Significance
Myosin-binding protein C (MyBP-C) is a critical component of the skeletal muscle sarcomere, muscle’s smallest contractile unit. MyBP-C’s importance is evident by genetic mutations leading to human myopathies, such as distal arthrogryposis (i.e., club foot). However, the molecular basis of MyBP-C’s functional impact on skeletal muscle contractility is far from certain. Complicating matters further is the expression of fast- and slow-type MyBP-C isoforms that depend on whether the muscle is fast- or slow-twitch. Using multiscale proteomic, biophysical, and mathematical modeling approaches, we defined the expression, localization, and modulatory capacities of these distinct skeletal MyBP-C isoforms in rat skeletal muscles. Each MyBP-C isoform modulates muscle contractility differentially, providing the capacity to fine-tune a muscle’s mechanical performance as physiological demands arise.
Keywords: muscle contraction, calcium regulation, myosin thick filament, in vitro motility, mass spectrometry
Abstract
Skeletal muscle myosin-binding protein C (MyBP-C) is a myosin thick filament-associated protein, localized through its C terminus to distinct regions (C-zones) of the sarcomere. MyBP-C modulates muscle contractility, presumably through its N terminus extending from the thick filament and interacting with either the myosin head region and/or the actin thin filament. Two isoforms of MyBP-C (fast- and slow-type) are expressed in a muscle type-specific manner. Are the expression, localization, and Ca2+-dependent modulatory capacities of these isoforms different in fast-twitch extensor digitorum longus (EDL) and slow-twitch soleus (SOL) muscles derived from Sprague–Dawley rats? By mass spectrometry, 4 MyBP-C isoforms (1 fast-type MyBP-C and 3 N-terminally spliced slow-type MyBP-C) were expressed in EDL, but only the 3 slow-type MyBP-C isoforms in SOL. Using EDL and SOL native thick filaments in which the MyBP-C stoichiometry and localization are preserved, native thin filament sliding over these thick filaments showed that, only in the C-zone, MyBP-C Ca2+ sensitizes the thin filament and slows thin filament velocity. These modulatory properties depended on MyBP-C’s N terminus as N-terminal proteolysis attenuated MyBP-C’s functional capacities. To determine each MyBP-C isoform’s contribution to thin filament Ca2+ sensitization and slowing in the C-zone, we used a combination of in vitro motility assays using expressed recombinant N-terminal fragments and in silico mechanistic modeling. Our results suggest that each skeletal MyBP-C isoform’s N terminus is functionally distinct and has modulatory capacities that depend on the muscle type in which they are expressed, providing the potential for molecular tuning of skeletal muscle performance through differential MyBP-C expression.
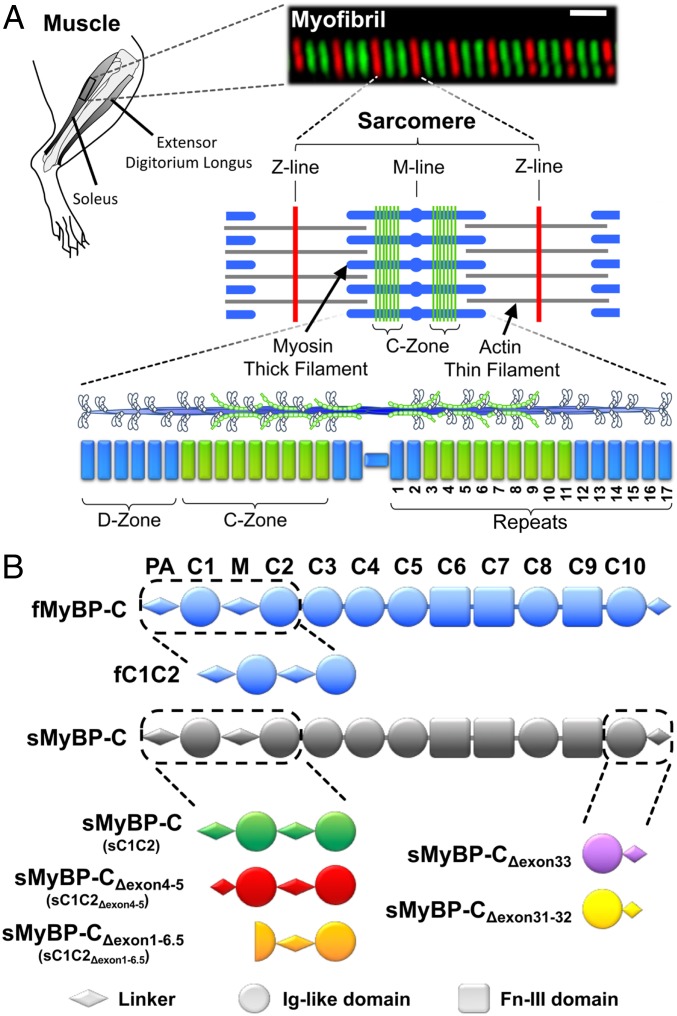
Skeletal muscle myosin-binding protein C (MyBP-C) is a ∼130-kDa, thick filament-associated protein and a key modulator of muscle contractility (1). Its importance is emphasized by mutations in its genes resulting in human skeletal muscle myopathies, such as distal arthrogryposis (2, 3). MyBP-C has a modular structure, consisting of 7 immunoglobulin (Ig) and 3 fibronectin-like (Fn) domains, C1 to C10 (Fig. 1B). Through its C-terminal domain interactions with the thick filament backbone (4), MyBP-C is reportedly localized to 7 to 9 stripes within a distinct central region (i.e., the C-zone) of the sarcomere (Fig. 1A), muscle’s smallest contractile unit (5). At its other end, the N terminus extends away from the thick filament, allowing for potential interactions with the actin thin filament (6). N-terminal interactions with actin filaments have been reported for the orthologous cardiac MyBP-C (7–10) although there is also evidence for binding to the myosin head region in cardiac muscle (11–14).
Fig. 1.
MyBP-C isoforms and their localization in rat skeletal muscle. (A) In this study, contractile protein preparations were obtained from the rat hind limb soleus and extensor digitorum longus. An immunofluorescent image of a soleus myofibril composed of 8 sarcomeres, whose boundaries are identified by antibodies to the Z-line protein, α-actinin (red). Antibodies against slow-type MyBP-C (green) highlight the distinct regions (C-Zones) in the sarcomere center where MyBP-C is localized. The sarcomere is comprised of myosin thick filaments (blue) and actin thin filaments (gray). Striated muscle myosin forms bipolar thick filaments where each half thick filament consists of 17 repeats that demarcate the 43-nm myosin head helical repeat. MyBP-C within the C-Zone occupies ∼9 repeats near the thick filament center where the ends of the thick filament are devoid of MyBP-C (D-Zone). (B) Skeletal MyBP-C consists of 7 Ig-like (Ig-like, circles) and 3 fibronectin-III (Fn-III, rectangles) domains named C1 through C10. A proline-alanine rich domain (PA) precedes the C1 domain and an M-domain linker (M) exists between the C1 and C2 domains. In skeletal muscle,1 fast-type MyBP-C and multiple slow-type MyBP-C splice isoforms are present. Slow-type MyBP-C is alternatively spliced at its N and C terminus. See SI Appendix, Figs. S1 and S2 for isoform-specific sequences.
The functional impact of MyBP-C was first demonstrated when partial extraction of the whole molecule in skinned skeletal muscle fibers resulted in both increased force production and shortening velocity at submaximal calcium levels (15, 16). Based on more recent in vitro data using expressed MyBP-C fragments, MyBP-C’s N-terminal domain interactions with the actin thin filament and/or myosin head may be the primary mechanistic modulators of muscle contractility (17, 18). Specifically, N-terminal fragment binding to the thin filament can sensitize the thin filament to calcium by shifting the position of tropomyosin out of the “blocked” state to facilitate myosin binding to actin (18). In addition, MyBP-C can act as a “brake” to reduce sarcomere shortening (19) and thin filament sliding velocity (17, 18) through its interaction with either the thin filament and/or myosin head region. Although extremely informative, the in vitro N-terminal fragment studies described above were performed with contractile proteins isolated from heterologous species or muscle tissue, creating nonnative simplified contractile model systems. To complicate matters further, 2 mammalian skeletal muscle MyBP-C isoforms exist that are encoded by separate genes, MYBPC1 and MYBPC2. These isoforms were historically named slow-type skeletal MyBP-C (MYBPC1) and fast-type skeletal MyBP-C (MYBPC2), based on the twitch speed of the muscle in which they were expressed (20). Recent studies suggest that the number of MyBP-C isoforms and their muscle type expression profiles are far more complex due to numerous slow-type MyBP-C isoforms that result from differential exon splicing (21, 22). Moreover, slow-type MyBP-C is also found in fast-twitch muscles as well, suggesting differential gene expression (23, 24). Therefore, these findings raise the intriguing possibility that a mixture of MyBP-C isoforms may exist in skeletal muscles, regardless of twitch speed, and their N termini may possess distinct functional capacities to modulate skeletal muscle contractility.
Here, using multiscale proteomic, biophysical, and in silico approaches, we define the expression, localization, and modulatory capacities of multiple skeletal MyBP-C isoforms in native skeletal muscle systems. To limit the inherent biological complexity associated with muscle-specific MyBP-C isoform expression (see above), we chose rat slow-twitch soleus (SOL) and fast-twitch extensor digitorum longus (EDL) muscles as model systems. These were the purest rat skeletal muscles with regard to fiber type composition identified in the literature (25). Regardless, we first identified and quantified the abundances and localization of the skeletal myosin and MyBP-C isoforms in the SOL and EDL muscle samples by mass spectrometry and immunofluorescence microscopy. Four different N-terminal MyBP-C isoforms were identified: one being the fast-type MyBP-C isoform, with the remaining 3 slow-type MyBP-C isoforms resulting from alternative N-terminal exon splicing (Fig. 1B). We then determined the Ca2+-dependent mechanical interactions between native thin and thick filaments isolated from the SOL and EDL muscles. Thin filament motion was sensitized to Ca2+, and maximal sliding velocities were slowed only within the thick filament C-zones. Next, we determined how each of the MyBP-C isoforms contributed to this modulation of actomyosin contractility at the molecular level. We combined in vitro motility assays using expressed recombinant N-terminal fragments and in silico mechanistic modeling. Our results suggest that each skeletal MyBP-C isoform is functionally distinct and, interestingly, their modulatory capacity depends on the muscle in which they are expressed. Therefore, these rat skeletal muscles use a combination of gene expression and alternative exon splicing in order to fine tune contractility and thus meet these muscles’ diverse physiological demands.
Results
Identification and Quantification of Myosin and MyBP-C Isoforms in Rat Soleus and Extensor Digitorum Longus Muscles by Mass Spectrometry.
To determine muscle myosin and MyBP-C protein compositions, myofibrillar protein preparations were isolated from ∼30-mg samples of rat slow-twitch SOL and fast-twitch EDL skeletal muscles (Materials and Methods). We digested these protein preparations with trypsin, analyzed the peptides by liquid chromatography mass spectrometry (LCMS), and quantified protein abundances by label-free analyses (26, 27). Unique peptides generated from the digestion of 5 myosin heavy chain isoforms were present at varying levels in the myofibrillar protein samples (Table 1). Slow-myosin heavy chains expressed from the MYH7 gene in the SOL samples and fast-myosin heavy chains expressed from the MYH4 and MYH1 genes in the EDL samples predominated (Table 1). The presence of multiple myosin heavy chain isoforms in both the rat SOL and EDL samples confirmed these muscles were composed of mixed fiber types at the proportions previously reported (28, 29).
Table 1.
Myosin heavy chain isoform abundance in rat slow-twitch soleus and fast-twitch extensor digitorum longus skeletal muscles determined by mass spectrometry
| Gene | Fiber type | SOL | EDL | ||
| Abundance, % | SD | Abundance, % | SD | ||
| MYH7 | I | 87.6 | 2.3 | 0.8 | 0.1 |
| MYH2 | IIA | 11.8 | 2.2 | 5.1 | 0.5 |
| MYH4 | IIB | 0.0 | 0.0 | 70.4 | 0.7 |
| MYH1 | IID/X | 0.2 | 0.1 | 23.7 | 0.5 |
| MYH3 | 0.5 | 0.8 | 0.2 | 0.3 | |
Abundance percentage determined as described in SI Appendix, Materials and Methods. SD determined from multiple muscle samples (SOL, n = 7; EDL, n = 7). MYH3 is an embryonic isoform, and not associated with any specific muscle fiber type.
To determine both the overall content and specific MyBP-C isoforms expressed in these muscles, we analyzed peptides originating from the tryptic digestion of the various MyBP-C isoforms (SI Appendix, Fig. S1). The total amount of MyBP-C relative to myosin heavy chain for each muscle was determined based on the abundance of a single, common peptide shared among all MyBP-C isoforms identified (SI Appendix, Fig. S1), relative to that of peptides shared among the myosin heavy chain isoforms expressed in that muscle. In the SOL muscle, we estimate 1 MyBP-C molecule per 11.4 ± 0.7 (SEM, n = 7) myosin heavy chain molecules, which is no different from the EDL with a ratio of 1 to 11.2 ± 0.6 (SEM, n = 7).
Unique peptides associated with multiple slow-type MyBP-C isoforms (MYBPC1 gene) were present in both the SOL and EDL samples (SI Appendix, Fig. S1). In contrast, unique peptides associated with fast-type MyBP-C (MYBPC2 gene) were present only in the EDL samples (SI Appendix, Fig. S1). The relative abundances of the slow- and fast-type peptides were indicative of 43.1 ± 3.9% (SEM, n = 7) expression of fast-type MyBP-C in the EDL samples. Next, in a more focused analysis of the MyBP-C isoform composition, we enhanced the detection of the various MyBP-C peptides by separating MyBP-C from the other myofibrillar proteins on sodium dodecyl sulfate (SDS) polyacrylamide gels and then trypsin-digested the 75- to 150-kDa gel region in preparation for mass spectrometry (n = 6 samples per group). Several unique peptides identified in these analyses were indicative of alternative MYBPC1 gene splicing resulting in both C- and N-terminal slow-type MyBP-C variants. The C-terminal splice variants were found only in the SOL samples (SI Appendix, Fig. S1). With the total MyBP-C content relative to myosin heavy chain content in the SOL and EDL samples being similar (see above), the slow-type MyBP-C C-terminal variants do not appear to affect the total number of MyBP-C molecules incorporated into the backbone of the thick filament.
Several peptides in both the SOL and EDL samples were indicative of alternative gene splicing in the 5′ region of the MYBPC1 gene, resulting in N-terminal slow-type MyBP-C variants (Fig. 1B). The longest of these variants (sMyBP-C) contained amino acids from all N-terminal exons (Table 2, Fig. 1B, and SI Appendix, Figs. S1 and S2). A shorter variant lacked exons 4 and 5 (amino acids 35 to 59, denoted sMyBP-CΔexon4-5) (Table 2, Fig. 1B, and SI Appendix, Figs. S1 and S2). Interestingly, the combined abundances of these N-terminal peptides associated with the sMyBP-C and sMyBP-CΔexon4-5 variants were consistently lower than the abundances of C-terminal peptides. This finding was consistent with the 40% expression of a third short N-terminal variant that uses an alternate start codon in the middle of exon 7, as described by Ackermann and Kontrogianni-Konstantopoulos (22). This variant lacks the first 125 amino acids originating from the first 6.5 exons (sMyBP-CΔexon1-6.5) (Table 2, Fig. 1B, and SI Appendix, Fig. S2), beginning midway through the C1 Ig-like domain (Fig. 1B and SI Appendix, Fig. S2), and, based on its sequence, would not be identified in our MS analyses. Therefore, the abundance of this variant was assumed to be that reported by Ackermann and Kontrogianni-Konstantopoulos (22) (Table 2) and then used to estimate by a mass-balance approach (26, 27) the relative abundances of the 2 longer N-terminal variants (Table 2). Collectively, these data demonstrate that both slow- and fast-twitch muscles utilize alternative exon splicing of the MYBPC1 gene to modify the structure and possibly the function of slow-type MyBP-C.
Table 2.
Slow-type and fast-type MyBP-C isoform abundance in rat slow-twitch soleus and fast-twitch extensor digitorum longus skeletal muscles
| Gene | Protein | SOL | EDL | SOL, high calpain-treated | ||
| Abundance, % | SEM | Abundance, % | SEM | Abundance, % | ||
| MYBPC2 | Fast-type MyBP-C | 41.7 | 4.9 | |||
| MYBPC1 | sMyBP-C | 35.9 | 2.2 | 7.7 | 2.6 | 7.3 |
| MYBPC1 | sMyBP-CΔexon4-5 | 24.1 | 2.2 | 21.8 | 2.6 | 14.5 |
| MYBPC1 | sMyBP-CΔexon1-6.5 | 40.0* | — | 28.5 | 2.1 | 14.6 |
Abundance percentage determined as described in SI Appendix, Materials and Methods. SEM determined from multiple muscle samples (SOL, n = 12; EDL, n = 12; SOL high calpain-treated, n = 2, therefore no SEM determined). Empty cells denote that fast-type MyBP-C was not detected in samples from soleus muscles.
Value estimated from Ackermann and Kontrogianni-Konstantopoulos (22), – denotes that SEM not calculated for this entry.
Spatial Distribution of Slow- and Fast-Type MyBP-C in the Sarcomere.
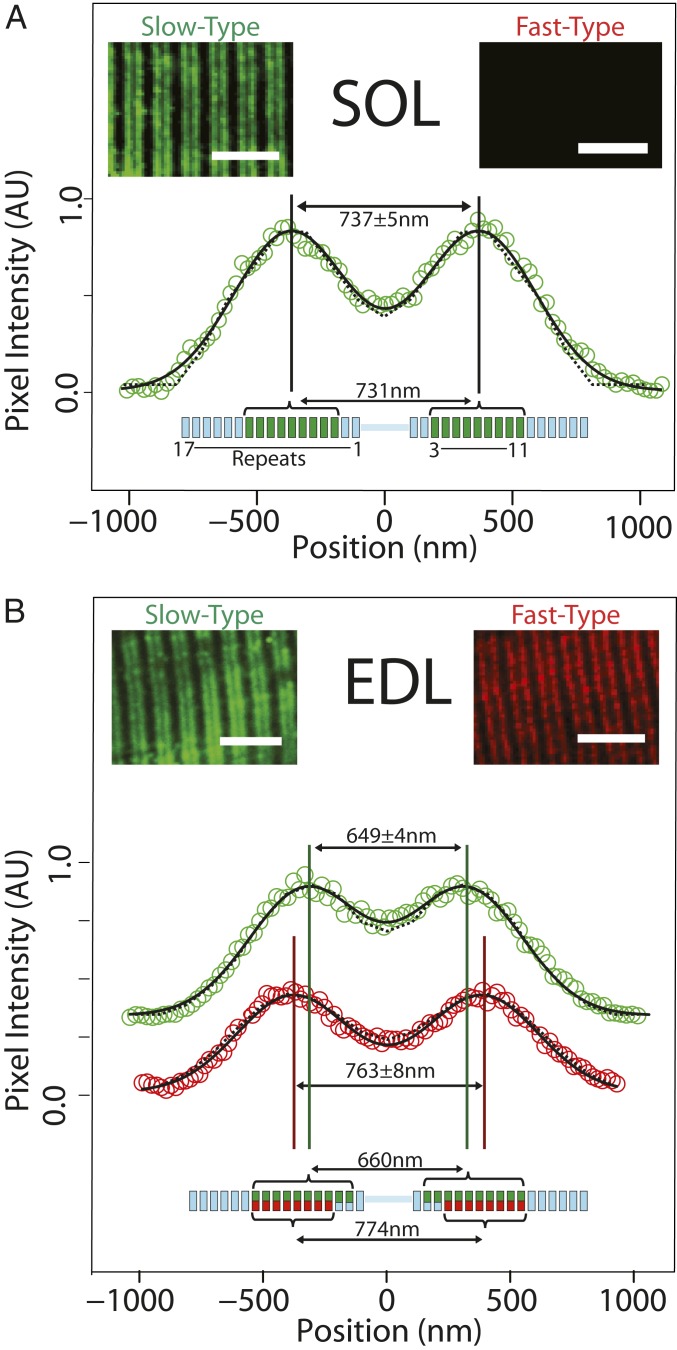
To determine if MyBP-C isoforms are differentially localized within the C-zones, we immunofluorescently labeled MyBP-C in cryo-sectioned SOL and EDL muscle samples and imaged them using confocal microscopy (Fig. 2, Insets). The slow-type antibody, capable of binding all slow-type MyBP-C isoforms, labeled both the SOL and EDL muscle samples while the fast-type MyBP-C antibody only labeled the EDL muscle samples. This was expected from the mass spectrometry data. Both antibodies resulted in 2 closely spaced, fluorescence bands in the sarcomere center, identifying the 2 C-zones (Fig. 2). To determine the spatial distribution of the slow- and fast-type MyBP-C isoforms within the C-zones, the signal-to-noise ratio of the fluorescence signal was improved by averaging multiple sarcomeres (Materials and Methods). The 2 fluorescence bands were fitted by the sum of 2 Gaussians, with the fits serving to constrain an analytical model that predicted the spatial distribution of slow- and fast-type MyBP-C molecules within each C-zone (see Materials and Methods and SI Appendix for details).
Fig. 2.
Immunofluorescence imaging and modeling of MyBP-C distribution in SOL and EDL muscle sections. (A) In slow-twitch SOL muscle, anti–slow-type MyBP-C antibodies label 2 bands in each sarcomere (Inset). The aligned and integrated intensity of these 2 bands (open circles) (SI Appendix, Materials and Methods) are well fit with two Gaussian peaks (solid line) with a separation of 737 ± 5 nm. Analytical modeling (dotted line) (SI Appendix, Materials and Methods) demonstrated that, if slow-type MyBP-C is distributed across thick filament repeats 3 to 11 (green bars in thick filament schematic below), the anticipated peak-to-peak separation would be 731 nm, coincident with the immunofluorescence data. Fast-type MyBP-C was not detected in SOL muscle (Inset). (B) For fast-twitch EDL muscle, doublet bands were apparent upon labeling with either slow-type or fast-type MyBP-C antibodies (Insets). Intriguingly, the separation distance of the two fitted Gaussian peaks differed for slow-type and fast-type MyBP-C. The spatial distribution for slow-type MyBP-C (649 ± 4 nm peak-to-peak separation, green) is closer toward the sarcomere midline than fast-type MyBP-C (763 ± 8 nm separation, red). Our model suggests that slow-type and fast-type MyBP-C cooccupy repeats 4 to 11 (green and red bars, respectively, in thick filament schematic below data); slow-type MyBP-C may also occupy repeats 2 and 3, although at reduced occupancy (i.e., 2 of 3 possible MyBP-C molecules per repeat, indicated with partial green bars in the thick filament schematic). This occupancy pattern would result in 660 nm peak-to-peak spacing for slow-type MyBP-C and 774 nm for fast-type MyBP-C. (Scale bars: 5.0 μm in all images.)
To determine how the MyBP-C molecules are distributed within the SOL sarcomeres (Fig. 2A), we developed an analytical model that generated a dual-Gaussian fluorescence image based on the point-spread function of the antibody’s fluorophores and their localization along the thick filament (see SI Appendix for model details). The model’s initial and unbiased assumption was that as many as 3 MyBP-C molecules could be located at any of 17 repeats, with each repeat spanning 43 nm (Fig. 2, schematics) along each half of the thick filament, with the 43 nm corresponding to the myosin helical repeat (5, 6). Based on there being 300 myosin heavy chain molecules per half thick filament and our LCMS ratio of 1:11.4 slow-type MyBP-C molecules per myosin heavy chain in the SOL muscle samples, the model assumed there were 27 slow-type MyBP-C molecules per half thick filament. The model then iteratively redistributed these 27 MyBP-C molecules in different arrangements among the 17 repeats, comparing the predicted fluorescence profile for each arrangement to the experimental data (Fig. 2A) (Kolmogorov–Smirnov test, P = 0.86, where P > 0.01 demonstrates significant overlap). This best fit was generated by 3 MyBP-C molecules in each of 9 consecutive repeats, with the position of the first repeat occupied by MyBP-C at 186 nm from the center of the thick filament. This localization corresponded to repeats 3 to 11 in the model (Fig. 2A).
Similar image analysis and modeling were performed on the EDL muscle samples. With both slow- and fast-type MyBP-C isoforms in the EDL, the model assumed that the 27 MyBP-C molecules per half thick filament were divided into 16 slow- and 11 fast-type molecules, based on the LCMS data (Table 2). The best fit distribution of these MyBP-C molecules (P = 0.83) suggested that the slow-type MyBP-C molecules in the EDL samples were distributed into 10 consecutive repeats, with the first repeat positioned at 143 nm from the center of the thick filament. This localization corresponded to repeats 2 to 11 (Fig. 2B, green) whereas the fast-type MyBP-C molecules were best distributed (P = 0.94) into 8 consecutive repeats, with the first repeat positioned at 229 nm from the thick filament center. This localization corresponded to repeats 4 to 11 in the model (Fig. 2B, red). Where the slow- and fast-type MyBP-C molecules occupied the same repeat, a total of 3 MyBP-C molecules were assumed to be present. Therefore, the C-zone spatial distribution of MyBP-C appeared to be both MyBP-C isoform and muscle-type specific.
MyBP-C Isoforms Differentially Modulate Native Thin Filament Motility in Native Thick Filament C-Zones.
We first set out to determine whether the endogenous complement of MyBP-C isoforms in the C-zones of thick filaments can modulate thin filament motility in a Ca2+-dependent manner. Therefore, we isolated native thin and thick filaments from both rat SOL and EDL muscles and observed the motility of Ca2+-regulated thin filaments over their corresponding native thick filaments to maintain physiological relevance.
Thin filament motility is both sensitized to Ca2+ and slowed in thick filament C-zones.
Native thick filaments present a unique opportunity to characterize how MyBP-C impacts thin filament motility in the C-zone since the thick filament serves as its own control with its tip being devoid of MyBP-C (i.e., D-zone) (Fig. 3A). Given that the D- and C-zones are each ∼350 nm in length (6), we ultrasonically shredded thin filaments into short ∼250-nm shards that were capable of interacting within each zone independently. These short thin filaments maintained their Ca2+ regulation and sensitivity (SI Appendix, Fig. S4) and traversed and sampled both the D- and C-zones (i.e., in the absence and presence of MyBP-C, respectively).
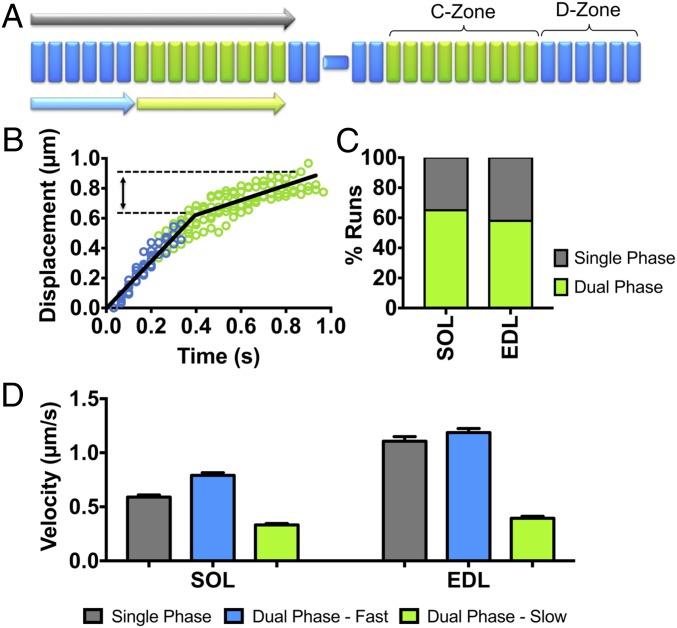
Fig. 3.
Native thin filament motility at pCa 5 over native thick filaments from SOL and EDL muscles. (A) Illustration of native thin filament trajectories observed over native thick filaments, which, as in B, are described as either a trajectory with a constant velocity (i.e., single phase; gray arrow) or having dual phases, where there is an initial fast velocity phase (light blue arrow) in the D-zone that is followed by a slower velocity phase (light green arrow) that corresponds to the length of the C-zone. (B) Representative EDL thin filament displacement vs. time trajectories, illustrating the behavior of dual-phase trajectories. The initial fast velocity phase (blue) persists for ∼350 to 400 nm, approximately the length of the D-zone, followed by a slow velocity phase that corresponds to the expected length of the C-zone (∼400 nm, green). (C) Percentage of observed thin filament trajectories that have single versus dual velocity phases. (D) The velocities that are associated with single phase and each of the dual velocity phase trajectories for the SOL and EDL thick filaments. The single-phase velocities of both SOL and EDL thin filaments are comparable to the fast velocity phase for the dual-phase trajectories. All data are presented as mean ± SEM. Data are summarized in Table 3.
At a Ca2+ concentration where thin filaments were fully “on” (−log[Ca2+] or pCa 5) (SI Appendix, Fig. S4) as determined in isolated myofibrils (SI Appendix, Fig. S4G), the majority (∼60%) of trajectories over both SOL and EDL thick filaments showed 2 distinct phases of velocity (Fig. 3 B and C): an initial fast velocity phase, with a value correlated to the rate of tension redevelopment in myofibrils and presumably the thick filament myosin type, which then transitioned to a ∼60% slower phase, lasting ∼400 nm (Fig. 3 B–D, SI Appendix, Fig. S4H, and Table 3). This distance corresponded to the established C-zone length (30, 31) and the values estimated from our immunofluorescence and modeling data (Fig. 2). The remaining 40% of trajectories over both SOL and EDL thick filaments were better described by a single velocity that was close to the fast velocity phase of the dual-phase trajectories (Fig. 3 C and D and Table 3). The presence of single-phase velocity trajectories may be due to the inability to identify the velocity transition with statistical certainty or physiological in nature (see modeling below).
Table 3.
Native thin filament motility over native thick filament motility under various experimental conditions
| pCa 7.5 | pCa 5 | |||||||||||
| Single-phase trajectories | Dual-phase trajectories | Dual-phase runs, % | ||||||||||
| Thick filament preparation | Velocity, µm/s | Displacement, µm | n | Velocity, µm/s | Displacement, µm | n | Fast-phase velocity, µm/s | Fast-phase displacement, µm | Slow-phase velocity, µm/s | Slow-phase displacement, µm | n | |
| Low calpain | ||||||||||||
| SOL | 0.39 ± 0.16 | 0.35 ± 0.14 | 205 | 0.59 ± 0.20 | 0.79 ± 0.04 | 106 | 0.79 ± 0.32 | 0.41 ± 0.15 | 0.33 ± 0.18 | 0.40 ± 0.14 | 197 | 65 |
| EDL | 0.46 ± 0.19 | 0.36 ± 0.12 | 135 | 1.11 ± 0.50 | 0.75 ± 0.09 | 133 | 1.19 ± 0.51 | 0.35 ± 0.16 | 0.39 ± 0.25 | 0.47 ± 0.15 | 187 | 58 |
| High calpain | ||||||||||||
| SOL | NA | NA | NA | 0.57 ± 0.24 | 0.78 ± 0.06 | 172 | 0.72 ± 0.26 | 0.35 ± 0.14 | 0.30 ± 0.13 | 0.50 ± 0.13 | 134 | 44 |
Native thin and thick filaments were isolated using low calpain treatment from rat slow-twitch soleus (SOL) and fast-twitch extensor digitorum longus (EDL) skeletal muscles. The motility of thin filament shards (Materials and Methods) over their respective thick filaments was observed at low Ca2+ (pCa 7.5) and high Ca2+ (pCa 5) concentrations. At pCa 7.5, only short displacements at a constant velocity were observed. At pCa 5, trajectories were described by a single velocity and displacement (single-phase trajectories) or by dual-phase trajectories having an initial fast velocity phase followed by a slow velocity phase, with the displacements for each phase reported. Following high calpain treatment that proteolytically cleaves only the slow-type MyBP-C N terminus, there was a reduction in the percentage of dual-phase trajectories over SOL native thick filaments. n, the number of thin filament trajectories. NA denotes that no trajectories were observed under the indicated experimental conditions.
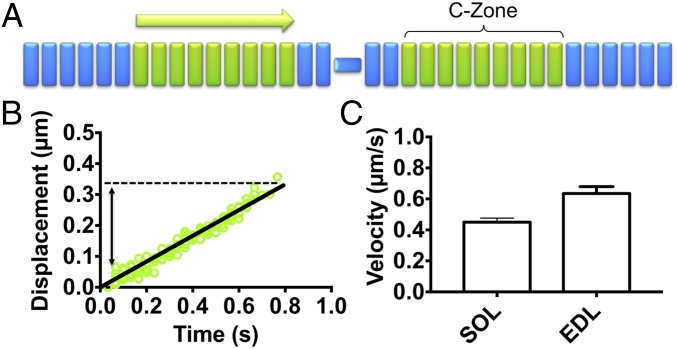
At a Ca2+ concentration where thin filaments should have been fully “off” as in myofibrils and thus motility should have been absent (pCa 7.5, SI Appendix, Fig. S4), we observed motile thin filaments on both SOL and EDL thick filaments that traveled short, ∼350-nm distances at a constant velocity (Fig. 4). Once again, the distances traveled were comparable to the C-zone lengths (Fig. 4B and Table 3), and the velocities were similar to that of the slow velocity phase of trajectories over the SOL and EDL thick filaments at pCa 5 (Fig. 3C and Table 3). Therefore, the complement of MyBP-C within the C-zones of both SOL and EDL thick filaments can both sensitize the thin filament to Ca2+ at low Ca2+ concentrations and slow motility at high Ca2+ concentrations.
Fig. 4.
Native thin filament motility at pCa 7.5 over native thick filaments from SOL and EDL muscles. (A) Illustration of native thin filament trajectories observed over native thick filaments, which, as in B, are described by a constant velocity (light green arrow) that corresponds to the length of the C-zone. (B) Representative SOL thin filament displacement vs. time trajectories that persist for ∼350 nm. (C) The velocities of the SOL and EDL thin filament trajectories are comparable to the slow velocity phase of the dual-phase trajectories at pCa 5 (Fig. 3D). All data are presented as mean ± SEM. Data are summarized in Table 3.
N-terminal domains of slow-type MyBP-C are required to modulate thin filament sliding.
To determine if MyBP-C’s N-terminal domains are the effectors of thin filament slowing within the C-zones of native thick filaments, we took advantage of a previously defined thrombin cleavage site in the M-domain that was identified in studies of bovine skeletal MyBP-C (32). Interestingly, this cleavage site target sequence, SAFK, that exists only in the slow-type MyBP-C variant isoforms (SI Appendix, Fig. S2), is identical to that of a calpain cleavage site in cardiac MyBP-C (27). Therefore, with the SOL thick filaments composed only of the slow-type MyBP-C, we treated these thick filaments with higher calpain concentrations and quantified the effect by LCMS. The abundance of all 3 slow-type MyBP-C N-terminal variants demonstrated reductions in abundance, with the total abundance of slow-type MyBP-C with intact N termini reduced by 64 ± 10% (Table 2).
Following N-terminal cleavage of SOL thick filaments, the proportion of thin filament trajectories at pCa 5 that were characterized by 2 phases of velocity were no longer the majority, falling from 65 to 44% (Table 3). All of the parameters that described these 2-phase velocity trajectories (i.e., fast- and slow-phase velocity values and displacements) were similar to those observed over thick filaments not subjected to high calpain treatment (Fig. 3 and Table 3). These results suggest that the N terminus of one or more of the slow-type MyBP-C variant isoforms is responsible for the thin filament slowing observed in the SOL C-zone.
Isoform-specific N-terminal MyBP-C fragments differentially modulate thin filament motility in a Ca2+-dependent manner.
To determine the functional contribution of individual MyBP-C N-terminal isoforms, we turned to the conventional in vitro motility assay to assess the impact of recombinant N-terminal MyBP-C fragments on SOL and EDL thin filament motility propelled by their respective monomeric myosins. Recombinant N-terminal fragments, up to and including the C2 domain, were bacterially expressed for the fast-type (fC1C2) and the 3 slow-type N-terminal variants (sC1C2, sC1C2Δexon4-5, and sC1C2Δexon1-6.5) (Fig. 1B). Unfortunately, both bacterial- and baculovirus-expressed sC1C2Δexon1-6.5 were insoluble under nondenaturing conditions, thereby precluding their characterization (SI Appendix, Fig. S5).
N-terminal fragments were added to the motility assay at pCa 7.5 and pCa 5 to assess each fragment’s ability to sensitize thin filaments to Ca2+ and to slow thin filament motility, respectively (Fig. 5 and SI Appendix, Fig. S6). By varying the fragment concentrations (0 to 2 µM), Hill activation (pCa 7.5) and noncompetitive inhibition (pCa 5) curves were generated and fitted to normalized motility data defined by the product of thin filament sliding velocity and the fraction of moving filaments (Fig. 5 and SI Appendix, Fig. S6 and Table S1). The product of velocity and fraction moving provides a means of taking into account the activation state of the thin filament (SI Appendix, Fig. S4).
Fig. 5.
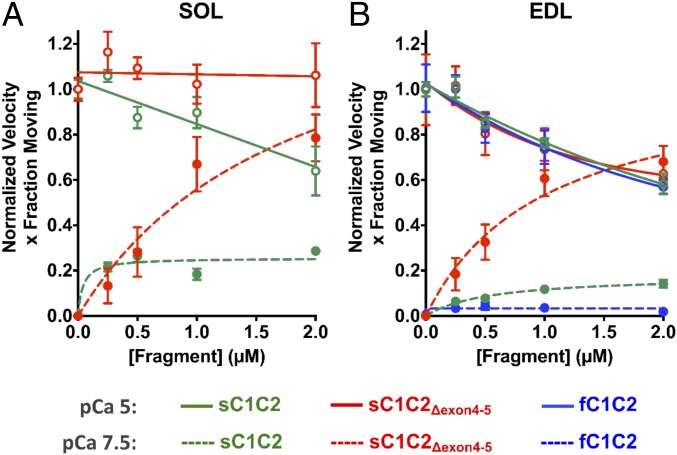
Isoform-specific N-terminal MyBP-C fragments differentially modulate SOL and EDL thin filament motility over monomeric myosin in a Ca2+-dependent manner. MyBP-C imparted concentration-dependent (0 to 2 µM) effects on thin filament sliding motility at low (pCa 7.5, closed symbols and dashed lines) and high (pCa 5, open symbols and solid lines) calcium concentrations. Plots are shown as velocity times the fraction of filaments moving, normalized to the value at pCa 5 for each MyBP-C fragment. Data were fitted with noncompetitive inhibition (Ki) and Hill activation (Ka) curves for pCa5 and pCa 7.5, respectively (see SI Appendix, Table S1 for Ki and Ka parameter fits). (A) For SOL thin filament motility at pCa 5, sC1C2 reduced motility by up to 36% with increasing fragment concentration (green solid line). sC1C2Δexon4-5 had negligible impact on thin filament motility (red solid line). At pCa 7.5, sC1C2Δexon4-5 (red dashed line) significantly enhanced thin filament motility, with increasing concentrations compared to sC1C2 (green dashed line). (B) For EDL motility, all 3 N-terminal fragments have comparable inhibitory effects on thin filament velocities at pCa 5 (solid lines). At pCa7.5, fC1C2, sC1C2, and sC1C2Δexon4-5 differentially sensitize the thin filaments to calcium with sC1C2Δexon4-5 (red dashed line) being the greatest sensitizer, compared to sC1C2 (green dashed line) and fC1C2 (blue dashed line). All data are presented as mean ± SEM. See SI Appendix, Fig. S6 for plots of individual velocity and fraction of moving filaments and SI Appendix, Table S1 for parameters of the fits to the data and the number of experiments for each condition.
For the SOL-based motility at pCa 5, where thin filaments are fully on, increasing concentrations of sC1C2 slowed normalized thin filament motility up to 36%, with a fragment concentration (inhibition constant [Ki]) of 3.8 ± 1.3 µM (Fig. 5A, green solid line). The shorter, sC1C2Δexon4-5 fragment had no effect on motility, as evidenced by a Ki (120 ± 644 µM) well beyond the 2-µM fragment concentration tested (Fig. 5A, red solid line). For the SOL-based motility at pCa 7.5, where thin filaments were normally off (SI Appendix, Fig. S4), the shorter sC1C2Δexon4-5 fragment had the greatest effect on sensitizing thin filaments to Ca2+ (Fig. 5A, red dashed line). At 2 µM sC1C2Δexon4-5, thin filament motility was nearly equal to that at pCa 5. In contrast, the sC1C2 was not as effective at sensitizing the thin filaments to Ca2+ (Fig. 5A, green dashed line).
In the EDL where both fast- and slow-type MyBP-C isoforms are expressed, we determined the effect of the fC1C2, sC1C2, and sC1C2Δexon4-5 N-terminal fragments on thin filament motility in the EDL-based assay at pCa 5. All of the fragments slowed thin filament motility up to 40%, with similar Ki ∼ 2.7 µM (Fig. 5B, solid lines and SI Appendix, Table S1). In contrast, at pCa7.5, each fragment differentially sensitized thin filament motility to Ca2+. Specifically, the sC1C2Δexon4-5 fragment was a potent Ca2+ sensitizer (Fig. 5B, red dashed line), which was not the case for both the sC1C2 (Fig. 5B, green dashed line) and fC1C2 fragments (Fig. 5B, blue dashed line). Therefore, all of the expressed N-terminal fragments tested imparted a concentration-dependent effect on sensitizing thin filaments to Ca2+ at pCa 7.5 and on slowing motility at pCa 5; however, the extent of these effects was muscle type-specific.
In Silico Model of Thin Filament Slowing in Thick Filament C-Zones.
Knowing how the individual N-terminal fragments modulate thin filament motility as described above, we implemented an in silico Monte Carlo-style simulation in an effort to understand how each N-terminal splice variant isoform contributes to the overall slowing of thin filament motility in the C-zones of native thick filaments at pCa 5. Another goal of this modeling effort was to infer the slowing capacity of the sMyBP-CΔexon1-6.5 splice variant, which could not be characterized in the in vitro motility assay due to the fragment’s insolubility. To simplify these modeling efforts, we focused only on SOL thick filaments due to the following: 1) fewer number of N-terminal isoforms (i.e., 3 for SOL vs. 4 for EDL) (Table 2); 2) profound difference in the slowing capacity for the characterized fragments (Fig. 5A); and 3) reduction in functional MyBP-C upon selective N-terminal proteolytic cleavage of slow-type MyBP-C by calpain.
The model and its simplifying assumptions are described in detail in SI Appendix but briefly are as follows:
-
1)
Each SOL half thick filament is comprised of 17 myosin helical repeats spaced 43 nm apart (Fig. 6A). Each repeat generates a maximal velocity of 0.79 ± 0.32 μm/s that equals the experimentally observed fast velocity phase of a thin filament trajectory (Fig. 5 and Table 3).
-
2)
The slow-type MyBP-C N-terminal splice variant isoforms are randomly distributed in C-zone repeats 3 to 11, as determined by our immunohistological imaging (Fig. 2), with the probability of a splice variant occupying a repeat determined by the LCMS abundance data (Table 2). An alternate model was considered in which a thick filament possesses only a single variant isoform and thus the variants are distributed by thick filaments (SI Appendix, Fig. S7) and not heterogeneously within a single C-zone (Fig. 6A).
-
3)
Each C-zone repeat has an inherent velocity that is slower than the maximal velocity based on the inhibition constant (Ki) (SI Appendix, Table S1 and Fig. 5) for the splice variant assigned to that repeat.
-
4)
A short thin filament (250 ± 50 nm) initiates a trajectory near the thick filament tip (repeat 17) and proceeds through the C-zone (Fig. 6A). As the thin filament is translocated over the thick filament, its effective velocity at any point is the mean of the velocities associated with the ∼6 repeats that the thin filament is in contact with. Therefore, the thin filament velocity is position-dependent.
-
5)
To simulate the experimental condition following high calpain treatment, N-terminally cleaved slow-type MyBP-C are assumed to no longer slow velocity. Therefore, the probability of these cleaved splice variants being in a C-zone repeat is determined by their relative abundance, measured by LCMS (Table 2). The repeat in which they are assigned thus generates the uninhibited maximal velocity.
-
6)
Simulated trajectories were parameterized to match the experimental frame rate (30 frames per second [fps]) and spatial resolution (14 nm) and analyzed for the number of velocity phases by the same approach employed experimentally (see SI Appendix for details). Once analyzed, velocity distributions for trajectories with a single constant velocity and for the fast and slow phases of the dual-phase trajectories were generated, and the model distributions were then compared to the experimentally obtained distributions (Fig. 6 D and E and SI Appendix, Fig. S7 B and C).
-
7)
The model has only 2 free parameters: 1) the effective total slow-type MyBP-C concentration and 2) the inhibition constant (Ki) for the uncharacterized slow-type MyBP-CΔexon1-6.5 splice variant. Therefore, trajectories were simulated for 2 experimental conditions (i.e., low and high N-terminal calpain cleavage) over a range of possible values for the 2 free parameters (Fig. 6B). For each condition and free parameter value, the simulated velocity distributions and fraction of trajectories that were 1 versus 2 phases were compared to the experimental data and assigned a “goodness of fit” (SI Appendix). The goodness of fit, over the entire parameter space explored, is presented as a color heat map (Fig. 6B).
Fig. 6.
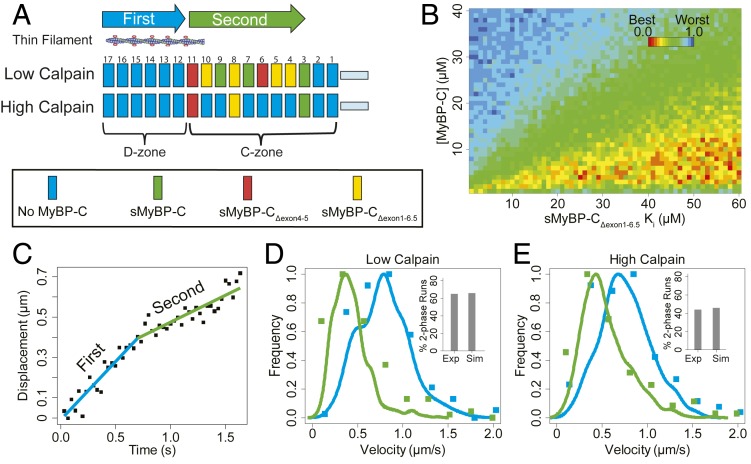
In silico modeling showing the slowing effects on thin filament motility at pCa 5 of the slow-type MyBP-C isoform complement in the C-zones of native SOL thick filaments. In addition, the model was used to extrapolate the functional influence of sMyBP-CΔexon1-6.5, which could not be characterized by in vitro motility (see main text and SI Appendix for model details). (A) Schematic illustrating the localization of slow-type MyBP-C molecules in repeats 3 to 11 within the C-zone of native thick filaments. The probability that a repeat is occupied by a specific MyBP-C isoform is related to the relative abundance for the isoform as determined by LCMS (Table 2). At low calpain treatment, slow-type MyBP-C N termini remain intact while high calpain treatment cleaves 64% of the N termini (Table 2), thereby reducing the intact MyBP-C repeat occupancy down to ∼3 repeats within the C-zone. (B) The model has two variable parameters: the effective total MyBP-C concentration and the Ki (inhibition constant) for the sMyBP-CΔexon1-6.5 isoform. The heat map shows the normalized goodness of fit for a range of modeled parameters that are used to generate model thin filament trajectories, as compared to the experimental data from Fig. 3 and Table 3. There is no unique best fit parameter set, but rather a range of best fit parameters for the effective total slow-type MyBP-C concentration of 3 to 12 µM. More importantly, the predicted Ki being >25 µM for the sMyBP-CΔexon1-6.5 isoform, which is well above the effective total MyBP-C concentration, suggests this isoform has little to no inhibitory effect on thin filament slowing. (C) Displacement vs. time trajectories generated by the model, showing the dual velocity phases. (D) Velocity distributions for the individual phases of the dual velocity phase trajectories (fast velocity phase, blue; slow velocity phase, green) for the simulated data (solid lines) compared to the experimental data (solid symbols), with the percentage of dual phase runs for the experimental (Exp) and simulated (Sim) data in the Insets. (E) As in D, but after high calpain treatment. All plots demonstrate that the simulated data generated using the modeled best fit values (4 µM total [MyBP-C], 26 µM Ki for MyBP-CΔexon1-6.5) closely resemble experimental native thick filament data.
Using the best fit parameters for the model where the slow-type MyBP-C isoforms are distributed within a single thick filament (4 µM total [MyBP-C], 26 µM Ki for MyBP-CΔexon1-6.5), the model effectively recapitulated the trajectory characteristics before and after N-terminal cleavage, specifically with respect to the proportion of dual-phase trajectories (Fig. 6 D and E, Insets) and the velocity distributions for the fast and slow phase of the trajectory (Fig. 6 D and E). The model suggests that the ∼40% of trajectories that have a single velocity phase can in part be due to the stochastic assignment of slow-type MyBP-C isoforms to the various repeats (Fig. 6A). Specifically, if the N-terminal variants that are minimally inhibitory to thin filament sliding velocity populate the repeats closest to the D- to C-zone transition, then the MyBP-C slowing effect would not occur until the thin filament is well into the C-zone. This would compromise our ability to statistically detect the transition between the fast and slow velocity phases of the trajectory. Although the simulation did not return a single unique parameter set, it did identify a range of parameters that gave equally good fits. Specifically, the effective total slow-type MyBP-C concentration was predicted to be 3 to 12 μM. This is in agreement with an ∼11-μM estimate, based on our LCMS ratio of slow-type MyBP-C to myosin heavy chain of 1:11.4 (Tables 1 and 2), assuming a myosin concentration of ∼120 μM in skeletal muscle (33). More importantly, the estimated Ki for the uncharacterized slow-type MyBP-CΔexon1-6.5 N-terminal variant isoform is no less than 25 µM (Fig. 6B), which would suggest that this isoform may have negligible impact on thin filament sliding velocity in the C-zone.
We also simulated an alternate model wherein individual thick filaments contain only a single slow-type MyBP-C N-terminal splice variant isoform (SI Appendix, Fig. S7A), instead of a mixture of isoforms as described above (Fig. 6A). These simulations estimated the total concentration of slow-type MyBP-C at 10 μM, with a Ki of the MyBP-CΔexon1-6.5 variant at 10 μM. However, these parameters did not adequately recapitulate the experimentally observed frequency of dual-phase trajectories (SI Appendix, Fig. S7 B and C, Insets) and the velocity distributions for the second, slower phase of the dual-phase trajectories (SI Appendix, Fig. S7 B and C, green curves). Therefore, our experimental results are most consistent with a model in which the slow-type MyBP-C isoforms are distributed within individual thick filaments.
Discussion
Using multiscale proteomic, biophysical, and in silico approaches, we defined the expression, localization, and modulatory capacities of MyBP-C isoforms in native myosin thick filaments from rat slow-twitch SOL and fast-twitch EDL skeletal muscle. Specifically, in SOL, at least 3 slow-type MyBP-C isoforms are expressed that differ in their N termini as a result of alternate exon splicing (Fig. 1B, Table 2, and SI Appendix, Fig. S2) whereas, in the EDL, these same slow-type MyBP-C isoforms are expressed with the addition of a fast-type MyBP-C (Fig. 1B, Table 2, and SI Appendix, Fig. S2). In native thick filaments, this mixed complement of MyBP-C isoforms both sensitizes the thin filament to Ca2+ while slowing thin filament velocity, only within the C-zones where MyBP-C is localized (Figs. 3 and 4). The major effector of this modulatory effect on filament sliding is the MyBP-C N terminus (Fig. 5, Table 3, and SI Appendix, Table S1), which presumably projects away from the thick filament backbone and interacts with the thin filament and/or the myosin head region (6).
Although, collectively, the assumed mixture of MyBP-C isoforms in the C-zone modulates thin filament motility, how each individual isoform contributes to these effects is far more complex. For example, in the EDL thick filament C-zone, where both slow- and fast-type MyBP-C isoforms are present, are these proteins spatially segregated? Are the fast-type and slow-type MyBP-C and its variants functionally identical? If not, then are functional differences dependent on the muscle-specific thick filament in which they exist? Only through the use of multiscale biophysical approaches described here have we been able to address these questions.
Expression and Spatial Distribution of MyBP-C Isoforms in the C-Zone.
The expression of slow-type MyBP-C isoforms in muscle has been observed in both slow-twitch, type-I and fast-twitch, type II muscle fibers (22) whereas fast-type MyBP-C is solely expressed in fast-twitch, type II muscle fibers (20). These general observations agree with our immunofluorescence data, where the slow-type MyBP-C is detected both in the rat slow-twitch SOL and fast-twitch EDL muscles, while the fast-type MyBP-C is only detected in the EDL (Fig. 2 and Table 2). However, fast-twitch, type II muscle fibers can be further categorized by their myosin heavy chain isoform expression (i.e., type IIA, IIB, and IIX). Therefore, the presence of 12% fast-twitch, type IIA myosin in the rat SOL, based on our own LCMS data (Table 1) and that of 20% reported in the literature (29), suggests that the assumed expression of fast-type MyBP-C in all fast-twitch, type II fibers may not hold. Specifically, the absence of any fast-type MyBP-C in the rat SOL (Table 2), even though fast-twitch, type IIA fibers do exist, suggests that the type IIA fibers in the SOL only express slow-type MyBP-C. Whether this applies to the type IIA fibers in the rat EDL, which only make up 5% of the fiber composition (Table 1), cannot be determined based on the existing data. Interestingly, type IIA fibers are the slowest of the fast-twitch fibers and thus closer in speed to type I fibers (34). This might imply that a muscle fiber’s speed dictates higher order transcriptional or translational regulation that determines the pattern of MyBP-C isoform expression.
Regardless of the MyBP-C isoform expression pattern, how are the slow- and fast-type MyBP-C isoforms spatially distributed in the C-zones of SOL and EDL muscle? Evidence from electron micrographs (5, 6) localizes MyBP-C within the C-zone to 7 to 9 transverse stripes separated by 43 nm, a separation distance that coincides with the myosin helical repeat (Figs. 1A and 2). Using a spatially explicit analytical model to help interpret our immunofluorescence data from tissue samples, the slow-type MyBP-C in the SOL appears to distribute into 9 repeats (Fig. 2A) coincident with stripes 3 to 11 within the C-zone, as observed in rabbit SOL by electron microscopy (5). Since the antibody epitope we used cannot distinguish between the various slow-type isoforms, the specific distribution of these isoforms among the 9 repeats cannot be determined. However, in the rat EDL, where both slow-and fast-type MyBP-C are expressed, the model does predict that these 2 isoforms are spatially distinct, which was previously reported for fast-twitch rabbit psoas muscle (5). Even without modeling, the raw immunofluorescence data imply that the peaks of the slow-type MyBP-C doublets compared to that of the fast-type MyBP-C are on average ∼60 nm closer to the sarcomere center (i.e., M-line) (Fig. 2B). More precisely, the model suggests that the fast-type MyBP-C is distributed into 8 repeats (repeats 4 to 11) within the C-zone whereas the slow-type MyBP-C is distributed into 10 repeats (repeats 2 to 11). This implies that the repeats closest to the M-line in the rat EDL are only occupied by the slow-type MyBP-C, as reported previously for rabbit psoas sarcomeres (5). Specifically, in the rabbit psoas, the slow-type MyBP-C was reported to occupy stripes 3 to 11 whereas the fast-type MyBP-C occupied 2 different stripe patterns of 4 to 11 and 5 to 11 (5). If we assume that our modeled repeat locations coincide with the immunolocalized stripe locations in electron micrographs (5), then there is remarkable agreement for the fast-type MyBP-C distribution in the rat EDL compared to the rabbit psoas. However, our modeled distribution of the slow-type into 10 consecutive repeats in the rat EDL versus 9 stripes in the rabbit psoas (5) could simply be due to species and muscle differences. Therefore, to our knowledge, this is the only report of the presence of MyBP-C within the thick filament repeat 2 for any muscle type. What governs this spatial segregation in the C-zone, not to mention the stoichiometry of the various MyBP-C isoforms? Regulated expression could be factor that determines stoichiometry. More interestingly, isoform differences in the MyBP-C C-terminal C10 domain, which occurs by alternate exon splicing in the slow-type MyBP-C (Results and SI Appendix, Figs. S1 and S3), were observed here and by others (22). Although these differences do not affect the overall stoichiometry, they may dictate an isoform’s binding affinity to particular locations along the thick filament through interactions with the backbone itself and/or titin (35). Regardless, the observed difference in the overall spatial distribution of MyBP-C isoforms in the C-zone might be related to functional differences between the isoforms, as described below.
Skeletal MyBP-C Isoforms Modulate Thin Filament Motility.
We demonstrated that the N-terminal domains of MyBP-C are the effector of MyBP-C’s actomyosin modulatory capacity (Fig. 5 and Table 3). Therefore, we used bacterially expressed N-terminal fragments (up to and including the C2 Ig-domain) of the various slow- and fast-type MyBP-C isoforms in a motility assay to define each isoform’s potential contribution to thin filament Ca2+ sensitization and slowing of velocity (Fig. 5). Based on the endogenous MyBP-C profile (Table 2), we expressed the fast-type MyBP-C N-terminal fragment (fC1C2) and the 3 slow-type MyBP-C N-terminal splice variants (sC1C2, sC1C2Δexon4-5, and sC1C2Δexon1-6.5) (Fig. 1B).
Isoform differences in thin filament Ca2+ sensitization.
In the absence of MyBP-C, Ca2+-regulated thin filaments are effectively off at pCa 7.5 (SI Appendix, Fig. S4), but, on native thick filaments from either SOL or EDL muscle samples, thin filament motility is apparent in the C-zone (Fig. 4). This is presumably due to MyBP-C’s Ca2+-sensitizing capacity. When the various N-terminal fragments were characterized in the conventional motility assay, the sC1C2Δexon4-5 fragment was the most effective Ca2+ sensitizer in both SOL- and EDL-based motility assays (Fig. 5 A and B, red dashed line) whereas the other fragments displayed more modest Ca2+ sensitization (Fig. 5A, green dashed line and B, green and blue dashed lines). Therefore, the sC1C2Δexon4-5 splice variant isoform may be the major contributor to thin filament Ca2+ sensitization in both the SOL and EDL thick filament C-zones (Figs. 4 and 5). In a previous study, we investigated how heterologous expression of skeletal MyBP-C isoforms in cardiac muscle during heart failure (18) might impact MyBP-C’s modulation of cardiac contractility. Interestingly, even within the context of a cardiac contractile protein system (i.e., cardiac myosin and thin filaments), the sC1C2Δexon4-5 fragment (referred to as ssC1C2 in that study) was more effective at Ca2+-sensitizing cardiac thin filaments than the fC1C2, suggesting that the enhanced Ca2+ sensitization by the sC1C2Δexon4-5 fragment is not muscle-specific (18). One mechanism by which the sC1C2Δexon4-5 fragment may Ca2+-sensitize the thin filament, as was the case for this fragment in the cardiac system (18), is its binding to the thin filament to shift tropomyosin from the “blocked” to the “closed” state, as observed for cardiac N-terminal fragments (36, 37), which, even at pCa 7.5, would then allow myosin to interact with the thin filament (38). From a structural standpoint, both the slow- and fast-type MyBP-C can Ca2+-sensitize the thin filament to varying degrees in the motility assay but do so without a C0 Ig-like domain that is present in cardiac MyBP-C. The lack of this extra C0 domain may limit the extent to which the slow- and fast-type MyBP-Cs sensitize the thin filament to Ca2+. Structural studies demonstrate that the cardiac C0 domain by itself can bind to the thin filament but is incapable of activating the cardiac thin filament (36). Therefore, the slow- and fast-type MyBP-C lacking the C0 domain would not have this additional weak point of contact with the thin filament, which, if present, may increase the probability that the other N-terminal domains stereospecifically interact with the thin filament and shift tropomyosin to a position more favorable for myosin binding.
Isoform differences in slowing of thin filament velocity.
At high Ca2+ concentrations (pCa 5), where thin filaments are effectively on (SI Appendix, Fig. S4), thin filament velocities slowed in both the SOL and EDL thick filament C-zones, thus defining MyBP-C’s second modulatory capacity (Figs. 3 and 5). This thin filament slowing upon entering the C-zone has recently been confirmed during fiber shortening in skinned rat SOL skeletal muscle fibers (19). Thin filament slowing appears to be inherent to the N termini of all slow- and fast-type MyBP-C fragments characterized in the motility assay, with 2 exceptions (Fig. 5, solid lines). The sC1C2Δexon1-6.5 variant, which could not be characterized due to its insolubility, was predicted to have little to no mechanical impact, based on our in silico modeling efforts (Results). If so, this may not be surprising since this variant’s sequence starts halfway through the C1 Ig-like domain (SI Appendix, Fig. S2). Previous studies with mouse cardiac N-terminal fragments showed that any fragment with slowing capacity required an intact C1 domain (8, 36). The other exception is the sC1C2Δexon4-5 variant, which is inhibitory in the EDL-based motility assay (Fig. 5B, red solid line), but not in the SOL-based motility assay (Fig. 5A, red solid line). Therefore, the observed slowing is context-specific in the sense that the same sC1C2Δexon4-5 N-terminal variant functions differently depending on the thin and thick filament proteins it interacts with: i.e., SOL versus EDL contractile proteins. Although Ackermann et al. (17) reported different degrees of bare actin filament slowing for several recombinant mouse slow-type MyBP-C N-terminal splice variant fragments (up to and including the M-domain), their motility assay used actin and myosin from heterologous species and muscle types. Therefore, interpretation of their results must be viewed in the context of our findings in which a MyBP-C fragment’s functional capacity can be dependent on the contractile proteins.
Potential mechanisms that may slow thin filament velocity are that the MyBP-C N-terminal domains act as internal load and/or as a competitive inhibitor that competes with myosin heads for binding to the thin filament. Evidence in the laser trap assay of cardiac MyBP-C N-terminal fragments binding transiently to actin (8) was interpreted as imparting a viscous-like load against which myosin motors must operate (39) although MyBP-C N-terminal binding to the myosin head region could also serve as an internal load, restricting myosin head movement and force production (16). Evidence from cardiac MyBP-C proteins also exists to support competition with myosin for actin binding based on solution cosedimentation (40), inhibition of ATPase activity (41), and recent direct visualization of reduced binding of S1-myosin to a single thin filament tightrope in the presence of N-terminal fragments (42). In fact, Walcott et al. combined the viscous load and competitive inhibition modes of MyBP-C interaction with the thin filament into a model that recapitulated the concentration dependence of N-terminal cardiac MyBP-C fragment-induced slowing of actin filament velocity in the motility assay (43). Why the slowing of thin filament velocity for the sC1C2Δexon4-5 variant is so different in the SOL- versus the EDL-based motility systems (Fig. 5) is a matter of speculation. For example, subtle differences may exist in the binding interface between this fragment and the protein isoforms that constitute the thin and thick filaments in these two muscle types.
If the shorter slow-type MyBPCΔexon1-6.5 N-terminal variant isoform is incapable of slowing thin filament velocity in the C-zone, as suggested by our modeling efforts, then what physiological purpose could it serve? A recent report suggests that, in addition to its modulatory role as defined here, slow-type MyBP-C may play a structural role in thick filament assembly within the sarcomere as knockdown of slow-type MyBP-C in mouse FDB skeletal muscle resulted in reduced abundance of myosin and myomesin (44). Therefore, the 1:11.4 ratio of MyBP-C to myosin, reported here, must be critical to the maintenance of thick filament stability. As such, significant expression of the slow-type MyBPCΔexon1-6.5 isoform in both the SOL and EDL thick filaments may allow its intact C-terminal domains to provide structural stability to the thick filament (4).
Conclusion.
Skeletal MyBP-C appears to sensitize the thin filament to Ca2+, while acting as an internal brake that slows thin filament velocity only within the C-zone, like cardiac MyBP-C. Interestingly, these two modulatory capacities are not equivalent among the multiple MyBP-C isoforms that populate the skeletal muscle C-zone. Rather, the various isoforms can Ca2+-sensitize or slow velocity to different extents, depending on the muscle fiber type in which they exist. As such, the differential expressions of slow- and fast-type MyBP-C isoforms may enable the muscle to fine-tune its mechanical performance, depending on the muscle’s physiological demands, such as exercise and potentially during development and aging. It is worth noting that, at least, the slow-type MyBP-C is posttranslationally modified by phosphorylation of its N-terminal domains (19, 45–47). Therefore, N-terminal phosphorylation of the slow-type MyBP-C may provide additional means of regulating its modulatory capacity both normally, under acute physiological stress, and abnormally, due to genetic mutations in MyBP-C that lead to skeletal myopathies (19, 45–47).
Materials and Methods
Recombinant protein isolation, production, and motility experiments were performed according to previously published protocols (18, 27). Comprehensive experimental details are provided in SI Appendix, Materials and Methods.
Proteins.
Myosin, native thick filaments, and native thin filaments (NTFs) were isolated from soleus (SOL) and extensor digitorium longus (EDL) muscles of Sprague–Dawley rats. F-actin was purified from chicken pectoralis muscle (48). Mouse MyBP-C N-terminal fragments (Fig. 1B) up to the C2 domain fC1C2 (1 to 337), sC1C2 (1 to 366), sC1C2Δexon4-5, (1 to 341), and sC1C2Δexon1-6.5 (1 to 241) were bacterially expressed as previously described (18) unless stated otherwise.
Thick Filament Isolations.
Thick filaments were isolated according to the procedures detailed in Previs et al. (27). Freshly isolated rat SOL or EDL were manually shredded in 0.5 mL of relaxing buffer (50 mM NaCl, 5 mM MgCl2, 2 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 1 mM dithiothreitol [DTT], 7 mM phosphate buffer (pH 7), 10 mM creatine phosphate, 2.5 mM adenosine 5′-triphosphate [ATP]) over the course of 30 min, while kept on ice. Tissue was further dissected in skinning solution (relaxing buffer containing 0.5% vol/vol Triton X-100) over 2 rounds of 30 min each, followed by 2 rinses (30 min each) with relaxing buffer. Finally, thick filaments were liberated by digestion of a small amount of skinned fibers over several minutes at room temperature, using Calpain-1 from porcine erythrocytes (Calbiochem), at either 0.05 U/μL (low calpain) or 0.25 U/μL (high calpain) concentrations in buffer containing 1 mM calcium acetate, 20 mM imidazole (pH 6.8), and 5 μM 2-mercaptoethanol. The slide was rinsed repeatedly with 10-μL aliquots of relaxing solution (supplemented to 75 mM NaCl total), and all washes were combined in a microcentrifuge tube. The total volume of the tube was raised to 250 μL, and the contents were clarified by centrifugation at 1,000 × g for 1 min.
Quantitative Mass Spectrometry.
Briefly, thick filament samples were isolated and digested in solution with trypsin, or MyBP-C was separated by SDS/PAGE and digested in-gel, as described in Results. Aliquots of each sample were separated by ultra-high pressure liquid chromatography, and the eluent was analyzed using a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were identified from the resultant spectra using SEQUEST in the Proteome Discoverer 2.2 (PD 2.2) software package (Thermo Fisher Scientific) and custom-built databases. Quantification was carried out using a label-free approach (26, 27). Details are described in SI Appendix, Materials and Methods.
Native Thin Filament In Vitro Motility.
Conventional in vitro motility assays were performed as previously described in ref. 18. Briefly, myosin was incubated onto a nitrocellulose-coated surface of a glass flow cell. Rhodamine-labeled NTFs were incubated in the flow cell prior to the addition of bacterially expressed fragments and activating buffer. The motion of NTFs was observed by epifluorescence microscopy. Myosin and NTFs were muscle type-matched in all experiments.
Native Thick Filament In Vitro Motility.
Native thick filament in vitro motility assays were performed as detailed previously (27). Sigmacote (Sigma Aldrich)-treated flow cells were initially incubated with freshly prepared native thick filaments and allowed to incubate at room temperature for 20 min. The flow-cell surface was then blocked with BSA (1 mg/mL). This was then followed by rhodamine-labeled thin filaments, which were sonicated (Model 300, Fisher) immediately prior to addition to the flow cell. Finally, motility buffer (25 mM KCl, 1 mM EGTA, 10 mM DTT, 25 mM imidazole (pH 7.4), 4 mM MgCl2, 100 μM ATP) with an oxygen scavenging system (0.1 μg/mL glucose oxidase, 0.018 μg/mL catalase, 2.3 μg/mL glucose) with freshly sonicated thin filaments and at the indicated pCa was introduced into the flow cell. The motion of the NTFs was captured under total internal reflection fluorescence imaging at 30 fps. Importantly, these motility experiments were performed with native thick filaments and NTFs derived from the same muscle type. Analysis details are provided in SI Appendix.
Myofibril Mechanics.
Myofibrils from rat SOL and EDL muscles were prepared as described in SI Appendix, Materials and Methods. In brief, myofibrils were attached between a rigid glass needle and a precalibrated atomic force cantilever that served as the force transducer within a microfluidic chamber in which calcium was varied between pCa 9.0 and 4.5 for tension:pCa measurements. The rate of force redevelopment following a large amplitude sarcomere length reduction (Ktr) at pCa 4.5 was determined as a measure of actomyosin kinetics.
Immunofluorescence.
Immunofluorescence staining was performed on isolated myofibrils and cryo-sectioned muscle tissue. All buffers were diluted into phosphate-buffered saline (PBS) at 4 °C unless stated otherwise. SOL and EDL muscles were sectioned at 10 µm thickness using a cryostat microtome. Cryostat sections were incubated with 0.1 M glycine (Sigma), followed by a 10% goat serum-based blocking buffer immediately prior to antibody labeling. Samples were incubated with primary antibodies against either slow (MYBPC1) or fast (MYBPC2) MyBP-C, and α-actinin (Sigma) for 2 h. The sections were then rinsed with PBS and incubated with species-specific Alexa Fluor secondary antibody for 2 h. After the secondary incubation step, the sections were rinsed with PBS, mounted in a fluorescence antifading medium, and sealed with nail polish. Imaging was performed on a Nikon Ti Eclipse inverted microscope equipped with an LU-N4/N4S 4-laser unit (405 nm, 488 nm, 561 nm, and 640 nm). The same protocol was used for myofibril staining. Myofibril isolation was performed as previously described in ref. 49. For more detail, see SI Appendix, Materials and Methods.
Immunofluorescence Image Analysis and Modeling.
Briefly, we decomposed confocal immunofluorescence images into a set of 1 pixel-wide intensity line scans and then aligned these to each other by maximizing the pairwise cross-correlation. The aligned intensity line scans were fitted with a double Gaussian to determine the peak-to-peak spacing of the fluorescence doublets. We then developed an analytical model to predict the molecular arrangements that resulted in the dual-Gaussian fluorescence profiles. The model accounted for the binding of MyBP-C at specific sites along each half of the thick filament, as described in Results , and the point spread function of the fluorophores. We compared the model results to the data using the Kolmogorov–Smirnov test, where P value > 0.01 demonstrates significant overlap, to maximize the goodness of fit. For more detail, see SI Appendix, Materials and Methods.
Monte Carlo Simulations of Native Thick Filament Motility.
Briefly, a Monte Carlo style simulation of thin filament motility over native thick filaments was used to generate 1,000 displacement-vs.-time trajectories for each set of possible model parameter values. Velocity distributions resulting from each of these simulations were then compared to experimental results to determine parameters that offer best agreement between simulation and experiment. For details, see SI Appendix.
Study Approval.
All protocols complied with the Guide for the Care and Use of Laboratory Animals (50) published by the NIH and were approved by the Institutional Animal Care and Use Committee at the University of Vermont Larner College of Medicine.
Supplementary Material
Acknowledgments
We thank Ms. Nicole Bouffard (Microscopy Imaging Center, University of Vermont) and Mr. Todd Clason (Imaging/Physiology Core Facility, University of Vermont) for expert assistance in cryo-section and confocal imaging. We thank Guy Kennedy from the Instrumentation and Model Facility at the University of Vermont for his microscopy expertise. We also thank Prof. Doug Taatjes (University of Vermont) for proficient advice on immunolabeling. This work was supported by NIH Grants HL059408, AR067279, and HL126909 (to D.M.W.), HL130356, HL139680, AR067279, and HL105826 (to S.S.), and HL124041 (to M.J.P.); by American Heart Association Grants 19TPA34830084 and 19UFEL34380251 (to S.S.) and 17POST33630095 (to J.W.M.); and by the Natural Sciences and Engineering Research Council of Canada (to D.E.R.). D.E.R. is a Canada Research Chair (Tier I) in Muscle Biophysics; and supported in part by a generous gift to D.M.W. from Arnold and Mariel Goran.
Footnotes
Competing interest statement: S.S. provided consulting and collaborative research services to the Leducq Foundation, AstraZeneca, Merck, Amgen, and MyoKardia unrelated to the content of this manuscript.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910549116/-/DCSupplemental.
References
- 1.Ackermann M. A., Kontrogianni-Konstantopoulos A., Myosin binding protein-C: A regulator of actomyosin interaction in striated muscle. J. Biomed. Biotechnol. 2011, 636403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayram Y., et al. , Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J. Clin. Invest. 126, 762–778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurnett C. A., et al. , Myosin binding protein C1: A novel gene for autosomal dominant distal arthrogryposis type 1. Hum. Mol. Genet. 19, 1165–1173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert R., Kelly M. G., Mikawa T., Fischman D. A., The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J. Cell Sci. 109, 101–111 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Bennett P., Craig R., Starr R., Offer G., The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J. Muscle Res. Cell Motil. 7, 550–567 (1986). [DOI] [PubMed] [Google Scholar]
- 6.Luther P. K., et al. , Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. U.S.A. 108, 11423–11428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer J. F., Kensler R. W., Harris S. P., The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 284, 12318–12327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weith A., et al. , Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 52, 219–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mun J. Y., et al. , Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C). J. Mol. Biol. 410, 214–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rybakova I. N., Greaser M. L., Moss R. L., Myosin binding protein C interaction with actin: Characterization and mapping of the binding site. J. Biol. Chem. 286, 2008–2016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampourakis T., Yan Z., Gautel M., Sun Y. B., Irving M., Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl. Acad. Sci. U.S.A. 111, 18763–18768 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Lange W. J., et al. , E258K HCM-causing mutation in cardiac MyBP-C reduces contractile force and accelerates twitch kinetics by disrupting the cMyBP-C and myosin S2 interaction. J. Gen. Physiol. 142, 241–255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruen M., Gautel M., Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 286, 933–949 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Bhuiyan M. S., et al. , In vivo definition of cardiac myosin-binding protein C’s critical interactions with myosin. Pflugers Arch. 468, 1685–1695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann P. A., Greaser M. L., Moss R. L., C-protein limits shortening velocity of rabbit skeletal muscle fibres at low levels of Ca2+ activation. J. Physiol. 439, 701–715 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann P. A., Hartzell H. C., Moss R. L., Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. J. Gen. Physiol. 97, 1141–1163 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackermann M. A., et al. , Loss of actomyosin regulation in distal arthrogryposis myopathy due to mutant myosin binding protein-C slow. FASEB J. 27, 3217–3228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin B. L., et al. , Skeletal myosin binding protein-C isoforms regulate thin filament activity in a Ca2+-dependent manner. Sci. Rep. 8, 2604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinett J. C., Hanft L. M., Geist J., Kontrogianni-Konstantopoulos A., McDonald K. S., Regulation of myofilament force and loaded shortening by skeletal myosin binding protein C. J. Gen. Physiol. 151, 645–659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber F. E., Vaughan K. T., Reinach F. C., Fischman D. A., Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur. J. Biochem. 216, 661–669 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Ackermann M. A., Kontrogianni-Konstantopoulos A., Myosin binding protein-C slow: An intricate subfamily of proteins. J. Biomed. Biotechnol. 2010, 652065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann M. A., Kontrogianni-Konstantopoulos A., Myosin binding protein-C slow: A multifaceted family of proteins with a complex expression profile in fast and slow twitch skeletal muscles. Front. Physiol. 4, 391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhoot G. K., Hales M. C., Grail B. M., Perry S. V., The isoforms of C protein and their distribution in mammalian skeletal muscle. J. Muscle Res. Cell Motil. 6, 487–505 (1985). [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto K., Moos C., The C-proteins of rabbit red, white, and cardiac muscles. J. Biol. Chem. 258, 8395–8401 (1983). [PubMed] [Google Scholar]
- 25.Bloemberg D., Quadrilatero J., Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One 7, e35273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Leary T. S., Snyder J., Sadayappan S., Day S. M., Previs M. J., MYBPC3 truncation mutations enhance actomyosin contractile mechanics in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 127, 165–173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Previs M. J., Beck Previs S., Gulick J., Robbins J., Warshaw D. M., Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337, 1215–1218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adala J. F., Campos G. E. R., Effects of long-term physical exercise in the skeletal muscles of rats. J. Morphol. Sci. 34, 226–231 (2017). [Google Scholar]
- 29.Eng C. M., et al. , Scaling of muscle architecture and fiber types in the rat hindlimb. J. Exp. Biol. 211, 2336–2345 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Craig R., Offer G., The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B Biol. Sci. 192, 451–461 (1976). [DOI] [PubMed] [Google Scholar]
- 31.Luther P. K., et al. , Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J. Mol. Biol. 384, 60–72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fürst D. O., Vinkemeier U., Weber K., Mammalian skeletal muscle C-protein: Purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. J. Cell Sci. 102, 769–778 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Marston S. B., Tregear R. T., Evidence for a complex between myosin and ADP in relaxed muscle fibres. Nat. New Biol. 235, 23–24 (1972). [DOI] [PubMed] [Google Scholar]
- 34.Schiaffino S., Reggiani C., Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Freiburg A., Gautel M., A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 235, 317–323 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Harris S. P., Belknap B., Van Sciver R. E., White H. D., Galkin V. E., C0 and C1 N-terminal Ig domains of myosin binding protein C exert different effects on thin filament activation. Proc. Natl. Acad. Sci. U.S.A. 113, 1558–1563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mun J. Y., et al. , Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 2170–2175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kad N. M., Kim S., Warshaw D. M., VanBuren P., Baker J. E., Single-myosin crossbridge interactions with actin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 102, 16990–16995 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weith A. E., et al. , The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. J. Muscle Res. Cell Motil. 33, 449–459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saber W., Begin K. J., Warshaw D. M., VanBuren P., Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. J. Mol. Cell. Cardiol. 44, 1053–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belknap B., Harris S. P., White H. D., Modulation of thin filament activation of myosin ATP hydrolysis by N-terminal domains of cardiac myosin binding protein-C. Biochemistry 53, 6717–6724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inchingolo A. V., Previs S. B., Previs M. J., Warshaw D. M., Kad N. M., Revealing the mechanism of how cardiac myosin-binding protein C N-terminal fragments sensitize thin filaments for myosin binding. Proc. Natl. Acad. Sci. U.S.A. 116, 6828–6835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walcott S., Docken S., Harris S. P., Effects of cardiac Myosin binding protein-C on actin motility are explained with a drag-activation-competition model. Biophys. J. 108, 10–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geist J., Ward C. W., Kontrogianni-Konstantopoulos A., Structure before function: Myosin binding protein-C slow is a structural protein with regulatory properties. FASEB J. 32, fj201800624R (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ackermann M. A., Kerr J. P., King B., W Ward C., Kontrogianni-Konstantopoulos A., The phosphorylation profile of myosin binding protein-C slow is dynamically regulated in slow-twitch muscles in health and disease. Sci. Rep. 5, 12637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackermann M. A., Kontrogianni-Konstantopoulos A., Myosin binding protein-C slow is a novel substrate for protein kinase A (PKA) and C (PKC) in skeletal muscle. J. Proteome Res. 10, 4547–4555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackermann M. A., Ward C. W., Gurnett C., Kontrogianni-Konstantopoulos A., Myosin binding protein-C slow phosphorylation is altered in duchenne dystrophy and arthrogryposis myopathy in fast-twitch skeletal muscles. Sci. Rep. 5, 13235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardee J. D., Spudich J. A., Purification of muscle actin. Methods Enzymol. 85, 164–181 (1982). [DOI] [PubMed] [Google Scholar]
- 49.Rassier D. E., Pre-power stroke cross bridges contribute to force during stretch of skeletal muscle myofibrils. Proc. Biol. Sci. 275, 2577–2586 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.