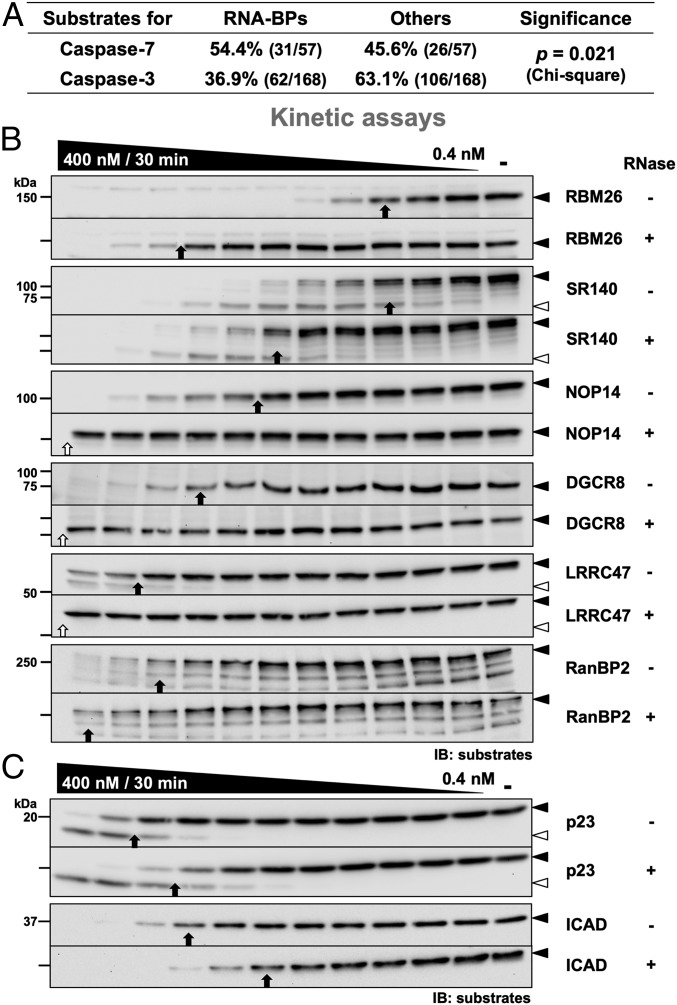
Significance
During apoptosis, hundreds of intracellular proteins are cleaved by caspases. In addition to recognizing optimized motifs in the primary structure of its substrates, we show that caspase-7 possesses an exosite to enhance poly(ADP-ribose) polymerase 1 (PARP-1) recognition via the mutual binding of RNA. We also demonstrate that caspase-7 binds RNA and that PARP-1, via 2 RNA-binding domains, likely binds the same RNA molecule, bringing itself near caspase-7 for prompt inactivation. This mechanism is conserved in the mouse ortholog. We have also validated the use of RNA for the efficient cleavage of 6 other RNA-BPs, demonstrating that this distinctive mode of proteolysis enhancement by caspase-7 is an essential feature of substrate recognition during apoptosis.
Keywords: caspase-7, poly(ADP-ribose) polymerase 1, apoptosis, exosite, RNA-BPs
Abstract
To achieve swift cell demise during apoptosis, caspases cleave essential proteins for cell survival and removal. In addition to the binding of preferred amino acid sequences to its substrate-binding pocket, caspase-7 also uses exosites to select specific substrates. 4 lysine residues (K38KKK) located in the N-terminal domain of caspase-7 form such an exosite and promote the rapid proteolysis of the poly(ADP-ribose) polymerase 1 (PARP-1), but the mechanism of recognition remains mostly unknown. In this study, we show that the overall positive charge of the exosite is the critical feature of this evolutionarily conserved binding site. Additionally, interaction with the caspase-7 exosite involves both the Zn3 and BRCT domains of PARP-1 and is mediated by RNA. Indeed, PARP-1 proteolysis efficacy is sensitive to RNase A and promoted by added RNA. Moreover, using affinity chromatography and gel shift assays, we demonstrate that caspase-7, but not caspase-3 or a caspase-7 with a mutated exosite, binds nucleic acids. Finally, we show that caspase-7 prefers RNA-binding proteins (RNA-BPs) as substrates compared to caspase-3 and that RNA enhances proteolysis by caspase-7 of many of these RNA-BPs. Thus, we have uncovered an unusual way by which caspase-7 selects and cleaves specific substrates.
Apoptosis is essential to the homeostasis of pluricellular organisms via the selective removal of unwanted cells during physiological and pathological conditions. This cell death process is without inflammation and is coordinated by caspases, a family of dimeric cysteinyl endopeptidases [clan CD, family C14 (1)]. Basically, apoptotic caspases are activated in a 3-step process in which initiator caspases (caspase-8, caspase-9, and caspase-10), activated by dimerization on multimeric platforms following internal and external cues, proteolyze executioner caspases (caspase-3 and caspase-7); then caspase-3 activates executioner caspase-6 (reviewed in refs. 2 and 3). Although several regulating and feedback mechanisms exist at many steps along the apoptosis-activating cascade, these straightforward steps ensure amplification of the death signal.
Together, executioner caspases cause cell demise by the cleavage of proteins essential to cell survival and removal. To date, up to 2,000 caspase substrates have been tallied (4, 5) encompassing all types of proteins, from structural to metabolic to signaling to nuclear. Although an individual caspase’s substrate repertoire overlaps with that of others, it has become clear that caspases must be specialized to achieve efficacy. This specialization is exemplified by caspase-6, which has a substrate preference that differs from that of caspase-3 and caspase-7, preferring amino acid sequences bearing short hydrophobic residues like valine in P4 (fourth residue preceding the scissile bond), whereas the other executioner caspases prefer substrates with an aspartate residue at that position (6, 7). However, despite almost identical substrate primary structure preference, caspase-3 and caspase-7 do not have a fully redundant death substrate list for reasons that are not fully understood. Some differences may be explained by the use of exosites, which are binding sites for specific substrates present outside the canonical substrate-binding pocket which enhance the cleavage rate by increasing the affinity for these substrates. Indeed, biochemical studies have revealed that caspase-7 uses determinants from its N-terminal domain (NTD) to assist in cleaving the HSP90 cochaperone p23 and poly(ADP-ribose) polymerase 1 (PARP-1) (8, 9). Also, caspase-6 has an exosite for the cleavage of substrates such as nuclear lamins (10, 11). The efficacious proteolysis of PARP-1 by caspase-7 relies on 4 lysine residues (K38KKK) that are exposed once the caspase is fully mature (8). Initial experiments showed that this exosite confers higher PARP-1 proteolysis efficacy (kcat/KM), rendering caspase-7 more efficacious than caspase-3 despite a lower intrinsic catalytic activity.
PARP-1 is a multidomain protein that plays critical roles in many processes such as regulation of chromatin structure, regulation of transcription, and DNA repair (12) in which it detects various types of DNA breaks via its many independently folded domains. The single- and double-strand breaks (SSBs/DSBs) in DNA are recognized by various combinations of the zinc finger 1 (Zn1), Zn2, Zn3, or WGR (tryptophane, glycine, arginine) domains, leading to domain reorganization specific to the type of DNA break (13, 14). In a process called PARylation, the newly structured PARP-1 uses nicotinamide adenine dinucleotide (NAD+) as a substrate to generate and transfer poly(ADP-ribose) (PAR) mostly on itself in the form of branched chains. The highly negatively charged PAR polymers are transferred on many residues of PARP-1 preferentially found in the linker following the BRCT (BRCA1 C terminus) domain (15). PARP-1 cleavage within a bipartite nuclear localization signal (NLS) is recognized as a hallmark of apoptosis (16) and prevents PARP-1-dependent energy collapse and ensuing necrosis (17). Thus, it is reasonable that cells have evolved a caspase with an exosite to quickly inactivate this necrosis-promoting enzyme during apoptosis.
Exosites in caspases remain poorly characterized for the substrates and motifs they bind, the details of their mechanism, and prevalence of use. In this study, we aimed at identifying the motif recognized by the exosite of caspase-7 and used a series of biochemical assays to reveal the mechanism by which it employs nucleic acids to enhance cleavage of PARP-1 and other RNA-binding proteins (RNA-BPs).
Results
Positive Charges Are a Critical Feature of the Exosite.
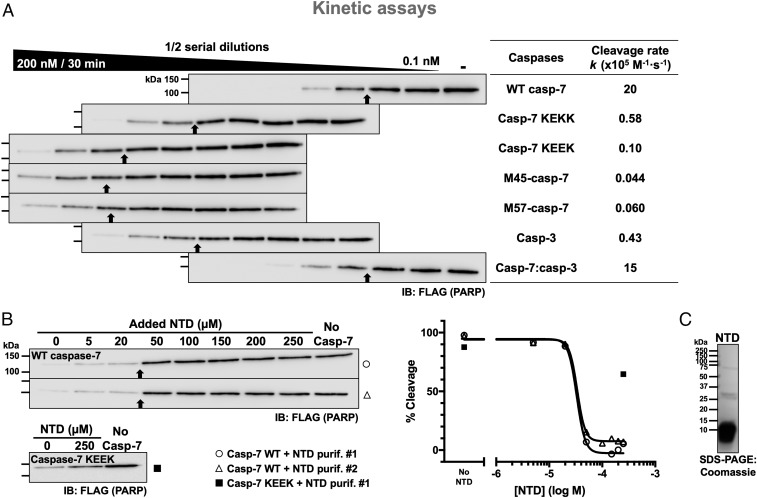
To study the interaction between the exosite of caspase-7 and PARP-1, we performed cleavage assays using cellular extracts from AD-293 cells transfected with a plasmid-encoding FLAG-tagged PARP-1 as a source of substrate. In these cells, caspase-7 expression was abolished using CRISPR/Cas9 gene editing (293C7KO; SI Appendix, Fig. S1A). Cell extracts mimic the natural environment in which the caspases operate, including potential posttranslational modifications. Immunoblotting for individual apoptotic caspases shows that they remain in their uncleaved forms (SI Appendix, Fig. S1B) and no activity was detected using a panel of preferred caspase substrates (SI Appendix, Fig. S1C), demonstrating the lack of caspase activation and activity in our cellular extracts. Because caspase-7 is inefficient at cleaving other caspases (18), no other endogenous caspase able to cleave PARP-1 was activated in our assays. First, we confirmed the ability of caspase-7 to cleave PARP-1 in optimized experimental conditions (Fig. 1A), and we calculated the PARP-1 cleavage rate (k; see Materials and Methods) as a mean of comparison between series of assays (9). The uncleaved PARP-1 band was quantified to determine the cleavage rate because the cleaved fragment is unstable and thus unreliably detected; serial dilutions of caspases were used to allow a better comparison of the different caspase-substrate pairs. As we previously reported (8), caspase-7 is more efficacious at cleaving PARP-1 than caspase-3 (higher k value: k = 20 × 105 M−1·s−1 vs. 0.43 × 105 M−1·s−1; Fig. 1A) despite its lower intrinsic activity measured on the small fluorogenic peptidic substrate Ac-DEVD-Afc (lower kcat/KM; SI Appendix, Table S1). The introduction of 1 (KEKK) or 2 (KEEK) negative charges in the form of glutamic acids in the exosite (wild type [WT] has K38KKK) reduces the cleavage rate of PARP-1 by up to 200-fold (k = 0.58 × 105 M−1·s−1 and 0.10 × 105 M−1·s−1, respectively). Importantly, mutation of 2 lysine residues neither altered its intrinsic activity (SI Appendix, Table S1) nor affected its inhibition by X-linked inhibitor of apoptosis protein (XIAP), an important endogenous inhibitor of caspase-3 and caspase-7 (19, 20) (SI Appendix, Fig. S2). The caspase-7 exosite mutants KAAK and AKKA, in which 2 lysines were substituted for alanine residues, have a slightly higher cleavage rate than caspase-7 bearing the KEEK motif (SI Appendix, Fig. S3A) and a similar cleavage to that of the KEKK exosite mutant, showing that the overall charge is key to the exosite. Furthermore, replacing all lysines for arginine residues resulted in the same cleavage rate of PARP-1 as with WT caspase-7 (SI Appendix, Fig. S3B). The M45 caspase-7 [caspase-7 starting at Met45 (18)] or a caspase-7 lacking the entire NTD (M57 mutant) showed similar cleavage rates (k = 0.04 × 105 M−1·s−1 or 0.06 × 105 M−1·s−1, respectively; Fig. 1A) and is also comparable to caspase-7 KEEK, meaning that the exosite is composed essentially of lysines 38–41 of caspase-7. Importantly, the NTD of caspase-7 can be added to the catalytic core of caspase-3 to generate a chimera which has a better cleavage rate than WT caspase-3 (k = 15 × 105 M−1·s−1 vs. 0.43 × 105 M−1·s−1), confirming that the exosite exerts its function independently of the catalytic core. All of the different caspases used have similar kinetic parameters on a small peptidic substrate (SI Appendix, Table S1), which implies that the differences seen in our assays come solely from the exosite. Because the murine caspase-7 ortholog has a similar positive patch (K38KKR) in its NTD, we tested its role in cleaving murine PARP-1. Lysates from murine RAW 264.7 macrophages were incubated with murine recombinant caspases (SI Appendix, Fig. S4) and, as with human proteins, murine caspase-7 is better at cleaving PARP-1 than caspase-3, albeit only 2-fold better whereas human caspase-7 is 32-fold better (Fig. 1A). Furthermore, the introduction of 2 negative charges (KEER) reduces the cleavage rate by murine caspase-7. Thus, the positively charged exosite of caspase-7 is conserved in mice. Of note, the murine caspase-7 expressed in this study showed lower intrinsic activity than the human ortholog (SI Appendix, Table S1).
Fig. 1.
Lysine residues 38–41 of caspase-7 promote the cleavage of PARP-1. (A) Efficient cleavage of PARP-1 requires the N-terminal domain. 293C7KO cell extracts expressing full-length FLAG-tagged PARP-1 were incubated with the indicated recombinant caspases at the different concentrations (2-fold serial dilutions of the caspase) for 30 min and analyzed using immunoblotting with an anti-FLAG antibody. The highest and lowest concentrations of caspases are indicated in and over the black wedge, respectively. Immunoblots are aligned to vertically position samples containing the same caspase concentration. The cleavage rates of PARP-1 (k values) are shown in the right-end table for comparison; higher k values signify a faster cleavage rate. As a reference throughout the manuscript, black arrows point to the sample with ∼50% cleavage; thus, the more rightward the arrow, the faster the cleavage. (B) Competition assays showing that the NTD of caspase-7 can prevent PARP-1 cleavage. Samples were treated as in A with a fixed amount of WT caspase-7 (0.78 nM) or caspase-7 KEEK (100 nM) and with varying concentrations of recombinant NTD (a.a. 24–65) from 2 different batches. Quantified data for the inhibition were plotted when possible. (C) Coomassie gel stain showing the integrity of the NTD protein used in B. Black arrows point to the sample with ∼50% cleavage based on densitometry analyses; repeated molecular weight marker values were omitted from subsequent immunoblots.
To demonstrate further the importance of the exosite for the cleavage of PARP-1, the recombinant mature NTD (residues 24–65) of caspase-7 was produced and added to cleavage assays with WT caspase-7 or the KEEK mutant (Fig. 1B). The added NTD almost completely inhibited the cleavage of PARP-1 by WT caspase-7 at a concentration of 50 µM, while the highest concentration of NTD used (250 µM) poorly inhibited cleavage by the caspase-7 KEEK mutant. This latter result demonstrates that the NTD competed only with the exosite and not with the catalytic core. Inhibition curves from those assays were plotted (Fig. 1B), allowing us to calculate IC50 values for 2 independent batches of NTD (33 and 34 µM). Furthermore, the steepness of the competition curve suggests that there is either positive cooperativity in the interaction or that the binding of the exosite to PARP-1 involves more than 1 binding site. Altogether, these results show that positive charges are the primary feature of the exosite and that its binding to PARP-1 involves multiple sites or uses an unusual binding mode.
Both the Zn3 and BRCT Domains of PARP-1 Participate in Interaction with the Caspase-7 Exosite.
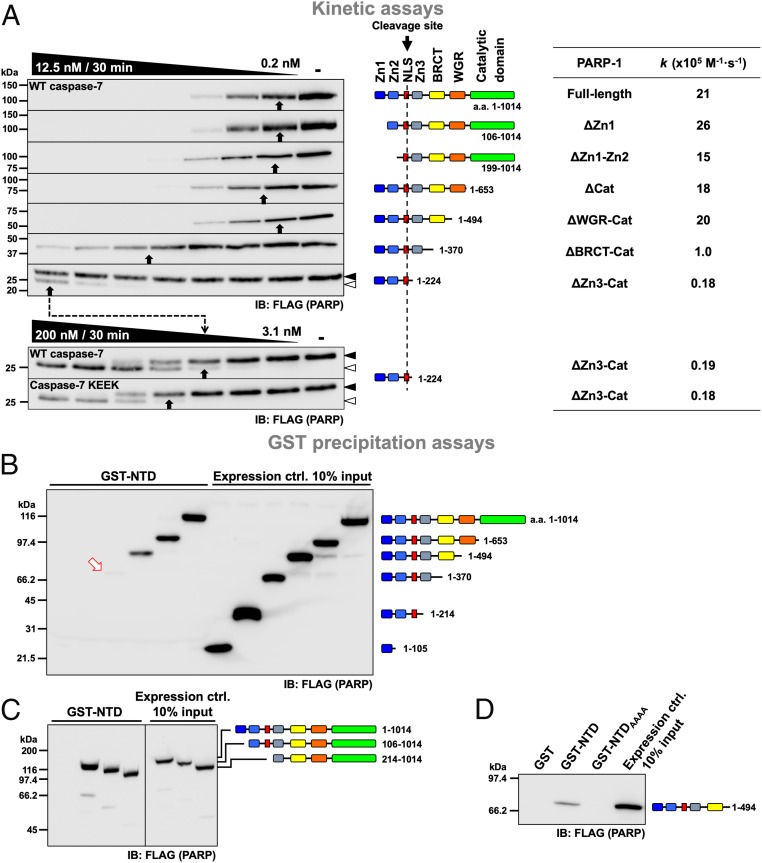
To identify the domain(s) the exosite binds to, PARP-1 truncation proteins were expressed in 293C7KO cells, and lysates were prepared and incubated with WT caspase-7 in cleavage assays; cleavage rates were calculated for each of them (Fig. 2A). Each truncated PARP-1 protein retained the cleavage site at Asp214 located in a linker between the Zn2 and Zn3 domains. There was no substantial difference in the cleavage rates of PARP-1 lacking either the Zn1 domain (ΔZn1) or both the Zn1 and Zn2 domains (ΔZn1-Zn2; k = 26 × 105 M−1·s−1 and 15 × 105 M−1·s−1, respectively; Fig. 2A) compared to the full-length PARP-1 (k = 21 × 105 M−1·s−1). Moreover, PARP-1 lacking either the catalytic domain (ΔCat) or the Cat and WGR domains (ΔWGR-Cat) was also cleaved with similar rates (k = 18 × 105 M−1·s−1 and 20 × 105 M−1·s−1, respectively). However, subsequent removal of the BRCT domain reduced the cleavage rate by 21-fold (ΔBRCT-Cat; k = 1 × 105 M−1·s−1), and removal of the Zn3 domain reduced the cleavage rate even further, by 6-fold (ΔZn3-Cat, k = 0.18 × 105 M−1·s−1). Because the cleavage site was preserved in all of the truncated PARP-1 proteins, we can conclude that the Zn3 and BRCT domains contribute to the interaction with the exosite of caspase-7. Additionally, cleavage of the ΔZn3-Cat PARP-1 by caspase-7 with a disabled exosite (KEEK) was no different from cleavage by WT caspase-7 (k = 0.18 × 105 M−1·s−1 and 0.19 × 105 M−1·s−1, respectively). This demonstrates that no additional interacting domain between the exosite and PARP-1 remained.
Fig. 2.
The Zn3 and BRCT domains of PARP-1 are critical to the caspase-7 exosite function. (A) Caspase-7 cleavage of various truncated PARP-1 constructs. Experiments were performed as described in Fig. 1A with cell extracts containing different PARP-1 proteins, which were analyzed by immunoblotting. The cleavage rates of PARP-1 (k values) are shown on the right-end table for comparison. The dashed arrow follows the cleavage range shift toward higher concentrations of caspases. (B–D) Caspase-7 NTD pull-down assays of various truncated PARP-1 constructs. WT NTD, mutated exosite (NTDAAAA), or GST alone was used in pull-down experiments. Cell extracts containing FLAG-tagged PARP-1 were incubated with GST proteins for 16 h at 4 °C. Pull-down proteins and input expression controls were analyzed by immunoblotting. Loss of cleavage and pull-down efficacy occurred when the Zn3 and BRCT domains were absent. In B, the red arrow points to a faint band; black and open arrowheads to the right of the immunoblots indicate uncleaved and cleaved proteins, respectively, when visible.
These results were confirmed using gluthathione S-transferase (GST) pull-down assays. In those assays, GST alone or linked to either the NTD (GST-NTD) or the NTD lacking the exosite (GST-NTDAAAA) bound to beads was used to precipitate truncated forms of PARP-1. In the first series of assays, PARP-1 domains were added one by one from the N terminus. Neither the Zn1 nor the Zn2 domain resulted in protein coprecipitation (Fig. 2B). However, a faint band was visible after the addition of the Zn3 domain (arrow), and clear precipitation was observed when the BRCT domain was present. The interaction remained with the further addition of the WGR and Cat domains. Additionally, pull-down assays in which only the Zn1 or Zn1 plus Zn2 domains were removed (Fig. 2C) did not show a decrease in interaction, confirming that they are dispensable for binding with the exosite. Similar assays with GST alone or with a GST-NTDAAAA did not show any interaction (Fig. 2D). In summary, all of the PARP-1 proteins that interacted with the exosite contained the Zn3 or the BRCT domain, confirming the cleavage assays and showing that these domains are important for interaction with the exosite.
To obtain more insight into the interaction between caspase-7 and PARP-1, we extensively mutated the BRCT domain because it is the one that contributed the most to the efficacy of cleavage in kinetic assays and is an established protein-protein interaction domain (21). We mutated negative patches on the surface of this domain with the expectation that they interacted with the positively charged exosite. Surprisingly, we did not identify any specific region that was important for the interaction with the exosite in the BRCT domain, but also in the following linker, or the Zn3 domain (SI Appendix, Fig. S5). Although we are confident that the exosite of caspase-7 interacts with both the Zn3 and BRCT domains, our results suggest that the interaction may not be a protein-protein interaction but may involve posttranslational modifications (PTMs) or an unknown intermediate molecule.
Interaction between the Exosite and PARP-1 Requires RNA.
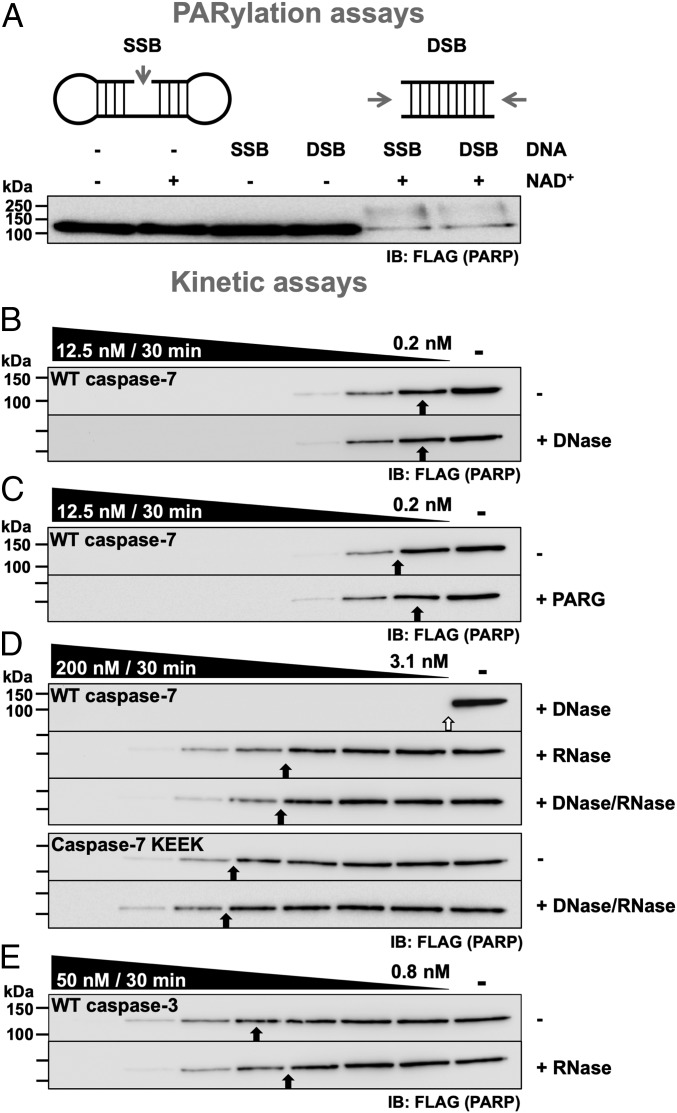
PARP-1 plays a key role in DNA repair, and this involves DNA binding, domain reorganization, and automodification by PARylation (i.e., the addition of PAR chains to itself). Consequently, we looked for the presence of PARylation and DNA as necessary to the interaction with the exosite. First, we forced PARP-1 PARylation with exogenous SSB or DSB DNA mimics in the presence of NAD+ (Fig. 3A). When both DNA and NAD+ were present, a smear that is characteristic of different lengths of polymers added on PARP-1 appeared. Therefore, the PARP-1 protein used does not seem to be modified compared with the positive control; also, even when NAD+ alone was added, no visible modification of PARP-1 was visible, suggesting that there was no damaged DNA from nuclei or mitochondria bound to PARP-1 in the cell extracts we used. Furthermore, adding DNase before the assays did not affect the cleavage of PARP-1 by caspase-7 (Fig. 3B). We also observed no significant difference in cleavage rate when PAR glycohydrolase (PARG) was added before the assays to remove any PAR polymers (Fig. 3C). Therefore, neither DNA nor PAR contributed to the interaction between the exosite of caspase-7 and PARP-1 under our experimental conditions.
Fig. 3.
The interaction between the exosite of caspase-7 and PARP-1 is mediated by RNA. (A) PARylation assays in which NAD+ and an SSB or a DSB mimic was added to cell extracts to detect the formation of PAR chains on PARP-1. (B–E) Effect of glycohydrolase and nucleases on PARP-1 cleavage. Experiments were performed as described in Fig. 1A, with cell extracts pretreated with various combinations of DNase, PAR glycohydrolase, and RNase; then samples were incubated with the indicated caspase. As with the caspase-7 KEEK mutant, the addition of RNase did not significantly affect the cleavage of PARP-1 by caspase-3. The open arrow indicates that the sample with ∼50% cleavage is out of the caspase concentration range.
Albeit less studied, PARP-1 also binds several types of RNA (22–25). To verify if RNA contributes to the interaction, RNase A was added to the extracts before performing cleavage assays. The addition of RNase reduced the cleavage of PARP-1 by WT caspase-7 to the same level as the KEEK exosite mutant (Fig. 3D); combining RNase and DNase had no greater effect than RNase alone. Importantly, the addition of RNase did not affect the cleavage of PARP-1 by caspase-3 (Fig. 3E), further showing the specificity of the interaction. Because caspase-7 KEEK is not affected by RNase, it is unlikely that the reduction in cleavage is due to an inhibition of caspase-7 by the RNase or that it induces a conformational change that renders the cleavage site less accessible. To confirm these results, an RNA rescue experiment was performed in which we sequentially added RNase to degrade RNA, RNasein to inhibit the RNase, and exogenous RNA (5 ng/µl) to replace the degraded RNA. This treatment resulted in enhanced cleavage of PARP-1 by caspase-7 to the untreated rates (SI Appendix, Fig. S6A); RNasein, inhibited RNase, and cleaved RNA showed no effect. Importantly, neither RNA, RNase, RNase-treated RNA (degraded RNA), nor bovine serum albumin (BSA) had any impact on the kinetics of caspase-7 on a small fluorogenic peptidic substrate (SI Appendix, Fig. S6B). Consequently, it seems that RNA acts as an intermediate bound by both PARP-1 and the caspase-7 exosite.
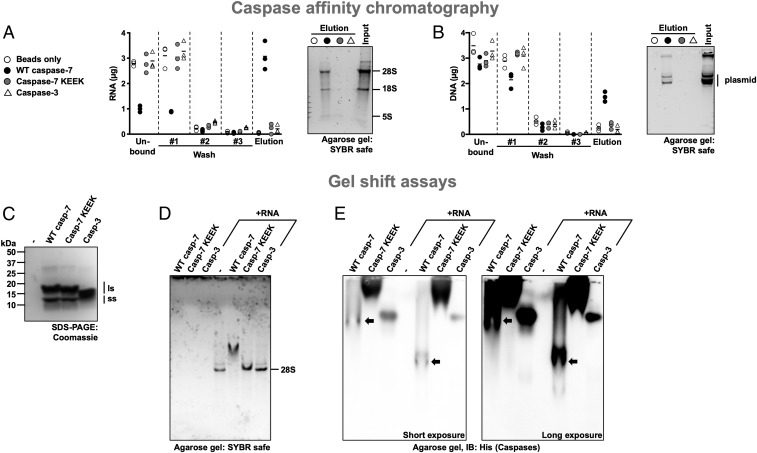
To date, no member of the caspase family has been shown to bind nucleic acids, although examples exist for other classes of peptidases (see Discussion). To investigate this further, we set up affinity columns containing immobilized metal affinity chromatography (IMAC) resin alone or resin saturated with His-tagged WT caspase-7, caspase-7 KEEK, or caspase-3. Each column was loaded with RNA and washed to remove unbound nucleic acids. The bound nucleic acids were eluted using sodium chloride, quantified by spectrophotometry, and separated by agarose gel. As seen in Fig. 4A, caspase-7 retained RNA after the washing steps (average of 3.1 µg out of 7.2 µg total RNA) while other caspases and resin alone had minimal RNA binding (<0.4 µg in all columns and assays). Accordingly, caspase-7 had less RNA coming out in the flow-through and washing steps. Following agarose gel migration, RNA was visible only for caspase-7. The same experiment was carried out with plasmid DNA, and similar results were obtained: WT caspase-7 retained DNA after the washing steps, although to a lesser extent (average of 1.5 µg out of 7.2 µg total DNA; Fig. 4B), while other caspases and resin alone did not (<0.5 µg in all columns and assays). Also, DNA was visible only in the WT caspase-7 eluate following agarose gel migration.
Fig. 4.
Caspase-7’s exosite binds nucleic acids. (A and B) Binding of RNA and DNA to caspase affinity columns. Purified His-tagged caspases were immobilized on 0.2 mL IMAC resin and incubated with 7.2 µg total RNA in phosphate-buffered saline (PBS) (A) or plasmid DNA (B), washed 3 times with PBS, and eluted using 1.25 M NaCl in PBS. Then nucleic acids were quantified at 260 nm, separated on a 1% agarose gel, and stained using SYBR Safe. WT caspase-7 retains nucleic acid more than the exosite mutant or caspase-3. (C) Coomassie gel stain of the caspases used in this figure showing the large (ls) and small (ss) subunits of the peptidase. (D and E) Caspase-7 forms a complex with RNA. Caspases alone or combined with RNA were analyzed on a nondenaturing agarose gel to assess alteration in RNA migration due to the interaction with a caspase (D). The agarose gel used in D was transferred onto a polyvinylidene fluoride membrane and analyzed by immunoblotting using an anti-His tag antibody (E). RNA alters the gel mobility of WT caspase-7 but not that of the exosite mutant or caspase-3. Short and long exposures of the membrane are shown; in E, black arrows point to bands containing WT caspase-7.
To further demonstrate the RNA-binding ability of the caspase-7 exosite, an electrophoretic mobility shift assay (EMSA) was employed. RNA was incubated with buffer, caspase-7, caspase-7 KEEK, or caspase-3 and run on a nondenaturing agarose gel (Fig. 4D). Neither caspase-3 nor caspase-7 KEEK had any effect on the migration of RNA. Conversely, the combination of RNA and caspase-7 resulted in a migration shift of the RNA that was slowed by the presence of the caspase. Following immunoblotting of those same gels, caspase-3 and caspase-7 KEEK migrated similarly with or without RNA while caspase-7 migrated further when combined with RNA (Fig. 4E). Based on the caspase affinity columns and the EMSA, we conclude that caspase-7 binds RNA and, more generally, nucleic acids, using its exosite. Because Guetg et al. (24) identified the Zn1 and Zn2 domains as RNA-binding domains of PARP-1, we wanted to rule them out as crucial for the interaction we studied here. Therefore, we performed cleavage assays in their absence or with those domains only. RNase hampered the cleavage of the PARP-1 construct lacking the Zn1 and Zn2 domains (SI Appendix, Fig. S7A) while no effect was observed when only those domains where present (SI Appendix, Fig. S7B), thus ruling them out as RNA-binding domains in the interaction with caspase-7. Considering all of the results obtained so far, we conclude that under our experimental conditions the exosite of caspase-7 and the Zn3 and BRCT domains of PARP-1 bind RNA and this facilitates proteolysis by bringing the protease and the substrate together.
RNA Enhances Cleavage of Many RNA-BPs by Caspase-7.
Since the interaction between caspase-7 and PARP-1 involves RNA, we tested other RNA-BPs to assess if this unusual mechanism is common to other substrates. We reanalyzed the caspase-3 and caspase-7 substrates identified by Agard et al. (26) to compare the proportion of RNA-binding substrates of each peptidase. From those data, 57 and 168 unique substrates for caspase-7 and caspase-3, respectively, were identified (SI Appendix). For caspase-7, 54.4% of the substrates were identified as RNA-binding (31/57) while the others were not classified as such based on their UniProtKB Gene Ontology molecular functions (Fig. 5A). For caspase-3, a smaller proportion of RNA-BPs was identified (36.9%, 62/168). Consequently, in this study, caspase-7 had significant RNA-binding protein preference over caspase-3 (P = 0.021, χ2 test). Unlike caspase-3, the 5 substrates cleaved the fastest for caspase-7 were all RNA-BPs. Importantly, similar analysis for DNA-binding protein preference was statistically nonsignificant (P = 0.326; SI Appendix, Table S2). To further demonstrate the importance of RNA for faster cleavage by caspase-7, we tested 6 RNA-BPs with cleavage rates above average: RNA-BP 26 (RBM26), U2-associated protein SR140 (SR140), nucleolar protein 14 (NOP14), microprocessor complex subunit DGCR8 (DGCR8), leucine-rich repeat-containing protein 47 (LRRC47), and E3 SUMO-protein ligase RanBP2 (RanBP2). As reported elsewhere (26), all of these proteins were cleaved by caspase-7 under our conditions (Fig. 5B). Interestingly, the addition of RNase reduced the cleavage rate of all of them by caspase-7 with different intensity (∼4-fold to >32-fold). However, treatment with RNase had limited, if any, effect on the cleavage rate of those same RNA-BPs by caspase-7 KEEK and caspase-3 (SI Appendix, Fig. S8). We also tested 2 substrates that do not bind RNA: the HSP90 cochaperone p23 (p23) and the inhibitor of caspase-activated DNase (ICAD). Unlike RNA-BPs, the cleavage of both p23 and ICAD was slightly faster when RNase was added (Fig. 5C). Consequently, RNA enhances cleavage by caspase-7 of many RNA-BPs.
Fig. 5.
RNA enhances caspase-7 cleavage of many RNA-BPs. (A) Analysis of caspase-7 and caspase-3 substrates previously identified by Agard et al. (26). Substrates were classified as RNA-BPs or as others based on their Gene Ontology molecular functions in the UniProtKB database. The RNA-BP preference of caspase-7 and caspase-3 was analyzed using a χ2 test. (B) Cleavage of various RNA-BPs by caspase-7. Experiments were performed as described in Fig. 1A using WT caspase-7 and cell lysates containing the indicated endogenous RNA-BPs treated or not with RNase. (C) Cleavage efficacy of non–RNA-BPs by caspase-7 is not decreased by RNase. Experiments were performed as in B.
Discussion
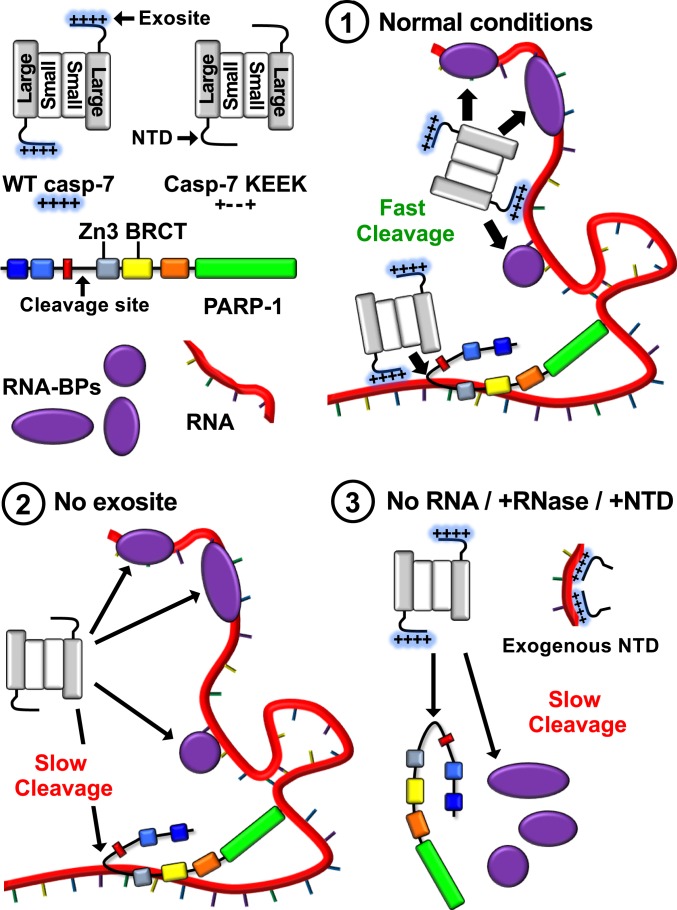
We initially assumed that the interaction between the exosite of caspase-7 and PARP-1 was of the protein-protein type, but we could not identify such direct interaction; instead, we found an interaction mediated by an intermediary RNA molecule. Fig. 6 presents a model based on our work: 1) During apoptosis, caspase-7, via a 4-lysine exosite located in its NTD, binds to RNA (or more generally to nucleic acids) on which several RNA-BPs are also attached; these include PARP-1 bound to the same RNA molecule as caspase-7 via its Zn3 and BRCT domains. The gathering of the peptidase and its substrates on RNA promotes proteolysis. 2) In the presence of a nonfunctional exosite, caspase-7 does not localize on RNA; thus, it is less exposed to nearby RNA-BPs, resulting in a slower cleavage of substrates. 3) Removing RNA using RNase or blocking binding sites on RNA using exogenous NTD also results in slower RNA-BP cleavage rates.
Fig. 6.
Schematic model of RNA-enhanced caspase-7 cleavage of RNA-BPs. See Discussion.
Germain et al. (27) identified PARylation as a modification that enhances PARP-1 cleavage and showed that caspase-7 binds PAR polymers. Because these polymers are negatively charged and the exosite of caspase-7 is of the opposite charge, PARylation was also assessed as a potential enhancer of proteolysis. Under our experimental conditions, no modification was apparent even when stimulated with DNA oligonucleotides or the addition of NAD+. Treatments with a PAR glycohydrolase before cleavage assays also did not affect cleavage. Finally, cleavage assays with truncated PARP-1 proteins lacking the catalytic domain (Cat) did not influence the proteolysis of PARP-1 by caspase-7. Because the modification of PARP-1 is essentially intramolecular (13), these results demonstrate that PARylation is not involved in interaction with the caspase-7 exosite under our experimental conditions. Additionally, it has been reported that PARylation of PARP-1 inhibits its RNA-binding properties, just as PARylation releases the enzyme from DNA strand breaks (28). Consequently, it is unlikely that PARP-1 was modified with PAR in our assays.
Degradation of RNA with RNase was the only condition we tested that recapitulated the loss of the exosite. Furthermore, because RNase treatment affected cleavage by caspase-7 but not by the exosite mutant or caspase-3, the inhibition cannot be the result of a structural rearrangement that renders the cleavage site less accessible; otherwise, this would have affected cleavage by any caspases. Thus, RNA is likely an intermediate between the exosite of caspase-7 and PARP-1. In hindsight, the inhibition of PARP-1 proteolysis by exogenous NTD and the steep drop in cleavage efficacy we observed (Fig. 1B) is well-explained by an RNA molecule bridging the caspase and its substrate. We suggest that in this experiment the NTD was titrating the free binding sites on RNA, and that the interaction worked until only a few RNA-binding sites remained. From these results, we conclude that the exosite and PARP-1 interact with the same RNA molecule to enhance the interaction and the ensuing cleavage. Although DNA played no part in the interaction we studied in vitro, we showed that caspase-7 can bind DNA. Thus, DNA may enhance the cleavage of some proteins in cells, especially if caspase-7 enters the nucleus during cell death or following the release of fragmented genomic DNA as part of the apoptotic process to act as a scaffold for protease-substrate rapprochement. However, current proteomic data (26) do not support a preference of caspase-7 for DNA-binding proteins.
In our study, we identified the Zn3 and BRCT domains of PARP-1 as crucial for indirect interaction with the exosite. It is already established that PARP-1 interacts with RNA (22, 23) via the Zn3 domain (24), thus supporting our hypothesis. In most studies, the BRCT domain was either omitted from experiments (25) or showed RNA binding in combination with the WGR and Cat domains (24). In one study (28), it was determined that the removal of specific domains had only limited or no effect on RNA binding, suggesting that PARP-1 uses at least 2 domains for RNA binding as our results suggest. Furthermore, in the same study the removal of the Zn3 domain resulted in the most considerable loss in RNA binding followed by Zn1 and Zn2 combined and BRCT alone, supporting the idea that the Zn3 and BRCT domains bind the RNA molecule. Because the Zn1 and Zn2 domains were previously identified as able to bind RNA in different studies, we tested them in cleavage assays with RNase but were unable to detect any effect on PARP-1 proteolysis. In addition, a protein with the Zn1 and Zn2 domains alone was cleaved at the same rate with or without RNase, again ruling them out as important for RNA binding. Finally, it was also shown that full-length PARP-1 binds preferentially to DNA (28) but prefers RNA in the absence of the Zn1 and Zn2 domains. This shows that the Zn3-Cat portion of PARP-1 contains domains that prefer RNA.
Using caspase-7 affinity columns and EMSA, we showed that caspase-7 is an RNA-BP and demonstrated that this requires the exosite. As the profile of bound RNA is similar to that of the input, it seems that no specific type of nucleic acid is preferred (e.g., ribosomal RNA or RNA of a specific size). Neither caspase-7 nor any other caspases had been identified as RNA-BP before this work. We can assume that the 4 positive lysine residues bind the negative phosphate backbone of RNA. Indeed, Castello et al. (23), who identified several mRNA-BPs, found that many of them have intrinsically disordered regions (IDRs) harboring many lysine residues. They proposed that the lysine patches bind the backbone of RNA similarly to some DNA-binding proteins with a basic IDR, and those characteristics are shared by the NTD of caspase-7 (9). Accordingly, Kreft and Nassal (29) demonstrated that the protein hRUL138 (human RNA-binding ubiquitin ligase of 138 kDa) binds RNA due to a lysine patch (K662KKTK), similar to the one found in caspase-7, and loses its RNA-binding ability upon mutation. Because of its simplicity and reliance solely on charges, we propose that the 4 lysine residues bind weakly using salt bridges to the phosphodiester backbone of nucleic acids without any preference for specific sequences. Similarly, hRUL138 does not show a significant preference for RNA sequences or structures (29). Furthermore, there is no necessity for caspase-7 to bind any specific RNA sequence because it is not involved in any particular RNA pathway, and the indiscriminate cleavage of RNA-BPs likely achieves the same goal as would selective targeting. Although beyond the scope of this study, in-depth biophysical characterization of the interaction between caspase-7 and nucleic acid will undoubtedly shed light on any sequence specificity, minimal or maximal nucleic acid length, structure, and affinity.
Although uncommon, the binding of nucleic acids by proteases is not unique to caspase-7; however, to our knowledge only a few human proteases bind RNA and DNA. Of those, neutrophil elastase and cathepsin G can bind nucleic acids, and their high affinity for DNA leads to their localization on neutrophil extracellular traps (NETs) which probably favors the cleavage of proteins from pathogens caught in those NETs (30). Closer in function to caspases, granzyme A and granzyme B, which can also induce cell death in a caspase-dependent (granzyme B) or independent (granzyme A) manner, bind RNA and DNA with nanomolar affinity and cleave RNA-BPs faster in the presence of nucleic acids (30). Our findings expand the body of evidence for the existence of regulatory sites on caspases, including by metallic ions (31) and small molecules like the nucleotide ATP (32). With our findings, these findings also increase the potential for design of caspase modulators targeting regions other than the canonical catalytic site and maybe achieve better selectivity.
A more distant analogy to our findings is thrombin and its inhibitory substrate antithrombin, which interact using the polysaccharide polymer heparin (33). Both antithrombin and thrombin bind to heparin, with the latter using a basic exosite. The interaction of both protease and substrate is enhanced when the 2 are bound to the same heparin molecule, bringing together the protease and its target to facilitate proteolysis. Thus, the binding of polymers seems an effective way of enhancing specific cleavage events in different biological processes.
Finally, all of the RNA-binding substrates tested so far have reduced cleavage rates upon RNA degradation, validating that the mechanism we uncovered for PARP-1 proteolysis may broadly apply to RNA-BPs. This further shows how the multiplication of caspases in higher animals has led to their specialization, even among 2 executioner caspases with highly similar primary structure preference in their substrates.
Materials and Methods
Enzymatic assays were performed using cellular extracts from FLAG-tagged PARP-1 transfected AD-293 cells (caspase-7 knockout using CRISPR/Cas9) and purified active site–titrated recombinant caspases. Data from unsaturated immunoblotting images acquired using an imaging system (VersaDoc 4000 MP) were used to determine proteolysis rates (k) using pseudofirst-order kinetics. Data for caspase substrates and relative cleavage rates determined by Agard et al. (26) were reanalyzed to compare the number of RNA-binding substrates for each caspase based on their Gene Ontology molecular functions listed in the UniProtKB database. A detailed description of materials and methods can be found in the SI Appendix.
Supplementary Material
Acknowledgments
We thank G. Poirier (Université Laval, Québec, QC) for the PARP-1 cDNA, J. Lafontaine (Université de Sherbrooke) for the caspase-7 KO cells, and R. Graham for the caspase-6 antibody. A.D. holds scholarships from Fonds de recherche du Québec-Santé (FRQS) and the Natural Science and Engineering Research Council of Canada (NSERC). This work was supported by an NSERC Discovery Grant (no. 2017-05988) to J.-B.D.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909283116/-/DCSupplemental.
References
- 1.Rawlings N. D., Morton F. R., Barrett A. J., MEROPS: The peptidase database. Nucleic Acids Res. 34, D270–D272 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guicciardi M. E., Gores G. J., Life and death by death receptors. FASEB J. 23, 1625–1637 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tait S. W., Green D. R., Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Dix M. M., Simon G. M., Cravatt B. F., Global identification of caspase substrates using PROTOMAP (protein topography and migration analysis platform). Methods Mol. Biol. 1133, 61–70 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Crawford E. D., et al. , The DegraBase: A database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thornberry N. A., et al. , A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S., Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 350, 563–568 (2000). [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher D., Blais V., Denault J. B., Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc. Natl. Acad. Sci. U.S.A. 109, 5669–5674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martini C., Bédard M., Lavigne P., Denault J. B., Characterization of Hsp90 co-chaperone p23 cleavage by caspase-7 uncovers a peptidase-substrate interaction involving intrinsically disordered regions. Biochemistry 56, 5099–5111 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Hill M. E., et al. , Reprogramming caspase-7 specificity by regio-specific mutations and selection provides alternate solutions for substrate recognition. ACS Chem. Biol. 11, 1603–1612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacPherson D. J., Mills C. L., Ondrechen M. J., Hardy J. A., Tri-arginine exosite patch of caspase-6 recruits substrates for hydrolysis. J. Biol. Chem. 294, 71–88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnakumar R., Kraus W. L., The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eustermann S., et al. , Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell 60, 742–754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langelier M. F., Planck J. L., Roy S., Pascal J. M., Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagné J. P., et al. , Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair (Amst.) 30, 68–79 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann S. H., Desnoyers S., Ottaviano Y., Davidson N. E., Poirier G. G., Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 53, 3976–3985 (1993). [PubMed] [Google Scholar]
- 17.Andrabi S. A., et al. , Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 10209–10214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denault J. B., Salvesen G. S., Human caspase-7 activity and regulation by its N-terminal peptide. J. Biol. Chem. 278, 34042–34050 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Takahashi R., et al. , A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 273, 7787–7790 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Deveraux Q. L., et al. , Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 18, 5242–5251 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masson M., et al. , XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18, 3563–3571 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltz A. G., et al. , The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Castello A., et al. , Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Guetg C., Scheifele F., Rosenthal F., Hottiger M. O., Santoro R., Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 45, 790–800 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Huambachano O., Herrera F., Rancourt A., Satoh M. S., Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 286, 7149–7160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agard N. J., et al. , Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc. Natl. Acad. Sci. U.S.A. 109, 1913–1918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Germain M., et al. , Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem. 274, 28379–28384 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Melikishvili M., Chariker J. H., Rouchka E. C., Fondufe-Mittendorf Y. N., Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov. 3, 17043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreft S. G., Nassal M., hRUL138, a novel human RNA-binding RING-H2 ubiquitin-protein ligase. J. Cell Sci. 116, 605–616 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Thomas M. P., et al. , Leukocyte protease binding to nucleic acids promotes nuclear localization and cleavage of nucleic acid binding proteins. J. Immunol. 192, 5390–5397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eron S. J., MacPherson D. J., Dagbay K. B., Hardy J. A., Multiple mechanisms of zinc-mediated inhibition for the apoptotic caspases-3, -6, -7, and -8. ACS Chem. Biol. 13, 1279–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okerberg E. S., et al. , Chemoproteomics using nucleotide acyl phosphates reveals an ATP binding site at the dimer interface of procaspase-6. Biochemistry, 10.1021/acs.biochem.9b00290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huntington J. A., Baglin T. P., Targeting thrombin–rational drug design from natural mechanisms. Trends Pharmacol. Sci. 24, 589–595 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.