Significance
Adherens junctions are specialized cell–cell adhesion complexes found in epithelial, endothelial, and neuronal tissues of multicellular organism. The cadherin–catenin complex is the core component of the adherens junction and transmits mechanical stress from cell to cell. This study reveals that the cadherin–catenin complex displays a wide spectrum of flexible structures, which suggests a dynamic mechanism for this complex in mechanotransduction for cell–cell adhesion.
Keywords: adherens junction, mechanotransduction, negative stain electron microscopy, small-angle X-ray scattering, small-angle neutron scattering
Abstract
The cadherin–catenin adhesion complex is the central component of the cell–cell adhesion adherens junctions that transmit mechanical stress from cell to cell. We have determined the nanoscale structure of the adherens junction complex formed by the α-catenin•β-catenin•epithelial cadherin cytoplasmic domain (ABE) using negative stain electron microscopy, small-angle X-ray scattering, and selective deuteration/small-angle neutron scattering. The ABE complex is highly pliable and displays a wide spectrum of flexible structures that are facilitated by protein-domain motions in α- and β-catenin. Moreover, the 107-residue intrinsically disordered N-terminal segment of β-catenin forms a flexible “tongue” that is inserted into α-catenin and participates in the assembly of the ABE complex. The unanticipated ensemble of flexible conformations of the ABE complex suggests a dynamic mechanism for sensitivity and reversibility when transducing mechanical signals, in addition to the catch/slip bond behavior displayed by the ABE complex under mechanical tension. Our results provide mechanistic insight into the structural dynamics for the cadherin–catenin adhesion complex in mechanotransduction.
Adherens junctions (AJs) are specialized cell–cell adhesion complexes found in epithelial, endothelial, and neuronal tissues of multicellular organisms (1). They are necessary for embryonic morphogenesis, tissue integrity and homeostasis, and wound healing (2). Disassembly of the AJs results in the loss of cell polarity and contact inhibition, and permits an epithelial-to-mesenchymal transition (3–5). Dynamic regulation of the AJs is needed so that moving or migrating cells can continually break and remake adhesive bonds to cell neighbors (2, 6). Dysfunction of the AJs has been implicated in cancer development and vascular disease (7, 8)
In AJs, the lateral plasma membranes of adjacent cells are held together by cadherin-associated macromolecular complexes (9, 10). The primary components of the cadherin-associated complex include the transmembrane protein cadherin, the intracellular protein β-catenin, and α-catenin proteins (1, 11). The cytoplasmic domain of cadherin binds to β-catenin inside the cell, and this β-catenin in turn interacts with the actin-binding protein α-catenin, forming the α-catenin•β-catenin•cadherin complex. The extracellular domain of a cadherin in one cell binds to the cadherin from a neighboring cell to form a trans homophilic interaction in a calcium-dependent manner (9). The α-catenin•β-catenin•cadherin complexes also associate with p120 catenin (which stabilizes the cadherin–catenin complex at the cell membrane), with cytoskeletal actin filaments, and with other actin-binding proteins, such as vinculin, eplin, and zyxin (1, 12).
The linkage of the α-catenin•β-catenin•cadherin complex to the actin cytoskeleton is necessary for cell–cell coupling and adhesion, and for providing mechanical strength to tissues (13, 14). Moreover, the α-catenin•β-catenin•cadherin complex senses and transmits mechanical tension between neighboring cells in quiescent tissues and during collective cell migration (6). These mechanical forces can be externally applied (such as via the shearing force of blood flow in blood vessels), or they can originate within cells from contractile motion of the actin cytoskeleton induced by myosin motor proteins (12). Mechanical tension is sensed and transmitted between neighboring cells through the α-catenin•β-catenin•cadherin complexes in the AJs (6, 15, 16). Cells respond to such mechanical tensions by strengthening cell–cell adhesion by recruiting larger AJ complexes and altering actin cytoskeleton dynamics (12). The α-catenin•β-catenin•cadherin complexes and the actin cytoskeleton thus form an integral part of the mechanosensing and mechanotransduction machinery, sensing and transducing mechanical stresses between neighboring cells. The response of cells to mechanical tension is believed to activate the transcription machinery that drives morphogenesis and tissue repair (17).
Because α-catenin alone binds to actin filaments, α-catenin was thought to be responsible for anchoring the α-catenin•β-catenin•cadherin complex to the actin cytoskeleton. However, contrary to results obtained from in-cell studies (13, 18), biochemical studies found that the reconstituted complex of the α-catenin•β-catenin•cadherin cytoplasmic domain complex does not bind to actin filaments as well as α-catenin does alone (19). This paradox has recently been explained by an optical trap experiment that showed that the α-catenin•β-catenin•cadherin cytoplasmic domain complex only binds strongly to actin filaments that are under tension, utilizing a mechanism called a catch bond (20). In this study, a 2-state catch bond model (21) was used to explain how an applied force shifts the equilibrium for the complex from a weakly bonding to a strongly bonding state. It has been hypothesized that this shift is made possible by α-catenin being able to adopt different conformations in the complex. However, the molecular structure of the whole α-catenin•β-catenin•cadherin complex has remained elusive despite considerable effort. Only the crystal structures of full-length α-catenin and β-catenin by themselves or of fragments forming binary complexes are available (18, 22–25), so that the molecular mechanisms that underlie the F-actin–binding behavior of the α-catenin•β-catenin•cadherin complex under equilibrium and under tension remain speculative.
Here, we report a structural study of the complex of α-catenin•β-catenin•epithelial cadherin cytoplasmic domain (ABE) utilizing negative stain electron microscopy (EM), small-angle X-ray scattering (SAXS), and small-angle neutron scattering (SANS) in combination with selective deuteration and contrast variation. Our study reveals that the complex exists as a surprisingly dynamic ensemble with a wide spectrum of flexible conformations. Notably, α-catenin adopts multiple conformational states within the complex. We further find that, within the ABE complex, the long and disordered N-terminal segment of β-catenin is inserted deep into α-catenin and interacts with the actin-binding domain (ABD) of α-catenin. This result suggests a mechanism by which the intrinsically disordered region of β-catenin modulates the assembly of the ABE complex. The revelation of an ensemble of flexible conformations adopted by the AJ complex provides insights into the mechanism governing its assembly and also, into the way in which the adherens complex functions as a mechanosensor.
Results
The ABE Complex Displays a Spectrum of Flexible Conformations.
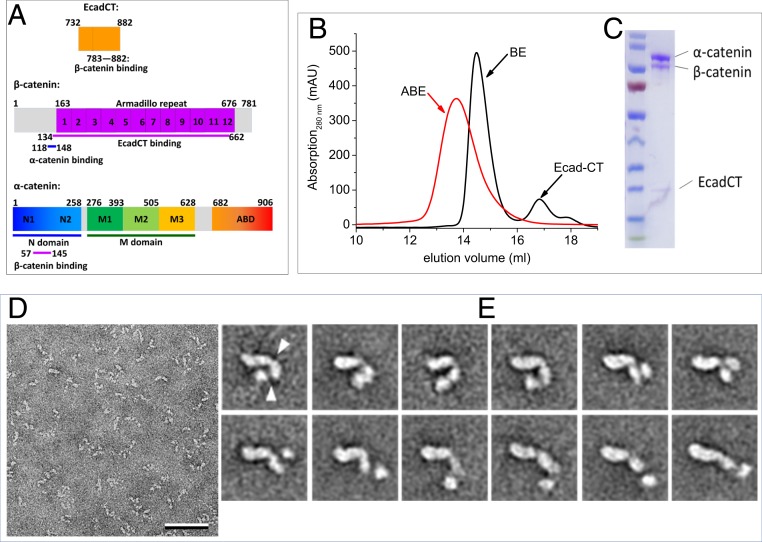
The domain organizations of α-catenin, β-catenin, and the epithelial cadherin cytoplasmic domain (EcadCT) are shown in Fig. 1A. α-Catenin is composed of the N-terminal (N) domain that consists of the N1 and N2 subdomains, the M domain that is subdivided into the M1 to M3 subdomains, and the ABD (22). The N1 domain is involved in the homodimerization of α-catenin, while the M1 domain binds to vinculin when α-catenin is under tension (26–29). β-Catenin is composed of a 12-armadillo repeat domain, a disordered N-terminal segment (residues 1 to 163), and a disordered C-terminal tail (residues 676 to 781) (30, 31). Previous studies showed that the armadillo repeat domain of β-catenin binds to the last 100 amino acid residues of EcadCT (23) and that residues 57 to 145 in the N1 domain of α-catenin bind to residues 118 to 148 of β-catenin (24). The region of the N1 subdomain of α-catenin that is involved in homodimerization overlaps with the region that binds to β-catenin so that the formation of an α-catenin/β-catenin heterodimer disrupts the α-catenin homodimer (24).
Fig. 1.
Conformational variability of the ABE complex revealed by negative stain EM. (A) Amino acid sequence and domain boundaries of EcadCT, α-catenin, and β-catenin. Previous studies showed that residues 783 to 882 in EcadCT bind to the armadillo repeat domain of β-catenin at residues 134 to 662 (23) and that residues 57 to 145 in the N domain of α-catenin bind to β-catenin at residues 118 to 148 (24). (B and C) Gel filtration profile (B) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel of the reconstituted ABE complex (C). (D) Electron micrograph area of negatively stained ABE complex. (Scale bar: 50 nm.) (E) Selected class averages of negatively stained ABE complex obtained with the ISAC procedure (SI Appendix, Fig. S1 shows all averages). Particles are shown from a more compact conformation starting in the upper left to a more extended conformation in the lower right. The arrowheads in the first average indicate the apparent hinge points around which the domains can move. Side length of individual averages: 31.2 nm.
The reconstituted and purified ABE complex elutes as a single peak in size exclusion chromatography (SEC) (Fig. 1 B and C). The inline size exclusion chromatography small-angle X-ray scattering (SEC-SAXS) analysis of the size and shape of the ABE complex will be described in the next paragraph. The peak fraction from SEC was imaged by negative stain EM. The raw images revealed a very heterogeneous population of poorly defined particles (Fig. 1D). However, many of the averages obtained with the iterative stable alignment and clustering approach (32) showed particles consisting of 3 domains: 1 larger comma-shaped domain and 2 smaller globular domains (Fig. 1E and SI Appendix, Fig. S1). The domains appear to be connected by flexible linkers, allowing the complexes to adopt a wide spectrum of conformations from a compact U shape to a fully extended rod-like conformation (Fig. 1E). The negative stain EM averages provide direct evidence that the ABE complex is very flexible and can adopt a wide spectrum of conformations.
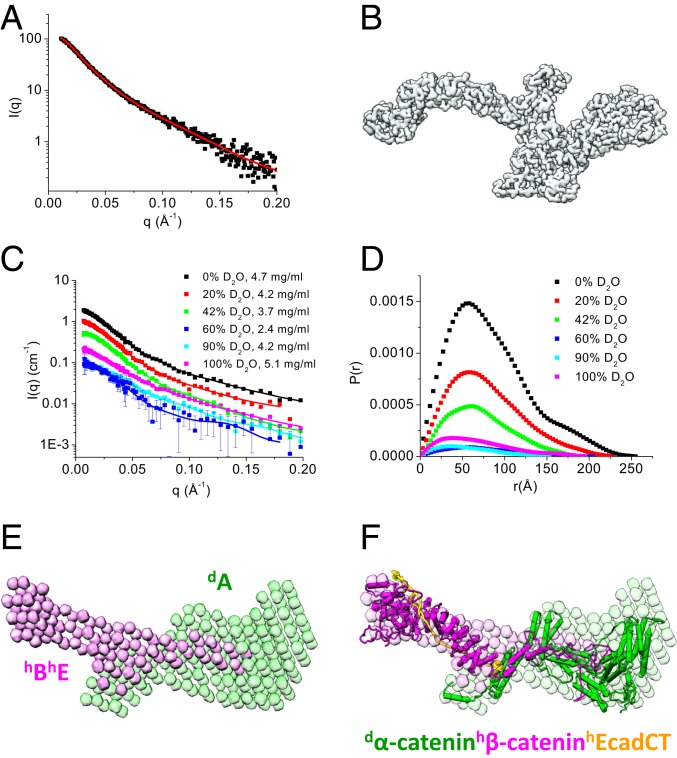
The ABE complex was analyzed by SEC-SAXS. The radius of gyration (Rg) measured from different frames across the single SEC peak of the ABE complex remained almost unchanged (SI Appendix, Fig. S2A), suggesting the absence of intermolecular interactions. Thus, for further analyses, we averaged 4 adjacent SAXS frames covering the center of the SEC peak (Fig. 2A). The asymmetric form of the length distribution function P(r) computed from the SAXS data indicates that the ensemble-averaged conformation of the overall ABE complex has an elongated shape (SI Appendix, Fig. S2C), with a radius of gyration Rg of 68.3 ± 2.2 Å and a maximum dimension Dmax of 265 Å (Table 1). We then produced an ab intio 3-dimensional (3D) structural shape of the ABE complex from the SAXS data (Fig. 2B). This 3D shape represents the average over an ensemble of molecular structures of the ABE complex. Interestingly, the 3D shape reconstructed from the SAXS data resembles some of the negative stain EM averages shown in Fig. 1E and SI Appendix, Fig. S1, with an elongated sickle-like lobe that is connected to 2 smaller compact domains.
Fig. 2.
Overall structure of the ABE complex. (A and B) SEC-SAXS and 3D shape of the ABE complex. Scattering intensity I(q) plot of the ABE complex (A). SI Appendix, Fig. S2 shows the SEC-SAXS profile, Guinier plot, and P(r) of the ABE complex. The 3D shape of the ABE complex reconstructed from the SAXS data using the program Gasbor (68) (B). The red line shown in A is the fit to generate the 3D shape. (C–F) Composite structure of the selectively deuterated dAhBhE complex from contrast variation SANS analysis. Contrast variation SANS data of dAhBhE in 0, 20, 42, 60, 90, and 100% (vol/vol) D2O buffer (C). P(r) functions of the dAhBhE complex at different contrasts (D); C shows quality of fit. Composite 3D shape of the ABE complex generated from the contrast variation SANS data using the program Monsa (37) (E). SI Appendix, Fig. S3A shows quality of fit to the scattering data when generating the 3D shape. Atomic models of α-catenin (from Monte Carlo simulation shown in Fig. 3G), β-catenin (from Monte Carlo simulation shown in Fig. 4F), and the cadherin cytoplasmic domain (PDB ID code 1I7W) were docked into the envelopes using the program UCSF Chimera (72). Green, deuterated α-catenin; magenta, hydrogenated β-catenin and EcadCT.
Table 1.
Summary of SEC-SAXS and SANS data
| Component measured | Method used | Rg (Å) | Dmax (Å) | C (mg/mL) |
| α-Catenin alone and in ABE complexes | ||||
| Monomer* | SEC-SAXS | 43.7 ± 1.1 | 147 | |
| Homodimer* | SEC-SAXS | 58.8 ± 1.6 | 205 | |
| dAhBhE | SANS in 42% D2O | 56.7 ± 0.9 | 200 | 4.2 |
| β-Catenin in BE and ABE complexes | ||||
| β-Catenin | SEC-SAXS | 50.2 ± 1.5 | 180 | Methods |
| hBdE | SANS in 100% D2O | 52.2 ± 2.1 | 220 | 3.2 |
| dAhBdE | SANS in 100% D2O | 51.9± 1.8 | 220 | 3.8 |
| dAhBhE | SANS in 100% D2O | 55.1 ± 1.9 | 230 | 4.2 |
| ABE complexes | ||||
| hAhBdE | SANS 100% D2O | 70.0 ± 1.5 | 250 | 5.4 |
| h(ABE) | SANS 100% D2O | 70.2 ± 1.0 | 251 | 4.5 |
| ABE | SEC-SAXS | 68.3 ± 2.2 | 260 | Methods |
| dAhBhE | SANS 0% D2O | 68.5 ± 1.1 | 257 | 4.7 |
From ref. 38.
To reveal the molecular conformation of α-catenin and β-catenin within the ABE complex, we performed contrast variation SANS on a selectively deuterated ABE complex consisting of deuterated α-catenin (dA), hydrogenated β-catenin (hB), and hydrogenated EcadCT (hE), dAhBhE. The parameters measured by SANS are similar to those measured by SAXS. With the aid of selective deuteration and contrast variation, SANS can further resolve the conformation of a component or a subunit within a multicomponent complex as well as the stoichiometry and the overall architecture of the complex (33–35). For the selectively deuterated dAhBhE complex, the scattering length density (SLD) of the buffer matches that of the hydrogenated components, hBhE, in 42% D2O buffer so that SANS provides information exclusively on the conformation of the deuterated component, dA. The SLD of 100% D2O buffer matches that of 75% deuterated component, dA, so that, in this case, SANS resolves the conformation of the hydrogenated components, hBhE in the complex.
SANS experiments were performed on this dAhBhE complex in 0, 20, 42, 60, 90, and 100% (vol/vol) D2O buffer solutions (Fig. 2C). Analysis of P(r) gives Rg, the forward scattering intensity I(0), and Dmax (36) at different contrasts (Fig. 2D). The molecular mass of the complex can then be determined from the slope of the normalized I(0) measured at different contrasts (SI Appendix, Eq. S1 and Fig. S3B), which was 202,338 ± 6,028 g/mol, close to the theoretical value of 207,505 Da of a selectively deuterated dAhBhE complex.
We then used the multiphase ab initio program Monsa (37) to reconstruct the 3D shape of the dAhBhE complex from the contrast variation SANS data by assuming a 2-phase complex that is composed of a deuterated dA phase and a hydrogenated hBhE phase. In the reconstructed 3D shape of the complex, the hBhE phase consists of 2 distinct lobes and adopts an elongated overall shape (magenta in Fig. 2E). One lobe of hBhE resembles the shape of a sickle, while the other lobe, which is about half the total length of the hBhE phase, overlaps with the dA phase and features a tip that extends into the center of the deuterated dA phase. The 3D shape of dA (green in Fig. 2E) is composed of an extruding arm that overlaps with the extruding tip of hBhE.
Placing the known crystal structure of the armadillo repeat domain of β-catenin in complex with EcadCT (Protein Data Bank [PDB] ID code 1I7X) (23) into the sickle-shaped hydrogenated phase (magenta in Fig. 2F) suggests that the tip or “tongue” that extends from the hBhE phase into the dα-catenin phase and forms a flexible mortise and tenon joint is not from EcadCT. In Fig. 2F, a structure of α-catenin in the deuterated dA phase generated by Monte Carlo simulation was docked into the green-colored shape representing the deuterated α-catenin phase. To gain further insights into the molecular conformation of α- and β-catenin in the ABE complex, we next analyzed the SANS data recorded at the contrast-matching points of the deuterated dA and hydrogenated hBhE phases, respectively.
A Dynamic Ensemble of Multiple α-Catenin Domain Configurations in the ABE Complex.
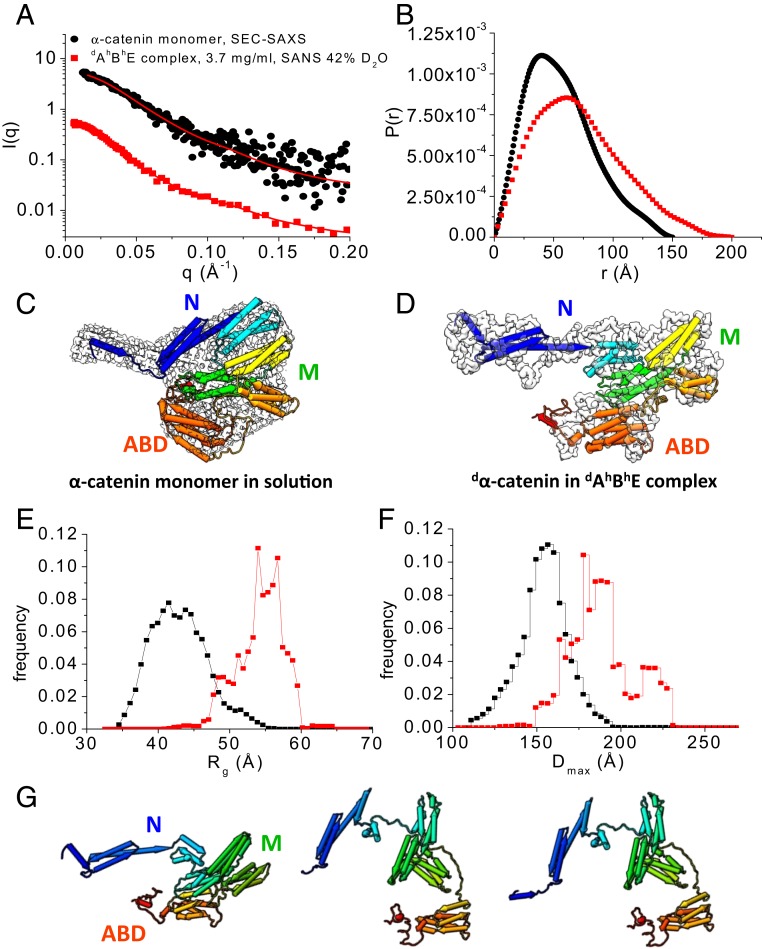
In 42% D2O buffer, the contrast-matching point for hydrogenated hBhE, the scattering of the dAhBhE complex comes exclusively from the deuterated dα-catenin in the complex (Fig. 3A). For comparison, Fig. 3 also shows the SEC-SAXS data of the α-catenin monomer by itself that we published earlier (38). A comparison of the P(r) functions suggests that the overall shape of dα-catenin within the dAhBhE complex is larger and more elongated than that of the α-catenin monomer by itself (Fig. 3B). In the complex, dα-catenin has a Dmax of 200 Å and an Rg of 56.7 ± 0.9 Å compared with the smaller α-catenin monomer by itself that has a Dmax of 147 Å and an Rg of 43.7 ± 1.1 Å (Table 1).
Fig. 3.
Conformation of α-catenin by itself and as part of the dAhBhE complex. (A) SEC-SAXS data of the α-catenin monomer alone in solution (black dots). Data are taken from ref. 38. SANS data of the dAhBhE complex, c = 3.7 mg/mL, in 42% D2O (red squares) at the contrast-matching point of hBhE, which only reveals the conformation of dα-catenin in the complex. Red lines are fits to the experimental scattering data for generating P(r). SI Appendix, Fig. S4 A and B shows Guinier plots and Kratky plots. (B) P(r) of the α-catenin monomer in solution (black) and of dα-catenin in the dAhBhE complex (red) generated from the scattering data in A. (C) The 3D shapes of the α-catenin monomer generated from SEC-SAXS data using the program DAMMIF/DAMMIN (73). (D) The 3D shape of dα-catenin within the dAhBhE complex generated from contrast-matching SANS data in 42% D2O using the program Gasbor (68). Docked into the 3D shapes are the α-catenin structures in solution and in the dAhBhE complex, which were generated by Monte Carlo simulations using the program SASSIE (43). (E and F) Rg distribution (E) and Dmax distribution (F) from EOM analysis (41, 42) of the SAXS and SANS data of the α-catenin monomer in solution (black) and of dα-catenin as a part of the dAhBhE complex (red). For the α-catenin monomer, Rflex = 75.3% (pool 84.5%) and Rσ = 0.68. For dα-catenin within the dAhBhE complex, Rflex = 72.9%, Pool 85.4%, and Rσ = 0.62. These values suggest that α-catenin is a flexible molecule. (G) Flexible structural models of dα-catenin in the dAhBhE complex obtained from Monte Carlo simulations show that the M domain and the ABD can adopt multiple configurations. The simulations were performed using the SANS data of dAhBhE in 42% D2O buffer as constraints. SI Appendix, Fig. S4 C and D shows quality of fit.
The Kratky plot analysis of SAXS or SANS data is an effective way to evaluate the compactness of a protein and the presence of disordered regions (39). For a compact single-domain protein, the Kratky plot shows a bell-shaped curve. For a multidomain protein connected by linkers but still adopting a compact overall conformation, the bell-shaped curve becomes asymmetrically stretched. As the protein becomes more disordered, the curve in the Kratky plot rises increasingly at large q values, where q is the magnitude of scattering vector. In the dimensionless Kratky plot, I(q) is normalized by I(0), and q is normalized by Rg, which allows the comparison of disorder and compactness of proteins of different size and molecular mass (40).
The dimensionless Kratky plots that we obtained from the SEC-SAXS data of the α-catenin monomer and the α-catenin homodimer in solution (38) show curves that are typical of a compact multidomain protein with domains connected by flexible linkers (39) (SI Appendix, Fig. S4B). However, the dimensionless Kratky plot suggests that dα-catenin as part of the dAhBhE complex is less compact than the α-catenin monomer or the homodimer in solution. The Kratky plot analysis thus agrees with the P(r) function that shows that α-catenin is more extended in the ABE complex than when it is free in solution.
The contrast-matching SANS data of the dAhBhE complex in 42% D2O was used to reconstruct the 3D shape of dα-catenin as part of the dAhBhE complex, and the SEC-SAXS data were used to reconstruct the 3D shape of the α-catenin monomer in solution. The α-catenin monomer by itself in solution adopts the shape of a relatively compact disk with a short handle (Fig. 3C), while the 3D shape of dα-catenin in the ABE complex resembles a hook that has an elongated extended N domain with the ABD forming the point of the hook (Fig. 3D). Analyses of the SAXS and SANS data thus show that α-catenin as part of the ABE complex adopts a more open structure compared with an α-catenin monomer by itself or with either protomer of an α-catenin dimer (38).
Our previous analysis of the SEC-SAXS data shows that, in solution, the α-catenin monomer and dimer are flexible, and the solution structures are thus more expanded than those seen in the crystal structures (38). To compare the flexibility of the α-catenin monomer by itself with that of dα-catenin as part the dAhBhE complex, we performed an ensemble analysis of the SEC-SAXS and SANS data using the Ensemble Optimization Method (EOM) program (41, 42). To generate a pool of 10,000 structures of full-length α-catenin, the N1, N2, and M domains and the ABD were taken from the available crystal structure of α-catenin (PDB ID code 4IGG) (22) and treated as rigid bodies. The unstructured linkers connecting N1 to N2, N2 to M, and M to ABD as well as the last 70 residues in the C-terminal tail were assumed to be flexible (Fig. 1A shows domain boundaries). The program Genetic Algorithm Judging Optimization of Ensemble (41, 42) was then used to select an ensemble of structures with average of computed scattering curves that fits the experimental data. This ensemble analysis suggests that the α-catenin monomer by itself and dα-catenin in the ABE complex are both flexible as indicated by the high Rflex and Rσ values (42) (Fig. 3 shows Rflex and Rσ values).
The ensemble analysis also suggests that the conformation distribution of dα-catenin in the ABE complex has 3 discrete populations (Fig. 3 E and F). Although the ensemble analysis for the α-catenin monomer by itself in solution yields 5 conformer populations, the difference in Dmax is less than 26 Å between the most compact fraction and the most expanded fraction. However, for dα-catenin residing in the dAhBhE complex, the difference in Dmax is large, namely 43 Å between the most compact and the most extended conformers (Fig. 3F). In 30% of the complexes, dα-catenin adopts compact conformations with a Dmax of 171.2 Å. In 20% of the complexes, dα-catenin adopts extended conformations with a Dmax of 214.9 Å. In the remaining 50% of the complexes, dα-catenin is in intermediate conformations with a Dmax of 187.7 Å.
Coincidently, the fraction of dα-catenin in the most extended conformation is comparable with the fraction of ABE complex that binds to F-actin (∼18%) as determined from sedimentation F-actin–binding assay and gel band intensity analysis (SI Appendix, Fig. S5). Thus, the extended conformers may correspond to states that bind F-actin strongly, while the compact populations of dα-catenin may represent inhibited states that do not bind F-actin. Additional contrast-matching SANS data show that ABE complexes adopt a more extended conformation when mixed with actin filaments than by themselves in solution (SI Appendix, Fig. S9).
To obtain structural models for α-catenin residing within the ABE complex, we used the structure of the α-catenin monomer in solution (38) as a starting point to perform Monte Carlo simulations using the program SASSIE (43). To generate a pool of α-catenin structures, the linkers between N1 and N2 (residues 164 to 168), between N and M (residues 259 to 275), between M and the ABD (residues 631 to 675), and the disordered C-terminal tail in the ABD (residues 836 to 906) were assumed to be flexible. The structures with computed scattering curves that fit the contrast-matching SANS data in 42% D2O with χ2 < 1.0 (SI Appendix, Fig. S4 C and D) were considered to be valid solutions. Fig. 3G shows representative conformations of dα-catenin as part of the dAhBhE complex, which range from compact to open with the M domain and ABD assuming different relative orientations.
Conformation of β-Catenin within the ABE Complex.
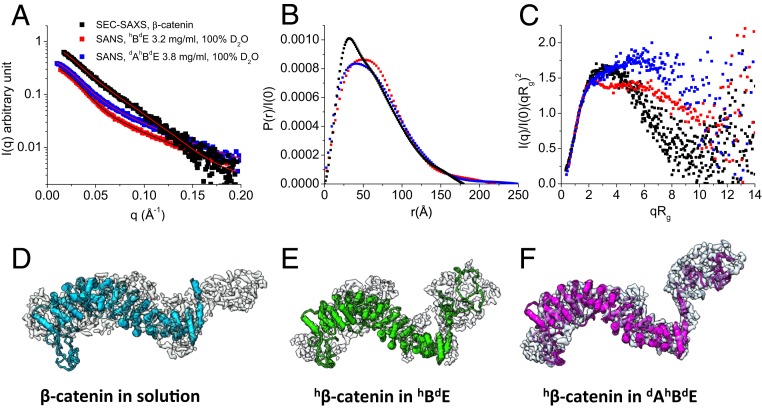
To compare the conformations of β-catenin by itself, in the binary complex with EcadCT (β-catenin•epithelial cadherin cytoplasmic domain [BE]), and as part of the ternary ABE complex, we performed SEC-SAXS experiments on β-catenin alone in solution (SI Appendix, Fig. S6), and contrast-matching SANS experiments on the complex of hydrogenated β-catenin with deuterated EcadCT (hBdE) and of hydrogenated β-catenin with deuterated EcadCT and deuterated α-catenin (dAhBdE) in 100% D2O buffer (Fig. 4A). These experiments revealed that, on binding to EcadCT and α-catenin, the 107-residue N-terminal disordered domain of β-catenin is released from its intramolecular interactions with the central armadillo domain.
Fig. 4.
Conformation of β-catenin by itself and as part of the hBdE or dAhBdE complex. (A) SEC-SAXS data of β-catenin (black squares; reveal the conformation of β-catenin alone in solution) and contrast-matching SANS data of the hBdE complex in 100% D2O buffer (red squares; reveal the conformation of hβ-catenin in hBdE) and of the dAhBdE complex in 100% D2O buffer (blue squares; reveal the conformation of hβ-catenin in the dAhBdE complex). The lines are fits to the experimental data for generating the P(r) functions. SEC-SAXS profile of β-catenin in solution is shown in SI Appendix, Fig. S6. (B) P(r) of β-catenin in solution (black) and as part of the hBdE complex (red) or the dAhBdE complex (blue). (C) Dimensionless Kratky plots of the SAXS and SANS data shown in A. (D–F) The 3D molecular shape of β-catenin in solution from SEC-SAXS (D) and β-catenin in the hBdE complex from SANS in 100% D2O (E) and in the dAhBdE complex (F) from SANS in 100% D2O buffer. The 3D shapes were generated using the program Gasbor (68).
For hβ-catenin as a part of the binary hBdE or the ternary dAhBdE complex, the P(r) function has an asymmetric shape and a long tail that extends to Dmax =∼240 to 250 Å (Fig. 4B). The long tail is more than one-third of the total length of β-catenin. A long P(r) tail is typical of a disordered or denatured protein. The P(r) function of β-catenin by itself in solution, with a Dmax of 180 Å, has a similar asymmetric shape but lacks the long tail. The dimensionless Kratky plots of β-catenin by itself in solution and as part of the hBdE and dAhBdE complexes all show features that are typical of a multidomain protein with disordered regions (Fig. 4C). However, the Kratky plot for β-catenin by itself shows an asymmetric bell-shaped curve, while the Kratky plots of β-catenin in the hBdE and dAhBdE complexes rise at qRg > 2. These comparisons indicate that β-catenin alone in solution is more compact than hβ-catenin as part of the hBdE or dAhBdE complexes. In the dAhBdE complex, β-catenin becomes even more open than in the hBdE complex as suggested by the higher rise in the Kratky plot at qRg > 2 than in that of the hBdE complex (Fig. 4C). Nevertheless, the Rg of β-catenin by itself is only slightly smaller than those of β-catenin in the hBdE and dAhBdE complexes (Table 1). The Kratky plot analysis thus shows that, as part of the ABE complex, β-catenin adopts a more open conformation with an increase in disordered regions.
The 3D shape of β-catenin, reconstructed from the scattering data, provides a real-space view of β-catenin by itself and as a part of the hBdE or dAhBdE complex. The 3D shape of β-catenin, whether by itself or as part of the hBdE or dAhBdE complex, has 2 distinct domains, with a larger sickle-shaped domain and a smaller globular domain (Fig. 4 D–F). Docking the crystal structure of the almost full-length β-catenin (PDB ID code 2Z6G) (31) into the envelopes of β-catenin by itself and as part of the hBdE and dAhBdE complexes suggests that the large sickle-shaped domain represents the armadillo repeat domain so that the smaller domain would represent about 107 residues at the N-terminal end of β-catenin. The structure of this N-terminal segment of β-catenin is not known but was predicted to be disordered using the program I-TASSER (44). In the 3D shape of β-catenin alone, the N-terminal lobe is smaller and less separated from the arched armadillo domain than those in the hBdE and dAhBdE complexes. This is likely due to the dynamic intramolecular interactions of the N-terminal disordered segment with the armadillo repeat domain in the apo form of β-catenin in solution. This observation supports a previous structural study of an almost full-length β-catenin, which found that the N- and C-terminal disordered regions in β-catenin interact with the central armadillo repeat domain (31).
The intramolecular interaction between the N-terminal disordered region and the armadillo repeat domain may be displaced on binding to EcadCT and α-catenin as suggested by a comparison of the 3D shapes of β-catenin by itself in solution and as part of the hBdE and dAhBdE complexes (Fig. 4 D–F). While the N-terminal disordered tail is already released from the armadillo domain in the hBdE complex (Fig. 4E), the separation is even more pronounced in the dAhBdE complex (Fig. 4F). The 3D shape comparison thus corroborates the Kratky plot analysis that shows a more open β-catenin structure in the dAhBdE complex than in the hBdE complex and in apo β-catenin in solution. Altogether, these analyses suggest that the N-terminal disordered segment of β-catenin is released from its intramolecular interaction with the armadillo repeat domain on forming the BE and ABE complexes, and that α-catenin likely stabilizes the separation of this N-terminal disordered segment from the armadillo repeat domain in the ABE complex.
In the 3D shapes of β-catenin by itself and as part of the hBdE and dAhBdE complexes, the C-terminal disordered tail of β-catenin is not distinguishable from the armadillo domain (Fig. 4D). The C-terminal tail of β-catenin thus likely remains bound to the armadillo domain in solution as well as in the complexes. Releasing the C-terminal disordered tail from the armadillo domain likely requires a signaling protein, such as a protein containing a PDZ domain that binds to the C-terminal tail of β-catenin (45).
We also performed contrast variation SANS experiments on the hBdE complex in 0, 42, and 100% D2O buffer (SI Appendix, Fig. S7). The 3D shape of the hBdE complex shows that an elongated dE is threaded through almost the entire armadillo repeat domain of β-catenin (SI Appendix, Fig. S8B). This result is consistent with a crystal structure of the β-catenin armadillo repeat domain bound to EcadCT (23).
The contrast-matching SANS experiment on the dAhBhE complex in 100% D2O revealed the conformation of hBhE in the complex (Fig. 2C). The Kratky plot suggests that the hBhE component in the dAhBhE complex has an even higher content of disordered regions than hB in the dAhBdE complex (SI Appendix, Fig. S8). This result is reasonable because of the presence of additional hE that is unstructured in the hBhE complex as seen in the crystal structure (23). The Rg of hBhE is larger than that of hB in the complex, but Dmax of hBhE in the dAhBhE complex is only slightly larger than that of hB in the dAhBdE complex (Table 1 and SI Appendix, Fig. S9B). The reasons for this are that the molecular mass of EcadCT is small (∼17 kDa) compared with that of the ABE complex (∼200 kDa) and that EcadCT is threaded inside the armadillo repeat domain of β-catenin as seen in the crystal structure.
The Intrinsically Disordered N-Terminal Segment of β-Catenin Modulates the Assembly of the ABE Complex.
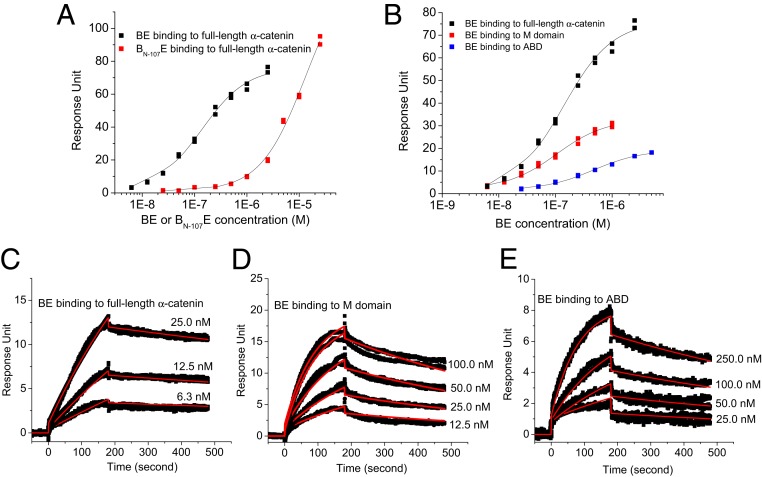
Fig. 4D indicates the separation of the N-terminal disordered segment of β-catenin from the armadillo repeat domain in the hBdE or dAhBdE complex. Fig. 2F shows a tongue that extends from the BE complex deep into α-catenin in the dAhBhE complex, forming a flexible mortise and tenon joint. In the crystal structure (PDB ID code 1I7X), EcadCT is threaded inside the armadillo repeat domain (23), which suggests that the extruding tongue of hBhE in the dAhBhE complex in Fig. 2F is not formed by EcadCT. Based on these observations, we hypothesize that, in the ABE complex, the long disordered N-terminal segment of β-catenin interacts with the M domain and with the ABD of α-catenin, forming interactions in addition to that formed by the N domain of α-catenin (residue 57 to 145) with a helix in β-catenin (residue 118 to 148) that was identified by an earlier crystallographic study (PDB ID code 1DOW) (24). To test this hypothesis, we performed surface-plasmon resonance (SPR) experiments using a β-catenin truncation mutant that lacks the N-terminal 107 residues (BN-107) (Table 2). This truncation mutant forms a stable complex with EcadCT (BN-107E) but aggregates in solution by itself. The SPR results showed that the BN-107E complex has reduced binding affinity for α-catenin with a dissociation constant Kd of 12,625.0 ± 758.2 nM compared with the full-length BE complex that has a Kd of 119.3 ± 6.3 nM (Fig. 5A). We also measured the binding of the BE complex to the M domain and to the ABD of α-catenin (Fig. 5B): BE complex binds to the M domain alone with a Kd of 103.5 ± 6.9 nM and to the ABD alone with a Kd of 485.8 ± 17.9 nM. Although the Kd for BE binding to M or to ABD is comparable with that of BE binding to full-length α-catenin, the SPR sensorgrams and kinetic fitting with a 1:1 binding model indicate that the binding of the BE complex to the M domain or to the ABD has a slower association rate constant ka and a faster dissociation rate constant kd than those for the binding of the BE complex to full-length α-catenin (Fig. 5 C–E and Table 2). Thus, the interactions of the disordered N-terminal segment with the M domain and the ABD are likely to be transient and dynamic. These binding results thus suggest that the long and intrinsically disordered N-terminal segment of β-catenin interacts with the M domain and the ABD of α-catenin, influencing the assembly of the ABE complex and thus, the AJ complex.
Table 2.
Summary of equilibrium and kinetic binding constants of BE to α-catenin
| Binding partners | Kd (nM) | ka (1/ms) | kd (1/s) |
| BE to full-length α-catenin | 119.3 ± 6.3 | (9.6 ± 2.2) × 104 | (4.0 ± 1.2) × 10−4 |
| BN-107E to full-length α-catenin | 12,625.0 ± 758.2 | (1.7 ± 1.1) × 103 | (2.0 ± 1.5) × 10−3 |
| BE to M domain | 103.5 ± 6.9 | (9.9 ± 1.7) × 104 | (1.3 ± 1.4) × 10−3 |
| BE to ABD | 465.8 ± 17.9 | (5.0 ± 0.5) × 104 | (1.0 ± 0.7) × 10−3 |
Fig. 5.
The intrinsically disordered N-terminal segment of β-catenin participates in ABE complex assembly. (A) The binding affinity of the BE complex for full-length α-catenin is reduced if β-catenin lacks the N-terminal 107 amino acid residues (BN-107E). (B) Comparing the binding affinities of the BE complex for the M domain, the ABD of α-catenin, and the full-length α-catenin (Kd values are in the text). (C–E) SPR sensorgrams for the binding of the BE complex to full-length α-catenin (C), to the M domain (D), and to the ABD (E) of α-catenin. The red lines are fit to a 1:1 kinetic binding model to obtain the association and dissociation rate constants ka and kd, respectively, shown in Table 2.
Other than being a component of the AJ, β-catenin is also a key player in the Wnt signaling pathway. The N- and C-terminal segments of β-catenin are known to harbor a number of phosphorylation sites, and they are the location of cancer-related mutations that influence the lifetime and concentration of β-catenin in the cytoplasm and thus, its subsequent translocation to the nucleus for the transcription activation of a set of target genes (46–48). The role of the N-terminal segment of β-catenin beyond the region shown in Fig. 1A in the assembly of the AJ complex has not been studied extensively. However, an earlier in-cell biochemical study reported that β-catenin fragments, with their N-terminal segments cleaved off by caspase-3, may not be able to assemble functional cadherin–catenin cell adhesion complexes (48).
Structural Models of the Entire ABE Complex.
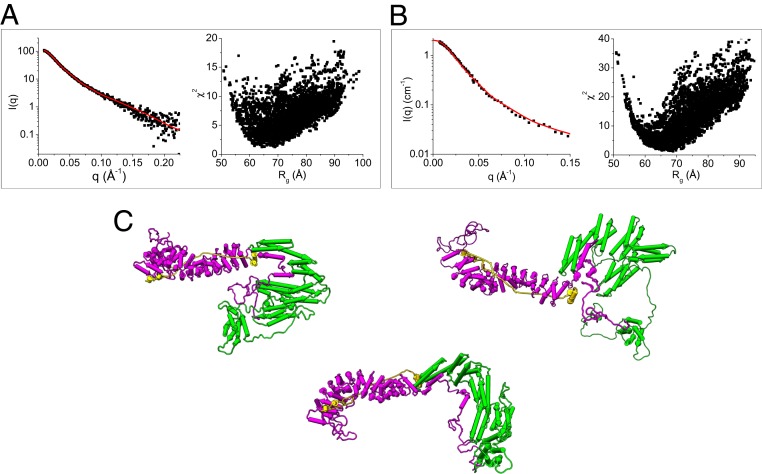
Using the structural models of α-catenin (Fig. 3D) and β-catenin (Fig. 4D) in the ABE complex generated from the contrast-matching SANS data, we assembled a composite structural model of the full-length ABE complex. Assuming that residues 151 to 162 in β-catenin and residues 260 to 275 and 629 to 677 in α-catenin form flexible linkers, this model was then subjected to Monte Carlo simulations to generate a pool of 5,000 plausible and representative ABE structures. The models that best fit both the SEC-SAXS data of ABE and the dAhBhE data obtained from SANS at 0% D2O were selected from this pool. The representative models are shown in Fig. 6. In the dynamic ABE complex, the flexible linkers in α-catenin and β-catenin allow the domains to sample different orientations relative to each other. Based on our finding that the N-terminal disordered domain of β-catenin binds to the M domain and the ABD, we derive a structural model of ABE in which the N-terminal disordered segment of β-catenin is in contact with the M domain and the ABD of α-catenin. Future studies will be aimed at pinpointing the exact residues in the N-terminal disordered region of β-catenin that mediate the binding to the M domain and the ABD of α-catenin.
Fig. 6.
Structural models of the ABE complex from SAXS and SANS. (A) Fit of the structural models generated by Monte Carlo simulations using SASSIE (43) to the SEC-SAXS data of the ABE complex and (B) to the SANS data of the dAhBhE complex in 0% D2O buffer; χ2 vs. Rg of the fits of the computed models to the experimental data are shown in A and B. (C) Representative structural models of the ABE complex that are consistent with the SAXS and SANS data. To comply with the binding data shown in Fig. 5, the N-terminal 107 amino acid residues have to be in contact with the M domain or ABD of α-catenin.
Discussion
The dynamic linkage of cadherin–catenin complexes to the actin cytoskeleton provides mechanical coupling between cells, which is necessary for cell–cell adhesion (1). An optical trap study found that the ABE complex only binds strongly to actin filaments under mechanical tension (20). This experiment mimicked the interactions of the cadherin–catenin complex with oscillating cytoskeletal actin under contraction motions caused by myosin motor proteins pulling on the filaments (15, 16). A 2-state catch bond model (21, 49) was used by these investigators to explain how force-stabilized ABE binds to F-actin. The 2-state catch bond model assumes that the ABE complex exists as an inactive binding state that binds to F-actin weakly and an active state that binds to F-actin strongly. In this model, mechanical force lowers the activation energy and as a result, shifts the ABE complex from a weakly binding low-affinity state to a strongly binding high-affinity state. By contrast, our results provide direct evidence that the ABE complex exists in a wide spectrum of multiple conformations that range from compact to extended conformers rather than occupying just 2 defined states. Thus, analysis of actin binding by the cadherin–catenin complex requires an expanded model that incorporates an ensemble of conformations rather than just the 2 states invoked by the current model. This scenario is similar to the model for the allosteric regulation of protein–ligand binding, which also progressed from an initial 2-state model to the realization of its multistate ensemble nature (50, 51).
The existence of multiple conformational states provides an explanation for why the ABE complex does not bind to F-actin well under equilibrium conditions (19) and why the ABE complex binds to F-actin under force (20). Extended conformations of α-catenin in the ABE complex are likely to be the ones that are competent for F-actin binding, because our contrast-matching SANS experiments show that the ABE complex is more extended when mixed with F-actin than it is when it is in solution (SI Appendix, Fig. S9). Our sedimentation experiments indicate that 18% of the ABE complex binds to F-actin (SI Appendix, Fig. S5). An ensemble analysis of the SANS data suggests that 20% of α-catenin in the ABE complex adopts extended conformations, while 30% adopts compact conformations, and 50% adopts conformations in between. The 80% of α-catenin that exists in somewhat compact conformations is thus likely the ones that are incompetent to bind actin filaments. Structurally, this may be explained by the fact that the more compact conformations of α-catenin may preserve some of the domain–domain interactions that autoinhibit the ABD (as shown in the crystal structure of α-catenin) (22), or else, the N-terminal disordered segment of β-catenin may inhibit the ABD of α-catenin in the ABE complex.
The spectrum of multiple dynamic ABE conformations also allows us to explain why ABE complexes form stable bonds with F-actin under force as observed in the optical trap study. The fact that the distribution of conformations adopted by the ABE complex is far more disperse and dynamic than those adopted by α-catenin or β-catenin by themselves implies the existence of a catch bond-binding mechanism. The ABE complex exists in an ensemble of states, few of which are competent to bind actin. The application of external force reduces this ensemble of states to a much smaller number of states that are competent to bind actin. Reducing the density of states implies a reduction of the entropy of the ABE complex, which requires the application of force. Alternatively, it is conceivable that the application of external force results in the exposure of a second binding site, which enhances actin binding. This possibility is less satisfying, however, as it fails to provide an explanation for the emergence of a large dynamic spectrum of the ABE complex and the subsequent collapse of the spectrum to a far smaller number of states on its force-assisted binding to actin.
It is important to recognize that it does not take a very large external force to substantially reduce the number of conformational states of the ABE complex. Indeed, the force of about 10 pN (of 2.43 kBT/nm) that is needed to strengthen the binding of the ABE complex to F-actin (20) is approximately the amount of force generated by myosin motors pulling on actin filaments (52–54). Thus, a relatively weak force, in cellular terms, is required to enhance the binding of ABE to F-actin. As a result, the catch bond-binding mechanism, created by the high flexibility of the ABE complex, produces a strong response to a small change in applied force. This correlates with high sensitivity. Additionally, the fact that the change in free energy is small implies that the reaction is nearly reversible. Thermodynamic reversibility is necessary for a sensory system to be capable of repeated acts of sensation without information loss. This notion follows from the second law of thermodynamics as expanded into Landauer’s principle (55, 56), which is the idea that any logically reversible transmission of information requires thermodynamic reversibility. High sensitivity and reversibility are likely required behaviors of a cellular mechanosensor and mechanotransducer (57).
Methods
Protein Purification and Complex Reconstitution.
The complementary DNAs encoding full-length human αE-catenin, human β-catenin, and the entire cytoplasmic domain of human E-cadherin (EcadCT; residues 731 to 882) were subcloned individually into the pET32a vector (EMD Biosciences, Inc) with an N-terminal tobacco etch virus (TEV) protease-cleavable 6xHis tag. The expression plasmids were transformed into competent Rosetta 2 (DE3) cells (EMD Biosciences, Inc.). The bacteria were grown in Luria–Bertani medium in the presence of 0.1 mg/mL ampicillin and 0.034 mg/mL chloramphenicol. When the culture reached an optical density at 600 nm of about 0.9, the cells were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside and grown overnight at 20 °C.
For protein purification, cells were pelleted by centrifugation at 5,000 × g for 20 min, and the cell pellet was resuspended in 30 mL/L cell culture of lysis/binding buffer containing 1× phosphate buffered saline (PBS) buffer, pH 7.4, 750 mM NaCl, 20 mM imidazole, 0.25 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The cell suspension was subjected to sonication cycles on ice for 12 cycles, 30 s for each cycle, using a Fisher Model 505 sonicator. The cell lysate was centrifuged at 13,000 × g for 20 min at 4 °C. The supernatant was loaded onto a HiTrap Chelating Sepharose HP column (GE Healthcare Life Sciences) precharged with Ni2+ and preequilibrated with lysis/binding buffer. The column was washed with 40 column volumes of lysis/binding buffer and eluted with 2 column volumes of elution buffer (150 mM phosphate buffer, pH 7.5, 500 mM NaCl, 500 mM imidazole, 0.25 mM TCEP, 0.1 mM PMSF). The protein was then subjected to SEC using a Superdex 200 Increase 10/30 column (GE Healthcare Life Sciences) equilibrated with SEC buffer (20 mM Tris⋅HCl, pH 7.5, 300 mM NaCl, 3 mM ethylenediaminetetraacetic acid [EDTA], 0.25 mM TCEP). Peak fractions were combined and incubated with 20 μM TEV protease on ice for 48 h to cleave off the 6xHis tag (after cleavage with TEV protease, only a glycine residue remains at the N terminus of the proteins). The protein was concentrated using a Vivaspin 20 centrifugal concentrator of 10 kDa molecular weight cut-off. A second round of SEC was used to remove the cleaved tag. Protein concentrations were determined by ultraviolet absorbance at 280 nm using extinction coefficients calculated using the ProtParam program (58) on the ExPASy Proteomics Server (https://www.expasy.org/).
Full-length β-catenin binds to EcadCT with a Kd of 104.4 ± 3.2 nM as determined by SPR. The BE complex was formed by incubating β-catenin with EcadCT at a molar ratio of 1:2 to saturate the binding site in β-catenin. The BE complex was isolated by SEC on a Superdex 200 Increase 10/30 column equilibrated with SEC buffer. The BE complex binds α-catenin with a Kd of 119.3 ± 6.3 nM. The ABE complex was reconstituted by incubating purified BE complex with α-catenin at a molar ratio of 2:1 with excess amount of BE. The reconstituted ABE complex was then isolated by SEC on a Superdex 200 Increase 10/30 column equilibrated with SEC buffer. ABE complex for analysis by EM or scattering experiments was run over SEC immediately before use.
Deuterated proteins were expressed in 85% D2O (vol/vol) M9 medium. Purification of the deuterated proteins and reconstitution of the selectively deuterated BE and ABE complexes were the same as those of the hydrogenated complexes. The online program MULCh (59) was used to compute the neutron SLD of the buffer at different D2O volume fractions and of the hydrogenated and deuterated proteins.
EM.
A 3.5-μL aliquot of ABE sample (0.01 mg/mL) was adsorbed for 1 min on a glow-discharged copper grid covered with a carbon-coated plastic film and negatively stained with 0.75% (wt/vol) uranyl formate solution as described (60). Specimens were imaged with a CM10 electron microscope (Philips) equipped with a tungsten filament and operated at an acceleration voltage of 100 kV. Micrographs were collected at a calibrated magnification of 41,513× (nominal magnification of 52,000×) with an XR16L-ActiveVu camera (AMT) at a defocus value of −1.5 μm.
EM Image Processing.
In total, 11,535 particles were interactively picked and windowed into 128 × 128-pixel images. After normalization and reduction of the particle images to 64 × 64 pixels, the particles were aligned and classified with the Interactive Stable Alignment and Clustering (ISAC) procedure (32) implemented in SPARX (61), specifying 50 particles per group and a pixel error threshold of 2. Three ISAC generations yielded 275 class averages.
SEC-SAXS.
The SEC-SAXS experiments were performed at the Stanford Synchrotron Radiation Light Source (SSRL) Bio-SAXS beamline 4–2. The experimental setup was as described previously (62). Two of the same SEC columns (Superdex 200 Increase 3.2/300 column; GE Healthcare Life Sciences) were used for high-throughput tandem SEC-SAXS data collection. The columns were equilibrated with SEC-SAXS running buffer (10 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid [HEPES], pH 7.5, 150 mM NaCl, 3 mM EDTA, 5 mM dithiothreitol). Before the SAXS experiments, the ABE complex was dialyzed overnight against the same SEC-SAXS running buffer; 10-μL aliquots of the ABE complex at 8.2 mg/mL or β-catenin at 17.4 mg/mL were applied to the column. A total of 500 images were recorded using 1-s exposures every 5 s at a flow rate of 0.05 mL/min.
The data were plotted as I(q) vs. q, where q = 4πsin(θ)/λ, with 2θ being the scattering angle and λ being the wavelength of the X-rays. The script hplcplots, available at SSRL beamline 4–2, was used for consecutive Guinier analysis implemented in the program AUTORG (63) and for assessment of data quality (e.g., radiation damage and cleanliness of the sample cell) by providing Rg, I(0), and an experimental intensity at a low q value. Since marginal interparticle interactions (concentration dependence) were observed over the peak, an average profile over images 285 to 289 was generated, scaled, and merged for further analyses.
SANS.
SANS data were collected at the EQ-SANS instrument at the Spallation Neutron Source (64) and the BIOSANS instrument at the High Flux Isotope Reactor (65), both at the Oak Ridge National Laboratory. The EQ-SANS used a single sample-to-detector distance setting of 4 m for all measurements. The beam was defined with a 25-mm-diameter source aperture and a 10-mm sample aperture. Two settings for the choppers were used for measuring the complex. In the first setting, the choppers ran at 30 Hz in frame-skipping mode, with the minimum wavelength set to 2 Å. This configuration provided a q range of 0.004 to 0.40 Å−1. In the second setting, the choppers ran at 60 Hz and were set to provide a minimum wavelength of 4 Å. This configuration spans a q range of 0.007 to 0.27 Å−1. A single configuration was used for the Bio-SANS that placed the main detector at 15.5 m from the sample, and the wing detector was fixed at 1.13 m from the sample. No guides were inserted, the source aperture was 40 mm in diameter, and the sample aperture was 10 mm. The wavelength was set to 6 Å. This configuration provides a q range of 0.003 to 0.80 Å−1. All SANS measurements were performed at 10 °C ± 1 °C.
The program Mantid (66) was used to reduce the data from the samples and from the backgrounds using standard procedures that correct for incident flux spectrum, sample transmission, and detector sensitivity as well as the detector dark current, which represents electronic noise and natural sources of radiation. Then, the data were azimuthally averaged to generate I(q) vs. q plots. Absolute intensity scaling for both configurations was done with a calibrated standard (67). The sample scattering was then corrected for solvent scattering by subtracting the 1-dimensional profiles to produce the final reduced data.
Before the SANS experiments, a PBS tablet (ThermoFisher) for making 500 mL 1× PBS buffer was soaked in 2 mL D2O and vacuum dried 5 times at 80 °C to exchange the H in the tablet into D. The D-exchanged PBS tablet was dissolved with 500 mL 99.9% D2O, which was then mixed with H-unexchanged 1× PBS buffer at different volume fractions. The protein complex of 0.4 mL was dialyzed 5 times, each for 6 to 8 h, against 5 mL of 0, 20, 42, 85, 90, and 100% PBS D2O (vol/vol) buffer.
Analysis of SAXS and SANS Data and Structural Modeling.
The length distribution function P(r) was generated using the program GNOM (36) to obtain Rg and Dmax. When computing P(r), the I(q) of q 0.2 Å−1 was selected for fitting. A series of Dmax values at which P(r) = 0 was tested and the optimum Dmax value with the minimum α-value and maximum total quality values was selected as the final solution.
The program Gasbor (68) was used to generate the ab initio shape of the ABE complex from the SAXS data. The program MONSA (37) was used to generate a multiphase ab initio shape for the dAh(BE) complex using the SANS data collected at different volume fractions of D2O buffer, with the input data of Rg values of dα-catenin in 42% D2O and hBhE in 100% D2O and the deuteration level of dα-catenin for neutron SLD contrast calculations. The EOM program (41, 42) was used to evaluate the flexibility of α-catenin and to estimate the distribution of the multiconformers by assuming that the unstructured linkers connecting the N, M, and ABDs are flexible.
The Monte Carlo simulation module in the program suite SASSIE (69) was used to generate the all-atomic models of α-catenin and β-catenin in the ABE complex, assuming that the linkers in α-catenin and the hinge region formed by amino acid residues 155 to 162 in β-catenin are flexible. The maximum angle that each torsion in each of the flexible regions could sample was 30°. Approximately 10,000 trial attempts were performed for each round of simulations. The SASCAL module in SASSIE was used to generate theoretical SAXS or SANS curves at different neutron SLD contrasts from the structural models, and the ANALYZE module was used to fit and compare the computed SAXS and SANS curves with the experimental SAXS and SANS data. Models with the minimum χ2 values were selected.
Before Monte Carlo simulations, the crystal structure of nearly full-length α-catenin (PDB ID code 4IGG) (22) was used as the starting structural model. The program SAXSTER (70) was used to build homology models for the disordered regions missing in the crystal structure, which are amino acid residues 18 to 81, amino acid residues 636 to 665 (the linker region between the M3 subdomain and the ABD), and amino acid residues 862 to 907 at the C-terminal end. The crystal structure of the N-terminal segment of mouse α-catenin (PDB ID code 4P9T) (71) served as a template for homology modeling. The starting model for β-catenin was the crystal structure of Danio rerio β-catenin (PDB ID code 2Z6G). Homology models for the N- and C-terminal disordered regions in β-catenin (amino acid residues 1 to 125 and 692 to 781) were built using the program SAXSTER (70).
SPR Experiments.
SPR experiments were performed on a Biacore X100 instrument (GE Healthcare Life Sciences). A Biacore CM5 Biosensor chip was activated by N-hydroxysuccinimide and N-ethyl-N′-[3-(diethylamino)propyl]carbodimide (GE Healthcare Life Sciences). The ligand (full-length α-catenin, the M domain, and ABD) was dissolved at 5 µg/mL in 10 mM sodium acetate, pH 4.9, and injected to coat the activated sensor chip surface in 1 of 2 flow cells. Noncross-linked ligand was washed away, and unoccupied sites were blocked with 1 M ethanolamine, pH 8.5. The control flow cell was activated and blocked without ligand injection. The analytes (BE or BN-107E) were dissolved in 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant polysorbate 20 (HBS-EP buffer) and injected in increasing concentrations over the ligand-coated surfaces at 30 µL/min for 180 s. The dissociation time was 800 s. At the end of each injection–dissociation cycle, the sensor chip was regenerated with 50 mM triethylamine, pH 9.15, 4.0 M MgCl2, and HBS-EP buffer. SPR experiments were performed at 15 °C. The BIA-evaluation software provided by the instrument manufacturer was used to obtain the equilibrium dissociation constant Kd and the kinetic association and dissociation rate constants (ka and kd, respectively) with a 1:1 binding model.
Supplementary Material
Acknowledgments
This research was funded by NSF Grant MCB-1817684 (to Z.B.) and National Center for Research Resources Grant 2G12 RR003060 (to City College of New York). A portion of the research conducted at Oak Ridge National Laboratory’s Spallation Neutron Source and High Flux Isotope Reactor was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy (DOE). The Bio-SANS of the Center for Structural Molecular Biology at the High Flux Isotope Reactor is supported by the Office of Biological and Environmental Research of the DOE. Use of the SSRL, Stanford Linear Accelerator Center’s is supported by DOE, Office of Science, Office of Basic Energy Sciences Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and NIH, National Institute of General Medical Sciences (NIGMS) Grant P41 GM103393. We thank Carrie Gao for technical support during the SANS experiments at Spallation Neutron Source. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911489116/-/DCSupplemental.
References
- 1.Takeichi M., Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15, 397–410 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner B. M., Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6, 622–634 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Kim N. G., Koh E., Chen X., Gumbiner B. M., E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U.S.A. 108, 11930–11935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendonsa A. M., Na T. Y., Gumbiner B. M., E-cadherin in contact inhibition and cancer. Oncogene 37, 4769–4780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery J. P., Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Mayor R., Etienne-Manneville S., The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17, 97–109 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jeanes A., Gottardi C. J., Yap A. S., Cadherins and cancer: How does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejana E., Orsenigo F., Endothelial adherens junctions at a glance. J. Cell Sci. 126, 2545–2549 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Shapiro L., Weis W. I., Structure and biochemistry of cadherins and catenins. Cold Spring Harb. Perspect. Biol. 1, a003053 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertocchi C., et al. , Nanoscale architecture of cadherin-based cell adhesions. Nat. Cell Biol. 19, 28–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia M. A., Nelson W. J., Chavez N., Cell-cell junctions organize structural and signaling networks. Cold Spring Harb. Perspect. Biol. 10, a029181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leckband D. E., de Rooij J., Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 30, 291–315 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Nagafuchi A., Takeichi M., Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 7, 3679–3684 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecuit T., Yap A. S., E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17, 533–539 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M., Alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Borghi N., et al. , E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U.S.A. 109, 12568–12573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladoux B., Nelson W. J., Yan J., Mège R. M., The mechanotransduction machinery at work at adherens junctions. Integr. Biol. 7, 1109–1119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai R., et al. , Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15, 261–273 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J., Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley C. D., et al. , Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346, 1254211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas W., et al. , Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 90, 753–764 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangarajan E. S., Izard T., Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber A. H., Weis W. I., The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105, 391–402 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Pokutta S., Weis W. I., Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol. Cell 5, 533–543 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ishiyama N., et al. , An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rangarajan E. S., Izard T., The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J. Biol. Chem. 287, 18492–18499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano Y., Amano Y., Yonemura S., Hakoshima T., The force-sensing device region of α-catenin is an intrinsically disordered segment in the absence of intramolecular stabilization of the autoinhibitory form. Genes Cells 23, 370–385 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Choi H. J., et al. , αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U.S.A. 109, 8576–8581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao M., et al. , Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5, 4525 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Huber A. H., Nelson W. J., Weis W. I., Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90, 871–882 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Xing Y., et al. , Crystal structure of a full-length beta-catenin. Structure 16, 478–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z., Fang J., Chittuluru J., Asturias F. J., Penczek P. A., Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaccaï G., Jacrot B., Small angle neutron scattering. Annu. Rev. Biophys. Bioeng. 12, 139–157 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Capel M. S., et al. , A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science 238, 1403–1406 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Whitten A. E., Trewhella J., Small-angle scattering and neutron contrast variation for studying bio-molecular complexes. Methods Mol. Biol. 544, 307–323 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Semenyuk A. V., Svergun D. I., GNOM–A program package for small-angle scattering data-processing. J. Appl. Crystallogr. 24, 537–540 (1991). [Google Scholar]
- 37.Svergun D. I., Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholl I. D., et al. , α-Catenin structure and nanoscale dynamics in solution and in complex with F-actin. Biophys. J. 115, 642–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Receveur-Brechot V., Durand D., How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durand D., et al. , NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Bernadó P., Svergun D. I., Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. Biosyst. 8, 151–167 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Tria G., Mertens H. D., Kachala M., Svergun D. I., Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2, 207–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis J. E., Raghunandan S., Nanda H., Krueger S., SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput. Phys. Commun. 183, 382–389 (2012). [Google Scholar]
- 44.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata T., Chuma M., Kokubu A., Sakamoto M., Hirohashi S., EBP50, a β-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology 38, 178–186 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Xu W., Kimelman D., Mechanistic insights from structural studies of beta-catenin and its binding partners. J. Cell Sci. 120, 3337–3344 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Mo R., et al. , The terminal region of beta-catenin promotes stability by shielding the Armadillo repeats from the axin-scaffold destruction complex. J. Biol. Chem. 284, 28222–28231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinhusen U., et al. , Apoptosis-induced cleavage of beta-catenin by caspase-3 results in proteolytic fragments with reduced transactivation potential. J. Biol. Chem. 275, 16345–16353 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Thomas W. E., Vogel V., Sokurenko E., Biophysics of catch bonds. Annu. Rev. Biophys. 37, 399–416 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boehr D. D., Nussinov R., Wright P. E., The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howard J., Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, Sunderland, MA, 2001). [Google Scholar]
- 53.Finer J. T., Simmons R. M., Spudich J. A., Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature 368, 113–119 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Veigel C., Bartoo M. L., White D. C., Sparrow J. C., Molloy J. E., The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys. J. 75, 1424–1438 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landauer R., Irreversibility and heat generation in the computing process. IBM J. Res. Dev. 5, 183–191 (1961). [Google Scholar]
- 56.Bennett C. H., Notes on Landauer’s principle, reversible computation, and Maxwell’s Demon. Stud. Hist. Philos. Mod. Phys. 34, 501–510 (2003). [Google Scholar]
- 57.Hoffman B. D., Grashoff C., Schwartz M. A., Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gasteiger E. H. C., et al. , “Protein identification and analysis tools on the ExPASy server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Humana Press, 2005), pp. 571–607. [Google Scholar]
- 59.Whitten A. E., Ca S. Z., Trewhella J., MULCh: Modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. Appl. Crystallogr. 41, 222–226 (2008). [Google Scholar]
- 60.Ohi M., Li Y., Cheng Y., Walz T., Negative staining and image classification–Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hohn M., et al. , SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Edwards A. L., Matsui T., Weiss T. M., Khosla C., Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol. 426, 2229–2245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I., ATSAS 2.1–towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40 (suppl. 1), s223–s228 (2007). [Google Scholar]
- 64.Zhao J. K., Gao C. Y., Liu D., The extended Q-range small-angle neutron scattering diffractometer at the SNS. J. Appl. Crystallogr. 43, 1068–1077 (2010). [Google Scholar]
- 65.Heller W. T., et al. , The suite of small-angle neutron scattering instruments at Oak Ridge National Laboratory. J. Appl. Crystallogr. 51, 242–248 (2018). [Google Scholar]
- 66.Arnold O., et al. , Mantid—Data analysis and visualization package for neutron scattering and μ SR experiments. Nucl. Instrum. Methods Phys. Res. Sect. A 764, 156–166 (2014). [Google Scholar]
- 67.Wignall G. D., Bates F. S., Absolute calibration of small-angle neutron-scattering data. J. Appl. Crystallogr. 20, 28–40 (1987). [Google Scholar]
- 68.Svergun D. I., Petoukhov M. V., Koch M. H., Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perkins S. J., et al. , Atomistic modelling of scattering data in the collaborative computational project for small angle scattering (CCP-SAS). J. Appl. Crystallogr. 49, 1861–1875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.dos Reis M. A., Aparicio R., Zhang Y., Improving protein template recognition by using small-angle x-ray scattering profiles. Biophys. J. 101, 2770–2781 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibahara T., Hirano Y., Hakoshima T., Structure of the free form of the N-terminal VH1 domain of monomeric α-catenin. FEBS Lett. 589, 1754–1760 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.