Significance
The spread of multidrug-resistant infections demands antibiotic development. Although new antibiotics targeting gram-positive bacteria have been developed, it has been more than 50 y since a new class of antibiotics targeting gram-negative bacteria has been approved for clinical use. The strong outer membrane permeability barrier makes it difficult for compounds to reach intracellular targets, contributing greatly to the lack of new antibiotics targeting this class of microbes. Here, we describe a compound, MRL-494, that inhibits the assembly of proteins into the outer membrane by targeting an essential member of β-barrel assembly machine. This compound appears to act at the cell surface, avoiding both the outer membrane permeability barrier and the action of multidrug-resistance efflux pumps.
Keywords: outer membrane biogenesis, antibiotic development, BAM complex, gram-negative bacteria, Escherichia coli
Abstract
The development of new antimicrobial drugs is a priority to combat the increasing spread of multidrug-resistant bacteria. This development is especially problematic in gram-negative bacteria due to the outer membrane (OM) permeability barrier and multidrug efflux pumps. Therefore, we screened for compounds that target essential, nonredundant, surface-exposed processes in gram-negative bacteria. We identified a compound, MRL-494, that inhibits assembly of OM proteins (OMPs) by the β-barrel assembly machine (BAM complex). The BAM complex contains one essential surface-exposed protein, BamA. We constructed a bamA mutagenesis library, screened for resistance to MRL-494, and identified the mutation bamAE470K. BamAE470K restores OMP biogenesis in the presence of MRL-494. The mutant protein has both altered conformation and activity, suggesting it could either inhibit MRL-494 binding or allow BamA to function in the presence of MRL-494. By cellular thermal shift assay (CETSA), we determined that MRL-494 stabilizes BamA and BamAE470K from thermally induced aggregation, indicating direct or proximal binding to both BamA and BamAE470K. Thus, it is the altered activity of BamAE470K responsible for resistance to MRL-494. Strikingly, MRL-494 possesses a second mechanism of action that kills gram-positive organisms. In microbes lacking an OM, MRL-494 lethally disrupts the cytoplasmic membrane. We suggest that the compound cannot disrupt the cytoplasmic membrane of gram-negative bacteria because it cannot penetrate the OM. Instead, MRL-494 inhibits OMP biogenesis from outside the OM by targeting BamA. The identification of a small molecule that inhibits OMP biogenesis at the cell surface represents a distinct class of antibacterial agents.
Antibiotic resistance is increasingly prevalent. In fact, rates of resistances in some countries for key pathogen–antibiotic pairs have risen above 90% (1), highlighting the critical need for the development of new antibiotics. Although recent efforts have led to the development of new antibiotics targeting gram-positive bacteria, antibiotic discovery efforts for gram-negative bacteria have been much slower due to the presence of the outer membrane (OM) (2). The OM is an asymmetric lipid bilayer composed of an outer leaflet of lipopolysaccharide (LPS) and an inner leaflet of phospholipids (3). Although small hydrophilic molecules permeate the OM through abundant porins, a class of OM β-barrel proteins (OMPs) (3, 4), its unique structure makes the OM impermeable to both large and hydrophobic molecules (5). Because of this barrier, gram-negative bacteria are protected from many antibiotics to which gram-positive bacteria are susceptible.
Beyond their intrinsic antibiotic impermeability, gram-negative bacteria can acquire high-level antibiotic resistance through several different genetic mechanisms. These include mutations or the acquisition of genes that alter or bypass the antibiotic target, inactivate or destroy the antibiotic, or cause the up-regulation of genes encoding multidrug-resistance (MDR) efflux pumps (6). Of these, MDR efflux pumps are not only the most frequently employed mechanisms of resistance, but they also confer protection from many antibiotics simultaneously in a manner independent of the target (7).
One strategy to avoid resistance provided by the MDR efflux pumps of gram-negative bacteria is to target essential processes that occur on the cell surface. This strategy would not only negate the activity of the pumps, but it would also avoid the intrinsic resistance posed by the OM permeability barrier. This requires the identification of antimicrobial agents that target the essential, surface-exposed proteins required for OM biogenesis: LptD, the surface-exposed component of the transenvelope complex, which translocates LPS to the cell surface, and BamA, the surface-exposed member of the β-barrel assembly machine (BAM complex) responsible for OMP assembly (3, 8).
Previous studies have validated this approach. Several antimicrobial peptides have been identified that target LptD and inhibit LPS transport to the cell surface, leading to cell death (9, 10). Moreover, an antibody binding to the extracellular region of BamA inhibits OMP biogenesis and kills cells (11). Recently, a small molecule inhibiting autotransporter assembly was described. Although this molecule may inhibit BAM function, the target has yet to be identified (12).
In order to identify new potential antibiotics targeting these surface-exposed essential OM proteins, we conducted a screen for small molecules that disrupted the OM barrier at sublethal concentrations and exhibited antimicrobial activity against Escherichia coli that was not reduced by efflux pump activity. Using these screens, we identified a compound, MRL-494, that inhibits the insertion of OMPs into the OM by targeting BamA.
Results
Identification of a Potential OM-Targeting Compound.
Previously, we described the identification of a collection of compounds that inhibited the growth of an E. coli strain deficient for efflux and OM integrity (13, 14). We hypothesized that this collection contained both compounds that are inhibited by the OM permeability barrier and/or efflux and compounds that can act on wild-type cells, bypassing these barriers. Therefore, we sought to identify bioactive molecules within this collection that had activity against efflux- and permeability-competent E. coli. These compounds are of particular interest since they appear to evade efflux and the OM permeability barrier due to either their chemical nature or the surface location of their target.
One such compound, MRL-494 (Fig. 1A), exhibited nearly equipotent activity against a wild-type strain as in a strain with a deletion of tolC, the gene for the efflux pump OM channel (15) (SI Appendix, Table S1). Strikingly, this compound also exhibited antimicrobial activity against a broad range of bacterial species (SI Appendix, Table S1). Interestingly, MRL-494 was an unintended byproduct created during the synthesis of an unrelated compound and therefore represented a unique chemotype within the screening collection.
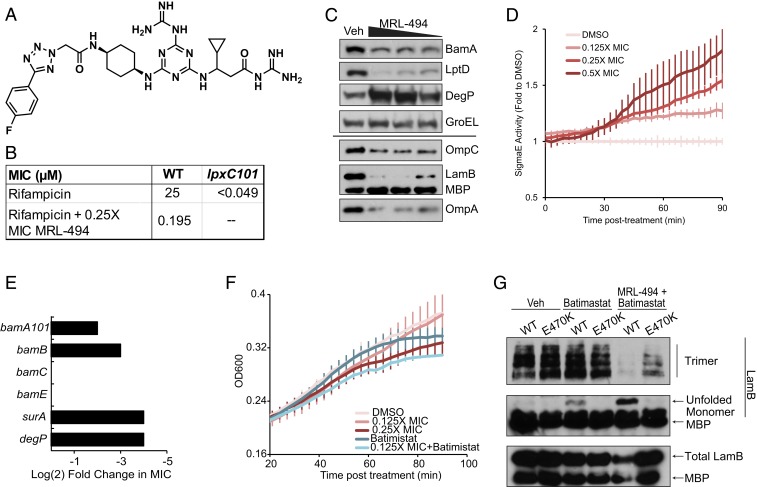
Fig. 1.
MRL-494 impairs biogenesis of OMPs. (A) Chemical structure of MRL-494. (B) MRL-494 potentiates rifampicin. MIC measurements for rifampicin with or without a sublethal dose of MRL-494 were performed. (C and D) Midlog cultures of TJS101 carrying a PmicA-gfp reporter were treated with MRL-494 at 1×, 0.5×, and 0.25× MIC or a DMSO control (vehicle [Veh]) and grown for 1.5 h. (C) MRL-494 treatment decreases OMP abundance. After 1.5 h of treatment, cells were harvested, normalized to OD, and assayed for protein abundance. The heavy line indicates separate biological replicates. Data are representative of 2 independent experiments. (D) MRL-494 treatment results in a dose-dependent increase in σE activity. GFP fluorescence posttreatment was measured as a reporter of σE activity and normalized to OD600. Data are shown as the average fold value to the DMSO treatment group ± SD of triplicate samples. (E) OMP-biogenesis mutants demonstrate increased susceptibility to MRL-494. The resistance to MRL-494 of strains with the indicated deletion or mutation in a tolC+ background was assayed by MIC. Log2 fold changes relative to the parent strain are shown. Data are representative of 2 independent experiments. (F) Batimastat treatment increases susceptibility to MRL-494. Cells were treated as in B and C and were additionally treated with 0.25× MIC batimastat where indicated. OD600 was assayed posttreatment. Data are shown as the average of triplicate samples ± SD. (G) OMP biogenesis is inhibited by MRL-494 and restored by bamAE470K. Midlog cultures of strains expressing wild-type (WT) bamA or bamAE470K in haploid were treated with DMSO (Veh), 0.25× MIC batimastat, or 0.25× MIC batimastat with 0.25× wild-type MIC MRL-494 for 1.5 h. The conformational state and total abundance of LamB were assayed. Data are representative of 2 independent experiments.
We then sought to determine whether MRL-494 1) was penetrating the cell envelope and was impervious to efflux or 2) was targeting an OM-related process. To this end, we tested the ability of MRL-494 to potentiate the activity of rifampicin, an antibiotic that is impervious to efflux but cannot easily traverse the OM of wild-type E. coli (13). We found the presence of sublethal concentrations of MRL-494 causes a large decrease in the minimum-inhibitory concentration (MIC) of rifampicin (Fig. 1B), similar to that observed with the allele envA101, a mutation within the lpxC gene that results in less LPS in the outer leaflet and a compensatory increase in phospholipid bilayer patches in the OM. These data demonstrate that MRL-494 disrupts the permeability barrier of the OM and allows for entry of rifampicin, suggesting that MRL-494 interdicts an essential OM-related pathway.
It is possible that MRL-494 disrupts OM permeability via a mechanism that perturbs the lipid composition of the cell. Bacteria carrying the envA101 mutation did not demonstrate a shift in the MIC of MRL-494 compared to wild type (SI Appendix, Table S1). Similarly, there was no shift in MIC in an Acinetobacter baumannii strain lacking LPS via a mutation in the lpxC gene. Together, these results suggest that MRL-494 leads to an increase in OM permeability, but this enhanced permeability does not potentiate the toxicity of MRL-494 or allow it to more readily penetrate the OM barrier. Therefore, MRL-494 might inhibit an essential OM-related process, the target of which could be accessible to the cell surface.
MRL-494 Inhibits OMP Biogenesis.
To investigate the possibility that MRL-494 acts by targeting an OM-related process, we looked for changes occurring to the OM during MRL-494 treatment and found that the abundance of major OMPs (BamA, LptD, OmpC, LamB, OmpA) decreased in a dose-dependent manner, whereas levels of cytoplasmic (GroEL) and periplasmic proteins (MBP) were maintained (Fig. 1C). Furthermore, we also observed a corresponding increase in the levels of the periplasmic protease DegP. DegP degrades unfolded OMPs that accumulate in the periplasm (16), and its expression is controlled by envelope stress responses (17, 18). This suggested that the σE stress response, which is activated by OMP- and LPS-biogenesis defects (19–23), might be activated by MRL-494 treatment. Using a GFP reporter driven by a σE-responsive promoter (24), we observed a dose-dependent increase in σE activity following MRL-494 treatment (Fig. 1D). Together, these data suggest that MRL-494 may target OMP biogenesis.
Newly translated OMPs are secreted into the periplasm by the Sec translocon (8, 25) (SI Appendix, Fig. S1). After secretion, the OMPs are bound by chaperones (e.g., SurA) (26) and transported through the aqueous periplasm to the heteropentomeric BAM complex (i.e., BamABCDE) (27–30). The BAM complex then folds and inserts the OMPs into the OM (8, 27–30). BamA and BamD are essential, while BamB, BamC, and BamE are not (28–30). To test whether MRL-494 might inhibit OMP biogenesis, we first investigated whether loss-of-function mutations in OMP biogenesis pathways synergized with MRL-494. Lowering the levels of BamA or deleting bamB caused decreases in the MIC of MRL-494 (Fig. 1E). Moreover, deletion of the gene for either the primary periplasmic chaperone responsible for delivery OMPs to the BAM complex (surA) (26) or the major protease responsible for degrading OMPs that fail to be assembled (degP) (16) caused even larger MIC shifts.
As previously mentioned, the σE system, which responds to unfolded OMPs in the periplasm (19, 22, 23), is activated by MRL-494. Induction of this stress response increases expression of BAM complex members, periplasmic chaperones, and periplasmic proteases, among others (19). Given the genetic interactions between MRL-494 and mutations in the genes for σE regulon members, we hypothesized that σE activation after MRL-494 treatment is protective. To test this possibility, we treated cells with sublethal doses of MRL-494 and batimastat, an inhibitor of σE activation (24). The cotreatment of cells with MRL-494 and batimastat caused a growth defect that was greater than an elevated dose of MRL-494 (Fig. 1F), demonstrating that σE activation in the context of MRL-494 treatment is protective.
During assembly, the commonly employed model OMP, LamB, transitions from an unfolded state to a transient monomeric β-barrel to a final trimeric β-barrel state in the OM (31). We examined the abundance of these states in the presence of a sublethal dose of MRL-494 and batimastat, which prevents the down-regulation of OMPs and up-regulation of periplasmic proteases that occurs upon σE activation (24). After cotreatment, the levels of functional LamB trimer were almost undetectable, whereas treatment with batimastat alone caused no change in trimer formation (Fig. 1G, see lanes marked “WT”), indicating that MRL-494 inhibits assembly of OMPs into the OM.
The current model for the primary cause of death following the inhibition of OMP biogenesis is a toxic accumulation of unfolded OMPs (24, 32, 33). Indeed, when we cotreat with MRL-494 and batimastat, we see an accumulation of unfolded LamB that is much greater than that observed with batimastat alone (Fig. 1G, see lanes marked “WT”). These data, taken as a whole, establish that MRL-494 acts by inhibiting OMP biogenesis.
BamAE470K Confers Resistance to MRL-494 by Restoring OMP Biogenesis.
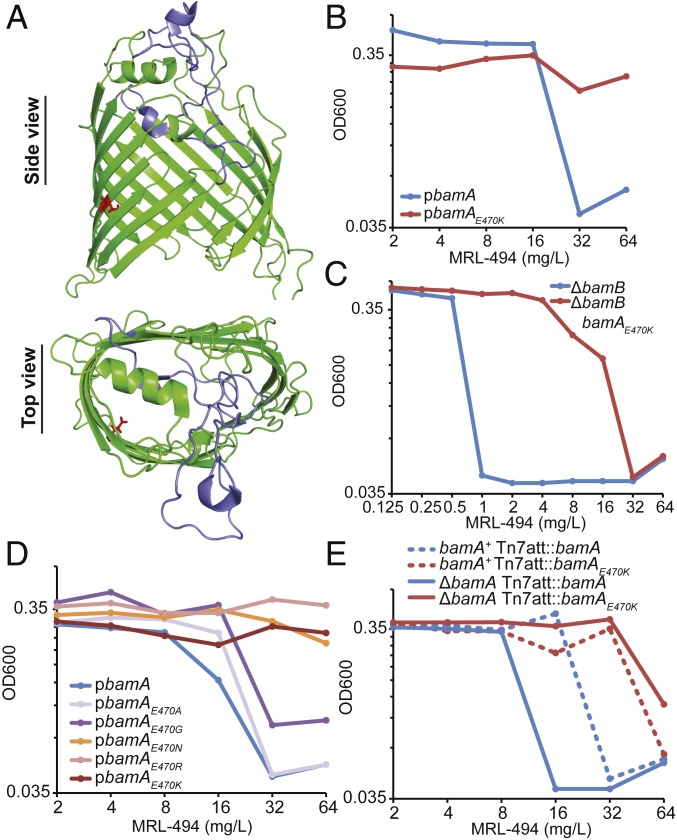
The BAM complex is composed of one essential β-barrel OMP (BamA), 1 essential lipoprotein (BamD), and 3 nonessential lipoproteins (BamBCE) that bind in equal stoichiometry (8). As MRL-494 is not a substrate for efflux and targets OMP biogenesis, we hypothesized that MRL-494 directly targets BamA, the only component of the OMP biogenesis pathway that is both essential and surface-exposed (28, 30). Therefore, we generated a pooled PCR mutagenesis library for bamA and screened for clones that exhibited enhanced resistance to MRL-494 as compared to a strain carrying wild-type bamA in liquid culture (SI Appendix, Fig. S2). We isolated one resistant clone encoding BamAE470K (Fig. 2A). BamAE470K was sufficient for growth in haploid when encoded chromosomally (SI Appendix, Fig. S3) and provided resistance to MRL-494 to the limit of MRL-494 solubility (Fig. 2B). During further investigation, we found that in a ΔbamB background, which greatly lowers the MIC of MRL-494 (Fig. 1D), BamAE470K provided 32-fold resistance to MRL-494 (Fig. 2C). The rate of spontaneous resistance in a ΔbamB background was found to be less than 3.5 × 10−10 using 1 to 3 times the MIC of MRL-494.
Fig. 2.
BamAE470K provides resistance to MRL-494 and restores OMP biogenesis. (A) A structure of BamA β-barrel domain (59) is shown a side view (from the plane of the OM) and a top view (from outside the OM). The E470 is indicated as red sticks, while extracellular loop 6 is indicated in blue. (B) Expression of bamAE470K results in increased resistance to MRL-494. BamA-depletion strains carrying the indicated plasmids were grown without arabinose to deplete BamA from the chromosomal copy of bamA, and MRL-494 resistance was then assayed by the MIC procedure. OD600 following overnight MRL-494 treatment is shown. Data are representative of 3 biological replicates. (C) Expression of bamAE470K results in a large increase in MRL-494 resistance in a ΔbamB background. ΔbamA ΔbamB with either wild-type bamA or bamAE470K at the Tn7 attachment site were assayed for MRL-494 resistance by the MIC procedure. OD600 following overnight MRL-494 treatment is shown. Data are representative of 2 independent experiments. (D) Resistance to MRL-494 correlates with the residue at BamAE470. Cells carrying plasmids with various alleles at bamAE470 were treated as in B and assayed for MRL-494 resistance. Data are representative of 2 independent experiments. (E) Expression of bamA in merodiploid increases resistance to MRL-494. Strains expressing bamA or bamAE470K in merodiploid or haploid were assayed for MRL-494 susceptibility through the MIC procedure. OD600 following overnight MRL-494 treatment is shown. Data are representative of 2 independent experiments.
We then wondered whether other residues at BamAE470 would also provide resistance. Remarkably, the degree of MRL-494 resistance correlated with the charge of the residue at E470 (Fig. 2D). Thus, the BamAE470 mutants with greater charge changes are increasingly resistant to MRL-494.
To test the effects of bamAE470K on OMP biogenesis directly, we monitored the assembly of the model OMP LamB in both the wild-type and the resistant strain in the presence and absence of MRL-494. Expression of bamAE470K does not change LamB assembly in the absence of MRL-494 (Fig. 1G, compare vehicle lanes). However, in the presence of MRL-494 and batimastat, assembly of LamB trimer was partially restored in strains carrying the bamAE470K mutation, demonstrating that BamAE470K acts to restore OMP assembly into the OM in the presence of MRL-494. Furthermore, BamAE470K prevented the toxic accumulation of unfolded OMPs in the presence of MRL-494 and batimastat (Fig. 1G, compare MRL-494 + batimastat lanes).
Two possible mechanisms could explain the restoration of OMP biogenesis in MRL-494 treated strains carrying bamAE470K: 1) MRL-494 inhibits BamA function and BamAE470K relieves this inhibition or 2) MRL-494 targets another component of the OMP biogenesis pathway and bamAE470K is a bypass suppressor restoring OMP biogenesis despite the inhibition of the targeted component. To investigate these possibilities, we changed the ratio of BamA to the rest of the BAM complex by expressing bamA in merodiploid. Expressing 2 wild-type copies of bamA resulted in a 2-fold increase in the MIC of MRL-494 (Fig. 2E), likely due to a portion of MRL-494 binding to BamA that is not part of an active BAM complex. These results also preclude the possibility that MRL-494 uses BamA as a channel to enter the cell, as a decrease in MIC would then be expected with bamA overexpression. In merodiploid, bamAE470K demonstrated incomplete dominance to wild-type bamA, because complete, functional BAM complexes were formed with both wild-type BamA and BamAE470K (Fig. 2E). Taken together, these data demonstrate that MRL-494 targets BamA function in order to inhibit the ability of the BAM complex to insert OMPs into the OM.
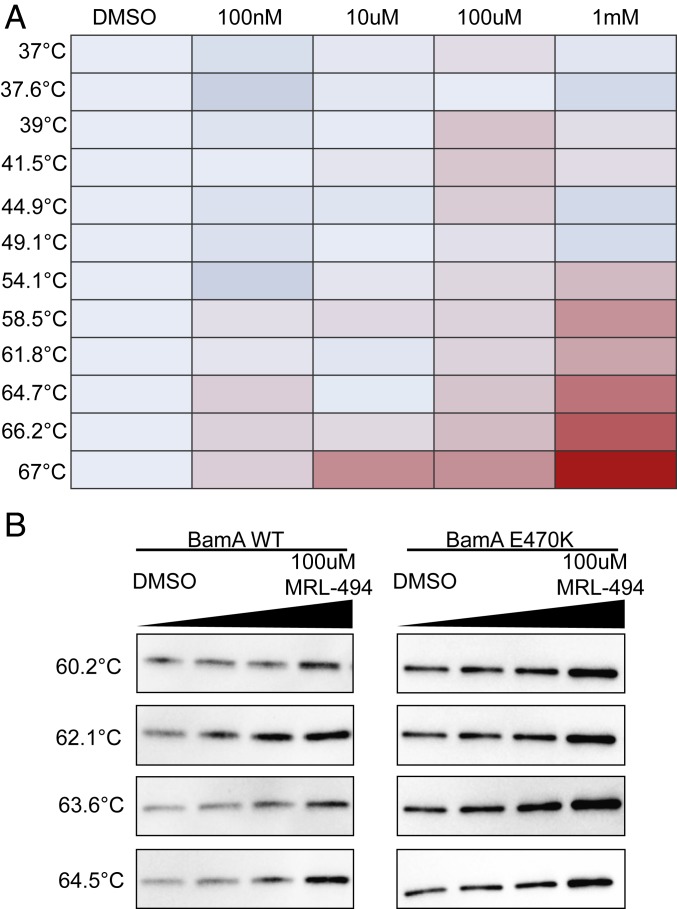
To support this conclusion, we carried out cellular thermal shift assay (CETSA) experiments with MRL-494 (Fig. 3) (34, 35). We found that MRL-494 stabilized BamA against thermally induced protein aggregation in vivo (Fig. 3A), a frequent indication of direct or proximal target engagement (36). We also measured the thermostability of other members of the BAM complex (BamB, BamC, and BamD) and other β-barrel OMP proteins, such as OmpA, but did not observe thermostabilization of these proteins by MRL-494 (SI Appendix, Fig. S4). These data do not support a biophysical effect on β-barrel proteins other than BamA or on a broader effect of MRL-494 on the BAM complex as a whole. Subsequently, we determined that the BamAE470K mutant was also thermally stabilized by MRL-494 treatment (Fig. 3B). This result raises the possibility that the BamAE470K mutant induces MRL-494 resistance through a mechanism other than blocking direct protein/compound binding.
Fig. 3.
MRL-494 binds directly or proximally to BamA. CETSA measures changes in thermostability of proteins in response to compounds in a cellular context. Direct or proximal binding of a protein to a compound can increase thermostability of the protein. (A) Two-dimensional CETSA mass spectrometry measurement of BamA in the presence of the indicated concentrations of MRL-494. MRL-494 stabilizes BamA at elevated temperatures but does not affect BamA abundance at 37 °C. Red indicates a log2 fold change of 1.2, light blue indicates a log2 fold change of 0, and dark blue indicates a log2 fold change of −0.2. Data are representative of 2 independent experiments. (B) bamA+ and bamAE470K cells were incubated with the indicated concentrations of MRL-494. An isothermal dose–response fingerprint was measured at a range of temperatures. Dose-dependent stabilization was observed in both bamA+ and bamAE470K cells. Data are representative of 2 biological replicates.
BamAE470K Retains the Ability to Assemble OMPs.
As the E470K mutation does not change MRL-494 binding to BamA, we characterized the bamAE470K allele in the absence of MRL-494 to determine whether the mutation alters BamA function. Cells expressing bamAE470K exhibited a similar pattern of growth to isogenic control cells (SI Appendix, Fig. S5A), indicating that bamAE470K does not cause a growth defect or alter the transition between different phases of growth. We then tested the strength of the OM permeability barrier and found that cells merodiploid or haploid for bamAE470K were not more sensitive to antibiotics than the wild type: a bamAE470K strain has an intact OM permeability barrier (SI Appendix, Fig. S5B).
We hypothesized that BamAE470K may activate the σE stress response and shift the cell’s expression profile to decrease the load on the inhibited BAM complex. We compared σE activation in untreated cells carrying bamA or bamAE470K and found that BamAE470K does not elevate σE signaling (SI Appendix, Fig. S5C). Furthermore, resistant cells exhibited no change in OMP levels (SI Appendix, Fig. S5D) and bamAE470K does not have synthetic effects with OMP biogenesis mutants (SI Appendix, Fig. S5 E and F), although we did observe partial suppression of the antibiotic sensitivity of a ΔsurA strain (SI Appendix, Fig. S5F). Thus, the function of BamAE470K is essentially indistinguishable from the wild-type protein.
BamAE470K Destabilizes the β-Barrel Domain.
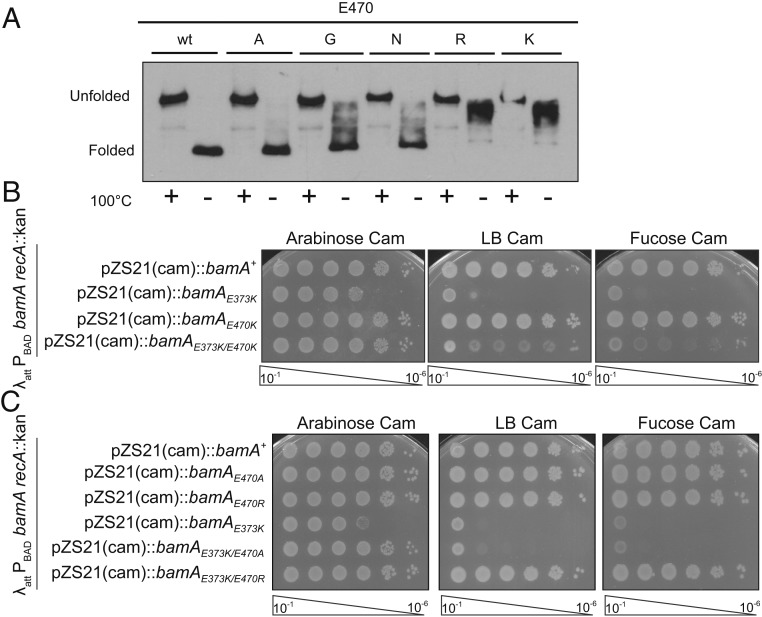
The bamAE470K mutation alters an amino acid in the middle of the fourth β-strand, and the affected side chain points into the β-barrel lumen (Fig. 2A). To determine if this mutation altered stability of the BamA β-barrel, we exploited the property of heat modifiability. β-Barrel proteins are so stable that they remain folded during sodium dodecyl sulfate (SDS) gel electrophoresis, unless the samples are heated (37). Thus, the stability of the β-barrel can be assayed through different migration of folded and unfolded proteins by gel electrophoresis.
We assayed the heat modifiability of BamAE470K and other BamAE470 residue changes to probe differences in the conformational flexibility of the proteins (Fig. 4A). Substitution of various amino acids at residue E470 influenced the stability of the BamA β-barrel, as indicated by changed migration of the nondenatured samples. Intriguingly, the stability was altered in a manner that correlated to the charge of the amino acid substitution. BamAE470K and BamAE470R, the most extreme substitutions, caused the largest shift in heat modifiability; most of the protein was unfolded before heat treatment. BamAE470A and BamAE470G, the least-charged substitutions, did not change β-barrel stability as compared to wild-type BamA. Remarkably, the changes in stability correlated well with the degree of MRL-494 resistance (compare Fig. 4A and Fig. 2D). However, β-barrel stability is not sufficient for resistance to MRL-494 as bamAG655D, a previously characterized mutation that decreases barrel stability (38), did not cause resistance to MRL-494 (SI Appendix, Fig. S6A). Thus, although BamAE470K retains wild-type function, it does have an altered conformation.
Fig. 4.
Charge changes at BamA E470 increase the activity of BamA. (A) Residues at BamAE470 affect the conformational stability of BamA. A BamA-depletion strain carrying plasmid-encoded bamAE470 alleles was grown to midlog in the absence of arabinose. Boiled (denatured) and folded samples were analyzed by gel electrophoresis. (B) BamAE373K/E470K is viable in haploid. The BamA depletion strain carrying the indicated plasmids was serially diluted onto LB media alone or LB containing arabinose or fucose to monitor dependence on the chromosomally encoded bamA. (C) Suppression of bamAE373K correlates with charge of bamAE470 allele. The indicated strains were analyzed for haploid viability as described in B.
Genetic Interactions of bamAE470K.
It is well established that BamA and BamD exist in multiple states and that OMP assembly requires these conformations to be properly coordinated. Proper interaction between the BamCDE and BamAB subcomplexes is necessary for each to be activated to fold and insert the OMP into the OM (39, 40). Mutations that alter the conformation of either BamA or BamD can interfere with this coordination and prevent OMP assembly (41). Suppressor mutations in bamA or bamD bypass the need for this coordination and restore OMP assembly (40, 41). Since the bamAE470K mutation allows for retained function despite the presence of MRL-494 (Fig. 2B), we tested how this mutant would affect the genetic interactions with our collection of bam mutations.
The bamAE373K mutation disrupts the interaction between BamA and BamD and is lethal in haploid due to a lack of proper coordination between the 2 proteins (41). This lethal phenotype can be suppressed by both intragenic suppressors in bamA (bamAK351E) and extragenic suppressors in bamD (bamDR197L) that bypass the need for proper coordination without restoring proper interaction between the 2 proteins (40, 41). In our genetic characterization of bamAE470K, we discovered that this mutation is an intragenic suppressor of bamAE373K: a strain carrying bamAE373K/E470K in haploid is viable, although sick (Fig. 4B). This demonstrates that, although BamAE470K is fully capable of assembling OMPs, the mutant protein bypasses checkpoints normally required for assembly.
Since altered conformation and degree of resistance to MRL-494 correlates with the amino acid substitution at codon 470, we tested whether intragenic suppression of the bamAE373K mutation correlates as well. Strikingly, bamAE373K/E470A was not viable in haploid, while bamAE373K/E470R was (Fig. 4C), demonstrating correlation between suppression of bamAE373K and MRL-494 resistance. The growth of bamAE373K/E470R appears to be more robust than that of bamAE373K/E470K, suggesting that the glutamate-to-arginine substitution is a stronger suppressor. However, neither an intragenic suppressor of bamAE373K, bamAK351E, nor an extragenic suppressor of bamAE373K, bamDR197L, caused MRL-494 resistance (SI Appendix, Fig. S6 C and D). The lack of resistance in cells expressing the gain-of-function allele bamDR197L decreases the plausibility of BamD as a potential target of MRL-494. Thus, while BamAE470K does not have altered function in the assembly of wild-type OMPs under laboratory conditions, it has both altered conformation and activity. Both of these correlate with resistance to MRL-494. However, neither the conformational change nor the altered activity seems to be sufficient to explain the MRL-494 resistance, although they may be necessary for resistance.
MRL-494 Inhibits Gram-Positive and -Negative Microbes through Distinct Mechanisms of Action.
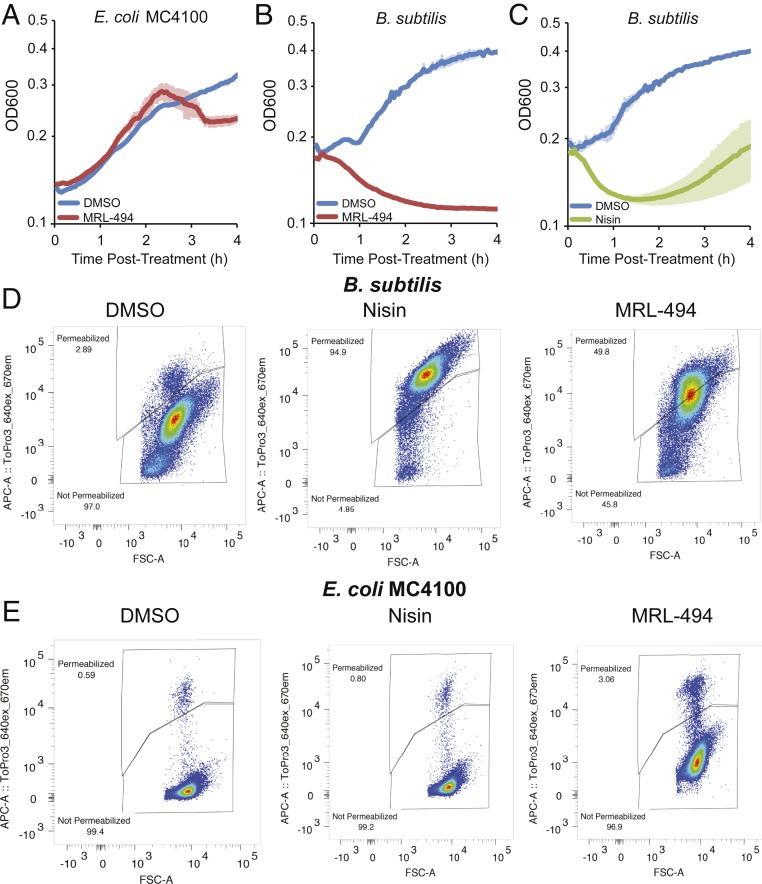
During initial screening, we found that MRL-494 inhibits the growth of gram-positive bacteria in addition to gram-negative bacteria (SI Appendix, Table S1). Since gram-positive species do not have an OM, we hypothesized that MRL-494 must have a second mechanism of action that kills gram-positive bacteria. To investigate this model, we compared cell death of E. coli and Bacillus subtilis cells after MRL-494 treatment. E. coli survive for several generations after treatment before decreasing in cell number (Fig. 5A). However, B. subtilis cells died immediately after treatment with MRL-494 (Fig. 5B), demonstrating different death kinetics than those observed in E. coli.
Fig. 5.
MRL-494 disrupts the cytoplasmic membrane of B. subtilis but not E. coli. (A–C) MRL-494 kills gram-positive cells in a manner similar to nisin. The indicated species was grown to midlog. The OD600 was monitored after treatment with DMSO or 1× MIC of nisin or MRL-494. (D and E) MRL-494 and nisin permeabilize B. subtilis cells (D) but not E. coli cells (E). Cells were treated with the indicated antibiotic and stained with TO-PRO-3 iodide. Permeabilization was monitored by flow cytometry. Data are representative of 2 independent experiments.
Given the cationic, amphiphilic, and peptidic structure of MRL-494, we sought to understand whether it could be mimicking the membrane-disrupting activity of antimicrobial peptides. Thus, we tested whether the kinetics of death we observed were similar to those of a membrane disrupting antimicrobial peptide, nisin. The sudden death of B. subtilis cells appeared strikingly similar to the rapid decline in cell number observed after exposure to the antimicrobial peptide nisin (compare Fig. 5 B and C) (42). These data suggested MRL-494 might kill B. subtilis in a similar manner to nisin.
Nisin is a class I pore-forming bacteriocin that inhibits gram-positive bacteria by binding to lipid II, a cytoplasmic membrane-associated precursor of peptidoglycan biosynthesis. Nisin then inserts into the cytoplasmic membrane, forms a pore, and permeabilizes the membrane (42, 43). To test if MRL-494 similarly disrupts the gram-positive cytoplasmic membrane, we treated cells and monitored membrane integrity at a sublethal compound concentration using a dye that detects membrane permeability, TO-PRO-3 iodide (44). TO-PRO-3 iodide is membrane-impermeable but is able to enter membrane-damaged cells and binds to double-stranded DNA (44). Exponentially growing E. coli and B. subtilis cells were stained with TO-PRO-3 iodide and treated with dimethyl sulfoxide (DMSO), MRL-494, or nisin. E. coli cells showed no fluorescent shift after exposure to nisin and only minimal permeabilization by MRL-494 (Fig. 5D and SI Appendix, Table S2). In contrast, B. subtilis exhibited extreme permeabilization after treatment with both nisin and MRL-494, indicating that MRL-494 permeabilizes the gram-positive cytoplasmic membrane (Fig. 5E and SI Appendix, Table S2).
Although MRL-494 permeabilizes the cytoplasmic membrane of gram-positive bacteria, it does not permeabilize the cytoplasmic membrane of gram-negative bacteria, suggesting the compound is physically excluded from reaching this membrane due to the presence of the OM. Similar prevention of activity on gram-negative bacteria is observed with nisin (Fig. 5D), where permeabilization is prevented unless the OM is severely compromised (45). We attempted to alter the OM by increasing phospholipids in the outer leaflet of the OM (mlaA* ΔpldA; ref. 46) and by expressing an open-pore mutant of OmpC (47) to allow MRL-494 to contact the cytoplasmic membrane of gram-negative bacteria. However, these attempts were unsuccessful (SI Appendix, Fig. S7).
Discussion
In this work, we characterize a small molecule that inhibits OMP assembly in gram-negative bacteria. MRL-494 exhibits the hallmark characteristics of an OMP biogenesis inhibitor, namely a decrease in steady-state OMP levels together with an increase in the unfolded form of these proteins, an increase in σE activation, and synergy with OMP-biogenesis mutations. We identify BamA as the target of MRL-494 with the isolation of a resistant mutant, bamAE470K, which retains wild-type function but has altered activity and conformation.
Additionally, we establish a second mechanism of action of MRL-494 against gram-positive bacteria. The structure of MRL-494, as a cationic amphiphile, suggests that the compound may disrupt membrane bilayers. Indeed, our work shows that MRL-494 rapidly kills gram-positive microbes in a manner similar to the antimicrobial peptide nisin, which is known to form pores in the cytoplasmic membrane (42). We conclude that the cytoplasmic membrane is the target of MRL-494 in gram-positive cells.
Strikingly, MRL-494 does not permeabilize the cytoplasmic membrane of E. coli. We suggest that the molecule cannot penetrate the OM, preventing the death of gram-negative cells by disruption of the inner membrane. This would also explain why MRL-494 acts in a tolC-independent manner; the OM permeability barrier prevents the compound from reaching a cellular compartment that is subject to efflux. Together, these data suggest MRL-494 acts at the cell surface to inhibit OMP biogenesis. Although monoclonal antibodies are known to inhibit BamA function by binding a surface-exposed epitope (38), MRL-494 is distinct as a small molecule that targets a surface-exposed moiety.
Our study also reveals that MRL-494 binds directly or proximally to BamA. Using CETSA, we found that in vivo addition of MRL-494 stabilizes thermally induced aggregation of wild-type BamA. Intriguingly, MRL-494 also stabilizes BamAE470K, suggesting that the resistant mutation does not change binding of the compound to its target. Thus, our data indicate that bamAE470K is a bypass suppressor that allows BamA function despite the presence of MRL-494. This mechanism of antimicrobial resistance has been previously observed. Notably, earlier studies characterized streptomycin-resistant E. coli mutants that continue protein synthesis despite maintained binding of streptomycin to the ribosome (48, 49). Similarly, MRL-494–treated cells expressing BamAE470K continue to assemble OMPs and support cell viability.
We have not determined where on the surface-exposed region BamA MRL-494 binds. Without more detailed information to support results obtained using CETSA, we cannot exclude the possibility that MRL-494 partitions into the OM and alters membrane properties in a manner that negatively impacts BamA function without directly interacting with BamA. Previous work has demonstrated the critical importance of the extracellular regions of the BamA for the protein’s function (11, 39, 50), and BamA activity has been linked to OM fluidity (51). Movement of extracellular loop 6 of BamA is an important part of the conformational cycling of BamA (39, 50). Additionally, as noted above, binding of monoclonal antibodies to extracellular loop 4 inhibits BamA function (11). The interaction of MRL-494 with the surface-exposed region of BamA, or the altered membrane properties caused by drug addition, may prevent BamA function by disrupting essential conformational changes that promote OMP assembly. Further studies to characterize this conformational change and whether MRL-494 affects BamA directly or indirectly are ongoing.
Activation of the BAM complex involves communication between BamA and BamD, resulting in complementary conformational states that allow OMP assembly to proceed (32, 39, 40). Previous studies have isolated activating mutations in both BamA and BamD that suppress assembly-defective conditions and bypass the requirement for conformational coordination between these 2 essential proteins (38, 39, 41). For instance, a bamAE373K mutation, which disrupts the essential communication between BamA and BamD (6), can be suppressed by activating mutations in BamA or BamD (40, 41).
In this work, we isolated bamAE470K as a mutant that is resistant to the BAM complex inhibitor MRL-494. Intriguingly, we found that bamAE470K acts as an intragenic suppressor of bamAE373K allowing viability of the haploid double mutant. The suppression of bamAE373K by bamAE470K strongly suggests that the bamAE470K allele allows bypass of conformational coordination within the BAM complex. Moreover, the stability of BamAE470K barrel domain is similar to the thermolability observed in other BamA mutant proteins with altered activity (38, 39). The altered conformation of BamAE470K seems to bias the protein toward a more assembly-competent state. This may allow OMP assembly to continue even in the presence of MRL-494. We are currently investigating this intriguing question.
The prevalence of antibiotic resistance necessitates the identification of new antibiotics that circumvent traditional mechanisms of resistance. In this work, we demonstrate that the newly identified small molecule MRL-494 inhibits a fundamental process by modulating BamA function. Furthermore, the ability of MRL-494 to act from outside the cell overcomes the intrinsic resistance of gram-negative bacteria and the activity of MDR efflux pumps, making it impervious to many common mechanisms of resistance.
Materials and Methods
Bacterial Strains.
The strains, plasmids, and oligonucleotides used in this study are listed in SI Appendix, Table S3. All strains were constructed using standard microbiological and molecular biology techniques, as previously described (52). All strains were grown at 37 °C in Lysogeny broth (LB) media, unless otherwise noted, which was supplemented with 50 mg/L kanamycin, 20 mg/L chloramphenicol, 0.2% arabinose, and 0.05% fucose when necessary. Deletion alleles originated from the Keio collection and Blattner collection (53, 54). FRT-flanked resistance cassettes were removed by use of the Flp recombinase system, as previously described (55).
Plasmid Construction.
Site-directed mutagenesis was performed on the indicated plasmids using the Q5 site-directed mutagenesis protocol (New England Biolabs [NEB]). Mach-1 cells (Thermo Fisher Scientific) were used for cloning purposes. bamAE470K and bamA+ under the control of the native bamA promoter was integrated at the Tn7 attachment site using pGRG25Modular::bamA, as previously described (56, 57).
Random mutagenesis of bamA was performed by a combination error-prone PCR of designed domains and seamless Golden Gate Assembly into the wild-type plasmid backbone. Error-prone PCR was performed using the GeneMorph II Random Mutagenesis Kit (Agilent). As mutagenesis rate is promotional to the gene size, the mutagenesis was performed for periplasmic Potra (P1 to P5) and barrel domains separately to maintain an overall low frequency of mutations (1 to 5) per gene. Briefly, the periplasmic and barrel domains were amplified using AK-379/AK-381 and AK-382/AK-383 primer pairs, respectively, from the pBAD18::bamA plasmid following manufacturer’ instructions for low-frequency mutagenesis. At least 10 independent reactions were performed. AK-382/AK-380 and AK-384/AK-381 primers were used to amplify the plasmid backbone together with the remaining bamA gene from the pZS21::bamA plasmid using high-fidelity Q5 polymerase (NEB). All PCR products were digested with DpnI to remove template plasmids, and the Golden Gate assembly was performed using BsaI (NEB) and T4 DNA ligase enzymes (NEB) following the manufacturer’ protocol. Assemblies were transformed into E. coli Mach-1 and plated on selective plates, giving rise to ∼10,000 colonies. The assembly fidelity and the mutagenesis frequency were confirmed by sequencing of 20 random colonies. The rest of the colonies were pooled together, and the isolated plasmid library was used for the downstream screening.
Minimum Inhibitory Concentration (MIC) Measurement.
Two-fold serial dilutions of MRL-494 were made in DMSO to encompass final treatment at 4-fold higher than the MIC (64 μg/mL) to 512-fold lower than the MIC (0.03125 μg/mL). The dilutions were made 50-fold higher than the desired treatment concentration to account for dilution upon treatment. Overnight cultures were diluted 1:50 and then further diluted 1:1,000 in fresh media. A 98-μL volume of the diluted cells was inoculated into a 96-well plate, and 2 μL of the serial MRL-494 dilutions were added. The plate was incubated overnight at 37 °C, and the optical density at 600 nm (OD600) was read at 16- and 20-h time points on a BioTek Synergy H1 plate reader. MICs for OMP biogenesis mutants were performed in Müeller Hinton II media.
Selection for MRL-494 Resistant Mutants.
JCM320 (the BamA-depletion strain) was transformed with bamA mutagenesis library plasmid pools and grown in the absence of arabinose to deplete chromosomal wild-type BamA. Cells transformed with a pZS21 plasmid containing wild-type bamA served as a control. These bamA mutants were screened against MRL-494 using the MIC assays described above with dilutions from 4-fold higher than the MIC to 32-fold lower than the MIC. From these screens, we harvested cells growing above MIC and identified the plasmid copy of bamA by sequencing.
Efficiency of Plating Assay.
Overnight cultures were normalized by OD600 and serially diluted into LB. Cells were spotted onto the indicated media and grown overnight at 37 °C.
Growth Curves and σE Reporter Assay.
Growth curves and reporter assays were performed as previously described (24) with minor modifications. Briefly, overnight cultures of the indicated strains were diluted 1:100 into LB media containing chloramphenicol for plasmid maintenance where appropriate. Cells were grown shaking in a 96-well plate for 1.5 h at 37 °C in at BioTek Synergy H1 plate reader. Then, DMSO or MRL-494 with and without batimastat (1.56 µM, 0.25× MIC) were added at a 1:50 dilution for a final volume of 100 µL/well. Cells were then returned to the plate reader for the indicated time. For growth curves without compound treatment, cells were grown for 12 h without interruption shaking in a 24-well plate with a 2-mL volume. For all assays, growth was determined based on OD600 measurements. For σE reporter assay, strains carrying the pACYC184-PmicA::GFP reporter plasmid (24) were grown in a clear-bottomed, black 96-well plate and treated as above. Fluorescence (excitation, 481 nm/emission, 507 nm) was measured and normalized to the OD600. For compound treatment experiments, fold values to untreated samples were then calculated. In some instances, growth curves, σE reporter assay, and immunoblots were performed on the same samples.
Sample Preparation for Immunoblot Analysis.
Samples were prepared for immunoblot analysis as previously described with minor modifications (24). For samples following compound treatment, samples were prepared as for growth curves and incubated 1.5 h after compound treatment. Samples were resuspended at an equivalent of 1 mL at an OD600 of 1 in 25 µL of Bugbuster (Millipore) with benzonase (1:100, Sigma-Aldrich) and incubated at room temperature 10 min. Samples were combined with 25 µL of Laemelli sample buffer (Bio-Rad) supplemented with 4% β-mercaptoethanol. For total OMP levels, samples were boiled for 10 min. For analysis of LamB folding, samples were prepared as above but were not boiled. For analysis of OMP abundance without MRL-494 treatment, cells were harvested from stationary phase and resuspended in protein sample buffer at an equivalent of 1 mL at an OD600 of 1 in 50 µL of sample buffer.
Immunoblot Analysis.
Immunoblot analysis was performed as previously described with minor modifications (28). Briefly, samples were loaded on 10% SDS/polyacrylamide gel electrophoresis (PAGE) gels and run at 120 V. Proteins were transferred to nitrocellulose membranes, and membranes were blocked in 5% nonfat dried milk for 2 h at 4 °C. Membranes were incubated in primary antibody in tris(hydroxymethyl)aminomethane (Tris)-buffered saline Tween 20 overnight at 4 °C (αBamA 1:20,000; αLptD 1:10,000; αDegP 1:30,000; αGroEL 1:10,000 [Sigma-Aldrich]; αOmpF [OmpC] 1:30,000; αLamB [OmpA, MBP] 1:5,000). Membranes were washed and incubated with donkey α-rabbit–HRP (Sigma-Aldrich) secondary antibody (1:20,000; 1 h at room temperature or 2 h at 4 °C). Membranes were washed and signal was detected using chemical luminescence (Millipore).
Heat Modifiability Assay.
Midlog cultures (OD600, ∼0.8 to 1.0) were normalized by OD600 and lysed in a mixture of Bugbuster (Millipore), protease inhibitor mixture (1:100; Sigma-Aldrich), benzonase (1:100; Sigma-Aldrich), and 1 M MgCl2 (1:100). Samples were divided in two, with one set incubated at room temperature for 10 min and the other boiled at 100 °C for 10 min. The samples were diluted 1:2 with Laemelli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol (Sigma-Aldrich). The samples were electrophoresed at 90 V at 4 °C and immunoblot analysis was carried out as described above.
TO-PRO-3 Staining and Flow Cytometric Analysis.
Midlog samples (OD600 ∼ 0.6) grown in Müeller Hinton II (MHII) media at 37 °C were diluted to ∼1 × 106 cells in 1 mL of filtered MHII media. Cells were treated with DMSO, nisin (to a final concentration of 5 μM), or MRL-494 (final concentration of 4 μg/mL, 0.25× MIC) for 5 min at room temperature. TO-PRO-3 iodide (Thermo Fisher Scientific) was added to a final concentration of 500 nM and incubated at room temperature shielded from light for 5 to 10 min. The samples were processed on a BD Biosciences LSRII flow cytometer and acquired using the BD FACSDiva 8.0.2 software. At least, 125,000 events were analyzed. The TO-PRO-3 iodide dye was excited at 640 nm, and emission was detected via a 670/30 bandpass filter. Populations were gated on forward-scatter area vs. side-scatter area to exclude debris, forward-scatter width vs. forward-scatter height to exclude aggregates, and finally on forward-scatter area vs. TO-PRO-3 signal to assess permeability. Quantification of flow cytometric analysis (SI Appendix, Table S2) represents the average of 2 biological replicates and flow cytometric plots (Fig. 4 D and E) are representative. Data were analyzed using FlowJo 10.5.3 software (FlowJo LLC).
Cellular Thermal Shift Assay (CETSA) Sample Preparation.
A starter culture of LB was inoculated with wild-type or bamAE470K mutant E. coli. This was grown overnight at 37 °C with shaking (180 rpm). The subsequent day, a 1:50 dilution of the starter culture was made into 200 mL of LB. Samples were grown for 2.5 h at 37 °C with shaking at 180 rpm. Cultures were subsequently transferred to 50-mL conical tubes and pelleted by centrifugation at 3,000 × g for 20 min at 4 °C. Samples were divided, treated with 0, 25, 50, or 100 µM MRL-494 in DMSO (final DMSO concentration 1% [vol/vol]), and incubated at 37 °C for 30 min with aeration; 80-µL samples were aliquoted into PCR tubes and heated to the indicated temperature for 3 min in a Bioer Genetouch thermocycler, followed by a 3-min incubation at 25 °C; 40 µL of 1.2% (vol/vol) IGEPAL CA-630 in phosphate-buffered saline were added to the samples and the samples were freeze-thawed 4 times alternating liquid nitrogen and 25 °C incubation in the thermocycler. Samples were centrifuged at 20,000 × g for 20 min at 4 °C and 40 µL of the supernatant were collected.
Samples were combined with Tris-glycine SDS loading buffer containing dithiothreitol and were analyzed by PAGE on 10% Tris glycine gels with Novex Tris-Glycine Native Running Buffer. Electrophoresis was performed at 20 mV for 5 h at 4 °C. Proteins were transferred to nitrocellulose membranes and membranes were blocked with LICOR blocking buffer for 30 min. Membranes were probed using α-BamA antibody and goat α-rabbit–HRP secondary antibody. Signal was detected using the SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and a Bio-Rad Chemidoc Imaging System.
CETSA Sample Preparation for Mass Spectrometry Analysis.
CETSA samples were prepared as described above; 60 μL of supernatant was collected and combined with 100 μL of 50 mM triethylammonium bicarbonate buffer, pH 8.0, and 16.8 μL of Proteasemax 1% wt/vol. Tris(2-carboxyethyl)phosphine hydrochloride and iodoacetamide were added to a final concentration of 10 and 15 mM, respectively. Samples were incubated at 55 °C for 30 min. After incubation, CaCl2 was added to a final concentration of 1 mM. Samples were digested with 2 μg of trypsin/LysC overnight at room temperature.
Following digestion, trifluoroacetic acid (TFA) was added to a concentration of 1% vol/vol and incubated for 15 min. The samples were pelleted at 2,000 × g, and the supernatant was collected. An Assaymap Bravo system with C18 desalting cartridges was used to desalt the samples. Peptides were eluted using 35% acetonitrile and 65% water (0.1% TFA). Samples were dried by speedvac, and the peptides were resuspended in 100 μL of 50 mM TEAB. Peptides were labeled with tandem mass tag (TMT) 10-plex reagents. Peptides from each treatment were mixed at every 2 adjacent temperatures. The peptide mixtures were then dried, resuspended in water containing 0.1% TFA, and purified using Pierce Detergent Removal Spin columns. The purified peptides were desalted again. The dried, purified peptides were resuspended in 20 μL of 95% water, 5% acetonitrile, and 0.1% formic acid.
Mass Spectrometry Analysis.
Samples were injected onto an Orbitrap Fusion Lumos (50 cm × 75 μm C18 column) and run on a 4-h linear gradient from 5% acetonitrile to 40% acetonitrile. TMT quantification was performed by MS3, as previously described (58). Spectra were searched against the E. coli K12 database downloaded from UniProt using Maxquant version 1.6.3.4 (tryptic peptides without missed cleavages). Methionine oxidation was excluded from quantification and the data were filtered to remove contaminant peptides. Using Perseus 1.6.0.7, the data were log2-transformed, and missing values were inputed. Proteins were normalized by the average TMT reporter tag per channel.
Supplementary Material
Acknowledgments
We thank members of the T.J.S. laboratory for productive discussion. This work was funded by a grant from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ (to T.J.S.), and by National Institute of General Medical Science Grant GM118024 (to T.J.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Christina DeCoste and Katherine Rittenbach at the Princeton University Flow Cytometry Resource Facility for their guidance and help with the flow cytometry analysis and Zemer Gitai (Princeton University) for sharing strains.
Footnotes
Competing interest statement: X.H., F.P.R.-R., A.G.S., J.C.M., C.J.B., S.B., Q.S., H.W., M.F.H., R.E.P., A.K.O., T.A.B., D.M.R., and S.S.W. are employed by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and may be shareholders in Merck & Co., Inc., Kenilworth, NJ, USA. H.S. and T.R. are employed by Prokaryotics, Inc., and may be shareholders in Prokaryotics, Inc., Union, NJ, USA. T.J.S. received research funds under a grant from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912345116/-/DCSupplemental.
References
- 1.World Health Organization , Antimicrobial Resistance: Global Report on Surveillance (World Health Organization, Geneva, Switzerland, 2014), pp. xxii–7. [Google Scholar]
- 2.Gajdács M., The concept of an ideal antibiotic: Implications for drug design. Molecules 24, 892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payne J. W., Gilvarg C., Size restriction on peptide utilization in Escherichia coli. J. Biol. Chem. 243, 6291–6299 (1968). [PubMed] [Google Scholar]
- 5.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munita J. M., Arias C. A., Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco P., et al. , Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 4, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konovalova A., Kahne D. E., Silhavy T. J., Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vetterli S. U., et al. , Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 4, eaau2634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivas N., et al. , Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Storek K. M., et al. , Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl. Acad. Sci. U.S.A. 115, 3692–3697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steenhuis M., et al. , Inhibition of autotransporter biogenesis by small molecules. Mol. Microbiol. 112, 81–98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodali S., et al. , Determination of selectivity and efficacy of fatty acid synthesis inhibitors. J. Biol. Chem. 280, 1669–1677 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Howe J. A., et al. , Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–677 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Hinchliffe P., Symmons M. F., Hughes C., Koronakis V., Structure and operation of bacterial tripartite pumps. Annu. Rev. Microbiol. 67, 221–242 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Misra R., Peterson A., Ferenci T., Silhavy T. J., A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J. Biol. Chem. 266, 13592–13597 (1991). [PubMed] [Google Scholar]
- 17.Lipinska B., Sharma S., Georgopoulos C., Sequence analysis and regulation of the htrA gene of Escherichia coli: A sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16, 10053–10067 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danese P. N., Snyder W. B., Cosma C. L., Davis L. J., Silhavy T. J., The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9, 387–398 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Grabowicz M., Silhavy T. J., Envelope stress responses: An interconnected safety net. Trends Biochem. Sci. 42, 232–242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein G., et al. , Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 291, 22999–23019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima S., Guo M. S., Chaba R., Gross C. A., Sauer R. T., Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340, 837–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mecsas J., Rouviere P. E., Erickson J. W., Donohue T. J., Gross C. A., The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T., OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113, 61–71 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Konovalova A., et al. , Inhibitor of intramembrane protease RseP blocks the σE response causing lethal accumulation of unfolded outer membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E6614–E6621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driessen A. J., Nouwen N., Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Sklar J. G., Wu T., Kahne D., Silhavy T. J., Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sklar J. G., et al. , Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 6400–6405 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T., et al. , Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Malinverni J. C., et al. , YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Ruiz N., Falcone B., Kahne D., Silhavy T. J., Chemical conditionality: A genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Ureta A. R., Endres R. G., Wingreen N. S., Silhavy T. J., Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 189, 446–454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabowicz M., Koren D., Silhavy T. J., The CpxQ sRNA negatively regulates skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. MBio 7, e00312-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell A. M., Silhavy T. J., Envelope stress responses: Balancing damage repair and toxicity. Nat. Rev. Microbiol. 17, 417–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez Molina D., et al. , Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Mateus A., et al. , Thermal proteome profiling in bacteria: Probing protein state in vivo. Mol. Syst. Biol. 14, e8242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan C. S. H., et al. , Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science 359, 1170–1177 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Noinaj N., Kuszak A. J., Buchanan S. K., Heat modifiability of outer membrane proteins from gram-negative bacteria. Methods Mol. Biol. 1329, 51–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tellez R. Jr, Misra R., Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J. Bacteriol. 194, 317–324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., et al. , Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc. Natl. Acad. Sci. U.S.A. 115, 2359–2364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe A. L., Ricci D., Adetunji M., Silhavy T. J., Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J. Bacteriol. 199, e00373-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricci D. P., Hagan C. L., Kahne D., Silhavy T. J., Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc. Natl. Acad. Sci. U.S.A. 109, 3487–3491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oppedijk S. F., Martin N. I., Breukink E., Hit ’em where it hurts: The growing and structurally diverse family of peptides that target lipid-II. Biochim. Biophys. Acta 1858, 947–957 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Prince A., et al. , Lipid-ii independent antimicrobial mechanism of nisin depends on its crowding and degree of oligomerization. Sci. Rep. 6, 37908 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortimer F. C., Mason D. J., Gant V. A., Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescent probes. Antimicrob. Agents Chemother. 44, 676–681 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan A., Vu K. D., Riedl B., Lacroix M., Optimization of the antimicrobial activity of nisin, Na-EDTA and pH against gram-negative and gram-positive bacteria. Lebensm. Wiss. Technol. 61, 124–129 (2015). [Google Scholar]
- 46.Sutterlin H. A., et al. , Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misra R., Benson S. A., Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 170, 3611–3617 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelberg H., Artman M., Studies on streptomycin-dependent bacteria: Effect of streptomycin on protein synthesis by streptomycin-sensitive, streptomycin-resistant and streptomycin-dependent, mutants of Escherichia coli. Biochim. Biophys. Acta 80, 256–268 (1964). [DOI] [PubMed] [Google Scholar]
- 49.Viceps D. I., Brownstein B. L., Streptomycin dependence in Escherichia coli: Effects of antibiotic deprivation on ribosomes. Antimicrob. Agents Chemother. 7, 271–280 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rigel N. W., Ricci D. P., Silhavy T. J., Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 5151–5156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storek K. M., et al. , The Escherichia coli β-barrel assembly machinery is sensitized to perturbations under high membrane fluidity. J. Bacteriol. 201, e00517-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silhavy T. J., Berman M. L., Enquist L. W., Experiments with Gene Fusions (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1984). [Google Scholar]
- 53.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang Y., et al. , Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186, 4921–4930 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenzie G. J., Craig N. L., Fast, easy and efficient: Site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 6, 39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hart E. M., Gupta M., Wühr M., Silhavy T. J., The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the bam complex by the lipoprotein RcsF. MBio 10, e00662-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAlister G. C., et al. , MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Y., et al. , Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.