Significance
The excitatory amino acid transporter 2 (EAAT2) is the predominant glutamate transporter in the mammalian brain, and its expression is reduced in aging and sporadic Alzheimer’s disease (AD) brains. The mechanisms through which reduced EAAT2 expression contributes to cognitive decline and AD-related neurodegeneration and the differential functional roles of its astrocytic and neuronal components remain largely unknown. This study uses behavioral analysis to show that astrocytic and neuronal EAAT2 deficiency in mice causes age-dependent cognitive dysfunction. Furthermore, we show that gene expression from human AD and aging datasets have a robust overlap with astrocytic EAAT2 deficiency. This study highlights the distinct signaling mechanisms mediated by neuronal and astrocytic EAAT2 and EAAT2 as a potential therapeutic target in aging and AD.
Keywords: Alzheimer’s disease, glutamate transporter, aging
Abstract
The excitatory amino acid transporter 2 (EAAT2) is the major glutamate transporter in the brain expressed predominantly in astrocytes and at low levels in neurons and axonal terminals. EAAT2 expression is reduced in aging and sporadic Alzheimer’s disease (AD) patients’ brains. The role EAAT2 plays in cognitive aging and its associated mechanisms remains largely unknown. Here, we show that conditional deletion of astrocytic and neuronal EAAT2 results in age-related cognitive deficits. Astrocytic, but not neuronal EAAT2, deletion leads to early deficits in short-term memory and in spatial reference learning and long-term memory. Neuronal EAAT2 loss results in late-onset spatial reference long-term memory deficit. Neuronal EAAT2 deletion leads to dysregulation of the kynurenine pathway, and astrocytic EAAT2 deficiency results in dysfunction of innate and adaptive immune pathways, which correlate with cognitive decline. Astrocytic EAAT2 deficiency also shows transcriptomic overlaps with human aging and AD. Overall, the present study shows that in addition to the widely recognized astrocytic EAAT2, neuronal EAAT2 plays a role in hippocampus-dependent memory. Furthermore, the gene expression profiles associated with astrocytic and neuronal EAAT2 deletion are substantially different, with the former associated with inflammation and synaptic function similar to changes observed in human AD and gene expression changes associated with inflammation similar to the aging human brain.
Aging is associated with cognitive decline in humans, nonhuman primates, and rodents (1–5). Aging is the major risk factor for development of Alzheimer’s disease (AD), and the prevalence of AD is expected to rise with increasing life expectancy (6). The same glutamatergic connections in the neocortex and hippocampus are affected in aging and AD (7, 8). Even though the same neural circuits are affected, in aging, synaptic changes occur with minimal neuronal death, whereas in AD, there is frank loss of neurons (8, 9). There is a compelling need to understand the mechanisms regulating the selective vulnerability of glutamatergic neurons in aging and AD to develop novel targets and more effective treatments.
Glutamate, the major excitatory neurotransmitter in the mammalian central nervous system (CNS), plays critical roles in neural functions, including cognition (10). Dysregulation of glutamate homeostasis and its spillover into extrasynaptic sites can cause long-term depression (LTD), excitotoxicity, and neuronal degeneration (10–13). The excitatory amino acid transporter 2 (EAAT2) is the predominant glutamate transporter in the mammalian brain (14, 15). Approximately 80 to 90% of EAAT2 protein is localized on astrocytes, and 5 to 10% is localized in the axonal terminals of neurons (16). Decreased EAAT2 expression is found in aging (17–19) and sporadic AD patients’ brains (20–23) and is associated with indices of neuronal death in postmortem AD brains (20). A number of studies have demonstrated a critical role for EAAT2 in neurodegeneration and synaptic plasticity. EAAT2 has been shown to associate with presenilin 1 in neurons and astrocytes and has important implications in maintenance of glutamate homeostasis and amyloid β (Aβ) pathology in AD (24). Aβ has been shown to prolong the extracellular lifetime of synaptically released glutamate by reducing surface expression in astrocytes (25), and soluble Aβ oligomers perturb synaptic plasticity by altering glutamate recycling at the synapse and promoting LTD (26). A recent study also showed that Aβ-dependent neuronal hyperactivity is initiated by the suppression of glutamate reuptake (27). In human brains, activated forms of astrocytes with increased EAAT2 expression may exert beneficial roles in preserving cognitive function, even in the presence of Aβ and neurofibrillary tangles (NFTs) (28). These findings indicate that glutamate transporters and, in particular, EAAT2 play a critical role in synaptic health and the pathogenesis of AD.
The mechanisms through which EAAT2 deficiency might contribute to age-related cognitive decline, AD-related neurodegeneration, and the differential functional roles of its astrocytic and neuronal components remain largely unknown. The present study employs behavioral analysis and RNA sequencing (RNA-Seq) to analyze cognitive dysfunction and identify transcriptome-wide alterations in the hippocampus resulting from the loss of astrocytic and neuronal EAAT2.
Results
Astrocytic EAAT2 Deficiency Results in Accelerated Age-Related Cognitive Decline.
The EAAT2 gene was constitutively deleted by synapsin I promoter-driven Cre expression in neurons (syn-Cre+ EAAT2−/−) and through the tamoxifen-inducible human GFAP/Cre-ERT2 driver in astrocytes (29). GFAP/Cre-ERT2 EAAT2Δ/+ mice (gfap-Cre+ EAAT2Δ/+) were used for all aging experiments as homozygous knockouts exhibit seizures and increased morbidity (15, 29–32), which could confound findings. The gfap-Cre+ EAAT2Δ/+ mice were injected with 4-hydroxy tamoxifen at 10 mo of age to induce conditional deletion of EAAT2. As a baseline cognitive measure, at 10 mo of age (before 4-hydroxytamoxifen injections), both syn-Cre+ EAAT2−/− and gfap-Cre+ EAAT2+/+ mice were subjected to a 2-trial spatial reference memory task in a Y-maze. Mice were tested again in a Y-maze task at 13 mo of age and in a Morris water maze (MWM) task for spatial reference learning and memory at 17 mo of age. EAAT2 deletion was quantified by immunoblotting on plasma membrane vesicles (PMVs) from the prefrontal cortex tissue of study mice after behavioral testing at ∼18 mo of age. The gfap-Cre+ EAAT2Δ/+ mice displayed a significant 22% reduction in EAAT2 expression, and the syn-Cre+ EAAT2−/− mice displayed a nonsignificant 14% reduction in EAAT2 expression, as previously reported (16, 29, 33), presumably because of the predominance of astrocytic EAAT2 (SI Appendix, Fig. S1).
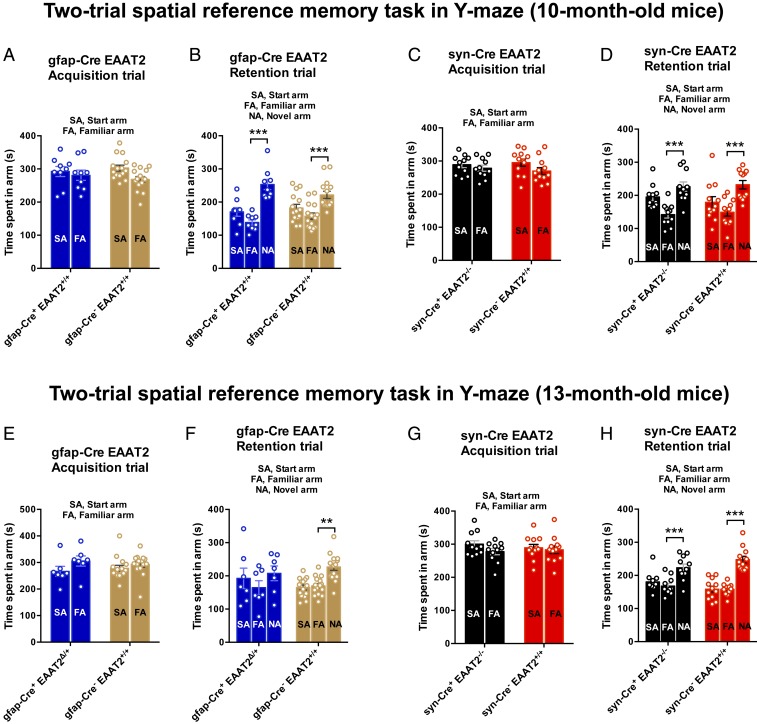
At 10 mo of age, syn-Cre+ EAAT2−/− and gfap-Cre+ EAAT2+/+ mice displayed no statistically significant differences in the spatial reference memory performance in the Y-maze (Fig. 1 A–D). In the acquisition trial, gfap-Cre+ EAAT2+/+ and gfap-Cre− EAAT2+/+ mice spent an equal amount of time in the start arm (SA) and familiar arm (FA) [Fig. 1A; F(1,44) = 0.9685, P = 0.3304]. In the retention trial, after a 1-h intertrial interval, gfap-Cre+ EAAT2+/+ and gfap-Cre− EAAT2+/+ mice spent a significantly longer time in the novel arm (NA) as compared with the FA (Fig. 1B; P < 0.0001 and P = 0.0002, respectively, Bonferroni’s post hoc test), suggesting an absence of any spatial memory deficit. The syn-Cre+ EAAT2−/− mice did not show any spatial memory deficit at 10 mo of age [Fig. 1 C and D; F(1,42) = 0.5658, P = 0.4561], as previously reported (34). Astrocytic EAAT2 deletion was induced in 10-mo-old gfap-Cre+ EAAT2+/+ mice with 4-hydroxy tamoxifen (intraperitoneally [i.p.]). The gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− mice were tested in the Y-maze at 13 mo of age. Remarkably, at 13 mo of age, gfap-Cre+ EAAT2Δ/+ mice showed memory impairment in the Y-maze task [Fig. 1F; F(1,40) = 0.6938, P = 0.4098 in the acquisition trial; P = 0.4823, P = 0.0062 in the retention trial]; however, the syn-Cre+ EAAT2−/− mice still did not show any memory deficit as compared with controls [Fig. 1H; F(1,42) = 0.6239, P = 0.4340 in the acquisition trial; P = 0.0003, P < 0.0001 in the retention trial]. The gfap-Cre+ control mice without the floxed EAAT2 allele did not display memory impairment in the Y-maze task at 13 mo of age (SI Appendix, Fig. S3B).
Fig. 1.
Conditional heterozygous astrocytic EAAT2 knockout mice exhibit accelerated spatial reference memory deficits. (A) In the acquisition trial of Y-maze evaluation at 10 mo of age, both gfap-Cre+ EAAT2+/+ and gfap-Cre− EAAT2+/+ mice explored the SA and FA equally. F(1,44) = 0.9685, P = 0.3304. (B) In the retention trial, both gfap-Cre+ EAAT2+/+ and gfap-Cre− EAAT2+/+ mice spent more time exploring the NA than the FA, suggesting an absence of any spatial reference memory deficit at baseline (prior to induction of conditional astrocytic EAAT2 knockout). P < 0.0001 and P = 0.0002, respectively. (C) In the acquisition trial of the Y-maze task at 10 mo of age, both syn-Cre+ EAAT2−/− and syn-Cre− EAAT2+/+ mice spent equal time in the SA and FA. F(1,42) = 0.5658, P = 0.4561. (D) In the retention trial, both syn-Cre+ EAAT2−/− and syn-Cre− EAAT2+/+ mice explored the NA significantly more than the FA, indicating a lack of spatial reference memory deficit in syn-Cre+ EAAT2−/− mice at 10 mo of age. (E) In the Y-maze task acquisition trial at 13 mo of age, both gfap-Cre+ EAAT2Δ/+ and gfap-Cre− EAAT2+/+ mice explored the SA and FA equally. F(1,40) = 0.6938, P = 0.4098. (F) In the retention trial (intertrial interval of 1 h), the gfap-Cre+ EAAT2Δ/+ mice failed to show a statistically significant preference for the NA, suggesting impairment in spatial reference memory in these mice. The gfap-Cre− EAAT2+/+ control mice explored the NA more than the FA, as expected. P = 0.4823 and P = 0.0062, respectively. (G) In the acquisition trial of the Y-maze task at 13 mo of age, both syn-Cre+ EAAT2−/− and syn-Cre− EAAT2+/+ mice spent equal time in the SA and FA. F(1,42) = 0.6239, P = 0.4340. (H) In the retention trial, both syn-Cre+ EAAT2−/− and syn-Cre− EAAT2+/+ mice spent more time exploring the NA than the FA, suggesting an absence of any spatial reference memory deficit in these mice at 13 mo of age. P = 0.0003 and P < 0.0001, respectively. The data in all panels are presented as mean ± SEM. (A–D ) Y-maze data for 10-mo-old mice are based on gfap-Cre+ EAAT2Δ/+, n = 9; gfap-Cre− EAAT2+/+, n = 15; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 mice. (E–H) Y-maze data for 13-mo-old mice are based on gfap-Cre+ EAAT2Δ/+, n = 7; gfap-Cre− EAAT2+/+, n = 15; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 mice.
Neuronal and Astrocytic EAAT2 Deficiency Results in Impairment of Spatial Reference Learning.
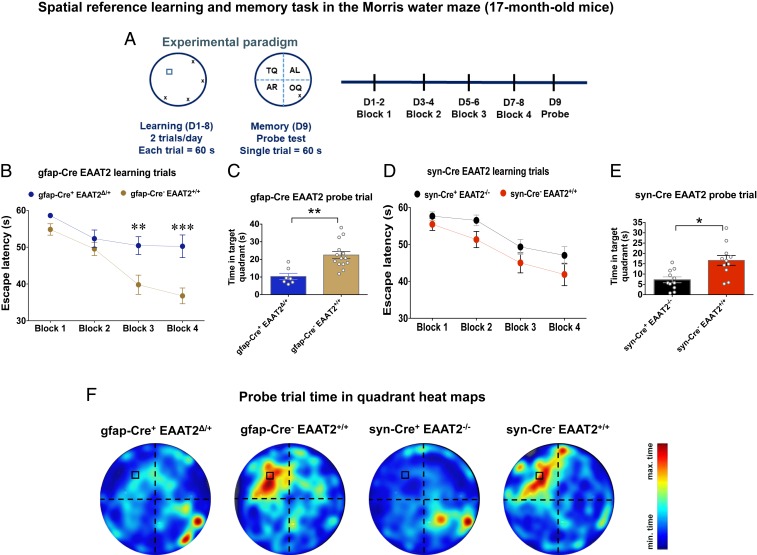
At 17 mo of age, mice were tested for spatial reference learning and memory using the MWM task (Fig. 2A). The gfap-Cre+ EAAT2Δ/+ mice displayed significantly longer latency to locate the submerged platform on the third and fourth blocks of training as compared with controls (Fig. 2B; P = 0.0096 for block 3 and P = 0.0005 for block 4, Sidak’s post hoc test). In contrast, syn-Cre+ EAAT2−/− mice did not show a statistically significant difference in spatial reference learning as compared with controls [Fig. 2D; F(3,80) = 0.2117, P = 0.8880, repeated measures 2-way ANOVA]. The study groups did not differ significantly in the latency(ies) to find the visible platform (P = 0.5268, Kruskal–Wallis test followed by Dunn’s multiple comparison test) (SI Appendix, Fig. S3). The gfap-Cre+ and control mice without the floxed EAAT2 allele did not display any significant differences in spatial reference learning at 17 mo of age (SI Appendix, Fig. S3C). These data showed impairment of spatial reference learning only in gfap-Cre+ EAAT2Δ/+ mice.
Fig. 2.
Conditional astrocytic EAAT2 knockout mice exhibit spatial reference learning deficits, and both astrocytic and neuronal EAAT2 knockout mice exhibit spatial reference memory deficits at 17 mo. (A) Experimental paradigm of the spatial reference learning and memory task in the MWM performed by mice at 17 mo of age. D, day. (B) Spatial learning curves of gfap-Cre EAAT2 mice in the MWM. The escape latency during the training phase (blocks 1 to 4, D1 to D8) of gfap-Cre EAAT2 mice is shown. Compared with gfap-Cre− EAAT2+/+ mice, the gfap-Cre+ EAAT2Δ/+ mice exhibited a significant delay in finding the hidden escape platform in blocks 3 and 4 (D5 to D8) of the learning trials. P = 0.0096 for block 3 and P = 0.0005 for block 4, Sidak’s post hoc test. (C) Spatial reference memory evaluation of gfap-Cre EAAT2 mice in the MWM. In the probe trial (D9, 24-h retention trial), gfap-Cre+ EAAT2Δ/+ mice spent significantly less time in the TQ as compared with the gfap-Cre− EAAT2+/+ control mice. P = 0.0019. (D) Spatial learning curves of syn-Cre EAAT2 mice in the MWM. The escape latency during the learning trials (blocks 1 to 4, D1 to D8) did not differ significantly between syn-Cre+ EAAT2−/− and syn-Cre− EAAT2+/+ mice. F(3,80) = 0.2117, P = 0.8880. (E) Spatial reference memory assessment of syn-Cre EAAT2 mice in the MWM. The syn-Cre+ EAAT2−/− mice spent significantly less time in the TQ as compared with syn-Cre− EAAT2+/+ mice in the probe trial (D9, 24-h retention trial). P = 0.0107. (F) Heatmaps of search intensity during probe trials conducted on D9. A high dwell time across the MWM pool area is indicated by colors close to red, whereas colors close to blue indicate a lower dwell time. Compared with control groups, both gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− mice did not show a preference for the TQ (where the escape platform was located during learning trials). max, maximum; min, minimum. The data in all panels are presented as mean ± SEM. The MWM data for 17-mo-old mice are based on gfap-Cre+ EAAT2Δ/+, n = 7; gfap-Cre− EAAT2+/+, n = 13; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 mice. *P < 0.05, **P < 0.01, ***P < 0.001.
However, during the probe trial on the ninth day, both gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− mice spent significantly less time in the target quadrant (TQ; platform location during training trials) as compared with controls (Fig. 2 C and E, respectively; P = 0.0019 and P = 0.0107, respectively), indicating age-related cognitive dysfunction with loss of astrocytic and neuronal EAAT2. Additionally, both gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− mice exhibited a preference for the opposite quadrant (OQ; opposite to TQ) as compared with controls (Fig. 2F; P = 0.0294 and P = 0.0009), suggesting impairment of spatial reference memory.
Astrocytic EAAT2 Deficiency Activates Innate and Adaptive Immune Pathways in the Hippocampus.
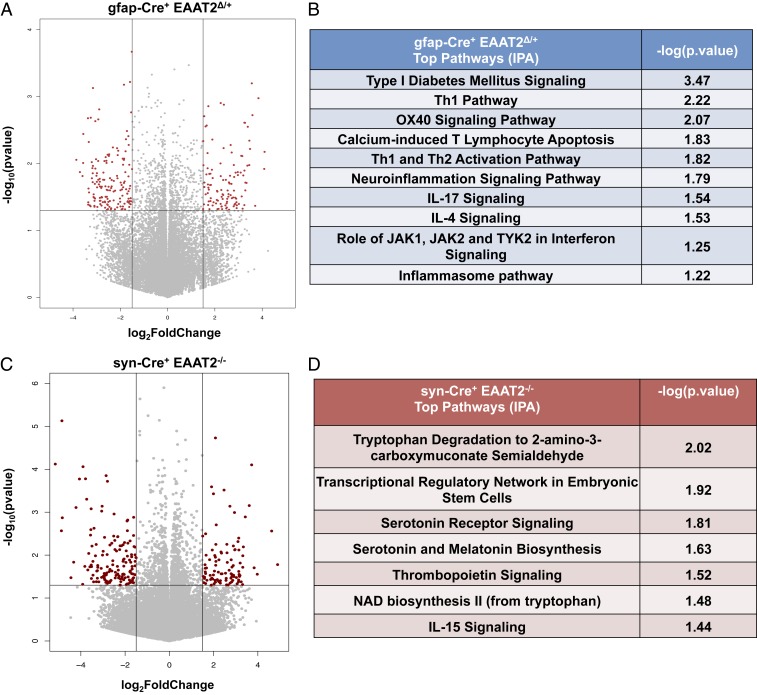
RNA-Seq and differential gene expression analysis were performed on hippocampal RNA of 18-mo-old mice. Astrocytic EAAT2 deficiency (gfap-Cre+ EAAT2Δ/+) altered the expression of 268 genes (P < 0.05, |fold-change| ≥ 1.5) (Fig. 3A), and neuronal EAAT2 deletion (syn-Cre+ EAAT2−/−) altered 246 genes (P < 0.05, |fold-change| ≥ 1.5) (Fig. 3C) compared with Cre− controls. Top pathways identified from differentially expressed genes (DEGs) (Ingenuity Pathway Analysis [IPA] tool; Qiagen) resulting from astrocytic EAAT2 deficiency were related to the immune system, including type I diabetes mellitus signaling, T helper 1 (Th1) and Th2 activation pathways, the neuroinflammation signaling pathway, the interleukin-4 (IL-4) signaling pathway, the IL-17 signaling pathway, and the inflammasome pathway (Fig. 3B). Recent evidence, including human genetics (35), strongly points to neuroinflammation and immune system activation in the early stages of AD pathology, which may also contribute to disease progression (36, 37).
Fig. 3.
Conditional heterozygous astrocytic EAAT2 knockout mice have dysregulated immune signaling, and constitutive neuronal EAAT2 knockout mice have dysregulated tryptophan metabolism in the hippocampus. (A) Volcano plot depicting DEGs in gfap-Cre+ EAAT2Δ/+ hippocampus. Horizontal and vertical lines represent filtering criteria (P < 0.05, |fold-change| > 1.5). Red dots represent transcripts expressed at significantly higher or lower levels compared with controls. (B) Canonical pathways derived from IPA gene ontology algorithms for significantly dysregulated genes in gfap-Cre+ EAAT2Δ/+ hippocampus. The table shows several innate and adaptive immune pathways that map to the canonical pathways from the IPA knowledgebase and P values generated from Fisher’s exact test. (C) Volcano plot depicting DEGs in syn-Cre+ EAAT2−/− hippocampus. Horizontal and vertical lines represent filtering criteria (P < 0.05, |fold-change| > 1.5). Red dots represent transcripts expressed at significantly higher or lower levels compared with controls. (D) Canonical pathways derived from IPA gene ontology algorithms for significantly dysregulated genes in syn-Cre+ EAAT2−/− hippocampus. The table shows tryptophan metabolism and kynurenine pathways that map to the canonical pathways from the IPA knowledgebase and P values generated from Fisher’s exact test. The data in this figure are based on gfap-Cre+ EAAT2Δ/+, n = 6; gfap-Cre− EAAT2+/+, n = 6; syn-Cre+ EAAT2−/−, n = 6; and syn-Cre− EAAT2+/+, n = 6.
Pathway analysis (IPA) on DEGs from loss of neuronal EAAT2 identified top pathways, including tryptophan degradation, the transcriptional regulatory network in embryonic stem cells, serotonin receptor signaling, serotonin and melatonin biosynthesis, nicotinamide adenine dinucleotide (NAD) biosynthesis, and IL-15 signaling (Fig. 3D). Serotonin is a neuromodulator that regulates glutamate transmission and suppresses long-term potentiation (LTP) in the hippocampus (38). Tryptophan, the precursor to biosynthesis of serotonin and melatonin, is metabolized through the kynurenine pathway (39).
These data therefore show that age-associated astrocytic EAAT2 deletion results in dysregulation of innate and adaptive immune response pathways, whereas age-associated neuronal EAAT2 deletion results in the disruption of tryptophan metabolism along the kynurenine pathway.
Expression of Inflammatory Genes Negatively Correlates with Behavior in Mice with Astrocytic EAAT2 Deficiency.
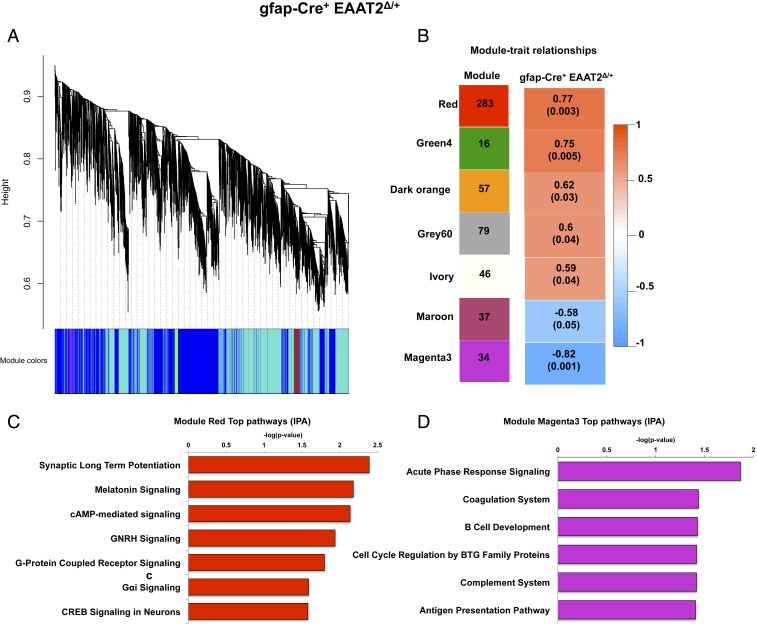
We then performed a weighted gene coexpression network analysis (WGCNA) (40) on the DEGs from gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− animals to identify the main gene coexpression modules that are disrupted from astrocytic and neuronal EAAT2 deficiency and investigate how groups of coexpressed genes were related to cognitive deficits observed in the MWM task at 17 mo (Fig. 4 A and B)
Fig. 4.
Dysregulation of inflammatory pathways in the hippocampus negatively correlates with cognitive dysfunction in astrocytic EAAT2 knockout mice by WGCNA. (A) Dendrogram of average hierarchical gene clustering from gfap-Cre+ EAAT2Δ/+ mice. The y axis corresponds to the distance determined by the extent of topological overlap, and significant branch points represent modules. The first band below the x axis shows automatically determined modules from correlation of gene expression without phenotypic considerations. (B) Module-cognitive dysfunction correlations in gfap-Cre+ EAAT2Δ/+ mice. (Left) Seven modules significantly correlated with MWM behavior and the number of their member genes. (Right) Color scale for module trait correlation from −1 to 1 with P values in parentheses. (C) Canonical pathways derived from IPA gene ontology algorithms for genes showing enrichment of pathways regulating synaptic plasticity in the module red. cAMP, adenosine 3′,5′-cyclic monophosphate; GNRH, gonadotropin releasing hormone. (D) Canonical pathways derived from IPA gene ontology algorithms for genes showing enrichment of immune modulatory pathways in the module magenta3. The data in this figure are based on gfap-Cre+ EAAT2Δ/+, n = 6; gfap-Cre− EAAT2+/+, n = 6; syn-Cre+ EAAT2−/−, n = 6; and syn-Cre− EAAT2+/+, n = 6.
We constructed a coexpression network of DEGs in Cre+ mice compared with Cre− mice, and the expression levels of each module were summarized by the first principal component and related to time spent by animals in the TQ in the probe trial (MWM). The eigengenes of 7 modules showed significant correlations (P ≤ 0.05) with age-related cognitive dysfunction in gfap-Cre+ EAAT2Δ/+ mice (Fig. 4C). Interestingly, the red module was significantly positively correlated with mouse behavior (r2 = 0.77, P = 0.003) (Fig. 4C), and was enriched for pathways regulating synaptic plasticity, including synaptic LTP, G protein-coupled receptor signaling, and CREB signaling (Fig. 4D).
Importantly, the magenta3 module was significantly negatively correlated with behavior (r2 = −0.82, P = 0.001) (Fig. 4C), and pathway analysis of genes in this module revealed enrichment in several immune function-related pathways, including acute-phase response signaling and the complement system (Fig. 4D).
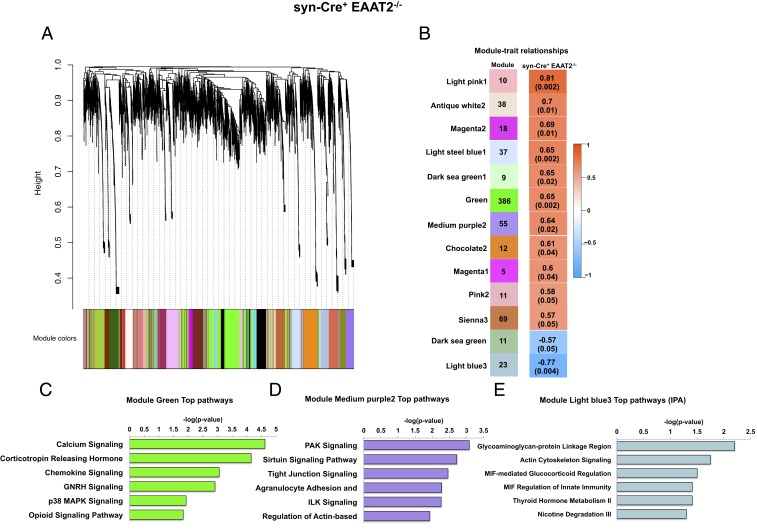
Thirteen eigengene modules were significantly correlated to cognitive dysfunction in syn-Cre+ EAAT2−/− mice (Fig. 5A). The green module (r2 = 0.65, P = 0.002) and the medium purple2 module (r2 = 0.64, P = 0.02) were positively significantly correlated with mouse behavior (Fig. 5B) and were enriched for several signaling pathways, including calcium signaling, mitogen-activated protein kinase signaling, and the sirtuin signaling pathway (Fig. 5 C and D, respectively). The light blue3 module was significantly negatively correlated with mouse behavior (r2 = −0.77, P = 0.004) (Fig. 5B), and was enriched for pathways, including actin/cytoskeleton signaling and the MIF-mediated glucocorticoid response (Fig. 5E).
Fig. 5.
Dysregulation of multiple signaling pathways in the hippocampus negatively correlates with cognitive dysfunction in neuronal EAAT2 knockout mice by WGCNA. (A) Dendrogram of average hierarchical gene clustering from syn-Cre+ EAAT2−/− mice. The y axis corresponds to the distance determined by the extent of topological overlap, and significant branch points represent modules. The first band below the x axis shows automatically determined modules from correlation of gene expression without phenotypic considerations. (B) Module-cognitive dysfunction correlations in syn-Cre+ EAAT2−/− mice. (Left) Thirteen modules significantly correlated with MWM behavior and the number of their member genes. (Right) Color scale for module trait correlation from −1 to 1 with P values in parentheses. (C) Canonical pathways derived from IPA gene ontology algorithms for genes in the module green. GNRH, gonadotropin releasing hormone. (D) Canonical pathways derived from IPA gene ontology algorithms for genes showing enrichment of pathways regulating synaptic plasticity in the module purple2. (E) Canonical pathways derived from IPA gene ontology algorithms for genes showing enrichment of pathways regulating metabolism in the module light blue3.
WGCNA analysis suggests that partial loss of astrocytic EAAT2 rather than neuronal EAAT2 results in gene expression/phenotype relationships more closely resembling aging and AD, with significant associations with synaptic plasticity and immune-related genes.
Gene Expression Profile from Astrocytic EAAT2-Deficient Hippocampus Overlaps with Expression Profiles in Human AD and Aging.
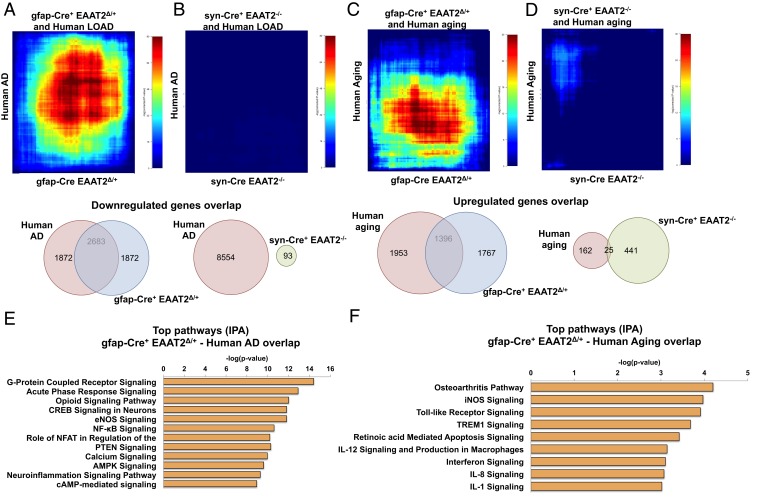
Next, we evaluated the extent of overlap between the gene expression data from our study mice and previously published human AD and aging datasets using rank-rank hypergeometric overlap (RRHO) (41) analysis to identify patterns of genome-wide expression overlap with the mouse data in a threshold-free manner. To generate relevant genes lists for comparison (hippocampus data from male brains), we performed data mining of GSE48350 from the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48350) to generate lists of DEGs in human AD and aging. We identified an overlap [-log (p.value) = 80] between the gfap-Cre+ EAAT2Δ/+ and the human AD gene list (Fig. 6A) and between the gfap-Cre+ EAAT2Δ/+ and the human aging gene list [-log (p.value) = 30] (Fig. 6C).
Fig. 6.
Transcriptome from conditional loss of astrocytic, not neuronal, EAAT2 overlaps with human AD and aging datasets in RRHO comparisons. (A) RRHO heatmap showing overlap between human AD and gfap-Cre+ EAAT2Δ/+ datasets. A map signal scale of log10-transformed hypergeometric P values is shown to the right of the heatmap. A Venn diagram below the heatmap represents the number of overlapping genes between GFAP/Cre and human AD datasets. LOAD, late-onset Alzheimer’s disease. (B) RRHO heatmap showing no overlap between human AD and syn-Cre+ EAAT2−/− datasets. A map signal scale of log10-transformed hypergeometric P values is shown to the right of the heatmap. A Venn diagram below the heatmap represents the number of overlapping genes between GFAP/Cre and human AD datasets. (C) RRHO heatmap showing overlap between human aging and gfap-Cre+ EAAT2Δ/+ datasets. A map signal scale of log10-transformed hypergeometric P values is shown to the right of the heatmap. A Venn diagram below the heatmap represents the number of overlapping genes between GFAP/Cre and human aging datasets. (D) RRHO heatmap showing minimal overlap between human aging and syn-Cre+ EAAT2−/− datasets. A map signal scale of log10-transformed hypergeometric P values is shown to the right of the heatmap. A Venn diagram below the heatmap represents the number of overlapping genes between Syn/Cre and human aging datasets. (E) Canonical pathways derived from IPA gene ontology algorithms of most codysregulated genes in gfap-Cre+ EAAT2Δ/+ versus human AD in the hippocampus. Pathways show enrichment of genes regulating neuroplasticity. AMPK, 5’ adenosine monophosphate-activated protein kinase; cAMP, adenosine 3′,5′-cyclic monophosphate; eNOS, endothelial nitric oxide synthase; NFAT, nuclear factor of activated T-cells; NF-κB, nuclear factor-κB. (F) Canonical pathways derived from IPA gene ontology algorithms of most codysregulated genes in gfap-Cre+ EAAT2Δ/+ versus human aging in the hippocampus. Pathways show enrichment of genes regulating inflammation. iNOS, inducible Nitric oxide synthase.
In contrast, there was little or no overlap between syn-Cre+ EAAT2−/− and human AD or human aging gene lists (Fig. 6 B and D, respectively). Top pathways identified from gene overlap of astrocytic partial loss of EAAT2 with human AD included pathways that have been implicated in the inflammatory response in AD (acute-phase response signaling, nuclear factor-κB signaling, and neuroinflammation signaling) and pathways that regulate synaptic function (opioid signaling pathway, CREB signaling pathway, and calcium signaling pathway) (42–44) (Fig. 6E). Pathway analysis of gene overlap with the human aging gene list included several inflammatory pathways (osteoarthritis pathway, Toll-like receptor signaling, TREM1 signaling, interferon [IFN] signaling, IL-12 signaling, and IL-1 signaling) (Fig. 6F).
RRHO analysis showed that partial loss of astrocytic EAAT2 drives gene expression changes in the hippocampus associated with inflammation and synaptic function, similar to human AD, and gene expression changes in the hippocampus associated with inflammation similar to the aging human brain. Furthermore, it points out that transcriptomic changes from astrocytic rather than neuronal EAAT2 deficiency in the hippocampus are more closely associated with AD and aging gene expression profiles.
Discussion
We report that astrocytic EAAT2 deficiency causes accelerated age-related cognitive dysfunction at 13 mo of age and that both astrocytic (gfap-Cre+ EAAT2Δ/+) and neuronal (syn-Cre+ EAAT2−/−) EAAT2-deficient mice exhibit age-dependent cognitive dysfunction at 17 mo of age. This study identifies cognitive deficits in mice lacking neuronal EAAT2. Furthermore, transcriptomic analysis of the hippocampus revealed dysregulation of several innate and adaptive immune response pathways in gfap-Cre+ EAAT2Δ/+ mice, whereas there was dysregulation of the kynurenine pathway in syn-Cre+ EAAT2−/− mice. Using an unbiased approach (WCGNA), we also identified a significant negative correlation between expression of inflammatory genes in the hippocampus and a hippocampus-dependent memory task in gfap-Cre+ EAAT2Δ/+ mice. Furthermore, we found that transcriptomic data from human AD and aging datasets have a robust overlap with our gfap-Cre+ EAAT2Δ/+ gene profile, but not with our syn-Cre+ EAAT2−/− gene expression profile.
Glutamate transport plays an essential role in LTP (45), a major phenomenon underlying synaptic plasticity, which has been proposed to be the cellular basis of learning and memory (46). An increase in glutamate transport via EAAT2 (abundant in the hippocampus and cortex), particularly astrocytic EAAT2, has been suggested to exert a pivotal role in the ability of hippocampal circuitry to code and store information (45). Previously, deletion of astrocytic EAAT2 has been shown to induce lower body weight and seizures (15, 29), and inhibition of glutamate uptake in rats was reported to induce impaired spatial reference memory (47). Additionally, loss of EAAT2 in a mouse model of AD resulted in accelerated onset of cognitive deficit (48). Furthermore, restoring EAAT2 expression in AD mice was found to improve cognitive function and restore synaptic integrity (49). Network hyperactivity and changes in expression of EAAT2 occur before amyloid plaque formation in mouse models of AD (50, 51). However, independent of EAAT2, astrocytic α7 nicotinic acetylcholine receptors have also been shown to induce glutamate release from astrocytes after associating with oligomeric Aβ (52). Glutamate dysfunction precedes cognitive decline in human AD, and decreased EAAT2 activity correlates with synaptic loss in aging (17) and neuronal loss in AD brains (20, 21). Taken together, these studies are in congruence with our data, where astrocytic EAAT2 deficiency in mice resulted in impaired short-term spatial reference memory in the Y-maze task at 13 mo of age and long-term spatial reference learning and memory deficit in the MWM task at 17 mo of age. In the probe trial for spatial memory in the MWM task, 17-mo-old gfap-Cre+ EAAT2Δ/+ mice performed below chance (defined as 15/60 s) (53) in time spent swimming in the platform quadrant, whereas the gfap-Cre− EAAT2+/+ controls exhibited a performance above chance level. These data indicate an overt cognitive deficit caused by astrocytic EAAT2 deficiency.
EAAT2 is expressed predominantly in astrocytes but is also present in neurons and excitatory axon terminals (16). Neuronal EAAT2 only constitutes 5 to 10% of all EAAT2 protein in the brain (16). Previously, neuronal EAAT2 knockout mice were found to have decreased glutamate uptake in synaptosomes (29); however, these mice did not display any behavioral phenotypes up to 1 y of age (29). Additionally, a recent study did not find any spatial reference learning and memory deficit in the MWM task or any impairment in cue- and context-dependent fear conditioning and novel object recognition tests in 12- to 16-wk-old neuronal EAAT2 conditional deletion mice (34). In congruence with these previous studies, we did not find impairment in spatial reference memory in the Y-maze task in 10-mo-old and 13-mo-old syn-Cre+ EAAT2−/− mice. However, interestingly, we found impairment in hippocampus-dependent spatial reference long-term memory in a probe trial in the MWM task in syn-Cre+ EAAT2−/− mice at 17 mo of age. In the probe trial, syn-Cre+ EAAT2−/− mice performed below chance level (53) in time spent in the platform quadrant and poorly as compared with syn-Cre− EAAT2+/+ controls. This is interesting because EAAT2 is highly expressed in neurons and axon terminals of the hippocampus (particularly the CA3 pyramidal neurons) (54, 55); however, previous studies in younger mice with neuronal EAAT2 deletion show no alteration in behavior dependent on hippocampal function (29, 34). A suggested possible explanation for the lack of cognitive deficits in younger mice with neuronal EAAT2 deletion was that EAAT2 expressed in neurons might not contribute significantly to glutamate clearance at the hippocampal synapses involved in learning and memory (34). While the overall functional significance of neuronal EAAT2 remains largely unknown, the present study points to a role of neuronal EAAT2 in cognitive function in aging, and possibly synaptic glutamatergic homeostasis, that needs to be further explored. Importantly, at the very least, our finding of late-onset cognitive impairment in mice with neuronal EAAT2 deletion challenges the long-held notion that the role of neuronal EAAT2 in glutamatergic synaptic homeostasis is minimal, given its lower expression in neurons relative to astrocytes.
We also report that the partial loss of astrocytic EAAT2 resulted in dysregulation of several innate and adaptive immune response pathways. Dyshomeostasis of the immune system also significantly negatively correlated with cognition in gfap-Cre+ EAAT2Δ/+ mice. Several recent genetic, bioinformatics, and preclinical data have shown that activation of the immune system and systemic inflammation contribute to the pathogenesis and progression of AD (36, 56–63). The peripheral immune system communicates with the brain through cytokines, acute-phase proteins, and complement factors (64), and causes activation of microglia, increased secretion of proinflammatory cytokines, and acceleration of disease progression in rodent models of AD (65–68). CNS resident microglia and T lymphocytes that migrate from the peripheral blood through the blood–brain barrier drive the inflammatory reaction in AD brains by secreting several immune modulators such as cytokines and chemokines (69, 70). Cluster of differentiation 3+ (CD3+) T cell numbers also significantly correlate with tau pathology in human AD brains (71). High glutamate levels have also been shown to stimulate IFN-γ secretion by CD3-activated T cells that can, in turn, activate resident microglia, resulting in synaptic alterations in multiple sclerosis (72). Systemic inflammation in humans and nonhuman primate brains has also been shown to increase the risk of developing neurological disorders (73–75). Future work will focus on the protein function of key genes that show a significant correlation with mouse behavior. Understanding the cross-talk between systemic inflammation and neuroinflammation has important implications for developing therapies for AD.
Our data also show dysregulation of diverse signaling pathways associated with cognitive dysfunction in syn-Cre+ EAAT2−/− mice that are distinct from gfap-Cre+ EAAT2Δ/+ mice. Interestingly, we found dysregulation of the kynurenine pathway in neuronal EAAT2 deletion mice. Previously, both in humans and animal models, alterations in the kynurenine pathway have been associated with cognitive impairment (76, 77). Tryptophan, an essential amino acid, is degraded primarily through the kynurenine pathway, which generates metabolites collectively referred to as the kynurenines (78). It has been suggested that the kynurenine pathway can be responsible for moderating the activity of 2 neurotransmitters fundamentally involved in cognition: glutamate and acetylcholine (77). Increased expression of indoleamine-pyrrole 2,3-dioxygenase, a key enzyme in the kynurenine pathway, and imbalances in the kynurenine pathway with an inverse correlation with cognitive decline have been observed in the serum and brains of AD patients (79–82). Reduced nicotinamide-adenine dinucleotide phosphate (NADPH) and oxidized nicotinamide-adenine dinucleotide (NAD), the primary end products of the kynurenine pathway, are also essential for mitochondrial function (83). EAAT2-mediated glutamate uptake in axon terminals in neurons has recently been shown to provide glutamate for mitochondrial energy metabolism (84). The sirtuin signaling pathway, a major regulator of mitochondrial function, is significantly positively correlated with cognition in syn-Cre+ EAAT2−/− mice. Additionally, the kynurenine pathway is induced by immunological activation and stress; thus, it also can mediate the effects of environmental factors on cognition and behavior (77). Thus, we suspect that the age-related cognitive deficit observed in neuronal EAAT2-deletion mice in our study could be driven by alterations in genes regulating the kynurenine pathway.
Furthermore, transcriptomic data from gfap-Cre+ EAAT2Δ/+ mice, but not syn-Cre+ EAAT2−/− mice, show a robust overlap with gene expression datasets of human AD and aging. Concordant gene expression from gfap-Cre+ EAAT2Δ/+ and human AD datasets was found to be enriched for genes regulating synaptic function and inflammatory responses. Synaptic dysfunction in excitatory neurons has been shown to be the best predictor of cognitive decline in AD (85, 86). Our data suggest that glutamate dyshomeostasis from partial loss of astrocytic EAAT2 in the aged hippocampus may potentially trigger inflammatory genes similar to the aging human hippocampus and may potentially suppress genes necessary for synaptic plasticity similar to human AD. Our previous work with riluzole, a known modulator of EAAT2 (18, 87–90), showed that riluzole induces neuroplastic changes in the brain (91) and rescues age- and AD-related gene expression changes related to neural transmission and synaptic plasticity (19). Further, riluzole reduces amyloid pathology in 5XFAD mice and reverses gene expression associated with disease-associated microglia (92). Future studies will further evaluate the direct and indirect mechanisms causing neuroinflammation and activation of the peripheral immune system from reduced EAAT2 expression. Recent evidence suggests activated forms of astrocytes in human brains with higher EAAT2 expression may exert beneficial effects to preserve cognitive function, even in the presence of Aβ and NFTs (28). The little or no overlap observed from syn-Cre+ EAAT2−/− and human AD datasets could potentially arise from the predominance of astrocytic EAAT2 in the brain. Loss of neuronal EAAT2 could also potentially be compounding gene expression effects from loss of astrocytic EAAT2 in the human brain.
Thus, astrocytic rather than neuronal EAAT2 deficiency in the hippocampus is transcriptionally more closely associated with human aging and AD. This study further implicates glutamatergic dyshomeostasis in inflammation and neural plasticity in aging and AD, highlights the distinct signaling mechanisms mediated by neuronal and astrocytic EAAT2, and brings out EAAT2 as a potential therapeutic target in aging and AD.
Methods
Animals.
To generate heterozygous astrocytic knockout of EAAT2(gfap-Cre+ EAAT2Δ/+), we bred male or female homozygous mice carrying the conditional EAAT2 knockout allele (29) (Mouse Genome Informatics identifier Slc1a2tm1.1Pros) obtained from the founder colony at Boston Children’s Hospital with male or female mice carrying the inducible human glial fibrillary acid protein human GFAP-CreERT2 (C57BL/6J) (29). To induce deletion, 10-mo-old mice were treated daily (i.p.) for 7 d with 33 mg/kg 4-hydroxy tamoxifen (H7904; Sigma). To generate homozygous neuronal knockout of EAAT2 (syn-Cre+ EAAT2−/−), we bred male mice that carried the conditional EAAT2 knockout allele (29) with female mice carrying the synapsin I promoter-driven cre recombinase allele (C57BL/6) (29). Genotypes were confirmed using PCR protocols from The Jackson Laboratory. All animal experiments were performed in accordance with NIH guidelines and were approved by the Mount Sinai Institutional Animal Care and Use Committee (IACUC) and The Rockefeller University IACUC. In consideration of sex as a biological variable, only male mice were used for all experiments in the present study.
Behavior.
Y-maze task for hippocampus-dependent spatial reference memory.
We used the 2-trial Y-maze task to assess the hippocampus-dependent spatial reference memory retention at 10 and 13 mo of age in study mice, as described previously (91). The 2-trial Y-maze experimental paradigm provides a sensitive and robust measure of spatial reference memory in rodents as it utilizes their natural tendency to explore novel environments (93).
A day prior to Y-maze testing, mice were habituated in an open-field arena (45 × 45 cm, ambient luminosity: ∼20 lux on edges and ∼40 lux in the center) in the testing room. Mice were acclimated to the testing room for 1 h prior to being placed in the open-field arena for 15 min. The open-field arena was cleaned between each mouse with 70% ethanol. Twenty-four hours after open-field habituation, mice were tested in the 2-trial Y-maze task. The task consisted of a 10-min acquisition trial, a 1-h intertrial interval, and a 10-min retention trial. During the acquisition trial, mice were placed facing away from the center of the maze in the SA and allowed to freely explore the SA and FA, while the NA was blocked. After a 1-h intertrial interval, mice were placed again in the SA for a 10-min retention trial and allowed to explore all 3 arms (i.e., SA, FA, NA). Each arm of the Y-maze was 40 cm long, 9 cm wide, and 21 cm high. Corncob bedding from the cage of the tested mouse was mixed with clean bedding and distributed evenly on the floor of the maze to minimize anxiety. Ambient luminosity was maintained at ∼25 lux in the arms and ∼40 lux in the center, believed to be nonanxiogenic for rodents. To eliminate the possibility of NA preference being based on external factors, the SA, FA, and NA were changed for each mouse. Black curtains surrounded the Y-maze to prevent the availability of noncontrolled distal cues. Visual cues of varying sizes and patterns were mounted on these curtains facing the 3 arms of Y-maze to facilitate mice in spatial orientation.
The time spent in each arm, frequency of arm entries, distance traveled, and velocity were automatically tracked and recorded using an Ethovision video-tracking system (Noldus Information Technology, Inc.). The data for time spent in each arm in the acquisition trial and retention trial (measure of spatial reference memory) were analyzed (gfap-Cre+ EAAT2Δ/+, n = 9; gfap-Cre− EAAT2+/+, n = 15; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 for 10-mo-old mice data and gfap-Cre+ EAAT2Δ/+, n = 7; gfap-Cre− EAAT2+/+, n = 15; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 for 13-mo-old mice data). Additionally, the Y-maze task was performed in age-matched gfap-Cre+ control mice (n = 9). In the 2-trial Y-maze task, a mouse with an unimpaired spatial reference memory is expected to spend a greater amount of time in the NA during the retention trial phase.
MWM task for hippocampus-dependent spatial reference learning and memory.
We conducted the MWM test at 17 mo of age in study mice to assess hippocampus-dependent spatial reference learning and memory. The experimental paradigm used was an adaptation of the protocol originally described by Morris et al. (94) in rats, and previously validated in mice (95–97). The MWM behavior task depends on the utilization of a spatial navigational strategy by rodents to find a fixed submerged escape platform. The hippocampal formation devises a spatial map based on distal environmental cues (94, 98) and plays a critical role in encoding, storage, consolidation, and retrieval of the spatial memory (94, 99, 100).
The learning trials were performed over 8 d, with 2 trials (60 s each; intertrial interval ∼ 20 min) each day. The test was conducted in a circular water tank, which had a diameter of 120 cm. The pool was filled with water (at room temperature, 23 ± 2 °C) rendered opaque with a small amount of nontoxic white paint to prevent animals from being able to see the location of the platform. Four start positions were marked around the perimeter of the circular tank to be used for successive trials. Extramaze visual cues were mounted on the black curtains surrounding the MWM tank. These cues were kept in the same position during all training trials and the probe trial. During the training trials, a square Plexiglas escape platform (25 cm2) was placed in the center of the TQ and submerged 1 cm beneath the surface of the water. The platform stayed in the same position throughout the learning trials and was removed from the tank only during the probe trial. Prior to the first trial, on the first training day, each mouse was placed on the platform for 30 s to allow orientation to extramaze cues. This was followed by a practice swim for 30 s and 3 practice climbs onto the platform. Subsequently, each mouse was placed in the desired start position in the pool facing the wall. The trial ended when a mouse reached the escape platform or the maximum trial duration of 60 s elapsed. If a mouse located the platform before 60 s had elapsed, it was immediately removed from the MWM pool. If a mouse failed to find the hidden escape platform within 60 s of a training trial, it was gently guided to it and allowed to stay on the platform for an additional 20 s to reorient to the distal visual cues. The mouse was then dried with a cloth towel before being returned to its home cage until the next trial. Each mouse was thoroughly inspected to ensure dryness. All testing was performed at approximately the same time each day to reduce variability in performance due to the time of day. With 2 trials per day for 8 consecutive days, each mouse performed a total of 16 spatial reference learning trials. A probe trial (i.e., a spatial reference memory retention trial) was conducted 24 h later. During the probe trial, the platform was removed from the pool; each mouse was placed in a quadrant opposite to the location of escape platform quadrant during learning trials and allowed to swim in the circular pool for 60 s. A single visible platform trial occurred on the last day of testing after the probe trial and served as a test for visual deficits. The platform was placed in the adjacent right (AR) quadrant and was made visible by a colorful flag and colored top that extended above the surface of the water. Latency to find the visible platform in the trial was used as a measure of visual ability.
The measure of spatial reference learning was the time taken to find the escape platform during the 16 training trials. The data were analyzed for 4 blocks, with each block consisting of 4 sequential trials over 2 d of the training period. For the probe trial, the circular MWM pool was divided into 4 imaginary quadrants (TQ, AR, adjacent left, and OQ), and a small zone where the escape platform had been located during the training trials (virtual platform). The measure of spatial reference memory was time spent in the TQ as compared with the other quadrants during the 60-s probe trial. The mouse behavior during the MWM task was automatically tracked and recorded using the Ethovision video-tracking system. Heatmaps of search intensity during the probe trial were also generated in Ethovision. The MWM data were analyzed from gfap-Cre+ EAAT2Δ/+, n = 7; gfap-Cre− EAAT2+/+, n = 13; syn-Cre+ EAAT2−/−, n = 11; and syn-Cre− EAAT2+/+, n = 12 mice. Additionally, the MWM task was performed in age-matched gfap-Cre+ control mice (n = 9).
RNA-Seq.
After completion of behavioral studies, ∼18-mo-old mice were killed by cervical dislocation, and brain tissue was harvested for RNA-Seq and biochemical studies. Each mouse brain tissue sample was immediately dissected into hippocampal and cortical regions, frozen on dry ice, and then stored at −80 °C until further evaluation. RNA was isolated (gfap-Cre+ EAAT2Δ/+, n = 6; gfap-Cre− EAAT2+/+, n = 6; syn-Cre+ EAAT2−/−, n = 6; syn-Cre− EAAT2+/+, n = 6 mice) using a RNeasy Lipid Tissue Mini Kit (Qiagen). Quality of RNA samples was determined using a bioanalyzer (Agilent), and samples for RNA-Seq with RNA integrity number values >9. 500 ng of purified RNA were used to prepare sequencing libraries according to the Illumina protocol and sequenced with an Illumina HiSeq 2500 system.
Raw sequencing reads were mapped to the mouse genome (mm10) using HISAT2. Reads were counted using HTSeq-count (v0.6.0) against ENCODE annotations, release 90. Differential gene expression between samples of different conditions was determined using the DESeq2 package (v1.6.3). For broad pattern identification, significance was set at an uncorrected P ≤ 0.05 and a threshold for absolute fold change at ≥1.5. IPA (Ingenuity Systems; Qiagen) was used to identify pathways from DEGs and upstream regulators from RRHO. The analysis used Fisher’s exact test to calculate P values.
WGCNA.
Analyses were carried out according to the online R WGCNA tutorial (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/). Normalized, variance stabilizing transformation-transformed counts data from mouse RNA-Seq samples were utilized to generate networks.
Separate unsigned networks were constructed for samples belonging to the gfap-Cre+ EAAT2Δ/+ and syn-Cre+ EAAT2−/− groups. A coexpression similarity matrix was generated using the absolute values of the correlation coefficient between the expression profiles of pairs of genes. The adjacency matrix was derived from the coexpression similarity matrix by raising all values to a power of 8 (soft-thresholding power determined using pickSoftThreshold). The adjacency matrix provides the measure of network interconnectedness for each pair of genes. Topological overlap-based dissimilarity was computed using the transformed adjacency matrix, which was further used to generate an average linkage hierarchical clustering. Modules were determined from the resulting clustered tree, where minModuleSize was set to 30. To summarize the profile in each module, the module eigengene was determined using the first principal component of the module matrix.
For each module, the module membership of a gene was defined as the correlation between the normalized gene expression and the module eigengene. Module eigengenes were correlated to traits representing the time spent by mice in respective TQs. Correlation between the individual genes with the trait of interest was found using the absolute value of correlation between the gene expression profile and trait.
Analysis of human microarray data.
GSE48350 contains microarray data from 4 regions of the forebrain. For our analysis of genes dysregulated in AD and aging, we chose the hippocampus from male subjects, which was relevant to our mouse data. AD-related gene expression changes were based on the comparisons between the male hippocampus of AD cases (n = 9, ages 76 to 94 y) versus age-matched male hippocampus controls (n = 13, ages 82 to 97 y). Aging-related gene expression changes were based on aged male hippocampus (n = 13, ages 82 to 97 y) versus young male hippocampus (n = 9, ages 20 to 52 y). All arrays were background-corrected and normalized (GCRMA algorithm) using the R Packages simpleaffy and limma. Probe annotation was carried out using the R Bioconductor package hgu133plus2.db. Differential gene expression between samples of different conditions was determined using limma at a raw P value cutoff of 0.05.
RRHO analysis.
Unfiltered differential gene expression lists were ranked by −log10(P value) * fold change. RRHO was used to evaluate the overlap of differential expression lists between astrocytic and neuronal EAAT2 knockout and human AD and between astrocytic and neuronal EAAT2 knockout and human aging. RRHO difference maps were generated for pairs of RRHO maps by calculating the normal approximation of difference in the log odds ratio and SE of overlap for each pixel. The Z score was then converted to a P value and corrected for multiple comparisons across pixels.
PMV preparation.
PMV fractions from the flash-frozen frontal cortex tissue were prepared for individual mice (gfap-Cre+ EAAT2Δ/+, n = 6; gfap-Cre− EAAT2+/+, n = 8; syn-Cre+ EAAT2−/−, n = 5; syn-Cre− EAAT2+/+, n = 5) using a modified version of previous protocols (29, 101–103). Briefly, the mouse cortical tissue was homogenized in a Teflon-glass homogenizer to generate 10% (wt/vol) homogenate. The prechilled homogenization buffer contained 10 mM Tris base (pH 7.4), 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), protease inhibitor mixture (catalog no. 78430; Thermo Fisher Scientific), and phosphatase inhibitor mixture (catalog no. 78428; Thermo Fisher Scientific), along with 320 mM sucrose. The homogenate was centrifuged at 800 × g for 10 min at 4 °C. The supernatant was then centrifuged at 20,000 × g for 20 min at 4 °C twice and resuspended. The supernatant was discarded each time. After the second centrifugation, the pellet was resuspended in homogenization buffer containing 35.6 mM sucrose with 6 up-and-down strokes of Teflon-glass homogenizer, followed by continuous mixing on a tube rotator for 30 min at 4 °C. Subsequently, this suspension was centrifuged at 20,000 × g for 30 min at 4 °C. The resultant pellet containing PMVs was resuspended in homogenization buffer containing 320 mM sucrose. The protein concentration in PMVs was determined using a Pierce BCA Protein Assay Kit (catalog no. 23227; Thermo Fisher Scientific).
Immunoblotting.
Western blotting was employed to evaluate the EAAT2 expression in the PMV fractions (gfap-Cre+ EAAT2Δ/+, n = 6; gfap-Cre− EAAT2+/+, n = 8; syn-Cre+ EAAT2−/−, n = 5; syn-Cre− EAAT2+/+, n = 5 mice). The samples were prepared in Laemmli buffer (catalog no. 1610747; Bio-Rad) and subjected to 10% polyacrylamide gel (catalog no. 4561035; Bio-Rad) electrophoresis, followed by transfer of separated proteins on poly(vinylidene difluoride) (PVDF) membrane (catalog no. 1620177; Bio-Rad). The immunoblot was developed with a C-terminal–directed rabbit anti-EAAT2 antibody (1:5,000; a kind gift from Jeffrey D. Rothstein, John Hopkins University School of Medicine, Baltimore, MD) (104). The corresponding horseradish peroxidase-conjugated goat–anti-rabbit secondary antibody was employed. For a loading control, blots were developed with mouse monoclonal antibody to β-actin (1:5,000; Thermo Fisher Scientific). The immunoreactive protein bands were visualized employing enhanced chemiluminescence reagents (catalog no. 1705060; Bio-Rad). The blot films were scanned and assessed using Multi Gauge software, version 3.0 (Fujifilm). For quantification purposes, each immunoreactive band was normalized to its corresponding β-actin band prior to group-wise comparisons.
Supplementary Material
Acknowledgments
This work was supported by an NIH grant (Paul B. Beeson Emerging Leaders Career Development Award in Aging 1 K76AG054772), the BrightFocus Foundation, the Dana Foundation, the Alzheimer’s Drug Discovery Foundation, the Alzheimer’s Association, and the Bernard L. Schwartz Award for Physician Scientists (to A.C.P.); the Hereditary Disease Foundation, Children’s Hospital Intellectual and Developmental Disabilities Research Center Core Grant HD 018655, NIH Grant RO1NS066019, and NIH Grant R21MH104318 (to P.A.R.); and by partial support of A.C.P. by Grant UL1TR001866 from the National Center for Advancing Translational Sciences and the NIH Clinical and Translational Science Award program.
Footnotes
Competing interest statement: P.A.R. and D.G.C. began a collaboration in August 2019 unrelated to this research.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903566116/-/DCSupplemental.
References
- 1.Burke S. N., Barnes C. A., Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 7, 30–40 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Coleman P. D., Flood D. G., Neuron numbers and dendritic extent in normal aging and Alzheimer’s disease. Neurobiol. Aging 8, 521–545 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Nagahara A. H., Bernot T., Tuszynski M. H., Age-related cognitive deficits in rhesus monkeys mirror human deficits on an automated test battery. Neurobiol. Aging 31, 1020–1031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster T. C., Defazio R. A., Bizon J. L., Characterizing cognitive aging of spatial and contextual memory in animal models. Front. Aging Neurosci. 4, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher M., Rapp P. R., The use of animal models to study the effects of aging on cognition. Annu. Rev. Psychol. 48, 339–370 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Subramaniam M., et al. , Prevalence of dementia in people aged 60 years and above: Results from the WiSE study. J. Alzheimers Dis. 45, 1127–1138 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Morrison J. H., Hof P. R., Life and death of neurons in the aging brain. Science 278, 412–419 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Morrison J. H., Hof P. R., Selective vulnerability of corticocortical and hippocampal circuits in aging and Alzheimer’s disease. Prog. Brain Res. 136, 467–486 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Morrison J. H., Hof P. R., Life and death of neurons in the aging cerebral cortex. Int. Rev. Neurobiol. 81, 41–57 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Hardingham G. E., Bading H., Synaptic versus extrasynaptic NMDA receptor signalling: Implications for neurodegenerative disorders. Nat. Rev. Neurosci. 11, 682–696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lüscher C., Malenka R. C., NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb. Perspect. Biol. 4, a005710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papouin T., Oliet S. H., Organization, control and function of extrasynaptic NMDA receptors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong G., Jahr C. E., Block of glutamate transporters potentiates postsynaptic excitation. Neuron 13, 1195–1203 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Furuta A., Rothstein J. D., Martin L. J., Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J. Neurosci. 17, 8363–8375 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K., et al. , Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 276, 1699–1702 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Furness D. N., et al. , A quantitative assessment of glutamate uptake into hippocampal synaptic terminals and astrocytes: New insights into a neuronal role for excitatory amino acid transporter 2 (EAAT2). Neuroscience 157, 80–94 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potier B., et al. , Reduction in glutamate uptake is associated with extrasynaptic NMDA and metabotropic glutamate receptor activation at the hippocampal CA1 synapse of aged rats. Aging Cell 9, 722–735 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Brothers H. M., et al. , Riluzole partially rescues age-associated, but not LPS-induced, loss of glutamate transporters and spatial memory. J. Neuroimmune Pharmacol. 8, 1098–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira A. C., et al. , Age and Alzheimer’s disease gene expression profiles reversed by the glutamate modulator riluzole. Mol. Psychiatry 22, 296–305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masliah E., Alford M., DeTeresa R., Mallory M., Hansen L., Deficient glutamate transport is associated with neurodegeneration in Alzheimer’s disease. Ann. Neurol. 40, 759–766 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Scott H. A., Gebhardt F. M., Mitrovic A. D., Vandenberg R. J., Dodd P. R., Glutamate transporter variants reduce glutamate uptake in Alzheimer’s disease. Neurobiol. Aging 32, 553.e1–533.e11 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Jacob C. P., et al. , Alterations in expression of glutamatergic transporters and receptors in sporadic Alzheimer’s disease. J. Alzheimers Dis. 11, 97–116 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Li S., Mallory M., Alford M., Tanaka S., Masliah E., Glutamate transporter alterations in Alzheimer disease are possibly associated with abnormal APP expression. J. Neuropathol. Exp. Neurol. 56, 901–911 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Zoltowska K. M., Maesako M., Meier J., Berezovska O., Novel interaction between Alzheimer’s disease-related protein presenilin 1 and glutamate transporter 1. Sci. Rep. 8, 8718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scimemi A., et al. , Amyloid-β1-42 slows clearance of synaptically released glutamate by mislocalizing astrocytic GLT-1. J. Neurosci. 33, 5312–5318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S., et al. , Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 62, 788–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zott B., et al. , A vicious cycle of β amyloid-dependent neuronal hyperactivation. Science 365, 559–565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi E., et al. , Activated forms of astrocytes with higher GLT-1 expression are associated with cognitive normal subjects with Alzheimer pathology in human brain. Sci. Rep. 8, 1712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petr G. T., et al. , Conditional deletion of the glutamate transporter GLT-1 reveals that astrocytic GLT-1 protects against fatal epilepsy while neuronal GLT-1 contributes significantly to glutamate uptake into synaptosomes. J. Neurosci. 35, 5187–5201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimoto J., et al. , Region-specific deletions of the glutamate transporter GLT1 differentially affect seizure activity and neurodegeneration in mice. Glia 66, 777–788 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kiryk A., et al. , Behavioral characterization of GLT1 (+/-) mice as a model of mild glutamatergic hyperfunction. Neurotox. Res. 13, 19–30 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Aida T., et al. , Astroglial glutamate transporter deficiency increases synaptic excitability and leads to pathological repetitive behaviors in mice. Neuropsychopharmacology 40, 1569–1579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y., Hassel B., Eid T., Danbolt N. C., Axon-terminals expressing EAAT2 (GLT-1; Slc1a2) are common in the forebrain and not limited to the hippocampus. Neurochem. Int. 123, 101–113 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Fischer K. D., et al. , Behavioral phenotyping and dopamine dynamics in mice with conditional deletion of the glutamate transporter GLT-1 in neurons: Resistance to the acute locomotor effects of amphetamine. Psychopharmacology (Berl.) 235, 1371–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertram L., McQueen M. B., Mullin K., Blacker D., Tanzi R. E., Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 39, 17–23 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Heppner F. L., Ransohoff R. M., Becher B., Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 16, 358–372 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Heneka M. T., et al. , Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brezun J. M., Daszuta A., Serotonergic reinnervation reverses lesion-induced decreases in PSA-NCAM labeling and proliferation of hippocampal cells in adult rats. Hippocampus 10, 37–46 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Bender D. A., Biochemistry of tryptophan in health and disease. Mol. Aspects Med. 6, 101–197 (1983). [DOI] [PubMed] [Google Scholar]
- 40.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plaisier S. B., Taschereau R., Wong J. A., Graeber T. G., Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie C. W., Lewis D. V., Endogenous opioids regulate long-term potentiation of synaptic inhibition in the dentate gyrus of rat hippocampus. J. Neurosci. 15, 3788–3795 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deisseroth K., Bito H., Tsien R. W., Signaling from synapse to nucleus: Postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron 16, 89–101 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Nanou E., Catterall W. A., Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron 98, 466–481 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Pita-Almenar J. D., Zou S., Colbert C. M., Eskin A., Relationship between increase in astrocytic GLT-1 glutamate transport and late-LTP. Learn. Mem. 19, 615–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neves G., Cooke S. F., Bliss T. V., Synaptic plasticity, memory and the hippocampus: A neural network approach to causality. Nat. Rev. Neurosci. 9, 65–75 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Bechtholt-Gompf A. J., et al. , Blockade of astrocytic glutamate uptake in rats induces signs of anhedonia and impaired spatial memory. Neuropsychopharmacology 35, 2049–2059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mookherjee P., et al. , GLT-1 loss accelerates cognitive deficit onset in an Alzheimer’s disease animal model. J. Alzheimers Dis. 26, 447–455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi K., et al. , Restored glial glutamate transporter EAAT2 function as a potential therapeutic approach for Alzheimer’s disease. J. Exp. Med. 212, 319–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schallier A., et al. , Region- and age-specific changes in glutamate transport in the AβPP23 mouse model for Alzheimer’s disease. J. Alzheimers Dis. 24, 287–300 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Busche M. A., et al. , Critical role of soluble amyloid-β for early hippocampal hyperactivity in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 109, 8740–8745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talantova M., et al. , Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proc. Natl. Acad. Sci. U.S.A. 110, E2518–E2527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terry A. V., Jr, “Spatial navigation (water maze) tasks” in Methods of Behavior Analysis in Neuroscience, Buccafusco J. J., Ed. (Frontiers in Neuroscience, CRC Press, Boca Raton, FL, ed. 2, 2009), chap. 13. [Google Scholar]
- 54.Berger U. V., Hediger M. A., Comparative analysis of glutamate transporter expression in rat brain using differential double in situ hybridization. Anat. Embryol. (Berl.) 198, 13–30 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Berger U. V., DeSilva T. M., Chen W., Rosenberg P. A., Cellular and subcellular mRNA localization of glutamate transporter isoforms GLT1a and GLT1b in rat brain by in situ hybridization. J. Comp. Neurol. 492, 78–89 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyss-Coray T., et al. , Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer’s mice. Proc. Natl. Acad. Sci. U.S.A. 99, 10837–10842 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakrabarty P., et al. , Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: Evidence against inflammation as a driving force for amyloid deposition. FASEB J. 24, 548–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meda L., et al. , Activation of microglial cells by beta-amyloid protein and interferon-gamma. Nature 374, 647–650 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Bhaskar K., et al. , Microglial derived tumor necrosis factor-α drives Alzheimer’s disease-related neuronal cell cycle events. Neurobiol. Dis. 62, 273–285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson T., et al. , Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abduljaleel Z., et al. , Evidence of trem2 variant associated with triple risk of Alzheimer’s disease. PLoS One 9, e92648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corder E. H., et al. , Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Bemiller S. M., et al. , TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol. Neurodegener. 12, 74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wrona D., Neural-immune interactions: An integrative view of the bidirectional relationship between the brain and immune systems. J. Neuroimmunol. 172, 38–58 (2006). [DOI] [PubMed] [Google Scholar]
- 65.McManus R. M., Mills K. H., Lynch M. A., T cells-protective or pathogenic in Alzheimer’s disease? J. Neuroimmune Pharmacol. 10, 547–560 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Rodrigues M. C., Sanberg P. R., Cruz L. E., Garbuzova-Davis S., The innate and adaptive immunological aspects in neurodegenerative diseases. J. Neuroimmunol. 269, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Schwartz M., Baruch K., Breaking peripheral immune tolerance to CNS antigens in neurodegenerative diseases: Boosting autoimmunity to fight-off chronic neuroinflammation. J. Autoimmun. 54, 8–14 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Holmes C., Review: Systemic inflammation and Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 39, 51–68 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Marsh S. E., et al. , The adaptive immune system restrains Alzheimer’s disease pathogenesis by modulating microglial function. Proc. Natl. Acad. Sci. U.S.A. 113, E1316–E1325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McManus R. M., Higgins S. C., Mills K. H., Lynch M. A., Respiratory infection promotes T cell infiltration and amyloid-β deposition in APP/PS1 mice. Neurobiol. Aging 35, 109–121 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Merlini M., Kirabali T., Kulic L., Nitsch R. M., Ferretti M. T., Extravascular CD3+ T cells in brains of Alzheimer disease patients correlate with tau but not with amyloid pathology: An immunohistochemical study. Neurodegener. Dis. 18, 49–56 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Centonze D., et al. , The link between inflammation, synaptic transmission and neurodegeneration in multiple sclerosis. Cell Death Differ. 17, 1083–1091 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Deleidi M., Isacson O., Viral and inflammatory triggers of neurodegenerative diseases. Sci. Transl. Med. 4, 121ps3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perry V. H., Holmes C., Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 10, 217–224 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Norden D. M., Muccigrosso M. M., Godbout J. P., Microglial priming and enhanced reactivity to secondary insult in aging, and traumatic CNS injury, and neurodegenerative disease. Neuropharmacology 96, 29–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koola M. M., Kynurenine pathway and cognitive impairments in schizophrenia: Pharmacogenetics of galantamine and memantine. Schizophr. Res. Cogn. 4, 4–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stone T. W., Darlington L. G., The kynurenine pathway as a therapeutic target in cognitive and neurodegenerative disorders. Br. J. Pharmacol. 169, 1211–1227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y., Guillemin G. J., Kynurenine pathway metabolites in humans: Disease and healthy states. Int. J. Tryptophan Res. 2, 1–19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Widner B., et al. , Tryptophan degradation and immune activation in Alzheimer’s disease. J. Neural Transm. (Vienna) 107, 343–353 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Majláth Z., Tajti J., Vécsei L., Kynurenines and other novel therapeutic strategies in the treatment of dementia. Ther. Adv. Neurol. Disorder. 6, 386–397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kincses Z. T., Toldi J., Vécsei L., Kynurenines, neurodegeneration and Alzheimer’s disease. J. Cell. Mol. Med. 14, 2045–2054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wennström M., et al. , Kynurenic acid levels in cerebrospinal fluid from patients with Alzheimer’s disease or dementia with lewy bodies. Int. J. Tryptophan Res. 7, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moroni F., Tryptophan metabolism and brain function: Focus on kynurenine and other indole metabolites. Eur. J. Pharmacol. 375, 87–100 (1999). [DOI] [PubMed] [Google Scholar]
- 84.McNair L. F., et al. , Deletion of neuronal GLT-1 in mice reveals its role in synaptic glutamate homeostasis and mitochondrial function. J. Neurosci. 39, 4847–4863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scheff S. W., Price D. A., Schmitt F. A., DeKosky S. T., Mufson E. J., Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology 68, 1501–1508 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Scheff S. W., Price D. A., Schmitt F. A., Mufson E. J., Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 27, 1372–1384 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Carbone M., Duty S., Rattray M., Riluzole elevates GLT-1 activity and levels in striatal astrocytes. Neurochem. Int. 60, 31–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banasr M., et al. , Glial pathology in an animal model of depression: Reversal of stress-induced cellular, metabolic and behavioral deficits by the glutamate-modulating drug riluzole. Mol. Psychiatry 15, 501–511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fumagalli E., Funicello M., Rauen T., Gobbi M., Mennini T., Riluzole enhances the activity of glutamate transporters GLAST, GLT1 and EAAC1. Eur. J. Pharmacol. 578, 171–176 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Hunsberger H. C., et al. , Riluzole rescues glutamate alterations, cognitive deficits, and tau pathology associated with P301L tau expression. J. Neurochem. 135, 381–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pereira A. C., et al. , Glutamatergic regulation prevents hippocampal-dependent age-related cognitive decline through dendritic spine clustering. Proc. Natl. Acad. Sci. U.S.A. 111, 18733–18738 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okamoto M., et al. , Riluzole reduces amyloid beta pathology, improves memory, and restores gene expression changes in a transgenic mouse model of early-onset Alzheimer’s disease. Transl. Psychiatry 8, 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dellu F., Mayo W., Cherkaoui J., Le Moal M., Simon H., A two-trial memory task with automated recording: Study in young and aged rats. Brain Res. 588, 132–139 (1992). [DOI] [PubMed] [Google Scholar]
- 94.Morris R. G., Garrud P., Rawlins J. N., O’Keefe J., Place navigation impaired in rats with hippocampal lesions. Nature 297, 681–683 (1982). [DOI] [PubMed] [Google Scholar]
- 95.Kazim S. F., Blanchard J., Bianchi R., Iqbal K., Early neurotrophic pharmacotherapy rescues developmental delay and Alzheimer’s-like memory deficits in the Ts65Dn mouse model of Down syndrome. Sci. Rep. 7, 45561 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kazim S. F., et al. , Disease modifying effect of chronic oral treatment with a neurotrophic peptidergic compound in a triple transgenic mouse model of Alzheimer’s disease. Neurobiol. Dis. 71, 110–130 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Lee T. T., Filipski S. B., Hill M. N., McEwen B. S., Morphological and behavioral evidence for impaired prefrontal cortical function in female CB1 receptor deficient mice. Behav. Brain Res. 271, 106–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Keefe J., Nadel L., The Hippocampus as a Cognitive Map (Oxford University Press, Oxford, UK, 1978). [Google Scholar]
- 99.Moser M. B., Moser E. I., Distributed encoding and retrieval of spatial memory in the hippocampus. J. Neurosci. 18, 7535–7542 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tonegawa S., Pignatelli M., Roy D. S., Ryan T. J., Memory engram storage and retrieval. Curr. Opin. Neurobiol. 35, 101–109 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Hallett P. J., Collins T. L., Standaert D. G., Dunah A. W., Biochemical fractionation of brain tissue for studies of receptor distribution and trafficking. Curr. Protoc. Neurosci. Chapter 1, Unit 1.16 (2008). [DOI] [PubMed] [Google Scholar]
- 102.Kamat P. K., Kalani A., Tyagi N., Method and validation of synaptosomal preparation for isolation of synaptic membrane proteins from rat brain. MethodsX 1, 102–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robinson M. B. “Examination of glutamate transporter heterogeneity using synaptosomal preparations” in Methods in Enzymology (Academic Press, 1998), vol. 296, pp. 189–202. [DOI] [PubMed] [Google Scholar]
- 104.Rothstein J. D., et al. , Localization of neuronal and glial glutamate transporters. Neuron 13, 713–725 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.