Significance
Houseplant enthusiasts know the power of plant regeneration—small cuttings can regrow into whole plants. Yet normally, plant tissues and cells maintain distinct and stable identities. A biochemical modification to histones (H3K27me3) is thought to lock in cell identity by preventing access to specific regions of DNA. Here, we investigated how H3K27me3 was used to maintain the identity of stomatal guard cells. We identified changes in H3K27me3 distribution and gene expression in normal guard cells and after we induced “reprogramming” into alternative cell identities. This linked the loss of H3K27me3 at stomatal precursor genes to a return to an earlier fate, but increased H3K27me3 at a wound-induced reprogramming gene indicated how plant cells may sense and resist inappropriate loss of identity.
Keywords: reprogramming, H3K27me3, stomata, Arabidopsis
Abstract
Plant cells maintain remarkable developmental plasticity, allowing them to clonally reproduce and to repair tissues following wounding; yet plant cells normally stably maintain consistent identities. Although this capacity was recognized long ago, our mechanistic understanding of the establishment, maintenance, and erasure of cellular identities in plants remains limited. Here, we develop a cell-type–specific reprogramming system that can be probed at the genome-wide scale for alterations in gene expression and histone modifications. We show that relationships among H3K27me3, H3K4me3, and gene expression in single cell types mirror trends from complex tissue, and that H3K27me3 dynamics regulate guard cell identity. Further, upon initiation of reprogramming, guard cells induce H3K27me3-mediated repression of a regulator of wound-induced callus formation, suggesting that cells in intact tissues may have mechanisms to sense and resist inappropriate dedifferentiation. The matched ChIP-sequencing (seq) and RNA-seq datasets created for this analysis also serve as a resource enabling inquiries into the dynamic and global-scale distribution of histone modifications in single cell types in plants.
Plants retain developmental plasticity in differentiated somatic cells while simultaneously maintaining these cells’ diverse and distinct fates. Extrinsic (cell-cell signaling) and intrinsic (regulation of gene expression) mechanisms have been shown to contribute to the generation of cell fates. Both extrinsic and intrinsic controls are likely also involved in cellular reprogramming, but little is known about how these layers of control function cooperatively and independently to define the potential and behaviors of individual Arabidopsis cells. Here, using a genetic manipulation in an epidermal cell type, we present a system that enables mechanistic dissection of intrinsic mechanisms of cell identity maintenance without disrupting tissue interactions.
Gene regulation via the Polycomb repressive complex 2 (PRC2) is required for intrinsic cell fate maintenance in many systems, including Arabidopsis. PRC2, which catalyzes trimethylation of lysine 27 of the histone H3 tail (H3K27me3), was first well-characterized in Drosophila; here, regional domain identity in the embryo is highly correlated with H3K27me3 distribution patterns (1). A failure to maintain H3K27me3 at Drosophila HOX genes through cell divisions leads to inappropriate HOX gene expression and misspecification of fate (2). The H3K27me3 histone itself, rather than PRC2, was shown to be critical for transcriptional regulation that prevents homeotic defects (3), suggesting that H3K27me3 is dynamically regulated at distinct targets to establish and abolish a cellular identity. Profiles of mammalian chromatin from individual cell types also support a connection between H3K27me3 and preservation of cellular identity, with domains of H3K27me3 enrichment greatly expanded in differentiated cells relative to embryonic stem cells (4). Monitoring H3K27me3 over the course of cellular reprogramming in tissue culture has shown this modification is dynamically remodeled at a small number of loci (5, 6). One possibility is that while H3K27me3 is important for fate maintenance, few loci enforce particular identities.
Plants globally lacking PRC2 have dramatic postembryonic developmental defects in cell identity maintenance, including the formation of somatic embryos from differentiated tissue (7). If PRC2 is lost only in postembryonic vegetative tissues, plants form undifferentiated masses of tissue called callus (8) and fully differentiated root hair cells can dedifferentiate to this state (9). These phenotypes support the hypothesis that plant cell identity maintenance requires transcriptional regulation via histone modifications (reviewed in ref. 10). The profundity and heterogeneity of these reprogramming events, however, obscures which changes to H3K27me3 are a cause and which are a consequence of reprogramming.
Genome-wide H3K27me3 profiles with finer spatial and temporal resolution are therefore required to address the role of chromatin modification in plant cell identities. Pioneering H3K27me3, H3K4me3, and gene expression profiling in Arabidopsis in root hair and nonhair epidermal cells established that genomic H3K27me3 distributions could exhibit cell type-specific behavior (11), and subsequent studies on other differentiated Arabidopsis tissues and cell types (12, 13) confirmed these findings. Further work improved mechanistic insight by observing changes in H3K27me3 during developmental transitions, although these studies were done in mixed tissues (14, 15). While these studies suggest H3K27me3 is important for cell identity maintenance, no study has observed dynamic regulation of this histone modification during cellular reprogramming. Ectopic expression of single transcription factors, typically PRC2 targets, can reprogram cells in Arabidopsis (16, 17), indicating expression changes at few loci can dramatically effect cell identity.
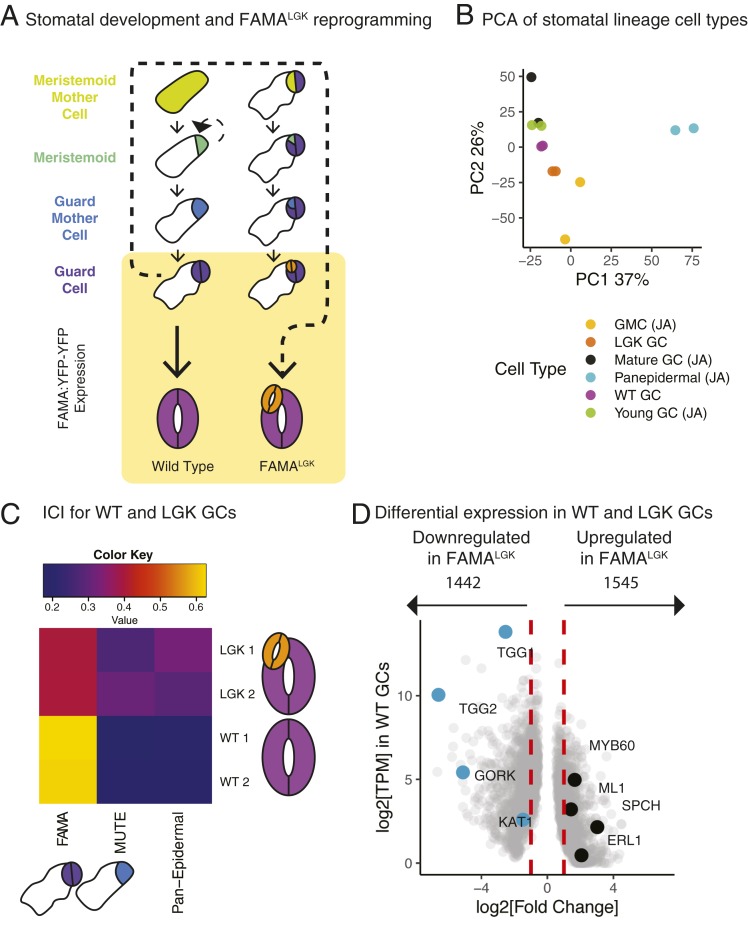
An ideal, but currently unavailable, dataset would capture the behavior of H3K27me3 in a single Arabidopsis cell type as it undergoes reprogramming to a progenitor cell type. If H3K27me3 locks in cell fates, must it be reorganized for such reprogramming to occur? If so, how and to what extent? The Arabidopsis stomatal lineage (Fig. 1A) provides a system to observe how histone modifications are reorganized over fine timescales during development of single cell types as well as during cellular reprogramming. Stomatal guard cells (GCs) can be isolated for transcriptional profiling (18) and reprogrammed back to a progenitor state in vivo (19) and are totipotent when cultured in isolation (20). GCs are the terminal products of a well-defined epidermal cell lineage. A basic helix–loop–helix transcription factor, FAMA, is necessary and sufficient for a cell to acquire and maintain GC identity (16). FAMA forms a complex with RETINOBLASTOMA RELATED (RBR), and when point mutations are introduced into FAMA to abrogate its interaction with RBR (referred to here as FAMALGK or LGK lines), GCs fail to maintain terminal identity (19). Strikingly, this leads to a reinitiation of early stomatal lineage behaviors. These behaviors include the expression of the stomatal lineage initiator SPEECHLESS (SPCH) and reinitiation of asymmetric cell divisions, followed by transition to MUTE-expressing Guard Mother Cell (GMC) identity and, ultimately, the production of new GCs in the footprint of the original GCs (Fig. 1A). RBR physically interacts with components of the PRC2 complex (21, 22), leading to the hypothesis that identity maintenance defects in LGK GCs was due to a failure to recruit PRC2 to the many FAMA targets (19). Independently, Lee et al. (23, 24) converged on a similar model of GC identity maintenance by different manipulations of FAMA and suppression of the GC reprogramming phenotype by overexpression of PRC2 components.
Fig. 1.
Single-cell type transcriptional profiles indicate that LGK GCs acquire early stomatal lineage identities. (A) Diagram of developmental trajectory of WT (Left) and reprogrammed (Right) GCs. The yellow box denotes cells marked by FAMA:YFP-YFP (young and maturing GCs) that were FACS-isolated for transcriptional and histone profiles. (B) PCA shows WT GCs cluster with previously characterized young and mature GCs, while LGK GCs cluster more closely with GMCs. Samples marked (JA) are from ref. 18. (C) ICI shows WT GCs are similar to transcriptomes previously generated from early FAMA-stage cells. LGK GCs lose early FAMA-stage identity and become more similar to early lineage cells. Columns are previously defined cell types. Rows are sorted WT and LGK GCs. Two replicates of each test cell type are shown. Color key indicates the ICI score and, therefore, the degree of similarity between known and unknown cell identities. (D) Differential expression analysis conducted with DESeq2 comparing WT and LGK GCs shows early lineage regulators (black) are up-regulated and genes associated with GC function (teal) are down-regulated.
In this work, to address intrinsic mechanisms behind cell identity, we report the generation of high-quality genome-wide maps of histone modifications in Arabidopsis GCs, along with corresponding transcriptome and histone modification profiles during in planta reprogramming of these cells. The reprogramming of GCs into early stomatal identities with scorable attributes occurs in an intact tissue, where extrinsic regulators of cell fate maintenance like niche signaling can be maintained. Furthermore, prior work establishing transcriptional signatures of stomatal lineage cell types (18) enabled us to define the extent of reprogramming and to identify potential regulators of this reprogramming. We find that genomic patterns of H3K27me3 deposition are clearly divergent between GCs and whole seedlings, revealing a strong correlation between cell identity and H3K27me3 distribution. GC reprogramming was not associated with widespread depletion of H3K27me3 at stomatal lineage regulatory genes, despite differential expression of nearly 3,000 genes. These data suggest H3K27me3 functions to establish cellular identity, but that previous models attributing GC reprogramming to global H3K27me3 depletion need to be reconsidered. We further use these datasets to show this in vivo reprogramming event may be negatively regulated by H3K27me3, via repression of a procallus gene expression program.
Results
Establishing Methods to Capture and Profile Normal GCs and Those Undergoing Reprogramming.
To capture differentiated stomatal GCs and those undergoing reprogramming, we generated a reporter (FAMAp:YFP-YFP) that in intact leaves and in fluoresence-activated cell sorting (FACS) analysis clearly distinguished WT and reprogramming GCs from autofluorescent nontarget cells (Fig. 1A and SI Appendix, Fig. S1 B and C). A FACS gating strategy and RNA extraction method described previously for the stomatal lineage (18, 25) was used to generate and analyze RNA-sequencing (seq) libraries (scheme laid out in SI Appendix, Fig. S1C). After completing general quality control analyses (SI Appendix, Fig. S2 A and B), we validated the specificity and effectiveness enabled by FAMAp:YFP-YFP expression to FACS-isolate GCs by comparing the resultant GC transcriptomes with published transcriptomes of stomatal lineage cell types (18). Principal component analysis (PCA) shows WT GCs profiled for this study cluster with previously profiled young GCs (18) and, to a lesser extent, with mature GCs, while LGK GCs cluster more closely with a precursor cell type, the GMC (Fig. 1B, [JA] indicates data from ref. 18). The identity of an unknown cell type can also be discerned by calculating an Index of Cellular Identity (ICI) where genes whose expression is most informative for individual known cell types are compiled and used as a metric to classify unknown cell types (26). When global measures of identity like ICI are applied to LGK GCs, we find that they lose FAMA-stage identity and become more similar to identities associated with the GMC stage (MUTE) and with samples containing all aerial epidermal cell types (Fig. 1C).
Having broadly defined the identities of our sorted cells, we sought a more granular understanding of the gene expression changes that occur during FAMALGK reprogramming. We identified 2,987 genes that are differentially expressed between WT and FAMALGK GCs (P < 0.01, log2 fold change threshold of 1); they are split nearly equally between increased or decreased expression (Fig. 1D and Dataset S1). Genes associated with GC function such as the ion channels KAT1 (27), and GORK (28), and the myrosinase enzyme TGG1 (29) are strongly down-regulated (Fig. 1D). Genes whose expression is normally high only in the early precursors of the stomatal lineage, including MERISTEM LAYER 1 (ATML1) and SPCH, are up-regulated in LGK cells (Fig. 1D).
Although reprogramming to intermediate fates is observed in animal systems (reviewed in ref. 30), studies of regeneration in Arabidopsis have focused on contexts in which identities are fully reset or where the reprogrammed state does not clearly correlate with an individual adult stem cell type (reviewed in ref. 31). Because LGK GCs are derived from a tissue that originates in the shoot apical meristem (SAM), we queried whether LGK GCs reprogram to a SAM identity. We calculated another ICI including microarray data from cells found in the SAM and leaf primordia (SI Appendix, Fig. S2D). Given a larger option of cell types, LGK cells are still much more transcriptionally similar to stomatal lineage cells than any other cell type, suggesting that LGK GCs reprogram but remain within the stomatal lineage of the leaf epidermis.
ChIP-Seq Data from FACS-Sorted GCs Is High Quality and Consistent with Tissue-Scale Data.
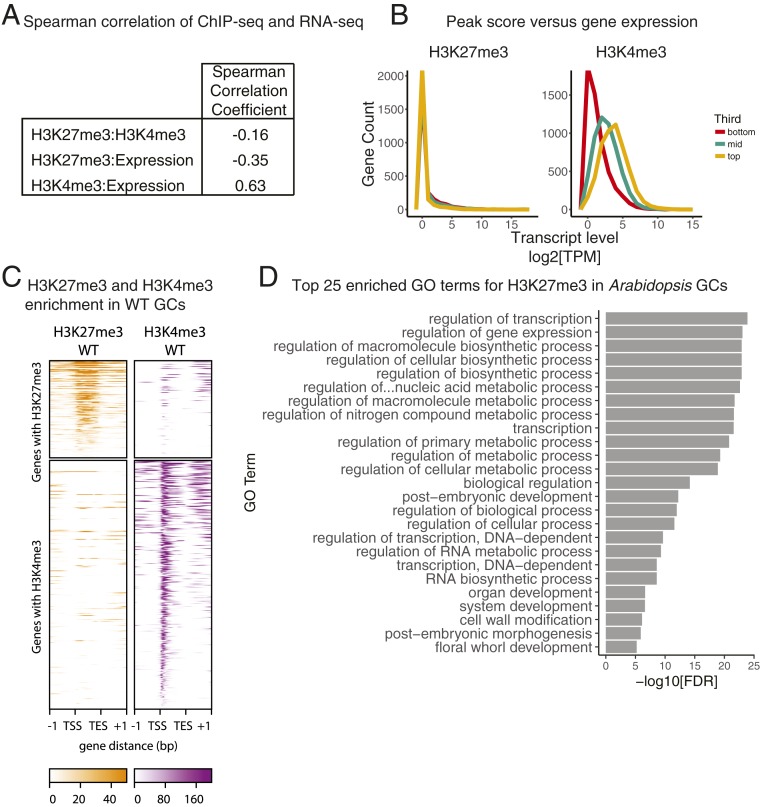
To investigate the relationship between histone modifications and cell identity, we then generated matched H3K27me3, H3K4me3, and transcriptional profiles for our pure (FAMAp:YFP-YFP expressing) cell populations. H3K4me3 was included in part because it has a positive correlation with gene expression and could serve as a control for linking ChIP and gene expression datasets. Despite the many experiments reporting H3K27me3 and H3K4me3 profiles, few are done on single cell types, and we therefore needed to optimize ChIP-seq protocols to accommodate low inputs (Materials and Methods). Using two replicates of 100,000 cells for each genotype and histone mark, our protocol enabled the generation of ChIP-seq data that met quality control standards based on metrics employed in ENCODE pipelines (SI Appendix, Fig. S3 A and B). As expected, H3K27me3 and H3K4me3 were primarily found associated with euchromatic, gene-rich regions (SI Appendix, Fig. S3D). We calculated the Spearman correlation of genome-wide tag distributions between our RNA-seq and ChIP-seq datasets and found a positive correlation between H3K4me3 and transcription and a negative correlation between H3K27me3 and transcription (Fig. 2A) as expected from studies on bulk tissue (32, 33).
Fig. 2.
High-quality H3K27me3 and H3K4me3 ChIP-seq datasets can be isolated from single cell types. (A) Spearman correlation coefficients calculated for H3K27me3, H3K4me3, and gene expression in isolated GCs. H3K27me3 is anticorrelated with gene expression, while H3K4me3 is correlated with gene expression in GCs. (B) Expression levels for genes associated with H3K27me3 and H3K4me3 peaks. Genes sets are divided into thirds based on peak score, and then the distribution of normalized expression levels (TPM) is plotted. (C) H3K27me3 and H3K4me3 enrichment within the gene body of target genes is represented as the signal P value, where the null hypothesis is that signal is present in the input control. Each row of the heatmaps represents a gene with a peak annotated to it. The top two heatmaps are composed of genes with H3K27me3 peaks. The bottom two heatmaps are composed of genes with H3K4me3 peaks. Genes are pseudolengthened to 1 kb, and 1-kb flank is shown on either side. (D) Top 25 enriched GO terms for genes with H3K27me3 peaks in WT GCs.
To assess the accuracy of our peak calling and subsequent assignment to genes, we examined transcript levels among genes carrying H3K27me3 and/or H3K4me3 peaks. Once identified, we subset genes carrying peaks into thirds based on peak score, and then generated histograms of transcript levels in these thirds (Fig. 2B). Genes in the top third of H3K27me3 peaks featured the lowest transcript levels, while the middle and bottom third show only modest increases in gene expression (Fig. 2B), a trend that mirrors findings from isolated SAMs (14). In contrast, gene expression level is well separated by H3K4me3 peak category (Fig. 2B). The top third of H3K4me3 peaks correlates with the highest gene expression levels, while the middle and bottom thirds correlate with their respective gene expression levels (Fig. 2B). This agreement between histone modification enrichments and gene expression state supports the conclusion that we have successfully generated high quality ChIP-seq profiles specifically for GCs.
Broadly, the single cell type H3K27me3 data were similar to data from bulk tissue: For example, H3K27me3 was abundant in the gene body (Fig. 2C), consistent with H3K27me3 datasets generated from whole Arabidopsis seedlings (32). We performed Singular Enrichment Analysis to identify overrepresented Gene Ontology (GO) terms for genes carrying H3K27me3. Among the top enriched (false discovery rate [FDR] < 0.05) GO terms for genes carrying H3K27me3 are regulation of transcription, postembryonic, organ, and system development (Fig. 2D and Dataset S2). The overrepresentation of developmental GO terms suggests that WT GCs, like other cell types in Arabidopsis, use H3K27me3 to repress developmental programs. This supports the hypothesis that H3K27me3 reinforces cell fates.
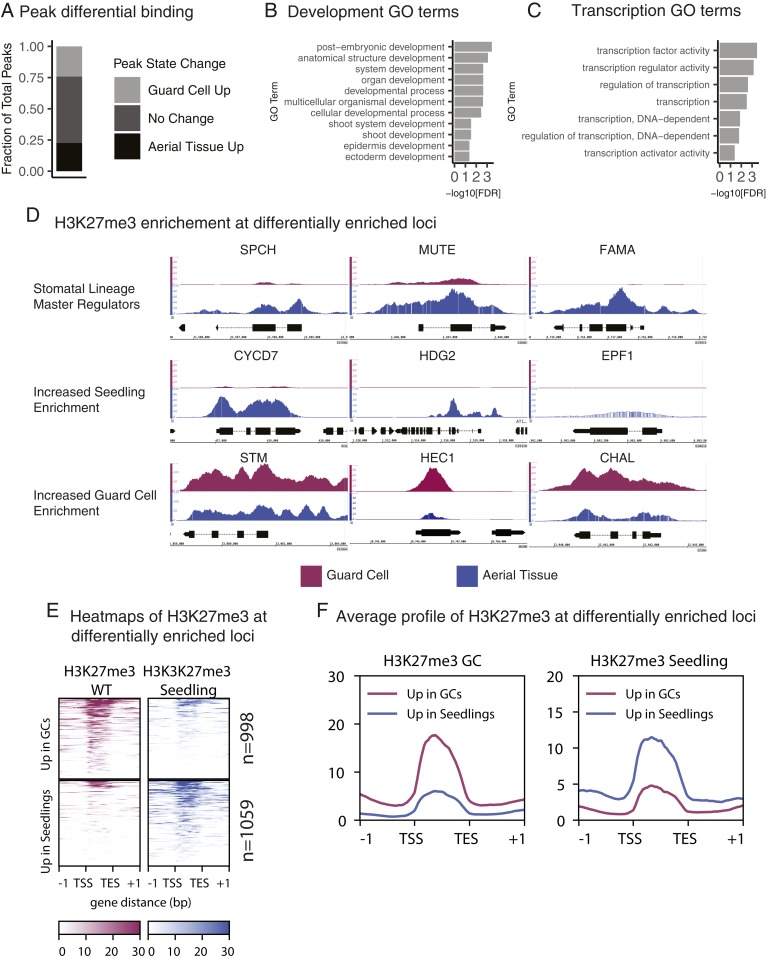
Differential Binding Analysis of H3K27me3 in WT GCs Versus Whole Aerial Tissue Reveals Tissue-Specific Enrichment Dynamics.
The ideal way to reveal tissue-specific H3K27me3 dynamics would be to compare our GC data with data from other single cell types, however, we found no published datasets whose platform or quality was suitable for this (SI Appendix, Fig. S4). We therefore compared the H3K27me3 profile from WT GCs to a high-quality published profile generated from whole Arabidopsis aerial tissue using the same H3K27me3 antibody (34). In a differential binding analysis of the H3K27me3 union peak set from WT GCs and aerial tissue (n = 8,480), one-quarter of the peaks have increased enrichment in aerial tissue, one-quarter have increased enrichment in WT GCs, and the remaining half are not called differential (Fig. 3A). Genes associated with peaks in the differential fraction are enriched for transcriptional and developmental regulators, including regulators of shoot and epidermal development (Fig. 3 B and C). H3K27me3 enrichment heat maps (Fig. 3E) and average H3K27me3 enrichment profiles (Fig. 3F) of these differentially enriched loci show a clear difference in enrichment levels across datasets.
Fig. 3.
Genome-wide distribution of H3K27me3 in GCs is unique relative to aerial tissue. (A) Differential binding analysis of WT GCs versus aerial tissue shows ∼50% of all H3K27me3 peaks across both datasets are differential. The differential set splits nearly evenly between Guard Cell Up (n = 2,052) and Aerial Tissue Up (n = 1,913). (B and C) Genes associated with the top 10 percent of the most differentially enriched H3K27me3 peaks identified in 3A are significantly enriched for GO terms related to development (B) or transcription (C). (D) H3K27me3 enrichment at representative loci. GC data are shown in purple. Aerial tissue data are shown in blue. All y axes are consistently scaled across data tracks at each gene. All loci shown are differentially enriched for H3K27me3 between WT GCs and aerial tissue. (E) Heatmap of enrichment of H3K27me3 at loci with differential enrichment in aerial tissue relative to GCs with a log2 fold change threshold of 1 (n = 2,057). Each row represents a gene with a differentially enriched H3K27me3 peak. The top two heatmaps show genes with increased H3K27me3 enrichment in GCs. The bottom two heatmaps show genes with increased H3K27me3 enrichment in aerial tissue. Enrichment in E and F is represented as the signal P value relative to the input control. (F) Average enrichment of H3K27me3 at the same loci shown in F. The plot at Left shows H3K27me3 enrichment in GCs. The plot at Right shows H3K27me3 enrichment in aerial tissue. Genes with increased enrichment in GCs are shown in purple. Genes with increased enrichment in seedlings are shown in blue.
As predicted, the master regulator transcription factor gene FAMA, normally expressed in GCs, completely lacks H3K27me3 in WT GCs (Fig. 3D). SPCH and MUTE are master regulators of the identities of GC precursor cells in which they are expressed, and those loci have reduced H3K27me3 in WT GCs relative to aerial tissue (Fig. 3D). The reduction, but not elimination of H3K27me3 at SPCH, a gene expressed only in GC precursors, may represent reestablishment of the H3K27me3 mark. This is consistent with previous observations that H3K27me3 is a “slow” mark that is reestablished over several cell divisions in human tissue culture (35). Lineage-expressed genes like CYCLIN D7;1 (CYCD7;1), HOMEODOMAIN GLABROUS 2 (HDG2), and EPIDERMAL PATTERNING FACTOR 1 (EPF1) also lack H3K27me3 repression in GCs (Fig. 3D). However, genes whose expression and activity fall outside of the stomatal lineage, such as the shoot meristem regulators SHOOTMERISTEMLESS (STM) (36), and HECATE1 (HEC1) (37), and the nonepidermally expressed peptide ligand gene CHALLAH/EPFL6 (38), have increased H3K27me3 in GCs (Fig. 3D).
Associated with the 25 H3K27me3 peaks most depleted in GCs relative to aerial tissue (Table 1) are GC-expressed genes such as FAMA and STOMATAL CARPENTER1 (SCAP1), which are required to establish correct GC morphology (16, 39) and members of the HD-ZIP IV family known to regulate stomatal fates including ATML1 and PROTODERMAL FACTOR 2 (PDF2) (40, 41), HDG2 (42), and HDG5 (43). Other genes in this list, such as GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE8 (GPAT8) and 3-KETOACYL-COA SYNTHASE 2 (KCS2), generate the epidermal-specific waxy cuticle (44, 45). This unbiased approach mirrors the cell type-specific trends we observe at the SPCH, MUTE, and FAMA loci (Fig. 3D), thereby validating that our FACS to ChIP-seq method provides high-quality data without perturbing GC identity.
Table 1.
Top 25 H3K27me3 peaks differentially depleted in GCs
| Gene ID | Gene name | FDR | Fold change |
| AT4G04890 | PDF2 | 1.65e-76 | −9.84 |
| AT3G24140 | FMA | 2.85e-72 | −9.66 |
| AT4G21750 | ATML1 | 8.14e-71 | −9.61 |
| AT5G46880 | HB-7 | 3.62e-67 | −9.44 |
| AT5G65590 | SCAP1 | 9.06e-65 | −9.36 |
| AT1G04220 | KCS2 | 1.68e-63 | −9.28 |
| AT1G75910 | EXL4 | 2.29e-63 | −9.28 |
| AT2G15840 | AT2G15840 | 3.52e-63 | −9.26 |
| AT5G62470 | MYB96 | 3.61e-62 | −9.22 |
| AT5G25830 | GATA12 | 5.16e-60 | −9.13 |
| AT4G35070 | AT4G35070 | 7.88e-58 | −9.02 |
| AT4G00400 | GPAT8 | 6.19e-57 | −8.98 |
| AT4G29020 | AT4G29020 | 1.78e-55 | −8.98 |
| AT5G53220 | AT5G53220 | 2.67e-53 | −8.84 |
| AT2G45970 | CYP86A8 | 2.67e-53 | −8.82 |
| AT4G01950 | GPAT3 | 4.67e-53 | −8.8 |
| AT1G33811 | AT1G33811 | 2.11e-52 | −8.77 |
| AT3G27400 | AT3G27400 | 2.74e-52 | −8.77 |
| AT1G75891 | AT1G75891 | 5.75e-50 | −8.66 |
| AT5G49334 | AT5G49334 | 1.56e-49 | −8.64 |
| AT1G70030 | AT1G70030 | 8.94e-48 | −8.57 |
| AT5G62470 | MYB96 | 3.15e-47 | −8.54 |
| AT1G05230 | HDG2 | 3.15e-47 | −8.51 |
| AT1G19620 | AT1G19620 | 5.06e-47 | −8.51 |
| AT1G65985 | AT1G65985 | 7.10e-47 | −8.51 |
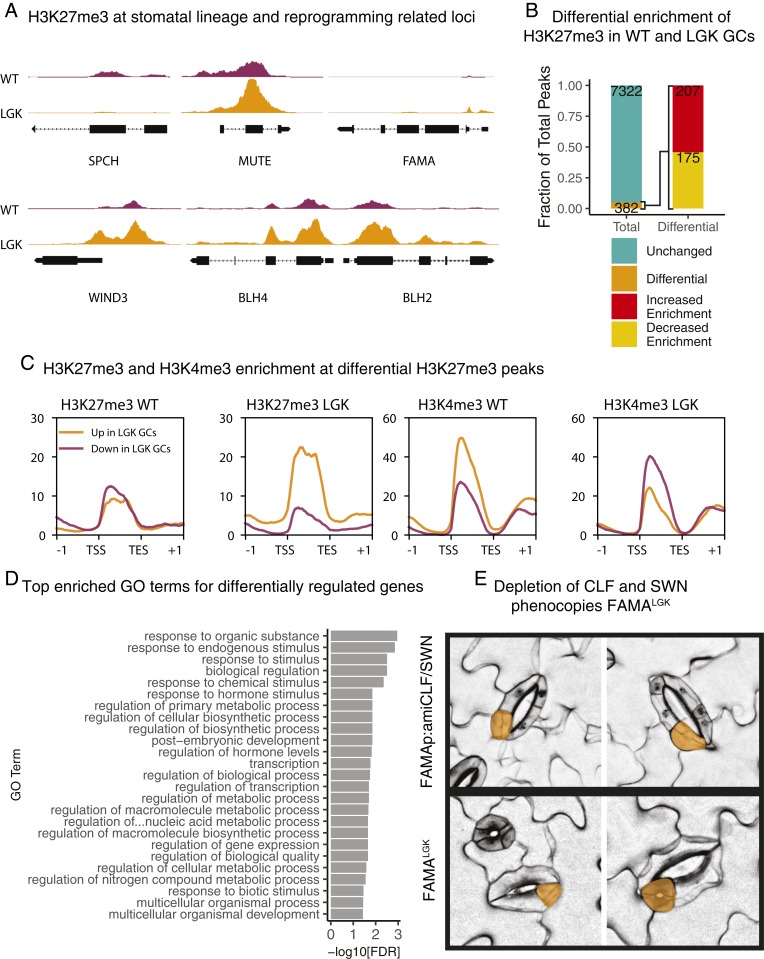
H3K27me3 Peaks Are Differentially Enriched at Several Regulatory Loci during GC Reprogramming.
From our comparison of WT GCs and aerial tissue, it was clear that H3K27me3 distribution varies widely in different tissues, corroborating previous observations (14, 15, 46). With this validation of our single-cell type methods and dataset, we could then generate profiles of H3K27me3 and H3K4me3 to compare WT GCs with reprogramming GCs. Transcriptional profiles of LGK GCs established that SPCH is strongly up-regulated (Fig. 1D); concomitantly, the SPCH locus loses all H3K27me3 in LGK GCs. H3K27me3 coverage of MUTE and FAMA, whose expression levels are not up-regulated in FAMALGK, remains unchanged based on a differential binding analysis (Fig. 4A). We identified overlapping and unique genomic intervals, or peaks, of enrichment for each modification in WT and LGK GCs. We found 15,288 common H3K4me3 peaks between WT and LGK GCs, with 635 and 563 unique to each cell type, respectively. For H3K27me3, we found 6,348 common peaks, 551 WT GC unique peaks, and 805 LGK GC unique peaks (SI Appendix, Fig. S3C). To assess H3K27me3 distribution more broadly, we performed differential enrichment analysis of the 7,704 H3K27me3 peaks found in WT and/or LGK GCs and found 382 peaks (367 genes) were differential between WT and reprogrammed GCs (Fig. 4B and Dataset S1). An earlier model of GC reprogramming was that it resulted from failure of FAMA to recruit RBR and the PRC2 complex to stomatal target genes (19). While H3K27me3 was depleted from SPCH in LGK GCs, as predicted by this model, the genome-wide result that roughly equal numbers of genes gained or lost H3K27me3 (Fig. 4B) was not expected.
Fig. 4.
Few, but critical, H3K27me3 peaks are differentially enriched in WT relative to LGK GCs. (A) SPCH and stemness regulators WIND3, BLH2, and BLH4 have differential H3K27me3 levels in WT and LGK GCs, but MUTE and FAMA are not differential by DiffBind analysis. The y axis is represented as the signal P value and is scaled consistently between tracks for each gene. (B) Differential binding analysis shows only ∼5% of peaks change in H3K27me3 enrichment during reprogramming. Those that change are split nearly equally between increased and decreased enrichment. (C) H3K27me3 enrichment at genes associated with differentially modified peaks. Intensity is represented as the signal P value. Genes with increased enrichment in WT GCs are shown in purple. Genes with increased enrichment in LGK GCs are shown in orange. (D) Top 25 enriched GO terms for genes with differentially enriched H3K27me3 peaks. (E) Confocal images of 9 dpg seedlings with stomata in stomata (reprogramming) phenotypes. The top two images are from seedlings expressing a FAMAp-driven artificial microRNA against CLF and SWN. The bottom two images are from FAMALGK seedlings. Stomata within stomata are false colored in orange.
When the average H3K27me3 enrichment levels at the loci that gained or lost H3K27me3 in WT and LGK GCs were plotted, both sets of loci had roughly equal amounts of H3K27me3 in WT GCs, but average enrichment levels diverged at these loci in LGK GCs (Fig. 4C). H3K4me3 behaves oppositely to H3K27me3 at these loci (Fig. 4C). The number of differentially enriched peaks between WT and LGK GCs (n = 382) is far fewer than those between WT GCs and aerial tissue (n = 3,965), and far fewer than the number of FAMA targets (47) and of genes differentially expressed during reprogramming (Fig. 1D). Differential H3K27me3 peaks identified do correlate well with the direction and magnitude of differential expression (SI Appendix, Fig. S5) and are enriched for genes involved in transcription and control of postembryonic development, suggesting the genes with altered H3K27me3 could represent regulators of cellular identity (Fig. 4D).
Given the modest changes to H3K27me3 we observed in reprogrammed cells, it was important to test whether H3K27me3 depletion in GCs could be causative for reprogramming at all. We generated a FAMA promoter-driven artificial microRNA that simultaneously targeted the two leaf-expressed SET domain-containing members of the PRC2 complex, CURLYLEAF (CLF) and SWINGER (SWN), to reduce H3K27me3 in GCs. Depleting PRC2 activity specifically in GCs caused a reprogramming phenotype qualitatively similar to the 1 observed in FAMALGK (Fig. 4E), but occurs at a lower frequency (SI Appendix, Fig. S6 A and B). This evidence suggests that the changes we observe to H3K27me3 at loci regulating transcription, although few in number, have a critical role in maintaining GC identity.
WIND3 Exhibits Differential H3K27me3 Enrichment and Modulates Reprogramming.
Genes that lost H3K27me3 could be linked to reestablishment of early stomatal lineage identity, but what about the genes that gained H3K27me3? Among the most interesting genes with this behavior were transcriptional regulators reported to act during reprogramming (Fig. 4A). For example, the WIND3 locus gains H3K27me3 marks and is transcriptionally down-regulated during GC reprogramming (Fig. 4A and SI Appendix, Fig. S8C). This is particularly intriguing because WIND3 is 1 of 4 AP2-ERF transcription factors that promotes wound-induced reprogramming, and the sole member of the group whose locus is clearly associated with H3K27me3 (9). Broad overexpression of WIND3 produces a dedifferentiation phenotype similar to H3K27me3 depletion in Arabidopsis roots (9).
Differential expression of WIND3 in LGK GCs could be caused by differential expression of this gene during stomatal development. However, in neither published RNA-seq nor microarray profiles was WIND3 lower in early stomatal lineage cells relative to GCs (SI Appendix, Fig. S7A) (18). Therefore, we hypothesized that differential expression of WIND3 was specific to LGK reprogramming rather than stomatal development. Because WIND3 has a role in wound-induced reprogramming, we next asked whether there were broader transcriptional patterns that implicated a wound response or other reprograming pathways in LGK GCs. We compared the transcriptome of LGK GCs to that of callus derived from cotyledons and petals (48); of the 1,490 genes up-regulated during callus formation in this study, however, only 162 were differentially expressed in WT v LGK GCs (P value of overlap = 1) (SI Appendix, Fig. S8A). Moreover, this small overlapping gene set is enriched for GO terms related to microtubule-based processes, ribosome biogenesis, and translation (SI Appendix, Fig. S8A), suggesting that any similarity between the two reprogramming events is cell cycle reentry, not shared regulatory pathways for reprogramming. That LGK GCs are not employing significant portions of known reprogramming pathways is reinforced by investigation of WIND3 and 102 other genes identified as inducers or repressors of callus formation (31, 49). Genes that act as callus inducers or repressors (31, 49) were equally likely to be up-regulated or down-regulated in LGK GCs relative to WT GCs (SI Appendix, Fig. S8C). Of genes associated with the “lateral root program” through which regenerating cells may transit (48), only SCARECROW (SCR) is weakly up-regulated in LGK GCs (SI Appendix, Fig. S8B), but SCR is also differentially expressed during stomatal development. Taken together, these analyses suggest that LGK GCs are not utilizing the known wound or hormone-based reprogramming pathways.
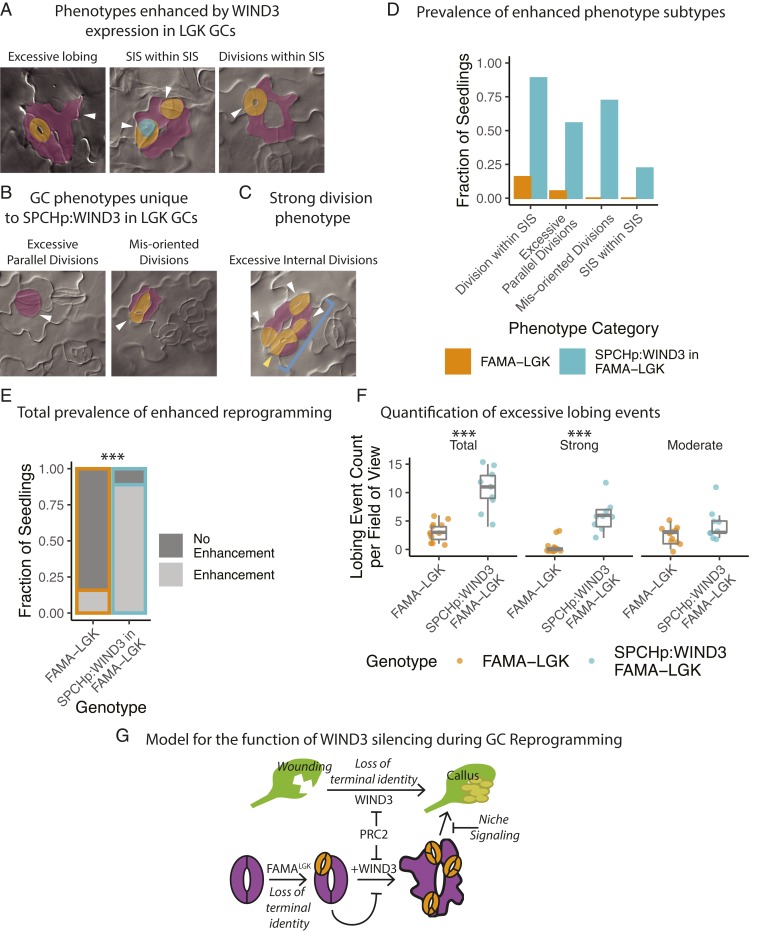
As a demonstrated positive regulator of reprogramming, WIND3 was expected to be more strongly expressed in reprogramming cells, but instead we observed a decrease in expression. We therefore considered whether LGK GCs were actually suppressing reprogramming via silencing of WIND3. We tested whether silencing WIND3 during LGK-mediated reprogramming was functionally relevant by reestablishing expression of WIND3 in LGK GCs by expressing WIND3 under the SPCH promoter, which is reactivated in LGK GCs upon reprogramming (19). The severity of LGK GC reprogramming was increased by this manipulation (Fig. 5A), manifesting as increased frequency of previously documented (19) reprogramming events, or as novel, more severe changes in GCs, overall comprising 5 classes of GC defects that serve as evidence of reprogramming enhancement (Fig. 5 A and B). We confirmed that the WIND3 overexpression levels in leaves directly correlated with phenotypic severity (SI Appendix, Fig. S7 B and C). When quantified, each of the phenotypic defect classes were present in almost all of the FAMALGK SPCHp:WIND3 plants scored (Fig. 5D), including the extreme case of stomata forming inside of stomata that had formed inside of stomata (SIS within SIS). Overall, there was a statistically significant increase in the number of seedlings displaying enhanced phenotypes in FAMALGK plants expressing SPCHp:WIND3 (Fig. 5E) (Fisher’s exact test, P = 1.006e-05). The strong reprogramming phenotype was dependent on the FAMALGK background as expression of SPCHp:WIND3 in the WT (Col-0) background had no discernable phenotypic effect (SI Appendix, Fig. S7D).
Fig. 5.
Reestablishing WIND3 expression in LGK GCs enhances the reprogramming phenotype relative to FAMALGK, suggesting plant cells sense and resist inappropriate reprogramming. (A and B) Representative DIC images of the different manifestations of the enhanced LGK phenotype (A) and the novel phenotypes (B) caused by SPCHp:WIND3 expression in FAMALGK GCs in 12 dpg cotyledons. White arrowheads denote instances of the referenced phenotype. SIS are false-colored in orange. Meristemoids in stomata are false-colored in green. Original stomatal complexes are false-colored in purple. (C) Image of excessive stomatal complex divisions. Here, white arrowheads indicate SIS, the yellow arrowhead indicates a division within an SIS, and the blue bracket points out the entire original complex. (D) Quantification of the prevalence of each class of enhanced phenotype among seedlings of the parental FAMALGK line (n = 19), and a T2 line expressing SPCHp:WIND3 in FAMALGK (n = 18). Values represent the number of seedlings having 1 or more instance of each enhanced phenotype. (E) Overall prevalence of phenotype enhancement per seedling. The total number of seedlings with enhanced phenotypes in each population are statistically different by Fisher’s exact test, P = 1.006e-05. (F) Quantification of GC lobing events per field of view (384 µm × 486 µm) in DIC images of 12 dpg cotyledons in the same lines used in 5A. There is a statistically significant increase in the total number of lobing events (P = 0.0002687) and the number of strong lobing events (P = 0.000256) in the line expressing SPCHp:WIND3 in FAMALGK by Welch’s t test. (G) Failure to silence WIND3 in LGK GCs causes reprogramming enhancement via increasing the number of cell divisions and the extent of dedifferentiation, which are also processes involved in callus formation. PRC2 regulates WIND3, limiting the extent of reprogramming. Asterisks indicates statistically significant P value, less than 0.0001.
Most of the enhanced GC defects we observe in FAMALGK plants expressing SPCHp:WIND3 involve increasing the number of cell divisions in LGK GCs (e.g., Fig. 5 A and C). It was formally possible that WIND3 expression in LGK GCs indirectly enhanced reprogramming events by increasing the number of cell divisions. We therefore scored another measure of reprogramming that was independent of cell division—cell lobing (19). When we compared FAMALGK and FAMALGK + SPCHp:WIND3 plants, the latter exhibited a significant increase in the number of stomatal complexes exhibiting lobes as well as the severity of ectopic lobes (Fig. 5F, strong lobe illustrated in Fig. 5A, Left and moderate lobe in SI Appendix, Fig. S1B, double arrow). This indicates that reprogramming occurs more profoundly (or more rapidly) when FAMALGK plants express SPCHp:WIND3 and supports the idea that LGK GCs suppress further reprogramming by silencing WIND3 (Fig. 5G).
Discussion
Understanding cell identity establishment and maintenance is critical to our understanding of developmental biology. To approach the mechanistic basis of cell identity maintenance and reprogramming in plants, we obtained transcript, H3K27me3, and H3K4me3 profiles from a single cell type, the Arabidopsis GC, and from GCs undergoing reprogramming to precursor stages in planta. We demonstrated that, although H3K27me3 distribution is cell type-specific, reprogramming can occur with minor H3K27me3 redistribution. H3K27me3 marks transcriptional and developmental regulators, so even minor remodeling can have major transcriptional consequences. For instance, the SPCH locus is depleted of H3K27me3 in reprogrammed GCs, and SPCH binds to nearly 6,000 genes (50). This can help to explain how we found 10 times more differentially expressed genes than genes with differential H3K27me3.
Loss of H3K27me3 in Arabidopsis by broad perturbation of PRC2 activity leads to cellular reprogramming (7–9). The FAMALGK manipulation, however, revealed that in GCs, H3K27me3 remodeling was nearly evenly split between increased and decreased H3K27me3 enrichment, allowing us to experimentally demonstrate that global depletion is not a necessary condition of reprogramming in vivo. Taken further, the increases to H3K27me3 at some loci suggest that “native” reprogramming events such as wound repair and lateral root initiation may even require active reorganization of H3K27me3. This is reminiscent of the directed conversion of mammalian ESCs into B cells that requires PRC2 and, therefore, active reestablishment of H3K27me3 marks (51). Given our observations of limited H3K27me3 changes in the GC context, as well as little overlap between the gene expression programs characterized for FAMALGK or wound-induced reprogramming, it also appears that Arabidopsis cells have access to previously unidentified reprogramming strategies.
Why might we observe H3K27me3-mediated repression of the proreprogramming factor WIND3 in FAMALGK GCs (Fig. 5G)? Parallel situations can be seen during animal regeneration, for example in Drosophila the expression of the transcription factor engrailed differs during wound repair and normal development (52). Engrailed is needed in both contexts, but failure to execute wounding-specific control of engrailed expression leads to ectopic cellular reprogramming. Thus, tissues have mechanisms by which they sense wounding to ensure that when repurposing developmental genes, they do so in a way that is appropriate for a tissue that has a history and existing pattern. We hypothesize that in plants, due to their constant creation of new organs and their exposure to wounding, it may be even more essential to build in molecular brakes to separate downstream progression of developmental programs. FAMALGK artificially triggered reprogramming without wounding; the response in GCs seems to indicate they may sense the occurrence of reprogramming itself. This mechanism, although speculative, may be relevant in contexts such as lateral root initiation or root apical meristem regeneration. In this case, roots need to sense whether they are rebuilding a tissue or generating a new one and regulate gene expression accordingly. In the case of our LGK reprogramming, loss of terminal guard cell identity is not accompanied by a wound signal, and therefore complete dedifferentiation is unproductive. The enhancement of reprogramming we observe upon reestablishment of WIND3 expression (Fig. 5 A–F) demonstrates that silencing this network is functionally relevant in this context. One hypothesis for this phenotypic enhancement is that reestablishment of WIND3 expression influences the expression of some of the 367 genes with differential H3K27me3 (Fig. 4B). We performed a search using algorithms in ref. 53 for the WIND3 binding motif derived from DAP-seq data (54) in the promoters of these 367 genes and found a total of 94 motif occurrences in 64 of the promoter sequences (Dataset S1). This does not constitute an overrepresentation of WIND3 motifs among these loci but does indicate that WIND3 may regulate a subset of these genes.
In addition to helping us understand cell identity, single-cell type datasets can inform the way we think about the behavior of histone modifications themselves. Much of our understanding of H3K27me3 establishment dynamics in Arabidopsis comes from close examination of the FLC locus in whole tissues (reviewed in ref. 55). Studies of FLC have the power to reveal fascinating chromatin dynamics, but to what extent these dynamics are employed at other loci remains an open question. The averaging effect of mixed cell populations are liable to mask locus-specific dynamics for genes that show tissue-specific behavior, as we show for H3K27me3 enrichment of meristem genes STM and HEC1 in GCs (Fig. 3D). Although the impetus of these studies was to evaluate the genome-wide changes in gene expression and histone modifications during a specific reprogramming transition, the high-quality single-cell type ChIP-seq datasets enable exploration of other genome-wide histone modification questions such as cooccurrence of different histone modifications (SI Appendix, Fig. S9).
The stomatal lineage has distinct phases of self-renewing precursors followed by committed precursors, and it is intriguing that the reprogramming proceeded back to the earliest lineage stage, but not beyond, even with enhanced WIND3 expression. It is interesting to contrast the reprogramming potential of GCs from intact Arabidopsis leaf tissue with those derived from Beta vulgaris epidermal peels where 100% of the GCs give rise to callus and then regenerate to form plantlets in culture (20). Inclusion of underlying vascular tissue, however, strongly or totally suppressed GC regeneration capacity, consistent with a block being set up by tissue context (20). Extrinsic cues, therefore, may still enforce epidermal fates in LGK GCs ectopically expressing WIND3. This is different from global overexpression of WIND3 (9), where all cells dedifferentiate and presumably normal cell-cell communication is lost in the process. As such, our findings support the conclusion that perturbations to intrinsic regulators of cell identity can remove some barriers to reprogramming but cannot alone override cues from extrinsic sources like tissue identity to fully unlock cell fate.
Materials and Methods
All lines used in this study were in the Col-0 ecotype. AGI codes: FAMA (AT3G24140), WIND3 (AT1G36060), CLF (AT2G23380), SWN (AT4G02020). Artificial microRNAs targeting CLF and SWN were designed using WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and amplified using the pRS300 vector as a template. FAMApro:YFP-YFP includes 2.5-kb FAMA promoter and tandem YFP sequences derived from R4pGWB440. SPCHpro:WIND3 was created by amplify a WIND3 from gDNA and combining with SPCHp in R4pGWB440. Sequences for all primers used in this work can be found in SI Appendix, Table S1. Detailed protocols for FACS-based cell isolation, preparation of RNA and chromatin, creation of libraries, and analysis of data are in SI Appendix, Materials and Methods. The raw data, bed files containing ChIP peaks, bigWig files showing ChIP fold change over input, and TPM-normalized RNA abundance measurements are deposited in GEO under accession no. GSE118138 (56).
Supplementary Material
Acknowledgments
We thank the members of the D.C.B. lab for constructive discussion and feedback and Dr. Ao Liu and Dr. Michael Raissig for detailed comments on the manuscript. L.R.L. was supported by an NIH Graduate Training Grant (NIH5T32GM007276) awarded to Stanford University. D.C.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov (accession no. GSE118138).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911400116/-/DCSupplemental.
References
- 1.Bowman S. K., et al. , H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3, e02833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman R. T., Struhl G., Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, eaai8236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pengelly A. R., Copur Ö., Jäckle H., Herzig A., Müller J., A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hawkins R. D., et al. , Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koche R. P., et al. , Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell 8, 96–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussein S. M. I., et al. , Genome-wide characterization of the routes to pluripotency. Nature 516, 198–206 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Bouyer D., et al. , Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7, e1002014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanvivattana Y., et al. , Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Ikeuchi M., et al. , PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1, 15089 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum K. D., Roudier F., Epigenetic memory and cell fate reprogramming in plants. Regeneration (Oxf.) 4, 15–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deal R. B., Henikoff S., A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lucas M., et al. , Transcriptional regulation of Arabidopsis polycomb repressive complex 2 coordinates cell-type proliferation and differentiation. Plant Cell 28, 2616–2631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno-Romero J., Jiang H., Santos-González J., Köhler C., Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35, 1298–1311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Y., et al. , Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nat. Commun. 8, 15120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafos M., et al. , Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7, e1002040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohashi-Ito K., Bergmann D. C., Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutilier K., et al. , Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adrian J., et al. , Transcriptome dynamics of the stomatal lineage: Birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matos J. L., et al. , Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 3, e03271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall R. D., et al. , Stomatal guard cells are totipotent. Plant Physiol. 112, 889–892 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosquna A., Katz A., Shochat S., Grafi G., Ohad N., Interaction of FIE, a polycomb protein, with pRb: A possible mechanism regulating endosperm development. Mol. Genet. Genomics 271, 651–657 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Jullien P. E., et al. , Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6, e194 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee E., Lucas J. R., Goodrich J., Sack F. D., Arabidopsis guard cell integrity involves the epigenetic stabilization of the FLP and FAMA transcription factor genes. Plant J. 78, 566–577 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Lee E., Lucas J. R., Sack F. D., Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 78, 555–565 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bargmann B. O. R., Birnbaum K. D., Fluorescence activated cell sorting of plant protoplasts. J. Vis. Exp. 1673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efroni I., Ip P.-L., Nawy T., Mello A., Birnbaum K. D., Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 16, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura R. L., et al. , Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109, 371–374 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ache P., et al. , GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K(+)-selective, K(+)-sensing ion channel. FEBS Lett. 486, 93–98 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Husebye H., Chadchawan S., Winge P., Thangstad O. P., Bones A. M., Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128, 1180–1188 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris S. A., Daley G. Q., A blueprint for engineering cell fate: Current technologies to reprogram cell identity. Cell Res. 23, 33–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeuchi M., et al. , Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Zhang X., et al. , Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, e129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Bernatavichute Y. V., Cokus S., Pellegrini M., Jacobsen S. E., Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., et al. , Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alabert C., et al. , Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long J. A., Moan E. I., Medford J. I., Barton M. K., A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Schuster C., et al. , A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438–449 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Abrash E. B., Bergmann D. C., Regional specification of stomatal production by the putative ligand CHALLAH. Development 137, 447–455 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Negi J., et al. , A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23, 479–484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe M., Katsumata H., Komeda Y., Takahashi T., Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–643 (2003). [DOI] [PubMed] [Google Scholar]
- 41.San-Bento R., Farcot E., Galletti R., Creff A., Ingram G., Epidermal identity is maintained by cell-cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J. 77, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Peterson K. M., et al. , Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140, 1924–1935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamata N., Okada H., Komeda Y., Takahashi T., Mutations in epidermis-specific HD-ZIP IV genes affect floral organ identity in Arabidopsis thaliana. Plant J. 75, 430–440 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Li Y., et al. , Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. U.S.A. 104, 18339–18344 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S. B., Suh M. C., Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56, 48–60 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C., H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6, e1001152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hachez C., Ohashi-Ito K., Dong J., Bergmann D. C., Differentiation of Arabidopsis guard cells: Analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 155, 1458–1472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto K., Jiao Y., Meyerowitz E. M., Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Ikeuchi M., Sugimoto K., Iwase A., Plant callus: Mechanisms of induction and repression. Plant Cell 25, 3159–3173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau O. S., et al. , Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira C. F., et al. , ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6, 547–556 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Schuster K. J., Smith-Bolton R. K., Taranis protects regenerating tissue from fate changes induced by the wound response in Drosophila. Dev. Cell 34, 119–128 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Malley R. C., et al. , Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whittaker C., Dean C., The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Lee L. R., Wengier D. L., Bergmann D. C., Cell-type–specific transcriptome and histone modification dynamics during cellular reprogramming in the Arabidopsis stomatal lineage. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118138. Deposited 5 August 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.