Significance
We report the development of a chemical proteomic platform, soluble activity-dependent proximity ligation (sADPL), which enables ultrasensitive and multiplexed quantification of endogenous active proteins in complex proteome samples from cells, fluids, and tissues. Single-plexed and multiplexed sADPL can be implemented to “write” and “read” barcoded oligonucleotide amplicons derived from specific active enzymes in extremely low levels of whole proteome. We apply sADPL to quantify in vivo protein–drug interactions from blood samples, as well as perform hundreds to thousands of parallel activity measurements in fresh or flash-frozen patient tumor samples in a matter of hours on a benchtop. Combined, these studies provide compelling proof-of-concept examples for future applications for the molecular analysis of biological and clinical samples.
Keywords: chemoproteomics, chemical probes, proteomics, diagnostics, proximity ligation
Abstract
Chemoproteomic methods can report directly on endogenous, active enzyme populations, which can differ greatly from measures of transcripts or protein abundance alone. Detection and quantification of family-wide probe engagement generally requires LC-MS/MS or gel-based detection methods, which suffer from low resolution, significant input proteome requirements, laborious sample preparation, and expensive equipment. Therefore, methods that can capitalize on the broad target profiling capacity of family-wide chemical probes but that enable specific, rapid, and ultrasensitive quantitation of protein activity in native samples would be useful for basic, translational, and clinical proteomic applications. Here we develop and apply a method that we call soluble activity-dependent proximity ligation (sADPL), which harnesses family-wide chemical probes to convert active enzyme levels into amplifiable barcoded oligonucleotide signals. We demonstrate that sADPL coupled to quantitative PCR signal detection enables multiplexed “writing” and “reading” of active enzyme levels across multiple protein families directly at picogram levels of whole, unfractionated proteome. sADPL profiling in a competitive format allows for highly sensitive detection of drug–protein interaction profiling, which allows for direct quantitative measurements of in vitro and in vivo on- and off-target drug engagement. Finally, we demonstrate that comparative sADPL profiling can be applied for high-throughput molecular phenotyping of primary human tumor samples, leading to the discovery of new connections between metabolic and proteolytic enzyme activity in specific tumor compartments and patient outcomes. We expect that this modular and multiplexed chemoproteomic platform will be a general approach for drug target engagement, as well as comparative enzyme activity profiling for basic and clinical applications.
The maturation of broad-scale “-omic” profiling methods and platforms has enabled the quantitative comparison of biological signal regulation at each step within the central dogma. In addition to providing unprecedented views of transcriptome-wide and proteome-wide regulation, these studies have unambiguously shown that mRNA transcript levels do not reliably predict protein abundance (1–3). Further complicating the situation, in principle the mere presence of a protein does not provide any information about the functional state of that protein, which can be impacted by posttranslational modifications, protein-protein or protein-metabolite binding, spatial compartmentalization, and other factors. Activity-based protein profiling methods utilize chemical probes upstream of common detection methods, such as liquid chromatography tandem mass spectrometry (LC-MS/MS) or gel-based protein separation, to specifically detect and quantify only active subpopulations of the proteome of interest (4, 5).
Family-wide chemical proteomic probes are particularly useful because they can report on the activity of large swaths of the active proteome simultaneously, with published examples available for serine hydrolases (6, 7), kinases (8, 9), metalloproteases (10), cysteine proteases (11), and other functionalities that regulate protein activity (12–14). Although large sections of the proteome can be interrogated using these probes, commonly used gel- and MS-based detection platforms place significant limitations on the biological contexts that can be efficiently studied. Gel-based profiling is readily accessible but is inherently low-throughput, suffers from poor resolution, and has a narrow dynamic range for low-abundance target proteins and input proteome. MS-based platforms, on the other hand, are multiplexed and target-agnostic, enabling profiling across entire families. However, these attributes are accompanied by several trade-offs, including laborious and expensive sample preparation (days per sample), the need for large quantities of input proteome (typically several milligrams) for a single analysis, and the capability of analyzing only one sample at a time per instrument, which limits parallel and reproducible sample analyses. Therefore, while ideal for discovery mode activity-based profiling, these strategies are poorly suited for rapid, ultrasensitive, and multifamily activity profiling that could be useful for complex and limited abundance samples like patient tissues, cells, and fluids. Indeed, chemical proteomic approaches have not been widely adopted for clinical samples, despite their significant potential to diagnose disease and ultimately track and guide patient treatment decisions.
To address the inherent shortcomings of existing chemical proteomic technologies and to more readily enable translational applications, we endeavored to develop a gel- and MS-free chemical proteomic platform that in principle can overcome limitations while providing 3 features: (1) broad and immediate compatibility with existing family-wide chemical proteomic probes to measure active proteins in native samples, rather than overall protein abundance; (2) signal deconvolution, superior dynamic range, and high sensitivity through signal amplification, to allow measurement of low abundance proteins and small quantities of input proteome; and (3) rapid, multiplexed, parallel, and simultaneous analysis of active targets within and between multiple protein families in diverse sample types.
We recently reported an imaging-based activity-dependent proximity ligation platform, termed ADPL imaging, which enables spatially resolved detection of active enzymes in single cells using family-wide probes (15). While the concept behind this approach addresses several shortcomings, the imaging-based format places limitations on samples types that can be interrogated (i.e., incompatible with biofluids), as well as low processing throughput involved with sample preparation and microscopy. Finally, the initial report of this approach was limited to single-plex measurements, which precludes profiling across multiple enzymes within a family or proteins among different families.
Here we report a platform that we call soluble activity-dependent proximity ligation (sADPL) and show that sADPL profiling can quantify active enzymes at picogram levels of whole proteome from cells, blood, and primary patient tissue samples. This multiplexed activity-based approach can be applied to directly quantify small molecule-protein target engagement in vivo, as well as molecular phenotyping of disease states through direct quantitative profiling of active enzyme biomarkers in tissue samples.
Results
sADPL Enables Ultrasensitive and Specific Activity Measurements for Diverse Enzyme Targets.
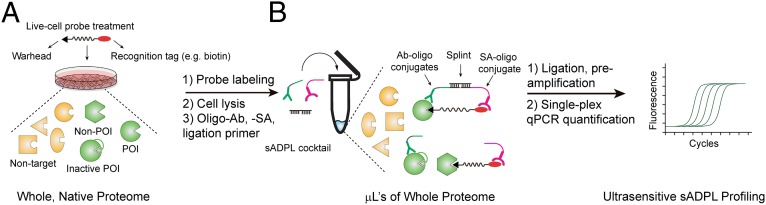
sADPL integrates the activity-dependent and family-wide tagging of endogenous, active enzymes using chemical probes with the specific and robust signal amplification afforded by barcoded oligonucleotide proximity ligation and amplification (Fig. 1) (15–18). In contrast to the majority of studies that only use chemical probes in homogenized cell lysates, we sought to label active enzymes in their native environment, and therefore performed sADPL by pulsing live cells with one probe or a combination of several family-wide probes (Fig. 1A). After cell lysis whole proteome is incubated with barcoded, protein of interest (POI)-specific antibody-oligonucleotide conjugates and streptavidin-oligonucleotide conjugates, which recognize the biotin recognition tag present on family-wide probes (Fig. 1B). The use of POI-directed antibodies allows for deconvolution of signals from several family-wide probes, which may have tagged hundreds of proteins, to the signal from a single POI. Subsequent incubation with complementary splint oligonucleotides permits ligation of proximal antibody- and streptavidin-conjugated oligonucleotides (i.e., on the same protein molecule), thereby forming unique barcoded amplicons that report on and amplify the activity of each POI. Finally, ligated amplicons are amplified and detected by PCR with a barcoded orthogonal primer binding site design, allowing for specific real-time quantitative PCR (qPCR) readout.
Fig. 1.
Schematic of the sADPL workflow. (A) Live cells pulsed with a combination of family-wide chemical probes label active proteins within their native environment. (B) Labeled proteome is incubated with a mixture of barcoded antibody-oligonucleotide conjugates that recognize the POIs and streptavidin-oligonucleotide conjugates directed to the probe detection handle (biotin), which enables specific templating of proximity ligation events and formation of unique barcoded amplicons. Ligated amplicons are preamplified by PCR and quantified by real-time qPCR with the barcoded primers.
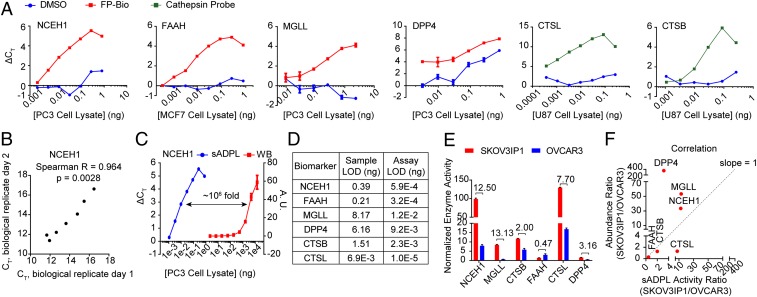
To pilot sADPL, we targeted proteins associated with cancer phenotypes in 2 well-characterized enzyme families, serine hydrolase and cathepsin cysteine protease enzymes, using widely available fluorophosphonate-biotin (FP-Bio) (15) and an acyloxymethylketone-based cathepsin probe (19), respectively. In parallel, we synthesized antibody-oligonucleotide (Ab-oligo) conjugates for specific proteins within each enzyme family, as well as probe-specific streptavidin-oligonucleotide (SA-oligo) conjugates (SI Appendix, Fig. S1) (20). Using monoacylglycerol lipase (MGLL) as a model enzyme, we optimized the absolute and relative concentrations of Ab-oligo and SA-oligo conjugates required for the sADPL signal and dynamic range in whole-cell proteome (SI Appendix, Fig. S2). We then tested the sensitivity and dynamic range of sADPL quantification for diverse protein targets, including the serine hydrolase enzymes neutral cholesterol ester hydrolase 1 (NCEH1), MGLL, fatty acid amide hydrolase 1 (FAAH), and dipeptidyl peptidase-4 (DPP4), as well as the cysteine protease enzymes cathepsin B (CTSB) and cathepsin L (CTSL). sADPL profiling and qPCR signal quantification of all 6 enzyme targets were successful, with ΔCT values of approximately 5 to 10 cycles between background and probe treatment, a linear dynamic range 3 to 4 orders of magnitude over proteome concentration, and high reproducibility among technical and biological replicates (Fig. 2 A and B). These data suggest that sADPL can be used for both comparative and competitive activity profiling of diverse targets within and between protein families while maintaining very low sample requirements. Indeed, side-by-side comparison of sADPL, which by design detects only active protein, and Western blot analysis for NCEH1 confirmed a limit of detection at least 6 orders of magnitude lower for sADPL, which could detect active protein in picogram quantities of whole proteome (Fig. 2 C and D).
Fig. 2.
Ultrasensitive, specific activity measurements for diverse protein targets in whole proteome with sADPL. (A) sADPL signal profiles for 6 enzyme targets across whole proteome dilutions from the indicated cancer cell lines. ΔCT, cycle threshold normalized to no lysate (PBS) control. (B) Correlation plot of representative sADPL runs for NCEH1 activity performed on different days. Each data point represents 3 technical replicates within each biological replicate. (C) Side-by-side comparison of sADPL with Western blot detection of active and total NCEH1 in PC3 cell proteome, respectively. (D) Total proteome concentration limit of detection values for each enzyme, expressed for the sample and assay, which also accounts for dilution during sample processing. (E) Comparative sADPL profiling of active enzyme levels between aggressive (SKOV3IP1) and nonaggressive (OVCAR3) ovarian cancer cell lines. The mean activity ratio between lines is shown above each enzyme. (F) Plot correlating SKOV3IP/OVCAR3 activity ratio measured by sADPL (x-axis) and protein abundance measured by LC-MS/MS (y-axis). Spearman’s r = 0.66. All data points are from triplicate technical replicates from representative biological replicates in 2 or more biological experiments. Values represent the mean, and error bars represent the SD.
sADPL Accurately Quantifies Differences in Endogenous Enzyme Activity.
Parallel, miniaturized, and rapid quantification of active enzymes from native biological samples could provide opportunities for accurate and efficient molecular phenotyping of cells, fluids, and tissues. To test whether sADPL could accurately quantify endogenous differences in enzyme activity between phenotypically distinct cells, we profiled the activity state of several enzymes in paired ovarian cancer cell lines SKOV3IP1 and OVCAR3 of high and low aggressiveness, for which both activity- and abundance-based proteomic profiling data have been published (15, 21–23). Each cell line was pulsed with either vehicle control (DMSO) or a combination of serine hydrolase and cathepsin protease probes, followed by sADPL profiling and relative activity quantification of 6 biomarker enzymes (Fig. 2E). NCEH1 and MGLL activities were 12.5- and 13.3-fold higher, respectively, in SKOV3IP1 cells relative to OVCAR3 cells, which matched previous reports using MS-based, gel-based, and imaging-based activity measurements in these cell lines (15, 21, 22). In contrast, FAAH activity was approximately 2-fold higher in the nonaggressive OVCAR3 cells relative to SKOV3IP1 cells, consistent with previously published bulk activity-based profiling data in several cancer cell lines (Fig. 2E) (24). Finally, the activity states of both cathepsin enzymes and the peptidase DPP4 were significantly increased in the more aggressive SKOV3IP1 cells (Fig. 2E).
To determine whether these high-throughput measurements of active enzyme were equivalent to or similar to overall protein abundance, we compared sADPL activity measurements with a previously published quantitative LC-MS/MS–based proteomics study in these cell lines (23). In general, there was a positive correlation between sADPL profiling and MS-based abundance measurements (Fig. 2F and SI Appendix, Fig. S3); however, closer inspection of these measurements revealed significant differences between relative enzyme abundance and activity in these cell lines. Cathepsin abundance and activity ratios between cell lines were almost identical for CTSB, but the CTSL activity ratio was moderately increased relative to abundance (Fig. 2F). Significant differences were found for NCEH1 and MGLL, with abundance ratios of 37.7-fold and 50.2-fold for each enzyme, respectively, compared with the more modest activity ratios of slightly above 10-fold (Fig. 2F). In contrast, the activity of FAAH was 2-fold lower in SKOV3IP1 cells compared with OVCAR3 cells, whereas abundance was nearly 4-fold lower in SKOV3IP1 cells, indicating that a higher percentage of FAAH is active in SKOV3IP1 cells relative to OVCAR3 cells. Finally, the correlation between MS-based abundance and activity of DPP4 was highly divergent, with nearly 350-fold higher protein levels observed in SKOV3IP1 cells by LC-MS/MS, contrasted with just a 3.2-fold increase in activity.
To further verify the differences observed for DPP4 and CTSL, we measured their protein levels by Western blot analysis. The DPP4 level was 3.7-fold higher in SKOV3IP1 cells, which closely matched the activity difference measured by sADPL (SI Appendix, Fig. S3). The CTSL level was 4.1-fold higher in SKOV3IP1 relative to OVCAR3; however, this measurement recognizes both precursors and mature CTSL, again supporting a significant difference between activity and abundance (SI Appendix, Fig. S3). The differences observed between MS-based and gel-based methods for these 2 proteins underscore the limitations in dynamic range accuracy and the caveat that MS can also detect protein fragments. Moreover, these results confirm that protein- and context-dependent differences exist between protein abundance and direct measures of active protein, which represents a fundamentally distinct and likely more meaningful assessment of proteome state in a biological system.
sADPL Enables Quantification of Small Molecule Target Engagement In Vitro and In Vivo.
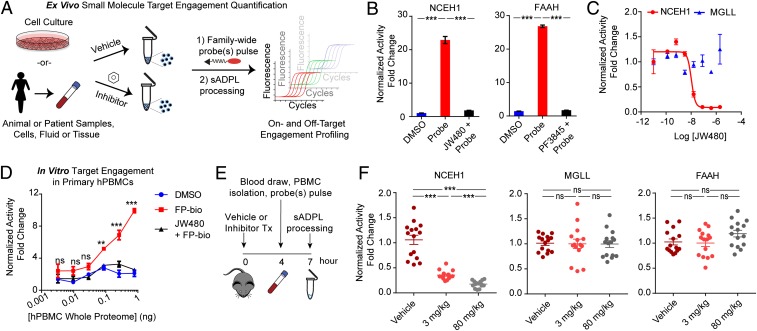
Due to the coupling of the enzyme activity state with the oligonucleotide signal, we hypothesized that sADPL could be a powerful platform for directly quantifying small molecule target engagement in a variety of biological sample types (Fig. 3A), which would be useful for both basic and translational studies (25–27). To test this, we first treated cultured cells with selective small molecule inhibitors of NCEH1 (JW480) (28) and FAAH (PF3845) (29), followed by FP-Bio probe treatment and sADPL processing. Each small molecule ablated the sADPL signal for its respective target protein (Fig. 3B and SI Appendix, Fig. S4). In addition, sADPL detected graded changes in NCEH1 activity levels in response to increased JW480 concentrations, an indication of direct occupancy of this enzyme by its small molecule inhibitor (IC50 = 10.5 nM; Fig. 3C). Parallel quantification of MGLL activity in response to JW480 treatment showed no effect (Fig. 3C), confirming the specificity of each sADPL signal within the serine hydrolase family. These results also suggested that both on- and off-target enzymes could be quantified from in vivo treatments and clinical samples. To test this possibility, we first asked whether our sADPL protocol was compatible with target enzyme profiling in patient-derived peripheral blood mononuclear cells (PBMCs). FP-Bio probe treatment and sADPL profiling of isolated PBMCs from human or mouse blood samples resulted in specific and inhibitor-sensitive signals from each enzyme tested (Fig. 3D and SI Appendix, Fig. S5).
Fig. 3.
Quantification of in vitro and in vivo small molecule target engagement by sADPL. (A) Schematic depiction of in vitro and in vivo proteome processing and sADPL target engagement profiling. (B) Enzyme-specific sADPL signals for NCEH1 and FAAH, which are completely abrogated following treatment of live cells with selective inhibitors. (C) Dose-dependent and target-specific engagement of the NCEH1 inhibitor JW480 in PC3 cancer cells quantified by sADPL quantification of both NCEH1 and MGLL activity. (D) In vitro target engagement in human patient-derived PBMCs across whole-cell proteome dilutions. Data points are from 3 technical replicates from 2 or more biological replicates. (E) Schematic timeline of in vivo target engagement experiments. (F) sADPL signals from on-target (NCEH1) and off-target (MGLL and FAAH) enzymes in isolated PBMCs from mice following the indicated in vivo compound treatments for 4 h. Data in F are derived from triplicate technical measurements from 5 mice dosed across 2 duplicate biological experiments. The whisker plot shows mean normalized activity signal, with error bars denoting SEM. ns, not significant; **P < 0.01; ***P < 0.001, Student’s t test.
Given these results, we next asked whether in vivo target engagement of a small molecule could be detected and quantified by sADPL profiling of PBMCs collected from live animals. C57BL/6 mice were treated with vehicle alone, an approximate ED50 dose of JW480 (3 mg/kg), or a supersaturating dose of JW480 (80 mg/kg) for 4 h, followed by PBMC collection and processing for sADPL (Fig. 3E). In agreement with previously published pharmacokinetic and pharmacodynamic data on JW480 (28), the NCEH1-dependent sADPL signal was reduced in a dose-dependent manner (Fig. 3F). Parallel quantification of the activity of FAAH and MGLL, 2 related off-target serine hydrolases, showed no significant inhibition of either target at either dose, consistent with previous gel-based profiling in mice treated with JW480 (28). These measurements proved to be extremely sensitive and statistically robust, confirming the potential to directly profile both on- and off-target engagement of small molecule inhibitors in vivo via sADPL.
Multiplexed sADPL Enables Parallel Biomarker Profiling of Patient Tumor Samples.
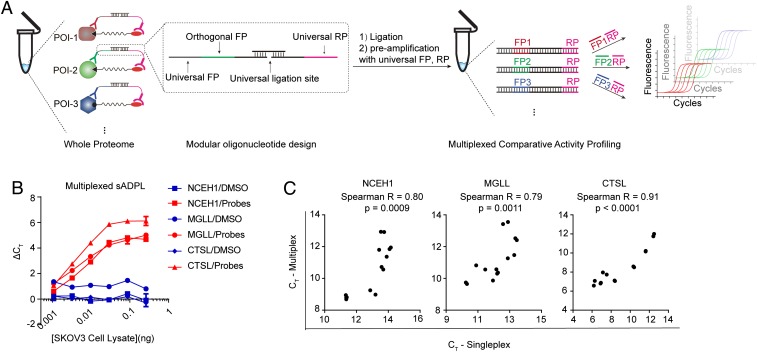
A central aspect of the sADPL design is the conversion of endogenous protein activity into an amplified, quantifiable oligonucleotide signal. The potential to barcode these signals enables quantification of multiple protein targets simultaneously via unique oligonucleotide identifiers and subsequent deconvolution with real-time PCR or other amplification/detection methods (30). Given this potential, and our intent to perform many measurements in parallel (e.g., profiling of many targets across many samples), we also tested whether sADPL could be reformatted for multiplexed signal “writing” and “reading” of many protein targets in the same sample. In the multiplexed format, single or multiple family-wide probes create a uniform biotin/desthiobiotin tag on all members of the protein family (or families) recognized by a streptavidin-oligo conjugate containing a universal identifier sequence, which also acts as reverse primer binding site (Fig. 4A and SI Appendix, Fig. S6). Specific active proteins of interest are recognized with antibody-oligo conjugates each harboring an orthogonal identifier sequence located adjacent to a universal forward primer site. Within this format, all probe-bound target proteins can be recognized by a universal splint oligonucleotide, which templates proximity ligation and sequence preamplification. Finally, each signal can be amplified and detected using a combination of orthogonal forward primers, universal reverse primer, and qPCR (Fig. 4A). We tested this multiplexed approach for the 6 active enzyme biomarkers using new oligonucleotide designs and barcoded reagents and observed equally robust signal and target fidelity compared with the single-plex format (Fig. 4 B and C and SI Appendix, Fig. S7).
Fig. 4.
Multiplexed sADPL enables simultaneous quantification of active enzymes. (A) Schematic outline of multiplexed “writing” and “reading” of active enzyme signals can be performed simultaneously with sADPL. FP, forward primer sequence; RP, reverse primer sequence. (B and C) Signal profiles for representative enzyme targets detected by multiplexed sADPL (B), which are highly correlated to single-plex measurements (C) and display comparable dynamic ranges. All data points are 3 technical replicates from 2 or more biological replicates. Values represent the mean, and error bars represent the SD.
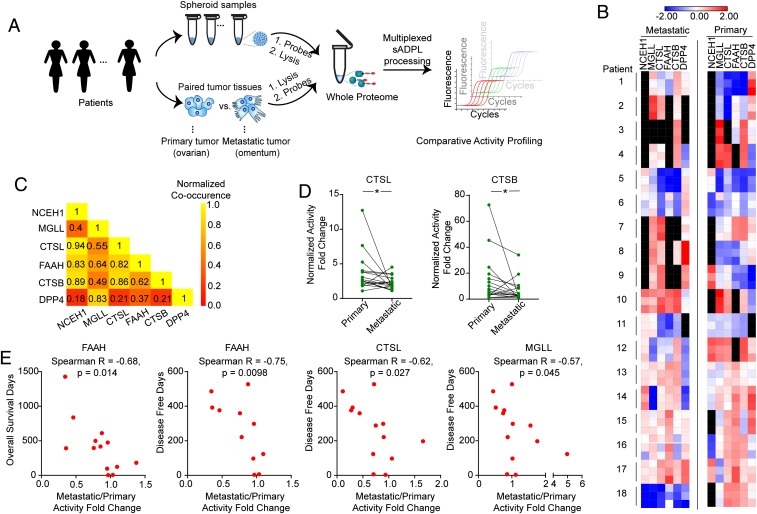
Having established the ability of sADPL to rapidly measure parallel target protein activities with very low input proteome, we next tested whether sADPL could be used to profile enzyme activities in human clinical samples. In particular, we sought to determine whether sADPL profiling of enzymes previously associated with cancer cell aggressiveness in ovarian cancer patient samples could provide insight into the biological mechanisms associated with disease state and progression. Toward this end, we performed a series of between-patient and within-patient sADPL profiling screens using banked, flash-frozen primary ovarian and corresponding metastatic tumors and ovarian cancer spheroids from patients with high-grade serous ovarian cancer (Fig. 5A). We first measured the relative activity of 4 serine hydrolases (NCEH1, MGLL, FAAH and DPP4) and 2 cathepsin proteases (CTSB and CTSL) in cancer spheroids collected from 15 patients during initial debulking surgery (SI Appendix, Table S1). In the same samples, we simultaneously measured the abundance of a loading control protein, GAPDH, using proximity ligation assay (PLA)-qPCR as a way to normalize for sample, processing, and amplification differences across patients (SI Appendix, Fig. S8) (20). The resulting activity profile confirmed that NCEH1, MGLL, CTSB, and CTSL activities were generally correlated within the same patient spheroids, whereas active FAAH levels were more variable and DPP4 activity was undetectable (SI Appendix, Fig. S9).
Fig. 5.
Multiplexed sADPL activity profiling in patient-derived tumor samples. (A) Schematic depiction of sADPL profiling of cancer spheroids and primary tumor and metastatic tumor tissues. (B) Heatmap depiction of a representative, multiplexed sADPL profiling experiment detecting 6 active enzymes from 2 tumor compartments in 18 patients. All data points are z-scores normalized from 3 technical replicates from 1 biological replicate. Individual activity measurements below the detection threshold are shown in black. (C) Metastatic/primary tumor activity ratio co-occurrence for each enzyme target measured across the 18-patient cohort. Active enzymes that show higher co-occurrence are yellow on the relative scale. (D) Within-patient activity measurements between primary and metastatic tumor sites reveals that CTSB and CTSL activities are significantly altered across the patient cohort. *P < 0.05, Wilcoxon signed-rank t test. (E) Correlation between patient survival and metastatic/primary tumor activity ratios demonstrates the presence of significant negative correlations for the enzymes shown. Each point represents an individual patient and corresponding mean activity ratio from 3 technical and 2 biological replicates.
These data confirm the potential to perform multiplexed activity profiling directly in primary patient samples with sADPL, which inspired us to next interrogate enzyme activities between unique tumor compartments in the same patients. To accomplish this, we profiled matched primary and metastatic tumor tissue from 18 patients with high-grade serous cancer (SI Appendix, Table S1). We hypothesized that comparing the relative enzyme activities between biologically distinct tumor tissues (i.e., primary tumors in hormone-producing ovaries and metastatic tumors in adipose tissue-rich omentum) (31) from the same patient could provide insight into the potential role of these enzymes in metastasis.
As a proof-of-concept example, we executed a multiplexed analysis of 6 biomarker activities in triplicate from 2 distinct tumor compartments in 18 ovarian cancer patients all in a single experiment that took approximately 5 to 6 h. This single biological replicate represents 648 individual quantitative activity measurements with minimal sample preparation and high sensitivity (e.g., from nanogram quantities of whole proteome) (Fig. 5 A and B). This analysis revealed a distinct pattern of activities between tumor sites and several general co-occurring enzyme activity trends in comparisons across all patients (Fig. 5 B and C). For example, the activities of both cathepsin proteases CTSB and CTSL were significantly higher in the primary tumor compared with the corresponding metastatic tumor (Fig. 5D). In contrast, several enzymes showed no quantitative difference in activity between the primary and metastatic tissues across the entire patient group (SI Appendix, Fig. S10A).
The notion that the relative protein activity between primary and metastatic sites could be informative prompted us to test for correlations between activity ratios and clinical outcomes, such as patient survival. Intriguingly, the activity ratio for several enzymes, including FAAH, CTSL, and MGLL, showed a significant negative correlation with disease-free survival across the patient cohort (Fig. 5E). In all cases, a larger metastatic/primary tumor activity ratio was correlated with decreased patient survival. The activity ratio for FAAH was also correlated with overall survival. Unsupervised k-means clustering of patients using a principal component analysis of all available clinical data (SI Appendix, Fig. S10B), segregated patients into 3 distinct groups with statistically significant differences in overall survival and disease-free survival. The mean FAAH activity ratio differed significantly among these groups (SI Appendix, Fig. S10 C and D). FAAH has been peripherally implicated in tumor metabolism, and thus these data present novel hypotheses for future testing. While conclusions about the prognostic potential of these enzyme activity ratios are premature given the small patient sample, these data demonstrate that hundreds to thousands of ultrasensitive parallel activity measurements can be made directly with fresh or flash-frozen patient samples in a matter of hours on a benchtop, which stands in stark contrast to activity-based readouts by existing methods.
Discussion
We have developed a general chemical proteomic platform that permits ultrasensitive, multiplexed, and activity-dependent quantification of endogenous proteins in complex biological samples. This platform is directly compatible with existing (or future) family-wide chemical probes provided that they have suitable recognition elements (e.g., biotin or desthiobiotin used here), thereby providing target-specific information without the need for dedicated probe development. Compared with PLA-based methods of detecting protein abundance alone, this method has the benefit of requiring only 1 polyclonal or monoclonal antibody and involves formation of the ADPL ternary complex in native binding conditions. This latter aspect carries the future possibility of interrogating the binding partners of active enzymes, but also the limitation that an active protein will not be recognized if its antibody-binding epitope is masked by neighboring proteins or other biomolecules. Future exploration of additional recognition moieties, both chemical and orthogonal receptor-based, is warranted to expand the sensitivity and multiplexing capacity of the approach.
Beyond direct compatibility with a wide range of probes, we sought to address several limitations associated with existing chemical proteomic platforms. Chief among these were the trade-offs between the breadth and depth of information procurable by gel-based or MS-based methods, their narrow dynamic range, and the general requirement for relatively large input proteome amounts. sADPL overcomes these issues through the coupling of probe labeling with specific and robust oligonucleotide amplification. Here we demonstrated that the proximity-dependent ligation and amplification of barcoded oligonucleotides permits measurement of active enzymes across families from samples that are several orders of magnitude smaller than required for Western blot analysis.
While we have shown that sADPL is uniquely suited for focused questions around a subset of target proteins, especially in samples with limited abundance or when higher-throughput profiling is needed, we do not believe that this method should be viewed as a replacement for LC-MS/MS–based activity profiling approaches. Rather, we view sADPL as complementary to traditional gel- and MS-based readouts of active enzymes.
Despite the minimal sample requirements, the resulting quantitative activity measurements by sADPL are highly consistent with bulk measurements made by gel- and LC-MS/MS–based methods. Direct comparison of protein abundance, measured by LC-MS/MS, and active protein levels, measured by sADPL, in 2 ovarian cancer cell lines provided further evidence that protein abundance alone is not a suitable surrogate for protein activity, as in several cases we observed large differences between abundance and activity. These results, along with accumulating evidence of poor correlations between mRNA and protein levels, confirm the need for methods like sADPL that can directly quantify the active proteome in complex biological samples. We envision that future integration of the sADPL workflow with mRNA and protein abundance measurements from the same samples (through, e.g., sequencing) could provide new insight into biological regulation across the central dogma.
Probe-dependent protein detection not only provides a way to specifically measure active proteins, but also provides opportunities for competitive profiling and detection of small molecule–protein interactions. Like gel- and MS-based activity profiling, sADPL is able to directly quantify specific and dose-dependent engagement of chemical probes with proteins of interest following in vitro and in situ cellular treatments. Here we extended this potential to directly measure pharmacodynamic target engagement following in vivo administration of a specific NCEH1 inhibitor in mice. sADPL profiling of an readily accessed cellular sample, PBMCs, showed highly consistent, dose-dependent target engagement of the intended target in mouse PBMCs. Parallel profiling of 2 related enzyme family members showed no effect of the NCEH1-specific inhibitor on their activity, confirming specific target engagement in vivo in a workflow that has minimal processing steps, is benchtop-compatible, and can be performed in a matter of hours.
Importantly, the solution-based ADPL approach described herein is distinct from our previously reported imaging ADPL readout for several reasons. First, compared with imaging ADPL, which involves the “writing” and “reading” of activity-dependent amplicons on the cell surface with iterative incubation and washing steps, it was not obvious that a “one-pot” approach would enable target-specific signal when profiling multiple targets with 1 or more family-wide probes. Indeed, we demonstrated that we can specifically measure the activity of multiple enzymes within and between families using barcoded reagents and a multiplexing scheme. This permits profiling of many types of biological samples with greater ease and higher throughput than would be possible with imaging-based detection and quantification alone. We anticipate that similar profiling studies could be performed from isolated cellular or tissue samples (e.g., fine-needle aspirates) as well as in biofluids for this and other target families, providing a more robust and accurate measure of drug action compared with serum concentrations alone.
Finally, the barcoding capacity of the oligonucleotide signals allows for multiplexed labeling, oligonucleotide amplification, and quantitative measurements in the same sample. These attributes are specifically attractive for complex, limited-abundance samples like clinical tissues and fluids, which are an active area of focus for ultrasensitive protein detection technologies (32–36). The high-throughput nature of this proof-of-principle application to clinical samples highlights the ability to perform hundreds to thousands of activity measurements with minimal sample preparation, high sensitivity, and good reproducibility. In comparison, sample preparation and analysis by LC-MS/MS detection would be considerably more expensive and take weeks to months for the same experiment.
Owing to the ability to quantify these marker activities across tumor sites and patients, we discovered several novel disease associations. For example, we found a significant decrease in the activity of the cathepsin proteases CTSL and CTSB in the metastatic tumor compartment compared with the primary tumor. This finding is consistent with previous studies implicating high CTSB and CTSL protein abundance in aggressive ovarian cancers; however, no previous studies have probed protease activity or tumor compartment-specific activity (37–39). The relatively high activity in the primary tumor site is in agreement with the roles of microenvironment remodeling factors and proteases during the epithelial-to-mesenchymal transition in primary metastatic tumors (40), and also could be related to microenvironmental conditions in the primary tumor that may promote the activity of these enzymes. Importantly, protein and mRNA abundance measurements alone could not detect this context-specific change in activity. Furthermore, we observed significant trends between the activity ratio in the metastatic tumors compared with primary tumors for the lipid-hydrolase enzyme FAAH. While the distinct biological mechanisms underlying these differences are the subject of future dedicated investigations, these results establish the potential for sADPL profiling in clinical samples to provide novel biological hypotheses as well as potential diagnostic correlations based on activity and not solely on protein expression. We expect that optimized protocols, automation, and contemporary methods to generate oligonucleotide-protein conjugates (20) will further augment these capabilities, and that sADPL will find many applications in the molecular analysis of biological and clinical samples.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix. Information includes banking patient-derived tumor tissue, spheroid samples, isolation of human and mouse PBMCs, antibody- and streptavidin-oligo conjugates, general single-plex sADPL, competitive sADPL profiling of small molecule target engagement, comparative multiplexed sADPL activity profiling in patient-derived spheroid cells and tumor tissues, and PLA assays. Patient-derived cells, tissues, and clinical information were collected with informed consent and protocol approval by the University of Chicago’s Institutional Review Board (IRB protocol 13372). All patient information was deidentified before use in this study.
Supplementary Material
Acknowledgments
We thank all the patients who kindly donated samples. We thank C. He for access to instrumentation, P. Dauer for assistance with mouse PBMC isolation, and S. Ahmadiantehrani for proofreading assistance. This work was supported by NIH Chemical Biology Interface Training Grant 2T32GM008720-16 ( to J.E.M.), the Marsha Rivkin Foundation (M.A.E.), National Cancer Institute Grant R01 CA111882 (to E.L.), NIH Grants R00 CA175399 and DP2 GM128199-01 (to R.E.M.), the Chicago Biomedical Consortium supported by Searle Family Funds (R.E.M.), University of Chicago Cancer Center Support Grant P30 CA014599, and the Duchossois Family Institute at the University of Chicago.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912934116/-/DCSupplemental.
References
- 1.Gygi S. P., Rochon Y., Franza B. R., Aebersold R., Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gry M., et al. , Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics 10, 365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghazalpour A., et al. , Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7, e1001393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadaghiani A. M., Verhelst S. H., Bogyo M., Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 11, 20–28 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Cravatt B. F., Wright A. T., Kozarich J. W., Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Patricelli M. P., Cravatt B. F., Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon G. M., Cravatt B. F., Activity-based proteomics of enzyme superfamilies: Serine hydrolases as a case study. J. Biol. Chem. 285, 11051–11055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patricelli M. P., et al. , Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 46, 350–358 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Zhao Q., et al. , Broad-spectrum kinase profiling in live cells with lysine-targeted sulfonyl fluoride probes. J. Am. Chem. Soc. 139, 680–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saghatelian A., Jessani N., Joseph A., Humphrey M., Cravatt B. F., Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. U.S.A. 101, 10000–10005 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato D., et al. , Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 1, 33–38 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Weerapana E., et al. , Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews M. L., et al. , Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat. Chem. 9, 234–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J. W., Montgomery J. E., Lee G., Moellering R. E., Chemoproteomic profiling of phosphoaspartate modifications in prokaryotes. Angew. Chem. Int. Ed. Engl. 57, 15712–15716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G., et al. , An activity-dependent proximity ligation platform for spatially resolved quantification of active enzymes in single cells. Nat. Commun. 8, 1775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredriksson S., et al. , Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Söderberg O., et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Gao X., Hannoush R. N., Single-cell in situ imaging of palmitoylation in fatty-acylated proteins. Nat. Protoc. 9, 2607–2623 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Okerberg E. S., et al. , High-resolution functional proteomics by active-site peptide profiling. Proc. Natl. Acad. Sci. U.S.A. 102, 4996–5001 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G., Moellering R. E., A concise, modular antibody-oligonucleotide conjugation strategy based on disuccinimidyl ester activation chemistry. ChemBioChem 20, 1599–1605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang K. P., Niessen S., Saghatelian A., Cravatt B. F., An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 13, 1041–1050 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Nomura D. K., et al. , Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140, 49–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coscia F., et al. , Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 7, 12645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessani N., Liu Y., Humphrey M., Cravatt B. F., Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci. U.S.A. 99, 10335–10340 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moellering R. E., Cravatt B. F., How chemoproteomics can enable drug discovery and development. Chem. Biol. 19, 11–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon G. M., Niphakis M. J., Cravatt B. F., Determining target engagement in living systems. Nat. Chem. Biol. 9, 200–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones L. H., Neubert H., Clinical chemoproteomics: Opportunities and obstacles. Sci. Transl. Med. 9, eaaf7951 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Chang J. W., Nomura D. K., Cravatt B. F., A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem. Biol. 18, 476–484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn K., et al. , Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem. Biol. 16, 411–420 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredriksson S., et al. , Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods 4, 327–329 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lengyel E., Makowski L., DiGiovanni J., Kolonin M. G., Cancer as a matter of fat: The crosstalk between adipose tissue and tumors. Trends Cancer 4, 374–384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson P. V., Tsai C. T., de Groot A. E., McKechnie J. L., Bertozzi C. R., Glyco-seek: Ultrasensitive detection of protein-specific glycosylation by proximity ligation polymerase chain reaction. J. Am. Chem. Soc. 138, 10722–10725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albayrak C., et al. , Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell 61, 914–924 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Ullal A. V., et al. , Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci. Transl. Med. 6, 219ra9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giedt R. J., et al. , Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens. Nat. Commun. 9, 4550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai C. T., et al. , Antibody detection by agglutination-PCR (ADAP) enables early diagnosis of HIV infection by oral fluid analysis. Proc. Natl. Acad. Sci. U.S.A. 115, 1250–1255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scambia G., et al. , Cathepsin D assay in ovarian cancer: Correlation with pathological features and receptors for oestrogen, progesterone and epidermal growth factor. Br. J. Cancer 64, 182–184 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lösch A., et al. , Cathepsin D in ovarian cancer: Prognostic value and correlation with p53 expression and microvessel density. Gynecol. Oncol. 92, 545–552 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Kolwijck E., et al. , Cathepsins B, L and cystatin C in cyst fluid of ovarian tumors. J. Cancer Res. Clin. Oncol. 136, 771–778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.