Significance
Immune receptor NLRC4 mediates caspase-1 activation upon stimulation. Mutations of NLRC4 and other cytoplasmic immune receptors cause familial cold autoinflammatory syndrome (FCAS). Individuals carrying the NLRC4-H443P mutation show exacerbated inflammation upon exposure to a cold environment due to caspase-1 hyperactivation. The mechanism of cold-induced hyperactivation of caspase-1 by none of the FCAS-causing mutants is known. We identified heat shock cognate protein 70 (HSC70) as an interacting partner of NLRC4-H443P that negatively regulates caspase-1 activation. Exposure to subnormal temperature reduces interaction of H443P with HSC70, causing caspase-1 hyperactivation. We provide a molecular mechanism for exacerbation of inflammation induced by cold temperature in individuals carrying the NLRC4-H443P mutation, which might have broader implications for temperature regulation of FCAS-causing mutations of other receptors.
Keywords: NLRC4, HSC70, caspase-1, cold hypersensitivity, inflammasome
Abstract
NLRC4 [nucleotide-binding domain and leucine-rich repeat (NLR) family, caspase recruitment domain (CARD) containing 4] is an innate immune receptor, which, upon detection of certain pathogens or internal distress signals, initiates caspase-1–mediated interleukin-1β maturation and an inflammatory response. A gain-of-function mutation, H443P in NLRC4, causes familial cold autoinflammatory syndrome (FCAS) characterized by cold-induced hyperactivation of caspase-1, enhanced interleukin-1β maturation, and inflammation. Although the H443P mutant shows constitutive activity, the mechanism involved in hyperactivation of caspase-1 by NLRC4-H443P upon exposure of cells to lower temperature is not known. Here, we show that heat shock cognate protein 70 (HSC70) complexes with NLRC4 and negatively regulates caspase-1 activation by NLRC4-H443P in human cells. Compared with NLRC4, the structurally altered NLRC4-H443P shows enhanced interaction with HSC70. Nucleotide binding- and leucine-rich repeat domains of NLRC4, but not its CARD, can engage in complex formation with HSC70. Knockdown of HSC70 enhances apoptosis-associated speck-like protein containing a CARD (ASC)-speck formation and caspase-1 activation by NLRC4-H443P. Exposure to subnormal temperature results in reduced interaction of NLRC4-H443P with HSC70, and an increase in its ability to form ASC specks and activate caspase-1. Unlike the NLRC4-H443P mutant, another constitutively active mutant (NLRC4-V341A) associated with autoinflammatory diseases, but not FCAS, showed neither enhanced interaction with HSC70 nor an increase in inflammasome formation upon exposure to subnormal temperature. Our results identify HSC70 as a negative regulator of caspase-1 activation by the temperature-sensitive NLRC4-H443P mutant. We also show that low-temperature–induced hyperactivation of caspase-1 by NLRC4-H443P is due to loss of inhibition by HSC70.
Cytoplasmic and membrane-bound receptors mediate an innate immune response upon sensing invading pathogens or intracellular danger signals (1–3). NLRC4 [nucleotide-binding domain and leucine-rich repeat (NLR) family, caspase recruitment domain (CARD) containing 4] is a member of the protein family of cytoplasmic immune receptors. It is expressed in immune cells such as monocytes, macrophages, and neutrophils. In addition, NLRC4 functions are known in nonhematopoietic cells such as lung and intestinal epithelial cells and brain cells (4–7). NLRC4 comprises an N-terminal CARD, a central nucleotide-binding and oligomerization domain (NBD), a winged helix domain (WHD), 2 helical domains (HD1 and HD2), and a C-terminal leucine-rich repeat (LRR) domain (8–12). Detection of bacterial proteins (flagellin, rod, and needle) by NAIPs leads to formation of the NLRC4 inflammasome, a multimolecular complex that activates caspase-1. There are 7 NAIPs in mice, which recognize different bacterial proteins; however, in human cells, a single NAIP is expressed, which recognizes type 3 secretion system needle and inner rod proteins (13–16). NLRC4 can engage and activate caspase-1 either directly through its CARD or through an adapter protein, apoptosis-associated speck-like protein containing a CARD (ASC) (11). Activated caspase-1 proteolytically cleaves pro–interleukin-1β (pro–IL-1β) and pro–IL-18 into mature cytokines that mediate an inflammatory response downstream of NLRC4 (11, 12, 14, 17–20). NLRC4 is maintained in an adenosine 5′-diphosphate (ADP)-bound autoinhibited state through intermolecular interactions between the NBD and WHD of NLRC4 and ADP, and the LRR domain folds over to inhibit its oligomerization and activation (8).
Mutations in NLRC4 (21–27) and some other cytoplasmic immune receptors like NLRP1 (11, 28–30), NLRP3 (31–34), and NLRP12 (35–37) cause autoinflammatory syndromes, which occur in the absence of any infection or autoimmunity. Certain dominant mutations in NLRP3, NLRP12, and NLRC4 cause familial cold autoinflammatory syndrome (FCAS), a mild form of autoinflammatory disorder characterized by arthralgia, intermittent fever, and skin rashes upon exposure of the individual to subnormal temperatures (21–27, 31–37). One of the mutants of NLRC4, H443P, causes FCAS in heterozygous individuals (21). NLRC4-H443P undergoes auto-oligomerization, leading to constitutive activation of caspase-1 and maturation of cytokine IL-1β. A hyperinflammatory response is seen in transgenic mice expressing NLRC4-H443P upon exposure to cold water at 4 °C (21). This inflammation in NLRC4-H443P transgenic mice is due to caspase-1–mediated IL-1β maturation. Overexpression of NLRC4-H443P in HEK293T cells causes hyperactivation of caspase-1 when exposed to a temperature of 32 °C (21). However, the mechanism by which exposure to lower temperature leads to hyperactivation of caspase-1 by the H443P mutant of NLRC4, or any FCAS-causing mutant of other NLR proteins, is not known.
Heat shock proteins (HSPs) are a family of molecular chaperones involved in protein homeostasis in a cell and are broadly conserved across all vertebrates (38). HSPs are involved in physiological processes like folding of newly synthesized polypeptides, refolding or degradation of misfolded proteins, protein sorting, transport and secretion, autophagy, apoptosis, stress response during hypothermia/hyperthermia, and an inflammatory immune response (39–41). HSPs have been divided into 2 broad categories based on their size and mode of function. Larger HSPs (i.e., HSP70, heat shock cognate protein 70 [HSC70], GRP78, HSP90) are 70- to 90-kDa proteins and require adenosine triphosphatase (ATPase) activity for most of their functions, while smaller HSPs (i.e., HSP27, HSP40) mostly act as cochaperones to larger HSPs and do not possess ATPase activity. HSC70, encoded by the HSPA8 gene in humans, is a constitutively and ubiquitously expressed member of the HSP family and constitutes 1 to 2% of total cellular proteins. Unlike HSP70, HSC70 is not induced in response to higher temperature. HSC70 possesses an N-terminal ATPase domain, a substrate-binding domain (SBD), and a C-terminal lid domain (42). It is involved in promoting protein folding and in processing of misfolded proteins (39, 41).
The chaperone function of HSC70 helps in proper folding of newly synthesized proteins, and this function involves transient interactions with short hydrophobic (or hydrophobic-basic) sequences in the partially folded/unfolded proteins. These interactions of HSC70 increase with increasing temperature in the physiological range of 30 to 37 °C. HSC70 undergoes temperature-dependent reversible conformational change beginning at about 30 °C, which increases its interaction with peptides and unfolded proteins, as well as its chaperoning activity (38, 43). Thus, HSC70 exists in active and inactive states, and the proportion of the active pool is determined by temperature-dependent conformational change. These properties of HSC70 indicate that it is a potential candidate to modulate temperature-dependent functional properties of client proteins in the physiological temperature range of 30 to 37 °C.
In this study, we attempted to understand how caspase-1 hyperactivation occurs downstream of the FCAS-causing mutant, NLRC4-H443P. We have identified HSC70 and HSP70 as interacting partners of NLRC4 and show that HSC70 negatively regulates the inflammasome function of NLRC4-H443P. Upon exposure to subnormal temperatures, NLRC4-H443P interaction with cellular HSC70 decreases, resulting in increased inflammasome formation and caspase-1 activation. Our results suggest that HSC70 plays an important role in modulating temperature-sensitive properties of NLRC4-H443P.
Results
HSC70 Forms a Complex with NLRC4 and Shows Enhanced Interaction with the H443P Mutant.
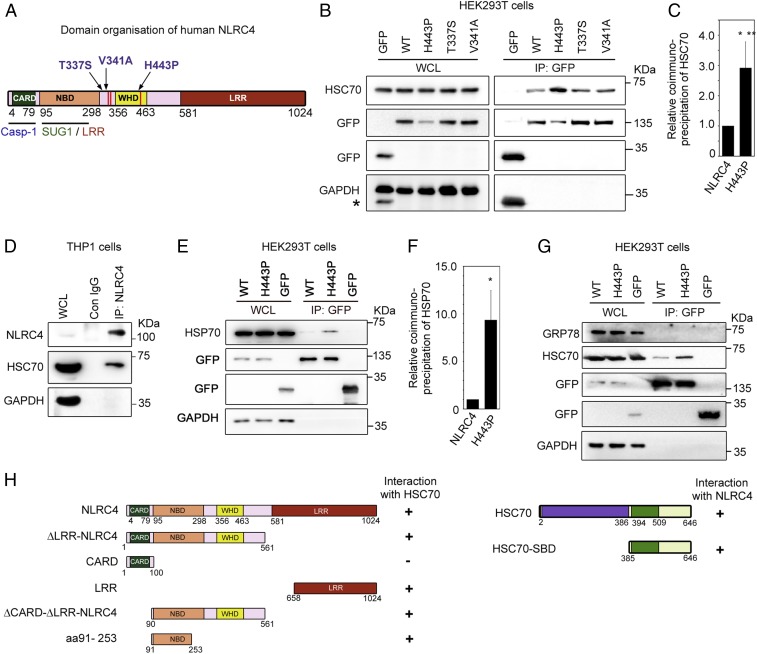
Gain-of-function mutations in NLRC4 cause autoinflammatory disorders in humans (27). Fig. 1A shows a schematic indicating the positions of various disease-associated mutations in the NLRC4 protein. A missense mutation, H443P in NLRC4, causes cold hypersensitivity in heterozygous individuals (21). While studying differential interaction of SUG1 with wild-type (WT)-NLRC4 and NLRC4-H443P by immunoprecipitation (IP) (44), we observed a prominent 70-kDa cellular polypeptide in NLRC4 immunoprecipitates from cells transiently expressing GFP-fusion proteins of WT-NLRC4 or NLRC4-H443P (SI Appendix, Fig. S1A). The prominence of this protein suggested that it could be one of the HSPs of 70 kDa, which play an important role in maintaining cellular homeostasis in response to abnormal temperatures. Western blot analysis of immunoprecipitates showed that cellular HSC70 forms a complex with NLRC4 (Fig. 1B and SI Appendix, Fig. S1B). Higher levels of HSC70 were seen repeatedly in immunoprecipitates of the H443P mutant compared with that of WT-NLRC4, suggesting that HSC70 complexes with the H443P mutant with higher affinity than with WT-NLRC4 (Fig. 1 B and C). We examined the interaction of HSC70 with 2 other disease-associated mutants of NLRC4, T337S and V341A, which also cause constitutive caspase-1 activation. The T337S and V341A mutants of NLRC4 do not cause FCAS but are involved in causing other autoinflammatory syndromes (22, 23). Interestingly, compared with WT-NLRC4, HSC70 showed stronger interaction only with the temperature-sensitive mutant NLRC4-H443P, but not with NLRC4-T337S or NLRC4-V341A (Fig. 1B). Interaction of endogenous NLRC4 with HSC70 was seen in differentiated THP1 cells by IP, indicating that complex formation was not due to forced expression of the proteins (Fig. 1D).
Fig. 1.
HSC70 and HSP70 are present in cellular complexes formed by NLRC4. (A) Schematic showing the domain organization of NLRC4. Sites of disease-associated mutations in NLRC4 are indicated. Interaction sites for caspase-1 (Casp-1) and the SUG1/LRR domain are also indicated (12, 55). (B) GFP-tagged NLRC4 and its mutants were transiently expressed in HEK293T cells, and lysates were subjected to IP using GFP antibody followed by Western blotting. WCL, whole-cell lysate. The asterisk indicates residual signal from bands of GFP. (C) Bar diagram shows quantitation of relative abundance of HSC70 in IP of the H443P mutant compared with WT-NLRC4 normalized with GFP signal from 8 independent experiments (n = 8). ***P < 0.0005. (D) Differentiated THP1 cells were lysed and subjected to IP using NLRC4 antibody or normal immunoglobulin G (IgG) as a control (Con) and analyzed by Western blotting. (E) HSP70 forms a complex with NLRC4 and shows enhanced interaction with NLRC4-H443P. Lysates of cells transfected with the indicated constructs were subjected to IP and Western blotting. (F) Bar diagram shows relative abundance of HSP70 in IP of WT-NLRC4 and NLRC4-H443P normalized with GFP signal (n = 3). *P < 0.05. (G) GRP78 is not present in the cellular complexes formed by NLRC4. (H) Identification of domains in NLRC4 and HSC70 required for complex formation. The schematic shows various deletion constructs of NLRC4 and HSC70. The plus symbol on the right-hand side indicates positivity for interaction, and the minus symbol indicates negativity for interaction.
In addition to HSC70, HSP70 was detected in the immunoprecipitates obtained from lysates of HEK293T cells transiently expressing WT-NLRC4 or NLRC4-H443P. Higher levels of HSP70 were seen in complexes with NLRC4-H443P compared with that of WT-NLRC4 (Fig. 1 E and F). GRP78, another HSP70 member, which is localized to the endoplasmic reticulum and is required for folding of newly synthesized proteins, was not detected in NLRC4 or NLRC4-H443P immunoprecipitates under similar experimental conditions (Fig. 1G). Endogenous NLRC4 did not show complex formation with GRP78 in differentiated THP1 cells (SI Appendix, Fig. S2A). HSP70 was not detectable in THP1 cells under our experimental conditions of Western blotting (SI Appendix, Fig. S2B). These results identified HSC70 and HSP70 as 2 interacting partners of NLRC4 and suggested a possible role for HSC70/HSP70 in signal transduction downstream of the temperature-sensitive mutant NLRC4-H443P.
Domain Requirements for Interaction between NLRC4 and HSC70.
To identify the domains in NLRC4 required for its interaction with HSC70, we transfected deletion constructs expressing GFP-fusion proteins of various domains of NLRC4 (Fig. 1H) in HEK293T cells and subjected the lysates to IP using GFP-antibody. HSC70 was present in the immunoprecipitates of deletion constructs expressing ΔLRR-NLRC4 (which lacks the LRR domain) as well as in ΔCARD-ΔLRR-NLRC4 (which lacks both the LRR domain and CARD) (SI Appendix, Fig. S3A). GFP-LRR and GFP-amino acids (aa) 91 to 253 interacted with endogenous HSC70, but GFP-CARD failed to coprecipitate HSC70 (SI Appendix, Fig. S3B). These results indicated that the NBD and LRR domain of NLRC4 can independently interact with HSC70, and the CARD is neither sufficient nor essential for interaction. In vitro glutathione S-transferase (GST) pulldown assays using GST-HSC70 showed that GFP-LRR and GFP-aa 91 to 253, but not GFP, can interact with HSC70 (SI Appendix, Fig. S3C). For GST pulldown assays, the binding reaction was carried out at 37 °C because no interaction of GST-HSC70 with NLRC4 or NLRC4-H443P was seen at 4 °C, a condition generally used for these assays. HSC70 is known to bind to its substrates primarily through its C-terminal SBD (aa 394 to 546) (shown schematically in Fig. 1H), which has affinity for extended hydrophobic motifs in the target protein (45, 46). GST-SBD interacted with NLRC4 and NLRC4-H443P in in vitro binding assays (SI Appendix, Fig. S3D).
Ubiquitination of NLRC4-H443P Enhances Interaction with HSC70.
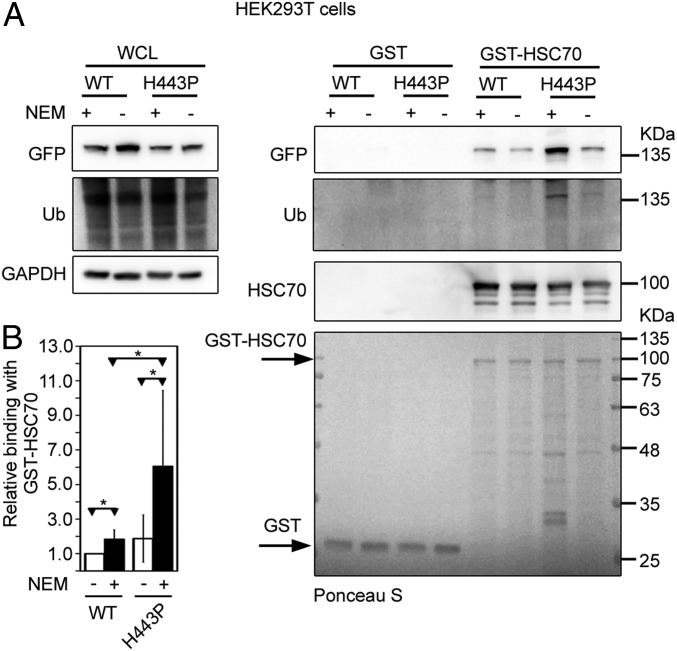
While a higher amount of HSC70 was present in cellular complexes formed by NLRC4-H443P compared with WT-NLRC4 (Fig. 1B), no difference was seen in interaction when in vitro binding assays were carried out (SI Appendix, Fig. S4). We hypothesized that the NLRC4-H443P mutant may be undergoing a posttranslational modification in cells that is lost or reduced in cell lysates. Previously, we have reported that NLRC4 undergoes ubiquitination and that a higher level of ubiquitination is seen on NLRC4-H443P (44) (SI Appendix, Fig. S1B). Therefore, we carried out cell lysis and GST pulldown assays in buffers containing N-ethylmaleimide (NEM), an inhibitor of cellular deubiquitinases. A higher amount of NLRC4, as well as NLRC4-H443P, complexed with GST-HSC70 when NEM was included in the cell lysis and binding assay buffers, compared with experiments without NEM (Fig. 2A). Compared with WT-NLRC4, NLRC4-H443P showed enhanced interaction with GST-HSC70 in the presence of NEM (Fig. 2B). Deprobing the blot, followed by reprobing with ubiquitin antibody, confirmed enhanced ubiquitination on polypeptide corresponding to NLRC4-H443P, compared with WT-NLRC4 in samples processed with NEM (Fig. 2A). These results suggest that HSC70 preferentially interacts with the ubiquitinated NLRC4-H443P mutant and enhanced binding of this mutant is likely due to higher ubiquitination of the NLRC4-H443P mutant compared with that of WT-NLRC4.
Fig. 2.
Ubiquitination (Ub) of NLRC4-H443P enhances interaction with HSC70. (A) Lysates of HEK293T expressing GFP-NLRC4 or GFP-NLRC4-H443P were prepared in buffer with or without NEM (10 mM) and incubated at 37 °C for 30 min with GST or GST-HSC70. After GST pulldown assays, bound proteins were analyzed by Western blotting with the indicated antibodies. (B) Bar diagram shows relative abundance of NLRC4 or NLRC4-H443P in pulldown samples of GST-HSC70 in the presence or absence of NEM, normalized with GFP signal in the lysates (n = 6). *P < 0.05.
HSC70 Negatively Regulates Caspase-1 Activation by NLRC4-H443P.
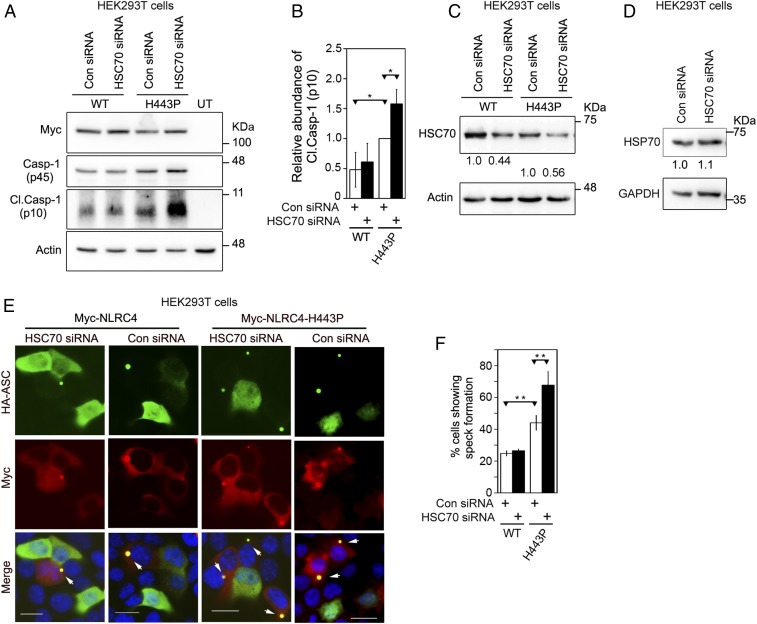
The FCAS-causing NLRC4-H443P constitutively activates caspase-1, leading to proinflammatory cytokine IL-1β maturation. To find out if HSC70 plays a role in caspase-1 activation by NLRC4-H443P, we examined the consequence of knockdown of HSC70 using short interfering RNAs (siRNAs). We observed increased caspase-1 activation by NLRC4-H443P compared with WT-NLRC4 in control siRNA-transfected samples (Fig. 3 A and B). Caspase-1 activation by NLRC4-H443P further increased upon HSC70 knockdown, suggesting that HSC70 negatively regulates caspase-1 activation downstream of NLRC4-H443P. There was only a marginal effect of HSC70 knockdown on caspase-1 activation by WT-NLRC4 (Fig. 3 A and B). We confirmed the efficacy of siRNA-mediated HSC70 knockdown by Western blot (Fig. 3C). HSP70 levels remained unaffected in these samples, indicating the specificity of siRNA for HSC70 (Fig. 3D).
Fig. 3.
HSC70 negatively regulates caspase-1 activation downstream of NLRC4-H443P. (A) HEK293T cells were transfected with control (Con) siRNA or siRNA targeted against HSC70 along with caspase-1 (Casp-1) and Myc-tagged NLRC4 or NLRC4-H443P. Whole-cell lysates were analyzed by Western blotting for the presence of cleaved caspase-1 (Cl. Casp-1 [p10]). (B) Bar diagram shows quantitation of relative abundance of Cl. Casp-1 in cells expressing NLRC4 or NLRC4-H443P normalized with caspase-1 p45 signal (n = 3). *P < 0.05. (C) Western blot shows efficacy of HSC70 knockdown by siRNA. (D) HSC70 siRNA does not affect cellular HSP70, indicating target specificity. (E) Representative images show the effect of siRNA-mediated knockdown of HSC70 on speck formation by the WT-NLRC4 or NLRC4-H443P mutant. Cells coexpressing HA-ASC and Myc-NLRC4 or Myc-NLRC4-H443P were scored for the presence or absence of specks. White arrows indicate ASC specks formed inside cells. (Scale bars, 20 μm.) (F) Quantitation of the effect of HSC70 knockdown on ASC-mediated speck formation by NLRC4 or NLRC4-H443P (n = 4). **P < 0.005.
NLRC4 coexpression enables ASC to form distinct specks in cells, an indicator of inflammasome assembly, which occurs due to oligomerization of ASC with NLRC4 (23, 47). NLRC4-H443P coexpression resulted in significantly higher number of cells showing ASC specks compared with those expressing NLRC4 (Fig. 3 E and F). ASC-mediated speck formation by NLRC4-H443P increased significantly upon HSC70 knockdown (Fig. 3 E and F). Importantly, reduction in levels of HSC70 did not affect speck formation by WT-NLRC4. Knockdown of HSP70 by siRNA did not enhance caspase-1 activation by NLRC4-H443P, although some effect on inflammasome formation by NLRC4-H443P was seen (SI Appendix, Fig. S5 A–C). Knockdown of HSP70 by siRNA was confirmed by Western blot analysis (SI Appendix, Fig. S5 D and E).
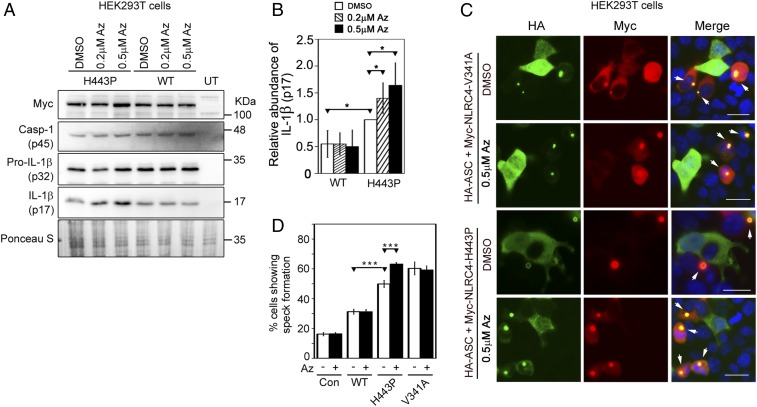
Apoptozole, an Inhibitor of HSC70/HSP70, Enhances Inflammasome Function of NLRC4-H443P.
HSC70 and HSP70 function through adenosine 5′-triphosphate–dependent cycles of substrate binding and release. Apoptozole (Az) is a chemical inhibitor of HSC70/HSP70 ATPase activity (48), and has been used as an inhibitor of HSC70 function (48–51). We tested the effect of Az on NLRC4-H443P–mediated caspase-1 activation by quantitating IL-1β maturation. HEK293T cells transiently expressing caspase-1 and IL-1β along with WT-NLRC4 or NLRC4-H443P were treated with dimethyl sulfoxide and 0.2 μM or 0.5 μM Az. Expression of NLRC4-H443P increased IL-1β maturation, compared with WT-NLRC4 (Fig. 4 A and B). Treatment with Az did not have a significant effect on WT-NLRC4–induced IL-1β maturation, but increased levels of mature IL-1β in the lysates of NLRC4-H443P–transfected cells (Fig. 4 A and B). The level of cleaved caspase-1 increased upon treatment with Az in H443P mutant-expressing cells but not in WT-NLRC4–expressing cells (SI Appendix, Fig. S6A). Az treatment also resulted in a significant increase in the frequency of ASC specks in NLRC4-H443P–expressing cells, while there was no effect on speck formation by WT-NLRC4 or NLRC4-V341A (Fig. 4 C and D and SI Appendix, Fig. S6 B and C). These results provide further support to our hypothesis that HSC70 regulates inflammasome formation and caspase-1 activation by NLRC4-H443P.
Fig. 4.
Effect of Az treatment on caspase-1 activation and IL-1β maturation downstream of NLRC4-H443P. (A) HEK293T cells were transfected with Myc-tagged NLRC4 or NLRC4-H443P along with caspase-1 (Casp-1) and pro–IL-1β. Whole-cell lysates were analyzed by Western blotting for the presence of mature IL-1β (p17). Az was added to the culture medium in concentrations as indicated. Dimethyl sulfoxide (DMSO) was used as a solvent control. (B) Bar diagram shows quantitation of relative levels of mature IL-1β normalized with pro–IL-1β signal in whole-cell lysates upon Az treatment (n = 3). *P < 0.05. (C) Representative immunofluorescence images showing the effect of Az treatment (0.5 μM for 6 h) on speck formation by Myc-NLRC4-V341A and Myc-NLRC4-H443P coexpressed with HA-ASC. White arrows indicate specks. (Scale bars, 20 μm.) (D) Quantitation of the effect of Az treatment (0.5 μM for 6 h, indicated as a plus symbol) on ASC-speck formation by NLRC4 and its mutants (n = 4). Minus symbol indicates treatment with DMSO used as a solvent control. ***P < 0.0005. HA-ASC plasmid was used alone without any NLRC4 plasmid as an additional control (Con).
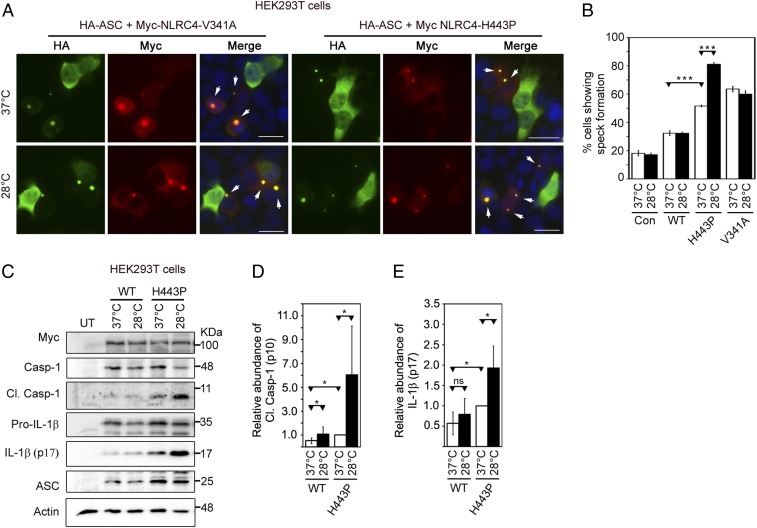
Effect of Subnormal Temperature on Inflammasome Formation and Caspase-1 Activation by NLRC4-H443P.
Caspase-1–mediated IL-1β maturation in NLRC4-H443P mutant–expressing cells increases upon exposure to subnormal temperature (21). This was validated by examining the effect of a subnormal temperature of 28 °C on ASC-mediated speck formation by the H443P mutant, the V341A mutant, and WT-NLRC4. We observed that NLRC4-H443P– and NLRC4-V341A–expressing cells showed a significantly higher percentage of cells with specks compared with WT-NLRC4–expressing cells. Upon exposure to 28 °C, NLRC4-H443P showed a further increase in speck formation, while there was no significant change in speck formation by WT-NLRC4 or NLRC4-V341A (Fig. 5 A and B and SI Appendix, Fig. S7). These results suggest that the NLRC4-H443P mutant shows enhanced inflammasome formation upon exposure to lower temperature, whereas the V341A mutant does not. As expected, we observed that NLRC4-H443P showed increased caspase-1 activation and IL-1β maturation, and NLRC4-H443P–mediated caspase-1 activation and IL-1β maturation further increased upon exposure to subnormal temperature (Fig. 5 C–E).
Fig. 5.
NLRC4-H443P–expressing cells exposed to subnormal temperature show an increase in inflammasome formation and caspase-1 activation. (A) Representative immunofluorescence images show the effect of exposure to subnormal temperature on ASC-speck formation in cells expressing HA-ASC along with NLRC4-V341A or NLRC4-H443P. One set of cells was shifted to 28 °C for 6 h after 12 h of transfection, while the other set remained at 37 °C. White arrows indicate specks. DAPI was used to stain the nucleus. (Scale bars, 20 μm.) (B) Quantitation of ASC-speck formation in response to subnormal temperature (n = 6). ***P < 0.0005. Con, control. (C) Western blot analysis of lysates of HEK293T cells expressing caspase-1 (Casp-1) and HA-ASC along with Myc-NLRC4, Myc-NLRC4-H443P, or Myc-NLRC4-V341A. One set of cells was exposed to 28 °C for 6 h after 18 h of expression, while the other set remained at 37 °C. Lysates were analyzed for the presence of cleaved (Cl.) Casp-1 (p10). UT, untransfected. The bar diagrams show quantitation of the effect of exposure to subnormal temperature on the relative abundance of Cl. Casp-1 normalized with caspase-1 p45 signal (D) and IL-1β normalized with pro–IL-1β signal (E) (n = 6). *P < 0.05. ns, not significant.
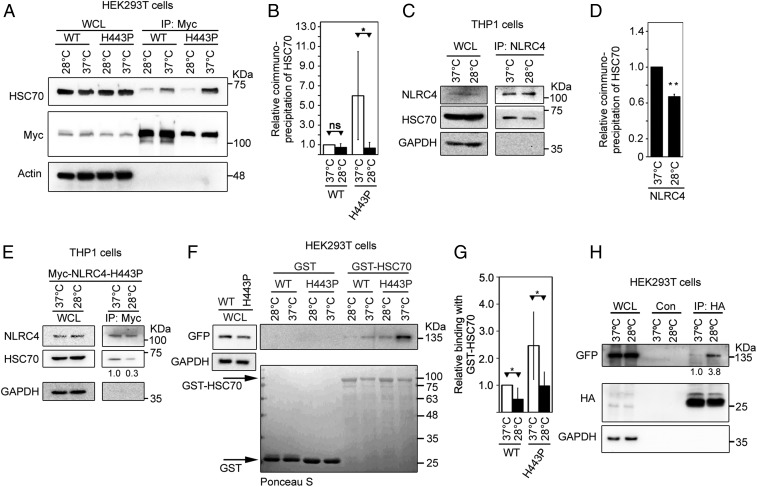
Effect of Subnormal Temperature on Interaction of NLRC4-H443P with HSC70 and ASC.
We examined the effect of exposure to subnormal temperature on the interaction of HSC70 with NLRC4 or NLRC4-H443P. HEK293T cells expressing Myc-tagged NLRC4 or NLRC4-H443P were maintained at 37 °C or exposed to 28 °C before lysis. Lysates were subjected to IP with Myc antibody and Western blotting. Significantly reduced levels of HSC70 complexed with NLRC4-H443P in cells exposed to subnormal temperature compared with that in cells at 37 °C (Fig. 6 A and B and SI Appendix, Fig. S8). The interaction of WT-NLRC4 with HSC70 showed a small reduction upon exposure to lower temperature (Fig. 6 A and B). The interaction of endogenous NLRC4 with HSC70 in differentiated THP1 cells was significantly reduced upon exposure to 28 °C (Fig. 6 C and D). Transiently expressed Myc-NLRC4-H443P in THP1 cells showed reduced interaction with endogenous HSC70 upon exposure to a subnormal temperature of 28 °C (Fig. 6E). In vitro binding assays also showed that GST-HSC70 binds weakly with NLRC4-H443P at 28 °C compared with that at 37 °C (Fig. 6 F and G). A significant effect of subnormal temperature on binding of GST-HSC70 with WT-NLRC4 was also seen (Fig. 6G). We did not see any significant difference in the extent of ubiquitination on the H443P mutant in response to temperature shift, although some decrease was observed (SI Appendix, Fig. S9). Western blot analysis of immunoprecipitates from lysates of cells expressing ASC along with NLRC4-H443P showed that NLRC4-H443P was more abundant in cellular complexes formed by ASC in cells exposed to a subnormal temperature of 28 °C compared with those grown at 37 °C (Fig. 6H). These results suggest that enhanced inflammasome formation and caspase-1 activation by the NLRC4-H443P mutant at lower temperature is likely to be due to reduced interaction with its negative regulator, HSC70.
Fig. 6.
Exposure to subnormal temperature alters the interaction of NLRC4-H443P with HSC70 and ASC. (A) HEK293T cells expressing Myc-NLRC4 or Myc-NLRC4-H443P were grown at 37 °C for 24 h or exposed to 28 °C for 6 h after 18 h of expression. Lysates were subjected to IP using agarose-conjugated Myc antibody, and immunoprecipitates were analyzed by Western blotting. WCL, whole-cell lysate. (B) Quantitation of the relative abundance of endogenous HSC70 in the immunoprecipitates normalized with Myc signal is shown (n = 4). *P < 0.05. ns, not significant. (C) THP1 cells were differentiated with 10 nM PMA for 66 h. One set was subjected to 28 °C for 6 h, while the other set was retained at 37 °C. WCLs were subjected to IP using NLRC4 antibody, and complexes were analyzed by Western blotting. (D) Quantitation of the relative binding of HSC70 with NLRC4 at 37 °C and 28 °C is shown after normalization with NLRC4 signal in the immunoprecipitates (n = 3). **P < 0.005. (E) THP1 cells were electroporated with Myc-NLRC4-H443P; one set was shifted to 28 °C for 6 h after 64 h of expression, while the other set remained at 37 °C. Cell lysates were subjected to IP using agarose-conjugated Myc antibody, and immunoprecipitates were analyzed by Western blotting. (F) Lysates of HEK293T cells expressing GFP-NLRC4 or GFP-NLRC4-H443P were incubated with GST or GST-HSC70 at the indicated temperatures, and bound proteins were analyzed by Western blotting. (G) Quantitation of relative abundance of NLRC4 or NLRC4-H443P in pulldown samples of GST-HSC70 shown after normalization with the corresponding GST-HSC70 signal (n = 4). *P < 0.05. (H) HEK293T cells were transfected with GFP-NLRC4-H443P along with HA-ASC, and exposed to 28 °C for 4 h after 12 h of transfection. Lysates were prepared 16 h posttransfection and subjected to IP using agarose-conjugated HA antibody. Western blot analysis shows an increase in binding of ASC with NLRC4-H443P upon exposure to subnormal temperature. Con, control.
Discussion
NLRC4 is generally activated by pathogen-associated molecular patterns (10), but certain point mutations result in its constitutive activation, inflammasome formation, and caspase-1 activation, leading to autoinflammatory diseases (27). Through this study, we show that HSC70 and HSP70 chaperone proteins form a complex with NLRC4. HSC70 keeps the constitutively active NLRC4-H443P mutant in check to suppress inflammasome formation and caspase-1 activation. Exposure of cells to subnormal temperature results in reduced interaction of HSC70 with the H443P mutant, which is accompanied by hyperactivation of caspase-1. Paradoxically, compared with WT-NLRC4, the constitutively active H443P mutant shows enhanced interaction with HSC70. The mutated histidine residue is at a location required for binding with the ADP molecule (8), which is crucial in maintaining the inactive configuration of NLRC4. We hypothesize that the H443P mutation, in addition to exposing oligomerization sites by inducing conformational changes in NLRC4, enables stronger interaction of HSC70 and HSP70 with the mutant protein. This mutation possibly induces very specific conformational changes in NLRC4 that are not seen in 2 other mutants, V341A and T337S, which do not show enhanced interaction with HSC70. It is possible that NLRC4-H443P is recognized as a misfolded protein by HSC70.
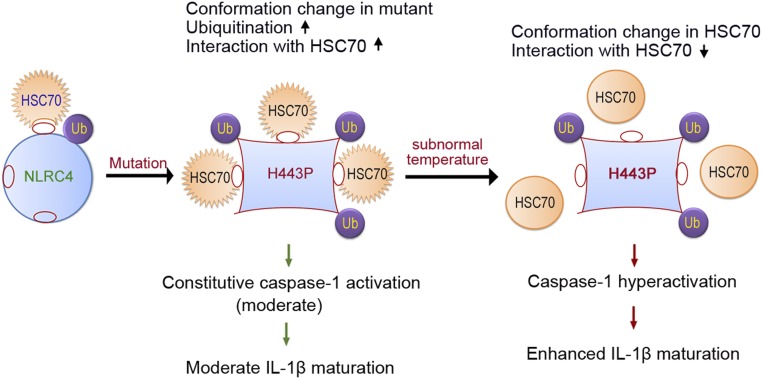
Binding of HSC70 with the H443P mutant may be preventing efficient oligomerization with ASC, as shown by the increase in speck formation in cells with reduced HSC70 levels. Lower temperature may be inducing a conformational change in HSC70 resulting in reduced interaction with the H443P mutant, leading to its hyperactivation. This is shown schematically in Fig. 7. Knockdown of HSC70 may have nonspecific effects on cells; thus, the effects observed on inflammasome formation and/or caspase-1 activation by NLRC4-H443P may not be due to loss of direct NLRC4 regulation. However, the proposed role of HSC70 in mediating low-temperature–induced hyperactivation of caspase-1 by the H443P mutant is supported by the following observations. HSC70 shows enhanced interaction with NLRC4-H443P, and this interaction is considerably reduced at lower temperature, which is accompanied by hyperactivation of inflammasome formation and caspase-1 activation. Unlike the NLRC4-H443P mutant, another constitutively active mutant (NLRC4-V341A) associated with autoinflammatory diseases, but not FCAS, showed neither enhanced interaction with HSC70 nor an increase in inflammasome formation upon exposure to subnormal temperature. Treatment with Az, an inhibitor of ATPase activity of HSC70/HSP70, increased inflammasome formation by NLRC4-H443P but not by NLRC4-V341A.
Fig. 7.
Proposed model for regulation of NLRC4-H443P by HSC70 upon exposure to lower temperature. NLRC4 is present in an inactive closed configuration with low levels of ubiquitination (Ub) and weak binding to HSC70. The mutation of H443 to proline causes a conformational change in NLRC4, which enables enhanced ubiquitination and more stable interaction with HSC70. The H443P mutant shows constitutive caspase-1 activation and moderate IL-1β maturation, causing mild inflammation. Upon exposure of cells to subnormal temperature, HSC70 undergoes a conformational change that lowers its ability to interact with H443P. This allows increased ASC-speck formation by H443P, and caspase-1 hyperactivation leading to enhanced IL-1β maturation and hyperinflammation. This mechanism of the differential interaction of HSC70 with H443P in response to subnormal temperature explains the hyperinflammation seen in FCAS patients carrying this mutation.
HSC70 performs several functions in the cell, including its role as a chaperone that helps in folding of newly synthesized proteins in the cytosol. The chaperone function involves transient interaction of HSC70 with hydrophobic patches in the native and partially folded/misfolded proteins (40). Based on biochemical and biophysical properties, it has been suggested that HSC70 may function as a thermal sensor in the physiological temperature range of 30 to 37 °C to adjust the chaperoning activity according to the requirement of the cell (38, 43). HSC70 undergoes an exothermic reversible transition beginning at about 30 °C, and it is more sensitive to protease chymotrypsin at 40 °C than at 20 °C, indicating a conformational change (43). These properties of HSC70 support our inference that the reduction of its interaction with the H443P mutant seen at lower temperature (28 °C) is likely to be due to a conformational change in HSC70.
Previously, we have shown that in comparison to WT-NLRC4, the H443P mutant undergoes a higher level of ubiquitination and interaction with ubiquitinated proteins (44). In comparison to WT-NLRC4, higher levels of the H443P mutant are seen in cellular complexes formed by HSC70. However, HSC70 did not show enhanced interaction with the H443P mutant in GST pulldown assays unless a deubiquitinase inhibitor was added in the lysis buffer. We suggest that HSC70 interacts with ubiquitinated NLRC4, and a higher level of ubiquitination of the H443P mutant contributes to enhanced interaction with HSC70. It is likely that ubiquitination induces a conformational change in the H443P mutant that results in exposure of HSC70-binding sites. An alternate possibility is that ubiquitination stabilizes an open conformation of the H443P mutant with exposed HSC70-binding sites. However, we cannot rule out the possibility of interaction of ubiquitin with HSC70, although the homologous protein HSP70 does not interact with ubiquitin (52).
The H443P mutation in NLRC4 results in enhanced interaction with HSC70 as well as HSP70, and these interactions are drastically reduced at lower temperature. Therefore, it is possible that in addition to HSC70, HSP70 may be involved in mediating the low-temperature–induced increase in inflammasome formation and caspase-1 hyperactivation. However, in comparison to HSC70, the constitutive expression of HSP70 in cells is very low. Furthermore, HSP70 generally mediates its effects upon induction by heat shock or other signals. Our results suggest that the contribution of HSP70 to cold-induced hyperactivation of inflammasome formation, if any, is likely to be small.
We conclude that HSC70 interacts with NLRC4 and the H443P mutation alters its conformation to favor a more stable complex with HSC70. Caspase-1 hyperactivation due to the H443P mutation is kept in check by HSC70, as its loss or reduction in interaction induced by exposure to cold temperature results in enhanced caspase-1 activation. Our results therefore provide an understanding of the molecular mechanism for the exacerbation of inflammation induced by cold temperature in individuals carrying the H443P mutation, causative of FCAS.
Methods
Cell Culture, Transfections, and Treatments.
Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS) was used to maintain HEK293T cells procured from the American Type Culture Collection. RPMI 1640 was supplemented with heat-inactivated 10% FBS and used to maintain the human macrophage cell line THP1. Cells were grown at 37 °C in a water-jacketed incubator, which maintained 5% CO2 and controlled humidity. For transient expression of proteins, plasmids were transfected using Lipofectamine 2000 or Lipofectamine 3000 (Invitrogen) as per the manufacturer’s protocol. In general, HEK293T cells showed about 80% transfection efficiency with control GFP, 40 to 50% with GFP-NLRC4, and 20 to 25% with Myc-NLRC4. Electroporation of THP1 cells was carried out in buffer 2M as described using a Lonza 4D-Nucleofector X Unit (53).
For siRNA-mediated knockdown of HSC70 or HSP70, HEK293T cells were seeded in a 24-well tissue culture plate or on coverslips and transfected with 100 pmol of control siRNA, HSC70, or HSP70-siRNA for 24 h. A second transfection was carried out after 24 h, with 100 pmol of control or HSC70-siRNA along with desired plasmids. Lysates were prepared 24 h after the second transfection for Western blot analysis. For immunofluorescence, cells on coverslips were fixed using 4% formaldehyde 18 h after the second transfection.
For inhibiting ATPase activity of HSC70, Az was added to the culture medium in desired concentrations for 12 h (unless indicated otherwise) after 12 h of transfection with desired plasmids.
For exposure to subnormal temperature, one set of cells expressing the desired plasmids was maintained at 37 °C, while another set was shifted to 28 °C for 6 h (unless indicated otherwise) after 18 h of transfection.
For indirect immunofluorescence and microscopy, immunostaining of cells fixed with formaldehyde was carried out as described previously (54). Immunostained cells were observed and images were captured with an automated AxioImager Z.2 (Zeiss) fluorescence microscope using Axiovision software under a 40×/0.75-numerical aperture dry objective.
Quantitation of Inflammasome Formation by Immunofluorescence.
HEK293T cells plated on coverslips were transfected with hemagglutinin (HA)-tagged ASC along with Myc-tagged NLRC4 and its desired mutants. Exposure to subnormal temperature or treatment with Az was for 6 h after 12 h of transfection. Cells were fixed 18 h posttransfection and immunostained with HA and Myc antibodies. In experiments involving siRNA-mediated HSC70 knockdown, transfections were carried out as described in the previous section, and cells were fixed 42 h posttransfection. Immunostained cells were observed under a fluorescence microscope and scored for the presence of specks in coexpressing cells. Data are presented as mean ± SD of the percentage of cells forming specks from at least 3 independent experiments done in duplicate. At least 300 expressing cells from each coverslip were examined.
Co-IP and Western Blotting.
For IP of Myc/GFP/HA-tagged proteins expressed in HEK293T cells, agarose-conjugated Myc/GFP/HA antibody was used as described earlier (44). For IP of endogenous NLRC4, THP1 cells were differentiated into macrophage-like cells by treatment with 10 nM phorbol 12-myristate 13-acetate (PMA) for 72 h. Differentiated cells were washed with ice-cold phosphate-buffered saline (PBS), followed by lysis in buffer containing 20 mM Tris⋅HCl (pH 7.5), 0.5% Nonidet P-40, 150 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1% bovine serum albumin, protease inhibitor mixture, and 10 mM NEM (a deubiquitinase inhibitor). Cells were scraped and collected in a precooled microfuge tube and allowed to undergo lysis at 4 °C for 30 min on a Roto-Torque. Lysate was centrifuged (10,000 × g for 10 min at 4 °C) to remove cellular debris. Two micrograms of NLRC4 antibody or control immunoglobulin G was incubated with agarose-conjugated protein A/G for 2 h at 4 °C before being added to the cell lysates and incubated for 8 h at 4 °C on a Roto-Torque. The bound proteins were washed 3 times with buffer containing 150 mM NaCl, 20 mM Tris⋅HCl (pH 7.5), 1 mM PMSF, 0.5 mM EDTA, protease inhibitor mixture, and 10 mM NEM. Immunoprecipitates were lysed in sodium dodecyl sulfate containing Laemmli sample buffer. The samples were then subjected to Western blot analysis as described by Shivakrupa et al. (54).
Quantification of Western Blots.
Bands were quantitated using ImageJ software (NIH). For quantification of IP blots, intensities of co-IP bands were normalized with the intensities of IP bands. For estimating caspase-1 activation, the amount of cleaved caspase-1 (p10) was normalized with full-length caspase-1 (p45) signal, and mature IL-1β was normalized with pro–IL-1β signal.
GST Pulldown Assays.
Escherichia coli BL-21 DE-3 cells transformed with plasmid expressing the desired GST-tagged protein were induced for protein expression by 1 mM isopropyl β-d-thiogalactoside for 15 h at 18 °C. Bacterial cells were lysed by sonication in chilled PBS containing 1 mM PMSF and protease inhibitors. Triton X-100 (1%) was added for solubilization and left for 30 min at 4 °C. Lysate was centrifuged for 10 min at 4 °C and 10,000 × g to remove the insoluble fraction. Glutathione-agarose beads (50% slurry) were added to the supernatant and incubated on a Roto-Torque at 4 °C for 1 h to pull down GST-fusion protein. The beads were pelleted (3,000 × g at 4 °C for 1 min), washed 3 times with chilled PBS (containing 1 mM PMSF and 0.1% Triton X-100), and incubated with lysates of HEK293T cells (prepared in buffer containing 20 mM Tris⋅HCl [pH 7.5], 0.5% Nonidet P-40, 150 mM NaCl, 0.5 mM EDTA, 1 mM PMSF, 10 mM NEM [optional], and protease inhibitor mixture) transiently expressing the desired proteins for 20 to 30 min at 37 °C, 28 °C, or 4 °C on a Roto-Torque. Unbound proteins were removed by washing 3 times at 4 °C, and bound proteins were boiled in Laemmli sample buffer and analyzed by Western blotting.
Statistical Analysis.
Quantitative data are represented as mean ± SD values. A two-tailed Student’s t test was used to calculate the significance of differences between 2 means, and a one-tailed t test was used to determine the significance of the relative difference of a test sample compared with control as 1.
Supplementary Material
Acknowledgments
We thank Dr. Barbara Kazmierczak (Yale University) for providing pCruzMycB-NLRC4 and pCruzMycB-NLRC4-V341A expression vectors. This work was carried out with support from the Department of Biotechnology, Government of India (Grant BT/PR14917/BRB/10/888/2010 to G.S. and V.R.). G.S. thanks the Department of Science and Technology, Government of India, for a J. C. Bose National Fellowship (Grant SR/S2/JCB-41/2010). A.K.R. thanks the Council for Scientific and Industrial Research, India, for a fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905261116/-/DCSupplemental.
References
- 1.Schroder K., Tschopp J., The inflammasomes. Cell 140, 821–832 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R., TLR-mediated innate immune recognition. Semin. Immunol. 19, 1–2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasselon T., Detmers P. A., Toll receptors: A central element in innate immune responses. Infect. Immun. 70, 1033–1041 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolle L., et al. , Redundant and cooperative interactions between TLR5 and NLRC4 in protective lung mucosal immunity against Pseudomonas aeruginosa. J. Innate Immun. 7, 177–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai S., Batra S., Wakamatsu N., Pacher P., Jeyaseelan S., NLRC4 inflammasome-mediated production of IL-1β modulates mucosal immunity in the lung against gram-negative bacterial infection. J. Immunol. 188, 5623–5635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellin M. E., et al. , Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16, 237–248 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Freeman L., et al. , NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J. Exp. Med. 214, 1351–1370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z., et al. , Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Vance R. E., The NAIP/NLRC4 inflammasomes. Curr. Opin. Immunol. 32, 84–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan J. A., Canna S. W., The NLRC4 inflammasome. Immunol. Rev. 281, 115–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma D., Kanneganti T. D., The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 213, 617–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poyet J. L., et al. , Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309–28313 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Yang J., Zhao Y., Shi J., Shao F., Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 110, 14408–14413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y., et al. , The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Kofoed E. M., Vance R. E., Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandjean T., et al. , The human NAIP-NLRC4-inflammasome senses the Pseudomonas aeruginosa T3SS inner-rod protein. Int. Immunol. 29, 377–384 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Sutterwala F. S., et al. , Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao E. A., et al. , Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107, 3076–3080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenthorey J. L., Kofoed E. M., Daugherty M. D., Malik H. S., Vance R. E., Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54, 17–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinon F., Burns K., Tschopp J., The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Kitamura A., Sasaki Y., Abe T., Kano H., Yasutomo K., An inherited mutation in NLRC4 causes autoinflammation in human and mice. J. Exp. Med. 211, 2385–2396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canna S. W., et al. , An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46, 1140–1146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romberg N., et al. , Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46, 1135–1139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang J., et al. , Novel NLRC4 mutation causes a syndrome of perinatal autoinflammation with hemophagocytic lymphohistiocytosis, hepatosplenomegaly, fetal thrombotic vasculopathy, and congenital anemia and ascites. Pediatr. Dev. Pathol. 20, 498–505 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Volker-Touw C. M., et al. , Erythematous nodes, urticarial rash and arthralgias in a large pedigree with NLRC4-related autoinflammatory disease, expansion of the phenotype. Br. J. Dermatol. 176, 244–248 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki Y., et al. , Identification of a high-frequency somatic NLRC4 mutation as a cause of autoinflammation by pluripotent cell-based phenotype dissection. Arthritis Rheumatol. 69, 447–459 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Romberg N., Vogel T. P., Canna S. W., NLRC4 inflammasomopathies. Curr. Opin. Allergy Clin. Immunol. 17, 398–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavarría-Smith J., Vance R. E., The NLRP1 inflammasomes. Immunol. Rev. 265, 22–34 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Jin Y., et al. , NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 356, 1216–1225 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Zurawek M., et al. , A coding variant in NLRP1 is associated with autoimmune Addison’s disease. Hum. Immunol. 71, 530–534 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D., Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301–305 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Torre-Minguela C., Mesa Del Castillo P., Pelegrín P., The NLRP3 and pyrin inflammasomes: Implications in the pathophysiology of autoinflammatory diseases. Front. Immunol. 8, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masters S. L., Simon A., Aksentijevich I., Kastner D. L., Horror autoinflammaticus: The molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 27, 621–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aksentijevich I., Kastner D. L., Genetics of monogenic autoinflammatory diseases: Past successes, future challenges. Nat. Rev. Rheumatol. 7, 469–478 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Jéru I., et al. , Mutations in NALP12 cause hereditary periodic fever syndromes. Proc. Natl. Acad. Sci. U.S.A. 105, 1614–1619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jéru I., et al. , Identification and functional consequences of a recurrent NLRP12 missense mutation in periodic fever syndromes. Arthritis Rheum. 63, 1459–1464 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Borghini S., et al. , Clinical presentation and pathogenesis of cold-induced autoinflammatory disease in a family with recurrence of an NLRP12 mutation. Arthritis Rheum. 63, 830–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuiderweg E. R., Hightower L. E., Gestwicki J. E., The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones 22, 173–189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu T., Daniels C. K., Cao S., Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol. Ther. 136, 354–374 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Young J. C., Agashe V. R., Siegers K., Hartl F. U., Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Sevin M., Girodon F., Garrido C., de Thonel A., HSP90 and HSP70: Implication in inflammation processes and therapeutic approaches for myeloproliferative neoplasms. Mediators Inflamm. 2015, 970242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stricher F., Macri C., Ruff M., Muller S., HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 9, 1937–1954 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Leung S. M., et al. , Thermal activation of the bovine Hsc70 molecular chaperone at physiological temperatures: Physical evidence of a molecular thermometer. Cell Stress Chaperones 1, 78–89 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghawan A. K., et al. , A disease-associated mutant of NLRC4 shows enhanced interaction with SUG1 leading to constitutive FADD-dependent caspase-8 activation and cell death. J. Biol. Chem. 292, 1218–1230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clerico E. M., Tilitsky J. M., Meng W., Gierasch L. M., How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young J. C., Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88, 291–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stutz A., Horvath G. L., Monks B. G., Latz E., ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040, 91–101 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Ko S. K., et al. , A small molecule inhibitor of ATPase activity of HSP70 induces apoptosis and has antitumor activities. Chem. Biol. 22, 391–403 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Evans L. E., Cheeseman M. D., Yahya N., Jones K., Investigating apoptozole as a chemical probe for HSP70 inhibition. PLoS One 10, e0140006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho H. J., et al. , Probing the effect of an inhibitor of an ATPase domain of Hsc70 on clathrin-mediated endocytosis. Mol. Biosyst. 11, 2763–2769 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Powers M. V., et al. , Targeting HSP70: The second potentially druggable heat shock protein and molecular chaperone? Cell Cycle 9, 1542–1550 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Parcellier A., et al. , HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol. Cell. Biol. 23, 5790–5802 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chicaybam L., et al. , An efficient electroporation protocol for the genetic modification of mammalian cells. Front. Bioeng. Biotechnol. 4, 99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shivakrupa R., Radha V., Sudhakar Ch., Swarup G., Physical and functional interaction between Hck tyrosine kinase and guanine nucleotide exchange factor C3G results in apoptosis, which is independent of C3G catalytic domain. J. Biol. Chem. 278, 52188–52194 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Kumar Y., Radha V., Swarup G., Interaction with Sug1 enables Ipaf ubiquitination leading to caspase 8 activation and cell death. Biochem. J. 427, 91–104 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.