pre-DCs exposed to HIV-1 R5GFP for 2 h fixed and stained for Siglec-1 with a mAb revealed by protein A gold coupled to 5 nm gold particles before embedding (Materials and Methods). An epon section is presented, with a magnified view. Arrowheads point to Siglec-1–specific staining at the interface between virions and plasma membrane invaginations. (Scale bar, 2 µm [Left image] and 0.5 µm for the magnified view.) *P < 0.05, **P < 0.01.
To evaluate the potential role of Siglec-1 on pre-DC infection, pre-DCs were exposed to Siglec-1–specific antibody prior to infection. This blockade prevented HIV-1 infection of pre-DCs by R5-tropic viruses to some extent (35% inhibition) but more extensively for X4-tropic ones (roughly 85% inhibition, Fig. 2C). Together these data suggested that Siglec-1 expression may endow pre-DCs to better capture HIV-1 particles and stabilize their interaction leading to better rates of infection.
Thus, we first compared the various DC subsets for their capacity to capture HIV-1. Sorted DC populations were exposed to HIV-1 for 2 h at 37 °C, washed, and stained for p24 before analysis by flow cytometry. Among the 4 subsets, only freshly isolated pre-DCs were able to capture HIV-1 (Fig. 2D). Pretreatment of pre-DCs with an anti–Siglec-1 antibody totally prevented capture of HIV-1 (Fig. 2 D and E). Moreover, electron microscopy (EM) ultrastructural analysis of pre-DCs exposed to HIV-1 for 2 h revealed the presence of HIV-1 particles tightly bound to microvilli at the pre-DC plasma membrane (Fig. 2F). Analysis of ultrathin immunolabeled sections revealed that Siglec-1 protein was localized at the ultrastructural level in the immediate vicinity of both viral particles and the plasma membrane (Fig. 2G). Given the very large size of the highly glycosylated Siglec-1 protein (1,709 amino acids), these images are highly suggestive of Siglec-1 bridging the plasma membrane of pre-DC to HIV-1 particles. Thus, among blood DCs, pre-DCs are uniquely equipped with Siglec-1 that mediates efficient HIV-1 capture and infection.
TLR Activation of Pre-DCs Induces RAB15 Up-Regulation and Prevents HIV-1 Fusion and Infection.
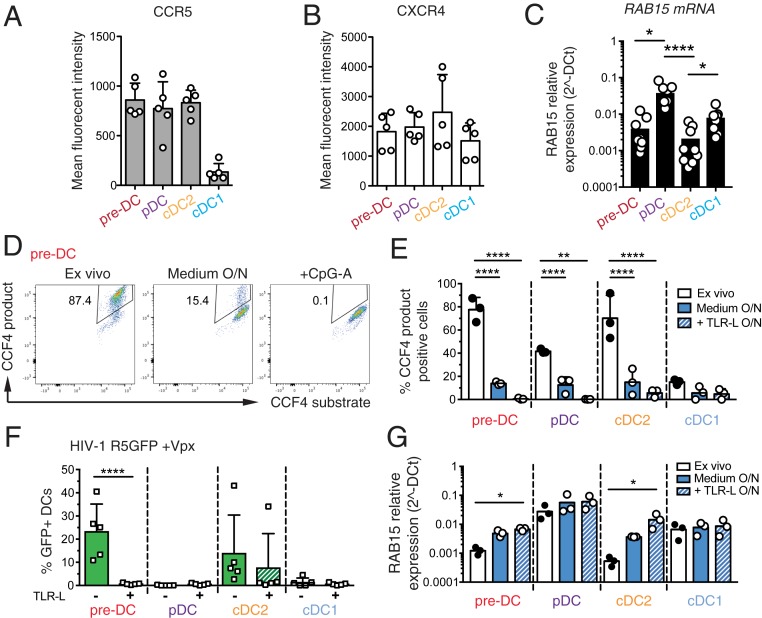
To identify other factors that may contribute to the preferential susceptibility of pre-DCs to HIV-1 as compared to other DC subsets, we first evaluated the expression levels of HIV-1 coreceptors on the various DC subsets. However, CCR5 and CXCR4 expression levels were similar among pre-DCs, cDC2s, and pDCs (Fig. 3 A and B). The molecular basis for the resistance of cDC1s and pDCs to HIV-1 infection at steady state has recently been attributed to their higher expression of the small GTPase RAB15 (11). Accordingly, we found that both HIV-1–suceptible subsets, pre-DC and cDC2, exhibited similar low levels of RAB15 mRNA, while these levels were higher in HIV-1–resistant cells, i.e., pDCs and cDC1s (Fig. 3C). Since RAB15 expression has been proposed to inhibit HIV-1 infection by blocking viral fusion (11), we measured viral fusion using a β-lactamase (BlaM)-Vpr assay in the 4 DC subsets purified ex vivo (Fig. 3 D and E). Strikingly, pre-DCs and cDC2s exhibited a high capacity to fuse with HIV-1 (close to 80% of the cells) while the 2 HIV-1–resistant DC subsets, pDC and cDC1, had lower capacity (40% and less than 20%, respectively). Thus, rates of viral fusion were higher than rates of infection for each of the 4 subsets purified ex vivo.
Fig. 3.
Pre-DCs high capacity to fuse with HIV-1 and to get infected is lost upon activation and is associated with increased expression levels of RAB15. (A) Surface expression levels of CCR5 and (B) CXCR4 on the 4 freshly sorted DC subsets determined by flow cytometry, n = 5 independent donors combined in 2 experiments. Individual donors are displayed with bars representing mean ± SD. (C) RAB15 mRNA expression levels among the 4 DC subsets freshly sorted, measured by qPCR, n = 9 independent donors combined in 3 experiments. Individual donors are displayed with bars representing mean ± SD. (D) Representative dot plot of viral fusion revealed by CCF4 fluorescence in sorted pre-DCs after infection with HIV-1 R5GFP containing a BlaM-Vpr fusion protein directly ex vivo or after an overnight culture with medium alone or in the presence of CpG-A. Fluorescence of the CCF4 product indicates viral fusion with target cells as a result of cleavage of the cell-loaded CCF4 substrate by the virus-contained BlaM. (E) Quantification of viral fusion revealed by CCF4 fluorescence after infection with HIV-1 R5GFP containing a BlaM-Vpr fusion protein directly ex vivo or after an overnight culture with medium alone or in the presence of TLR-L (CpG-A for pre-DCs and pDCs, or CL264 for cDC2s and cDC1s) for the 4 sorted DC populations, n = 3 donors combined from 2 independent experiments. (F) Quantification of DC infection with HIV R5GFP + Vpx directly ex vivo or following overnight (O/N) incubation with TLR ligands (CpG-A for pre-DCs and pDCs; CL264 for cDC2s and cDC1s). GFP expression was measured at 48 h postinfection, n = 5 from 3 independent experiments. Individual donors are displayed with bars representing mean ± SD. (G) RAB15 mRNA levels among the 4 DC subsets freshly sorted, measured by pPCR in DC subsets, ex vivo or after an overnight culture in medium alone or supplemented with TLR-L as in E, n = 3. *P < 0.05, **P < 0.01, ****P < 0.0001.
We next asked whether TLR-mediated activation of DCs could impact their susceptibility to viral fusion. Strikingly, while overnight culture already substantially reduced the fusion rates, overnight TLR activation induced a total block in HIV-1 fusion for all DC subsets (Fig. 3E). Indeed, overnight TLR activation abolished infection of pre-DCs and of cDC2s (in 4 out of 5 patients), while pDCs and cDC1s remained resistant to HIV-1 (Fig. 3F). Accordingly, overnight culture induced the expression of RAB15 mRNA in both pre-DCs and cDC2s, which was further increased following TLR activation.
We concluded that TLR activation completely switches pre-DC phenotype from highly susceptible to total resistance to HIV-1 infection. This phenotypic change is accompanied by an increased expression of RAB15 and a complete resistance to HIV-1 fusion.
HIV-1 Replicates in Pre-DCs in Apparently Internal Compartments.
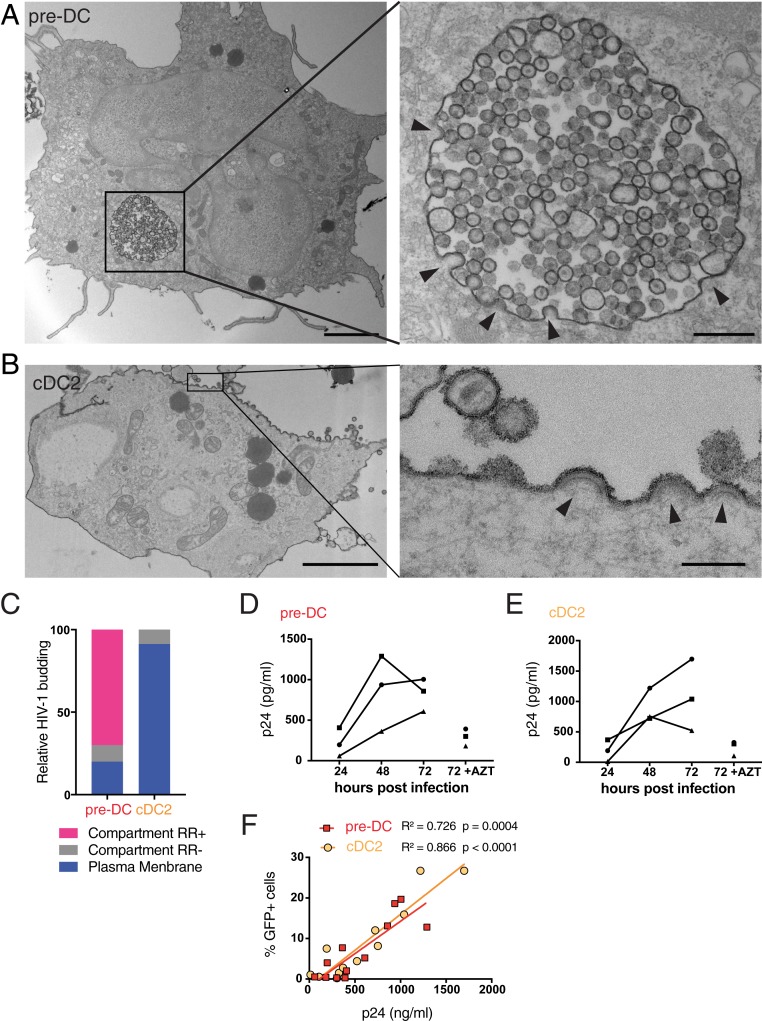
Siglec-1 has been proposed to contribute to the particular localization of viral assembly and storage in HIV-1–infected macrophages that take place in apparently internal compartments termed virus-containing compartments (VCCs) (27). To evaluate whether Siglec-1 could have a similar role in pre-DCs, pre-DCs and cDC2s infected for 48 h with HIV-1 R5 + Vpx were analyzed by EM (Fig. 4 A and B and SI Appendix, Fig. S3). Strikingly, pre-DCs exhibited typical VCCs similar to the ones observed in HIV-1–infected MDMs (28–30). Viral budding profiles were mostly absent from the plasma membrane but were clearly observed at the limiting membrane of the compartment, indicating de novo production of viral particles. These profiles exhibited a dark molecular coat typical of Gag precursor assembly (31, 32), that budded toward the lumen of the compartments in which numerous immature and mature viral particles accumulated (SI Appendix, Fig. S3A). A specific feature of the VCCs from macrophages is their accessibility to the external medium that can be evaluated by adding ruthenium red (RR), a membrane impermeant dye, during fixation (33, 34). Ruthenium red was visible in pre-DC VCCs as a dark and fuzzy shade at the limiting membrane of the VCCs and on the viral membranes (Fig. 4A and SI Appendix, Fig. S3A), demonstrating that these compartments are connected to the external medium. Interestingly, similar internal compartments preexist to the infection in MDMs (29, 35) and are hijacked by HIV-1 for its assembly, leading to their conversion into VCCs.
Fig. 4.
HIV-1–infected pre-DCs, but not cDC2s, produce new virions in apparently intracellular compartments. (A) Epon sections from sorted pre-DCs and (B) cDC2s infected with HIV-1 R5GFP + Vpx for 48 h and analyzed by EM. Cells were fixed in the presence of ruthenium red to label surface and surface-connected compartments. Arrowheads point to nascent viral buds. (Scale bar, 2 µm [Left images] and 0.15 µm for magnified views.) (C) Quantification of the location of viral budding profiles observed in HIV-1–infected pre-DCs (n = 11) and cDC2s (n = 22). Of note, some internal compartments containing viruses were observed in infected cDC2s; they were however unlabeled by RR and did not exhibit viral budding profiles at their limiting membranes. Rather than VCCs, they probably represent endosomes having internalized viral particles secreted by neighboring cells. (D) Kinetics of production of p24 by sorted pre-DCs and (E) cDC2s infected with HIV-1 R5GFP + Vpx measured by CBA (Materials and Methods). The RT inhibitor AZT was used as a control for the viral input remaining at the end of the assay, n = 3 independent donors combined in 2 experiments. Individual donors are displayed. (F) Correlation between the levels of infection of pre-DCs (red) and cDC2s (yellow) measured by GFP expression and the quantification of p24 in supernatants as in D and E.
In sharp contrast, infected cDC2s were free of VCCs, and viral budding only occurred in a polarized manner at the plasma membrane (Fig. 4B and SI Appendix, Fig. S3B), as previously observed in infected T lymphocytes and cell lines (36). In cDC2s, ruthenium red stained only the plasma membrane and the released viral particles. Quantification of compartments accessible to ruthenium red that exhibited viral buds and particles confirmed the striking differences between the 2 subsets (Fig. 3C). Thus, our results further support the idea that Siglec-1 plays a role in the VCC formation (27). Moreover, pre-DCs and cDC2s, although related through their precursor-to-progeny relationship as pre-DCs, can give rise to cDC2s (4, 5), exhibit different susceptibility to infection and completely different locations of viral assembly.
The EM profiles indicated that active viral replication can take place in both cell types. We compared their production after pre-DC and cDC2 exposure to HIV-1 R5GFP. Given the low numbers of pre-DC purification yield, we complemented the virus with Vpx to maximize the viral replication and be able to detect their eventual production. Both infected DC subsets released in their culture supernatant increasing amounts of p24 overtime (Fig. 4 D and E) that correlated with the rates of infection (Fig. 4F), indicating that HIV-1 can replicate in both subsets.
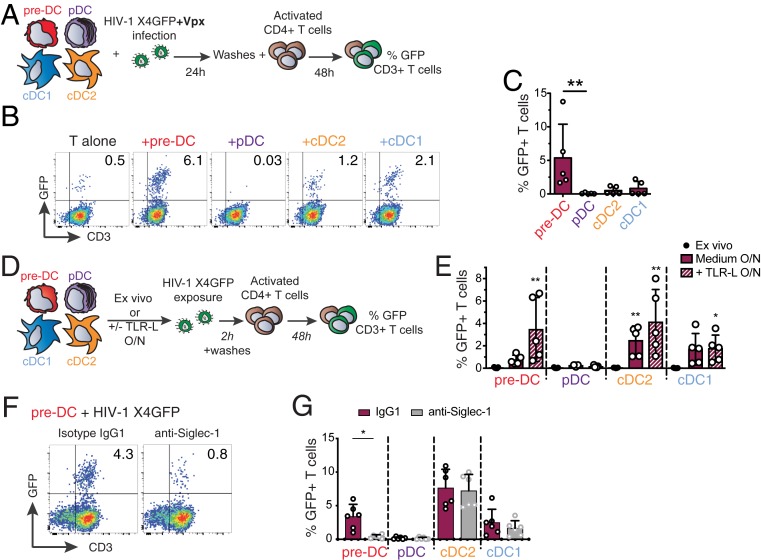
Pre-DCs Can Transmit HIV-1 to CD4+T Cells In Cis or Trans Depending on Their Activation State.
The infectious capacity of the viral particles produced by HIV-1–infected pre-DCs was evaluated on primary activated CD4+ T lymphocytes (Fig. 5A). In line with their preferential infection with X4-tropic virus, HIV-1–infected pre-DCs were the most efficient subset in transmitting the infection in cis to activated primary CD4+ T cells (Fig. 5 B and C). These results establish that the viral progeny of infected pre-DCs is infectious.
Fig. 5.
Infected pre-DCs and activated noninfected pre-DCs can both transmit HIV-1 to activated CD4+ T cells. (A) Cis-infection of T cells, outline of the experiment. (B) GFP expression in CD3+ T cells cocultured with HIV-1–infected DC subsets or not, as explained in A. Representative dot plots are presented. (C) Quantification of GFP expression in CD3+ T cells as in B, n = 5 independent donors combined from 2 experiments. Individual donors are displayed with bars representing mean ± SD. (D) Trans-infection of T cells, outline of the experiment. (E) GFP expression in activated T cells cocultured directly with ex vivo DCs or following overnight culture in medium or in the presence of TLR-L (CpG-A for pre-DCs and pDCs, CL264 for cDCs). Prior to CD4+ T cell coculture, DC populations were exposed to HIV-1 X4GFP (without Vpx) for 2 h and washed. Quantification of GFP+ T cells (CD3+) was performed by flow cytometry after a 48-h coculture, n = 5 independent donors combined in 2 experiments. Individual donors are displayed with bars representing mean ± SD. (F) GFP expression by CD3+ T cells. TLR-activated pre-DCs were preincubated with anti–Siglec-1 antibody or with an isotype control antibody (IgG1) before performing the trans-infection assay as in E. (G) Quantification as in F with the indicated DC populations. (n = 6 independent donors combined in 3 experiments). Individual donors are displayed with bars representing mean ± SD. *P < 0.05, **P < 0.01.
Given that TLR stimulation induced a state of resistance to HIV-1 infection, we evaluated the capacity of DC populations activated or not to perform HIV-1 trans-infection of T cells (Fig. 5D). First, none of the ex vivo sorted DC populations, including pre-DC, was able to transmit HIV-1 to activated CD4+ T cells (Fig. 5E). However, after overnight culture, pre-DCs, cDC1s, and cDC2s performed trans-infection to CD4+ T cells, which was further increased upon TLR activation (Fig. 5E). Similarly, MDDCs cultured overnight in the presence or absence of lipopolysaccharides (LPS) performed trans-infection of T cells (SI Appendix, Fig. S4A). Thus, following TLR activation, both pre-DCs and cDC2s performed HIV-1 trans-infection to CD4+ T cells to similar levels, while cDC1s did poorly and pDCs did not (Fig. 5E). Finally, preincubation of TLR-activated DC subsets with anti–Siglec-1 antibody strongly inhibited pre-DCs’ but not cDC2s’ capacity to perform trans-infection (Fig. 5 F and G). Anti-Siglec-1 treatment also reduced HIV-1 trans-infection to CD4+ T cells by MDDCs exposed to LPS (SI Appendix, Fig. S4B). We concluded that TLR-activated pre-DCs can perform genuine trans-infection, capturing HIV-1 without being infected (Fig. 3F) and transmitting the virus efficiently to activated CD4+ T cells. The fact that ex vivo-sorted unstimulated pre-DCs are unable to perform trans-infection to T cells, despite their high levels of surface Siglec-1, indicates that activation of pre-DCs induces the acquisition of this capacity through a still unknown mechanism.
Discussion
The recent identification of the human blood pre-DC population was based on single-cell data analysis using unsupervised clustering approaches (4, 5). Here we show that pre-DCs, as compared to the other blood DC subsets, exhibit a unique set of properties regarding HIV-1 that are linked to their constitutive Siglec-1 expression (SI Appendix, Fig. S5). Indeed, we show first that Siglec-1 is pivotal for the unique capacity of pre-DCs to capture HIV-1. In addition, among freshly sorted blood DC subsets, pre-DC stands out as the only subset susceptible to infection by both types of HIV-1: CCR5-tropic and CXCR4-tropic viruses. The CXCR4-tropic viruses are unable to infect any other DC subtype, even when the SAMHD1 restriction factor is counteracted (ref. 11 and Fig. 1). Despite their low frequency among blood DCs, pre-DCs were able to outcompete other DCs and get preferentially infected in a Siglec-1–dependent manner.
Stabilization of the interaction between HIV-1 and pre-DCs via Siglec-1 may increase the probability of the Env-mediated fusion to occur, a rather slow process (37, 38). It may also account for pre-DCs’ preferential susceptibility to X4 viruses as compared to other blood DC populations that do not express Siglec-1, despite their common low levels of expression of the coreceptor CXCR4. Regarding CCR5-tropic viruses, we observed that both pre-DCs and cDC2s had a high capacity to fuse with this type of virus (around 80%). However, cDC2s exhibited a much lower infection rate than pre-DCs, even when the SAMHD1 restriction factor was counteracted (roughly 20% versus 50%). On the same line, while pDCs are totally resistant to infection by R5 HIV-1, 40% of them were susceptible to viral fusion as judged using the BLaM-Vpr assay. The discrepancies between the rates of fusion and infection, even when SAMHD1 was counteracted, suggest that additional unknown mechanisms are at work in cDC2s and pDCs.
RAB15 high expression has been associated to resistance to viral fusion of cDC1s and thus to infection (11). We confirmed and extended these data documenting high levels of RAB15 transcripts in pDCs and cDC1s and low levels in cDC2s and pre-DCs at the steady state. Importantly, we further established that TLR activation up-regulates these levels in all DC subsets and confers to all of them full resistance to fusion and infection. More work is needed to understand the molecular mechanism involved but the comparison of the various DC subsets may represent a valuable approach for this purpose.
Our data also indicate that surface Siglec-1 on pre-DCs facilitates pre-DC infection, allowing for production of infectious particles. Infection leads to viral production and accumulation in apparently intracellular compartments termed VCCs. Of note, Siglec-1 is also present at the VCC limiting membrane of infected MDMs (27) and thus in the HIV-1 particles they produce (39). Therefore, Siglec-1 expression by pre-DCs might be implicated in HIV-1 assembly at the VCC level. VCCs have been initially observed in tissue macrophages from infected patients in a variety of tissues, see ref. 31, and were then essentially studied in in vitro-derived and infected MDMs (30). A recent study identified urethral macrophages from an individual under combined antiretroviral therapy containing infectious particles in VCCs, suggesting that such tissue macrophages can represent a viral reservoir (40). In contrast, HIV assembly at the plasma membrane, as observed here in primary cDC2s, has been largely documented in activated CD4+ T cells. The different location of viral assembly between pre-DCs and cDC2s is compelling since pre-DCs can give rise to cDC2s. Comparison between the 2 DC subtypes should help to approach the molecular basis for VCC formation and the potential role of Siglec-1 in this process, as previously proposed (41).
When activated, pre-DCs are no longer susceptible to infection but can perform efficient trans-infection to CD4+ T cells, again in a fully Siglec-1–dependent manner. We thus confirm with primary pre-DCs what has been established with LPS-stimulated MDDCs, i.e., the existence of this mode of transmission to T cells via non-infected DCs. The fact that ex vivo purified pre-DCs were unable to perform HIV-1 trans-infection suggests that DC activation is necessary. In contrast to a previous study performed with LPS-stimulated cDC2s (15), we observed that TLR7-activated cDC2s performed trans-infection independently of Siglec-1. The impact of different TLR stimulations of DCs on the HIV-1 trans-infection process remains to be established. In addition, innate immune responses, including to TLR ligands, can be modulated by Axl (42, 43), a receptor tyrosine kinase well expressed by pre-DCs. Axl has also been implicated in the susceptibility to viral infections (44, 45), opening the possibility that Axl on pre-DCs plays a role in their susceptibility and response to HIV-1.
Our results indicate that, stimulated or not, pre-DCs could contribute to viral spreading through trans- or cis-infection, respectively. How pre-DCs integrate in the highly organized share of work between the various DC subsets to face or contribute to the HIV infection remains to be established. Recent work of Silvin et al. (11) points out that cDC2s get infected by HIV-1, whereas cDC1s are in a resistant state that allows them to cross-present viral antigens from dead infected cDC2 cells. As pre-DCs can secrete IL-12 and support mixed-lymphocyte reactions (4, 5), it will be of interest to determine whether pre-DCs possess the hybrid capacity of both cDC1s and cDC2s to initiate an anti–HIV-1 response.
Interestingly, Siglec-1+ macrophages from the spleen marginal zone have been shown in the vesicular stomatitis virus (VSV) mouse model to support a locally restricted enforced viral replication by down-modulating type I IFN responsiveness thus providing a source of viral antigen required to elicit a protective immune response (46). Moreover, Siglec-1+ macrophages can transfer viral antigens to cDC1s which in turn cross-prime CD8+ T cell responses (47). Importantly, this antigen transfer was mediated by Siglec-1 on the macrophages interacting with sialylated proteins present on cDC1 cell surface (47). Thus, Siglec-1 expression may also confer blood pre-DCs the capacity to recruit and instruct the help of cross-presenting cDC1s. Future studies will certainly address this important hypothesis, since manipulation of pre-DCs response or protection against HIV-1 may represent new approaches to promote a better immune control of the infection.
Materials and Methods
Isolation of Cells.
Peripheral blood mononuclear cells were isolated from buffy coats from healthy human donors (approved by the Institut National de la Santé et de la Recherche Médicale ethics committee and the Health Sciences Authorities [Singapore]) with Ficoll-Paque PLUS (GE). Informed consent was obtained from all donors, and samples were deidentified prior to use in the study. Total blood DCs were enriched with EasySep human pan-DC pre-enrichment kit (Stemcell Technologies). DC-enriched fractions were stained with antibodies specific for HLA-DR APCeFluor780, CD1c PerCPeFluor710 (eBioscience), CD123 Viogreen, CD141 Vioblue (Miltenyi), CD33 APC, CD45RA PE (BD), and with a mixture of antibodies against lineage markers CD19 (Miltenyi), CD3, CD14, CD16, and CD34 (BD) in the FITC channel. Alternatively, DC-enriched fractions were stained with HLA-DR APCeFluor780, CD1c PerCPeFluor710, CD123 Viogreen, CD45RA Vioblue (Miltenyi), CD33 PE-CF594, and Clec-9A PE (BD) with lineage markers in the FITC channel. Pre-DCs were sorted as Lin− HLADR+ CD33int CD45RAint CD123+. pDCs were sorted as Lin− HLADR+ CD33− CD45RA+ CD123+. cDC2s were sorted as Lin− HLADR+ CD33+ CD45RA− CD1c+. cDC1s were sorted as Lin− HLADR+ CD33+ CD45RA− CD141+/Clec9A+.
For pre-DC depletion, DC-enriched fractions were stained with antibodies specific for HLA-DR APCeFluor780 with a mixture of antibodies against lineage markers CD19, CD3, CD14, CD16, and CD34 in the FITC channel or with the addition of CD123 Viogreen, Siglec-1 PE-Vio770 (Miltenyi), and Axl PE (clone 108724, R&D Systems). Total DCs were sorted as Lin− HLADR+. Alternatively, Pre-DCs (Lin− HLADR+ CD123+ Siglec-1+ Axl+) were excluded from Lin− HLADR+.
All cells were sorted on a FACSAria (BD) using Diva software (BD) in 5-mL polypropylene round-bottom tubes containing 1 mL X-VIVO-15 media (Lonza BE04-418F). Postsort cell purity after gating on live cells by FCS/SSC was routinely between 92 and 99%.
CD4+ T cells were isolated from PBMCs by negative selection using the CD4+ T Cell Isolation Kit (Miltenyi 130-096-533). CD14+ monocytes were obtained by positive selection from PBMCs using CD14 Microbeads (Miltenyi 130-050-20).
DC Phenotype by Flow Cytometry.
Enriched DCs obtained as described were stained with HLA-DR APCeFluor780, CD1c PerCPeFluor710, CD123 Viogreen, CD45RA Vioblue, CD33 PE-CF594, CD141 BV711 (BD), Axl PE, and Siglec-1 PE-Vio770 with lineage markers in the FITC channel. Cells were acquired on a FACS Verse and data analyzed using FlowJo 10 (Tree Star, Inc.). After compensation, tSNE representation of gated Lin− HLADR+ cells were generated. Siglec-1 and Axl expression were evaluated on gated pre-DCs, pDCs, cDC1s, and cDC2s as described (5).
Alternatively, PBMCs were stained as described previously (5) with DC lineage markers in addition to Siglec-1 (BD), CXCR4 (Biolegend), and CCCR5 (BD) and analyzed by flow cytometry. FCS files compensated for spillover between channels were exported using FlowJo v10 (Tree Star, Inc.) and CXCR4 and CCR5 expressions were measured on gated pre-DCs, pDCs, cDC1s, and cDC2s as described.
Cell Culture.
For lentiviral infections of sorted DCs, cells were cultured in X-VIVO-15 complemented with penicillin/streptomycin (Thermo Fisher 10378-016). For capture experiments, DCs were cultured in RPMI medium 1640, GlutaMAX (Thermo Fisher 61870-010) complemented with FBS 10% (Thermo Fisher 10270-106), and penicillin/streptomycin.
T cells were cultured in RPMI medium 1640, GlutaMAX complemented with FBS 10% and penicillin/streptomycin at 106 cells/mL in the presence of 5 µg/mL PHA (lectin from Phaseolus vulgaris leucoagglutinin; Sigma L2769) and 50 U/mL of IL-2 (eBioscience). On day 2 of culture, cells were washed and additionally cultured with IL-2.
Purified CD14+ cells were cultured at 0.8 106 cells/mL in complete RPMI supplemented with GM-CSF at 100 ng/mL and IL-4 (Miltenyi) at 50 ng/mL for 4 d to obtain MDDCs.
HEK293FT cells were cultured in DMEM, GlutaMAX (Thermo Fisher 61965-026) complemented with FBS 10%, and penicillin/streptomycin. GHOST X4R5 cells were cultured in DMEM, GlutaMAX complemented with FBS 10%, and penicillin/streptomycin. All cells were cultured at 37 °C with 5% CO2 atmosphere.
Plasmids.
The proviral plasmid HIV-1 R5GFP was derived from NL4-3 with BaL env, Δnef, and GFP in nef (48, 49). HIV-1 X4GFP corresponded to NL4-3, Δnef, and GFP in nef. pIRES2EGFP-VPXanyVPR plasmid was used to complement some of the viral preparations with Vpx (50) and pMM310 (BlaM-Vpr) to complement the viral preparations with BlaM to measure the fusion of virions (51).
HIV-1 Production and Titration.
Viral particles were produced by transfection of HEK293FT cells in 6-well plates with 3 µg DNA and 8 µL TransIT-293 (Mirus Bio) per well. For HIV-1 R5GFP and HIV-1 X4GFP, 3 µg of HIV plasmid was used. For HIV-1 virus containing Vpx and BlaM, 0.75 µg pIRES2EGFP-VPXanyVPR or pMM310, respectively, and 2.25 μg HIV plasmid were used. Sixteen hours after transfection, media were removed, and fresh X-VIVO-15 or RPMI medium was added. Viral supernatants were harvested 36 h later, filtered at 0.45 μM, used freshly or aliquoted, and frozen at −80 °C. Viral titers were determined on GHOST X4R5 cells as described (50) and multiplicity of infection (MOI) ranged from 0.04 to 1.99, mean = 0.99, SD = 0.49 for DC infection displayed in Fig. 1.
HIV Infection of DCs and Stimulations.
Sorted cells were pelleted and resuspended in complete X-VIVO-15 or RPMI media at 0.4 106 cells/mL, and 50 μL was seeded in round-bottom 96-well plates. In some experiments anti–Siglec-1 mAb (clone 7-239) or mIgG1 isotype control (Miltenyi) were added at 20 µg/mL and cells were incubated for 30 min at 37 °C before adding the virus. Azidothymidine (AZT; Sigma) was used at 25-μM final concentration, and nevirapin (Sigma) was used at 5 μM. CpG-A (ODN2216, Invivogen) was used at 5 µg/mL and CL264 (Invivogen) at 10 μg/mL. For infections, 150 μL of media or dilutions (150 µL or 50 µL) of viral supernatants were added. In the experiments depicted in Fig. 1, infections were spinoculated for 2 h at 800 × g at 25 °C.
For total DCs or pre-DC-depleted DCs, cells were resuspended at 2.106 cells/mL and 2.105 cells were cultured in the presence of 450 µL mock medium or HIV-1 R5GFP viruses in X-VIVO-15.
Forty-eight hours after infection, cell culture supernatants were harvested and cells were fixed in 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS prior to analysis on a FACSVerse flow cytometer (BD). Data were analyzed using FlowJo v10 and Prism v7 for Mac (GraphPad).
Electron Microscopy.
For routine embedding into Epon, cells were fixed in 2.5% glutaraldehyde in 0.1 M Na-cacodylate buffer pH 7.4 for 1 h, postfixed for 1 h with 1% buffered osmium tetroxide, dehydrated in a graded series of ethanol solution, and then embedded in epoxy resin as described (34). Electron micrographs were acquired on a Tecnai Spirit electron microscope (FEI, Eindhoven, The Netherlands) equipped with a 4k CCD camera (EMSIS GmbH, Münster, Germany). For pre-embedding immunogold labeling, cells were fixed in 4% paraformaldehyde for 1 h at room temperature (RT), quenched in PBS-glycine, and blocked with 1% BSA-PBS. Primary antibody anti–Siglec-1 mAb (Miltenyi) was diluted to 1:50 in 1% BSA-PBS and incubated with cells for 1 h. Cells were extensively washed prior to addition of protein A gold (EM laboratory, Utrecht University) and routine embedding in Epon.
For RR stain, cells were fixed with 2.5% glutaraldehyde in 0.1 M ice-cold Na-cacodylate buffer (pH 7.4) containing 0.15% RR overnight at 4 °C. Cells were then washed with 0.1 M Na-cacodylate buffer (pH 7.2), postfixed with 1.3% OsO4 in the same buffer containing 0.15% RR for 2 h at room temperature, dehydrated in a graded series of ethanol solution, and then embedded in epoxy resin.
HIV Capture and Trans-Infection of Activated CD4+ T Cells.
Sorted cells were pelleted and resuspended in complete X-VIVO-15 media at 0.4 106 cells/mL and 50 μL were seeded in round-bottom 96-well plates. In some experiments anti–Siglec-1 mAb or mIgG1 isotype control were added at 20 µg/mL and cells were incubated for 30 min at 37 °C before adding the virus. HIV-1 X4GFP was added onto the cells (150 µL/well of HEK293FT culture supernatant) and incubated for 2 h at 37 °C. Cells were washed extensively and fixed in 4% PFA in PBS. p24 staining was performed using KC-57 RD1 mAb (Beckman Coulter, 6604667). For trans-infection experiments, sorted DCs were washed extensively after the 2-h culture with HIV-1 X4GFP and activated CD4+ T cells were added at a ratio of 1:1. Alternatively, CpG-A was added at 5 µg/mL, CL264 was added at 10 μg/mL onto DCs, and cells were incubated overnight before the addition of HIV-1 X4GFP. Cells were then washed and activated CD4+ T cells added. After 48 h, cells were fixed in 2% PFA in PBS. Cells were then stained with PE-Cy7 anti-CD3 (BD) and analyzed on a FACS Verse (BD). Data were analyzed using FlowJo v10 and Prism v7 for Mac (GraphPad).
HIV-1 Fusion Assay.
DC infection was performed as described with 150 µL BlaM-containing viral supernantant and cells were spinoculated for 2 h at 800 × g, 25 °C. Cells were additionally incubated for 3 h at 37 °C, washed 3 times with CO2 independent medium (Thermo Fisher 18045-054), loaded with CCF4-AM dye (dilution 1/400) in CO2 independent medium complemented with solution B (dilution 1/125) for 1 h at RT. Cells were washed 2 times with CO2 independent medium and resuspended in 100 μL of CO2 independent medium complemented with 10% FBS and 2.5 mM probenicid. Cells were incubated for 12 h at RT, washed with CO2 independent medium, and fixed with 1% PFA in PBS before acquisition on a FACS Verse. Data were analyzed using FlowJo v10 and Prism v7 for Mac.
Microarray Analysis.
Total RNA was isolated from FACS-sorted blood pre-DC and DC subsets using a RNeasy Micro kit (Qiagen). Total RNA integrity was assessed using an Agilent Bioanalyzer (Agilent) and the RNA integrity number (RIN) was calculated. All RNA samples had a RIN ≥7.1. Biotinylated cRNA was prepared using an Epicentre TargetAmp 2-Round Biotin-aRNA Amplification Kit 3.0 according to the manufacturer’s instructions, using 500 pg of total RNA starting material. Hybridization of the cRNA was performed on an Illumina Human-HT12 Version 4 chip set (Illumina). Microrarray data were exported from GenomeStudio (Illumina) without background subtraction. Probes with detection P values >0.05 were considered as not being detected in the sample, and were filtered out. Expression values for the remaining probes were log2 transformed and quantile normalized. The microarray dataset is deposited in the Gene Expression Omnibus under accession no. GSE80171.
RAB15 Gene Expression.
A total of 4.105 sorted DCs were used ex vivo or treated as described. Cells were lysed, RNA was extracted using an RNeasy micro kit (Qiagen), and reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) by following the manufacturer’s instructions. Real-time quantitative PCR was performed using SYBR Green I Master (Roche) with the following primers (Eurogentec): RAB15-forward: 5′-CAGCAGCTGGCGAAGGAG-3′ and RAB15-reverse: 5′-GTTGGTGCAGGCACTTCTTTC-3′. RPS18 gene was used as a housekeeping gene with the following primers: RPS18-forward: 5′-CTGCCATTAAGGGTGTGG-3′, RPS18-reverse: 5′-TCAATGTCTGCTTTCCTCAAC-3′. The relative quantity of RAB15 mRNAs was calculated between RAB15 RNA Cp and RPS18 Cp by the 2-∆Ct method.
Measure of p24 by Cytometric Bead Assay.
In-house p24 cytometric bead assay (CBA) was used to quantify p24 concentration in the supernatant of HIV-1–infected DCs. Dilutions of virus-like particles with known concentration of p24 were used to establish a standard curve. A total of 300 beads/sample were acquired on a FACS Verse. Data were analyzed using FlowJo v10 and Prism v7 for Mac.
Statistical Analysis.
Data were analyzed using Prism v7 for Mac (GraphPad). A Friedman test was used followed by Dunn’s multiple comparisons tests, and a P value lower than 0.05 was considered as significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Supplementary Material
Acknowledgments
We acknowledge Nicolas Manel at Institut Curie for fruitful discussions, reagents, and critical reading of the manuscript. We also thank Ana-Maria Lennon-Dumenil, François-Xavier Gobert, Julie Helft, Constance Delaugerre, and Sebastian Amigorena for discussions. We acknowledge the “plateforme d’imagerie cellulaire et tissulaire BioImaging de l’Institut Curie” (PICT-IBiSA) for access to the imaging facilities and the flow cytometry facilities at Institut Curie and Institut Cochin (“plateforme Cytométrie et Immunobiologie”; CYBIO). This work was supported by grants from Agence Nationale de Recherche contre le SIDA et les hépatites virales (ANRS), Ensemble contre le SIDA (Sidaction), Laboratoire d’Excellence (Labex) DCBIOL (ANR-10-IDEX-0001-02 PSL and ANR-11-LABX-0043) to P.B., and Singapore Immunology Network (SIgN) core funding to F.G. Our work also benefitted from France-BioImaging, ANR-10-INSB-04, and CelTisPhyBio Labex (ANR-10-LBX-0038) part of the Initiative D’EXcellence (Idex) PSL (ANR-10-IDEX-0001-02 PSL). N.R. and E.G.-M. were supported by fellowships from ANRS, Sidaction, and Fondation pour la Recherche Médicale.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray dataset is deposited in the Gene Expression Omnibus under accession no. GSE80171.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911007116/-/DCSupplemental.
References
- 1.Schlitzer A., McGovern N., Ginhoux F., Dendritic cells and monocyte-derived cells: Two complementary and integrated functional systems. Semin. Cell Dev. Biol. 41, 9–22 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Guilliams M., et al. , Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlitzer A., et al. , Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Villani A.-C., et al. , Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.See P., et al. , Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, eaag3009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breton G., et al. , Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c +or CD141 +DCs. J. Exp. Med. 213, 2861–2870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granelli-Piperno A., Delgado E., Finkel V., Paxton W., Steinman R. M., Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J. Virol. 72, 2733–2737 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIlroy D., et al. , Infection frequency of dendritic cells and CD4+ T lymphocytes in spleens of human immunodeficiency virus-positive patients. J. Virol. 69, 4737–4745 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hladik F., McElrath M. J., Setting the stage: Host invasion by HIV. Nat. Rev. Immunol. 8, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L., KewalRamani V. N., Dendritic-cell interactions with HIV: Infection and viral dissemination. Nat. Rev. Immunol. 6, 859–868 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silvin A., et al. , Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol. 2, eaai8071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller-Trutwin M., Hosmalin A., Role for plasmacytoid dendritic cells in anti-HIV innate immunity. Immunol. Cell Biol. 83, 578–583 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Dutertre C.-A., et al. , Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity, S1074-7613(19)30334-6 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Garcia E., et al. , HIV-1 trafficking to the dendritic cell-T-cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic 6, 488–501 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Izquierdo-Useros N., et al. , Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol. 10, e1001448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izquierdo-Useros N., et al. , Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood 113, 2732–2741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puryear W. B., Yu X., Ramirez N. P., Reinhard B. M., Gummuluru S., HIV-1 incorporation of host-cell-derived glycosphingolipid GM3 allows for capture by mature dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 109, 7475–7480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puryear W. B., et al. , Interferon-inducible mechanism of dendritic cell-mediated HIV-1 dissemination is dependent on Siglec-1/CD169. PLoS Pathog. 9, e1003291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sewald X., et al. , Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science 350, 563–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrecka K., et al. , Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laguette N., et al. , SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cribier A., Descours B., Valadão A. L. C., Laguette N., Benkirane M., Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Arnold L. H., et al. , Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog. 11, e1005194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahouassa H., et al. , SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakri Y., et al. , The maturation of dendritic cells results in postintegration inhibition of HIV-1 replication. J. Immunol. 166, 3780–3788 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Cameron P. U., et al. , Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257, 383–387 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Hammonds J. E., et al. , Siglec-1 initiates formation of the virus-containing compartment and enhances macrophage-to-T cell transmission of HIV-1. PLoS Pathog. 13, e1006181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan J., Sattentau Q. J., The HIV-1-containing macrophage compartment: A perfect cellular niche? Trends Microbiol. 21, 405–412 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Mlcochova P., Pelchen-Matthews A., Marsh M., Organization and regulation of intracellular plasma membrane-connected HIV-1 assembly compartments in macrophages. BMC Biol. 11, 89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues V., Ruffin N., San-Roman M., Benaroch P., Myeloid cell interaction with HIV: A complex relationship. Front. Immunol. 8, 1698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orenstein J. M., Meltzer M. S., Phipps T., Gendelman H. E., Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: An ultrastructural study. J. Virol. 62, 2578–2586 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raposo G., et al. , Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3, 718–729 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Deneka M., Pelchen-Matthews A., Byland R., Ruiz-Mateos E., Marsh M., In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177, 329–341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jouve M., Sol-Foulon N., Watson S., Schwartz O., Benaroch P., HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe 2, 85–95 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Bèrre S., et al. , CD36-specific antibodies block release of HIV-1 from infected primary macrophages and its transmission to T cells. J. Exp. Med. 210, 2523–2538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freed E. O., HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 13, 484–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones D. M., Padilla-Parra S., Imaging real-time HIV-1 virion fusion with FRET-based biosensors. Sci. Rep. 5, 13449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B., HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chertova E., et al. , Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80, 9039–9052 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganor Y., et al. , HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4, 633–644 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Akiyama H., Ramirez N.-G. P., Gudheti M. V., Gummuluru S., CD169-mediated trafficking of HIV to plasma membrane invaginations in dendritic cells attenuates efficacy of anti-gp120 broadly neutralizing antibodies. PLoS Pathog. 11, e1004751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothlin C. V., Ghosh S., Zuniga E. I., Oldstone M. B. A., Lemke G., TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Strange D. P., et al. , Axl promotes zika virus entry and modulates the antiviral state of human sertoli cells. MBio 10, e01372-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid E. T., et al. , AXL receptor tyrosine kinase is required for T cell priming and antiviral immunity. elife 5, e12414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meertens L., et al. , Axl mediates ZIKA virus entry in human glial cells and modulates innate immune responses. Cell Rep. 18, 324–333 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Honke N., et al. , Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat. Immunol. 13, 51–57 (2011). [DOI] [PubMed] [Google Scholar]
- 47.van Dinther D., et al. , Functional CD169 on macrophages mediates interaction with dendritic cells for CD8+ T cell cross-priming. Cell Rep. 22, 1484–1495 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Oswald-Richter K., et al. , HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol. 2, E198 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oswald-Richter K., et al. , Identification of a CCR5-expressing T cell subset that is resistant to R5-tropic HIV infection. PLoS Pathog. 3, e58 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manel N., et al. , A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467, 214–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavrois M., De Noronha C., Greene W. C., A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20, 1151–1154 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.