Significance
The assembly of localized static structures is a culminating feature of developmental programs. Construction of the spore coat, a shell that surrounds bacterial spores, is a model for understanding how proteins localize and assemble at the correct place and time during morphogenesis. Using data derived from in vivo and in vitro single-molecule imaging, we propose a computational model in which coat assembly initiates via an interplay between 2 proteins: a protein that localizes preferentially to micron-scale convex membranes (the surface of the developing spore) and a recruited structural protein that polymerizes to hold the first protein in place. This ratchet model therefore describes how a static structure can be built specifically atop the 2-dimensional surface of a developing organelle.
Keywords: nuclear lamina, membrane curvature, DivIVA, MreB, septins
Abstract
Bacterial spores are dormant cells that are encased in a thick protein shell, the “coat,” which participates in protecting the organism’s DNA from environmental insults. The coat is composed of dozens of proteins that assemble in an orchestrated fashion during sporulation. In Bacillus subtilis, 2 proteins initiate coat assembly: SpoVM, which preferentially binds to micron-scale convex membranes and marks the surface of the developing spore as the site for coat assembly; and SpoIVA, a structural protein recruited by SpoVM that uses ATP hydrolysis to drive its irreversible polymerization around the developing spore. Here, we describe the initiation of coat assembly by SpoVM and SpoIVA. Using single-molecule fluorescence microscopy in vivo in sporulating cells and in vitro on synthetic spores, we report that SpoVM’s localization is primarily driven by a lower off-rate on membranes of preferred curvature in the absence of other coat proteins. Recruitment and polymerization of SpoIVA results in the entrapment of SpoVM on the forespore surface. Using experimentally derived reaction parameters, we show that a 2-dimensional ratchet model can describe the interdependent localization dynamics of SpoVM and SpoIVA, wherein SpoVM displays a longer residence time on the forespore surface, which favors recruitment of SpoIVA to that location. Localized SpoIVA polymerization in turn prevents further sampling of other membranes by prelocalized SpoVM molecules. Our model therefore describes the dynamics of structural proteins as they localize and assemble at the correct place and time within a cell to form a supramolecular complex.
The construction of static supramolecular structures such as flagella, teeth, and eggshells is an important endpoint in developmental programs (1–3). Bacterial endospores (“spores”) are encased in a thick proteinaceous shell termed the spore “coat” that provides an attractive model for understanding how large complex cellular structures localize and assemble (4–6). Bacillus subtilis is a rod-shaped, gram-positive bacterium. As nutrients in the environment are depleted, B. subtilis initiates a massive stress response termed “sporulation” that results in the metamorphosis of the rod-shaped, actively growing cell into a dormant endospore (7, 8). In the presence of nutrients, B. subtilis divides by binary fission to produce 2 identical daughter cells; at the onset of sporulation, B. subtilis instead divides asymmetrically to produce 2 dissimilar progeny: a larger “mother cell” and a “smaller” forespore that eventually display different cell fates (Fig. 1A). The initially straight polar-division septum then curves as the mother cell swallows the forespore in a process termed “engulfment.” Ultimately, the leading edges of the engulfing membrane undergo membrane fission to produce a double membrane-bound, topologically isolated forespore in the mother cell cytosol.
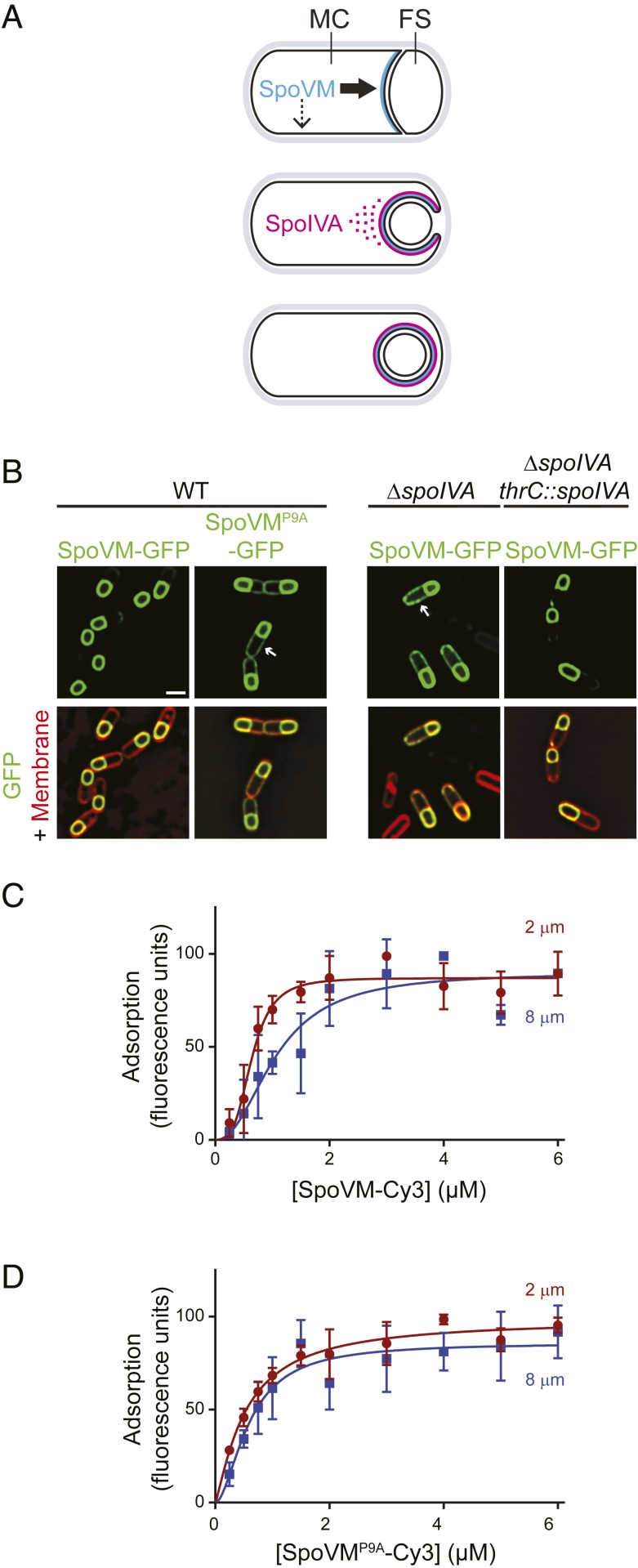
Fig. 1.
Preferential adsorption of SpoVM-Cy3 onto increasingly convex membranes. (A) Schematic of SpoVM and SpoIVA localization within a sporulating B. subtilis cell. Asymmetric division produces a larger mother cell (MC) and a smaller forespore (FS). SpoVM (blue) is produced in the MC and localizes to the engulfing membrane (Top). Thick and thin arrows indicate preferred and less-preferred sites of SpoVM localization, respectively. SpoIVA (magenta) is recruited to the engulfing membrane by SpoVM (Middle). For simplicity, SpoVM and SpoIVA are depicted at different stages of engulfment, but both proteins are produced in the MC after asymmetric division. ATP hydrolysis by SpoIVA drives polymerization on the FS surface (Bottom). (B) Localization of SpoVM-GFP (green; strain CVO1195) or SpoVMP9A-GFP (CVO1395) in wild-type (WT) sporulating B. subtilis cells; SpoVM-GFP localization in the absence of SpoIVA (KR128); or in the absence of SpoIVA complemented with spoIVA expressed from an ectopic chromosomal locus (KR209) 3 h after induction of sporulation. Membranes (red) visualized with FM4-64. Arrows indicate mislocalized GFP signal on the MC membrane. (Scale bar: 1 µm.) (C and D) Adsorption of SpoVM-Cy3 (C) and SpoVMP9A-Cy3 (D) onto differently curved membranes in vitro. Either SpoVM-Cy3 or SpoVMP9A-Cy3 were incubated with a mixed population of 2-μm (maroon) and 8-μm (blue) SSLBs. SSLBs were separated, and fluorescence intensity on each SSLB was measured using flow cytometry. Data were fit with a specific binding with allosteric sigmoidal model: Y = Bmax*Xh/(Kdh + Xh). Data points: median values; error bars: SD (n = ∼20,000).
Proteins that make up the coat are produced in the mother cell after asymmetric division. Of the ∼80 proteins that comprise the coat (4, 9), 2 of the proteins that are produced soon after elaboration of the polar septum are SpoVM and SpoIVA, which form the “basement layer” of the coat: a platform atop which the other coat proteins assemble (4). SpoVM is a 26 amino acid-long protein encoded by a small open reading frame (10) and localizes to the surface of the developing forespore (ref. 11 and Fig. 1B). Preferential localization of SpoVM to the forespore surface is driven by the protein’s intrinsic affinity toward membranes that are positively curved (convex) (12, 13). SpoVM, an amphipathic α-helix (14), spontaneously inserts into the membrane such that it is parallel to the plane of the lipid bilayer (15, 16). The NH2 terminus of SpoVM assumes a highly flexible structure (15) that is dependent on a conserved proline at position 9 (11). Substitution of Pro-9 with Ala results in an entirely α-helical structure (15) that fails to localize properly in vivo due to its inability to discriminate between convex and concave membranes (ref. 11 and Fig. 1B). SpoVM recruits (16) the unusual cytoskeletal protein SpoIVA (17), an ATPase that harnesses the energy released from ATP hydrolysis to drive a conformational change that places the protein in a polymerization-competent form (18–20). Recruitment of SpoIVA to the forespore surface increases the local concentration of SpoIVA, which permits the localized and irreversible assembly of SpoIVA polymers around the forespore. In the absence of SpoVM, SpoIVA fails to uniformly assemble around the forespore (16, 21), consistent with a recruitment and anchoring function for SpoVM. Interestingly, in the absence of SpoIVA, SpoVM promiscuously localizes to all available membranes in the developing sporangium (ref. 16 and Fig. 1B), suggesting a complex interdependence for subcellular localization of both proteins that initiate coat assembly.
Here, we describe how the protein pair, SpoVM and SpoIVA, function as reciprocal dependents to form the basement layer of the bacterial spore coat and therefore drive the developmental process of sporulation toward the maturation of a viable spore. Utilizing single-molecule fluorescence microscopy, we examine the behavior of SpoVM molecules in vitro on synthetic curved membranes and in vivo during sporulation, in the presence and absence of SpoIVA polymers. We demonstrate that SpoVM has an intrinsic affinity toward membranes that are positively curved and that this is driven primarily by a higher residence time, or lower off-rate, on membranes of preferred curvature. We also show that recruitment, followed by polymerization, of SpoIVA atop membrane-bound SpoVM effectively eliminates the off-rate of SpoVM, thereby trapping correctly localized SpoVM molecules on the forespore surface and preventing them from sampling other membranes. Using experimentally derived parameters, we show computationally that coat basement-layer assembly can initiate with just SpoVM and SpoIVA via a 2-dimensional ratchet model. First, a longer residence time of SpoVM on preferred membranes results in the preferential recruitment of SpoIVA molecules to the forespore. Second, recruitment of SpoIVA molecules permits the localized polymerization of adjacent molecules of SpoIVA. Third, SpoIVA polymerization locks prelocalized SpoVM molecules in place. Polymerization of SpoIVA therefore effectively eliminates the backward reaction of SpoVM coming off the membrane and drives a net-forward reaction in which the surface of the forespore is ultimately covered with SpoVM anchoring polymerized SpoIVA, similar to the manner in which a ratcheting socket wrench tightens a bolt when turned in one direction but does not engage and loosen the bolt when turned in the opposite direction when repositioning the wrench. Our results therefore provide a mechanism by which a complex protein structure can localize and assemble to initiate assembly of a supramolecular structure at a specific subcellular location.
Results
Preferential Adsorption of SpoVM-Cy3 onto More Convex Membranes.
To study the preferential adsorption of SpoVM onto convex membranes, we previously established an in vitro assay that utilized spherical supported lipid bilayers (SSLBs) (22, 23), consisting of a phospholipid bilayer encasing a silica bead of specified radius to generate membranes of defined positive (convex) curvature. We previously observed that purified SpoVM-GFP (using fluorescence microscopy; ref. 15) or synthesized SpoVM-FITC (using flow cytometry; ref. 13) preferentially bound to 2-μm diameter SSLBs (which display membrane curvature similar to that of the ∼1-μm diameter forespore) as compared to 8-μm diameter SSLBs (which display membranes with a 4-fold reduction in curvature). To test if SpoVM fused to the fluorescent dye Cy3, that we planned to use in single-molecule imaging, behaved similarly to SpoVM-GFP and SpoVM-FITC, we generated a saturation-binding curve by incubating increasing concentrations of synthesized SpoVM-Cy3 with a mix of 2- and 8-μm SSLBs present in a molar ratio that provided equal total surface area. After the reaction achieved equilibrium, we employed flow cytometry to distinguish between the 2 bead sizes based on forward and side scatter and quantified the Cy3 fluorescence present on each bead in the population. We then plotted the median fluorescence for each population as a function of SpoVM-Cy3 concentration and fitted the data to an allosteric sigmoidal model to discern differences in 3 binding parameters. As we reported previously (13, 15), the fitted Bmax values were similar for the adsorption of SpoVM-Cy3 onto the 2- and 8-μm SSLBs, indicating that the density of SpoVM-Cy3 binding sites were similar on both surfaces (Fig. 1C and SI Appendix, Table S1). However, SpoVM-Cy3 bound to 2-μm SSLBs with a half-maximum concentration of 0.64 (± 0.04) µM, compared to 1.1 (± 0.17) µM for the 8-µm SSLBs, indicating a ∼2-fold increase in affinity for the more convex surface (where numbers in parentheses indicate SDs). Additionally, whereas SpoVM-Cy3 binding to 2-µm SSLBs exhibited a Hill coefficient of 3.5 (± 0.8), binding to 8-µm SSLBs displayed a Hill coefficient of only 2.2 (± 0.7), suggesting increased cooperativity on the more convex membrane. In contrast, SpoVMP9A-Cy3 adsorbed similarly on all SSLBs regardless of curvature (Fig. 1D and SI Appendix, Table S1), consistent with the promiscuous in vivo localization of SpoVMP9A-GFP (ref. 11 and Fig. 1B) and its reported inability to distinguish membrane curvature (12). The affinity/cooperativity-driven adsorption of SpoVM-Cy3 is similar to our previous reports on SpoVM-FITC and SpoVM-GFP, suggesting that the addition of Cy3 does not affect the behavior of SpoVM.
SpoVM Displays Longer Residence Times on More Convex Membranes in Vitro and in Vivo.
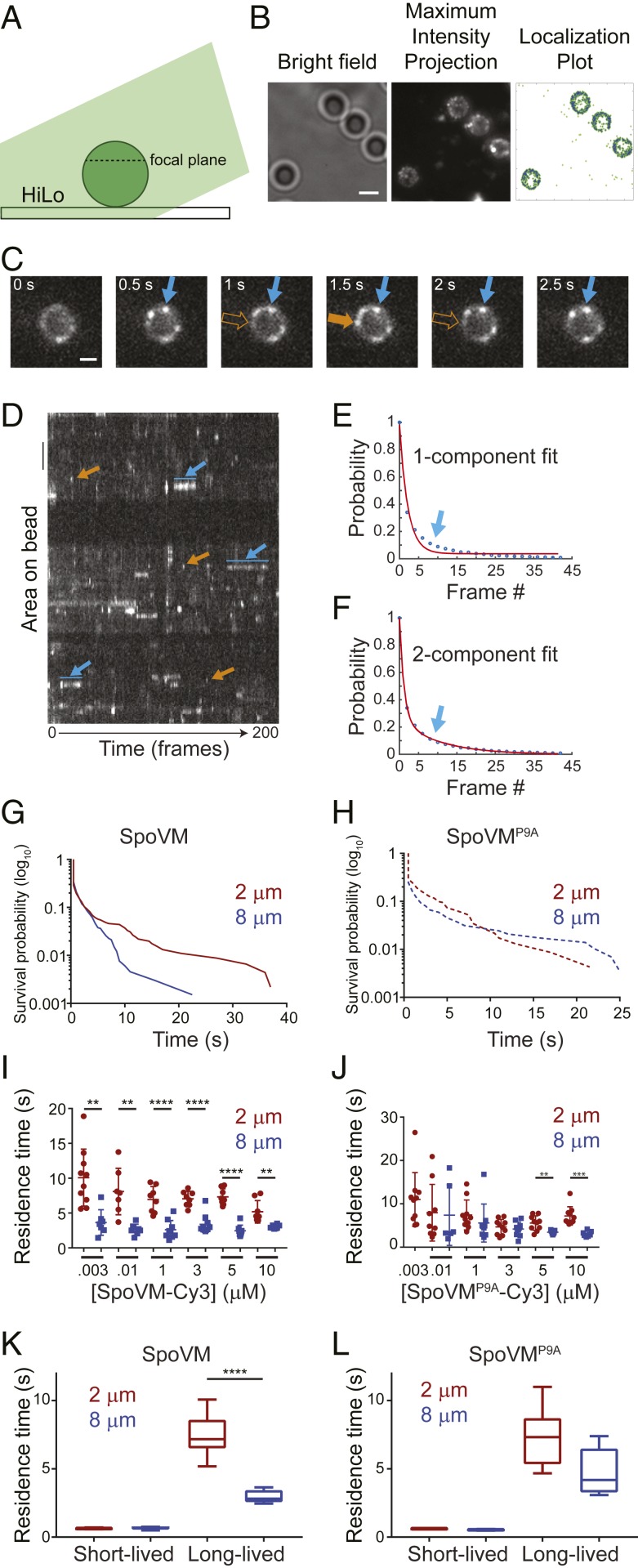
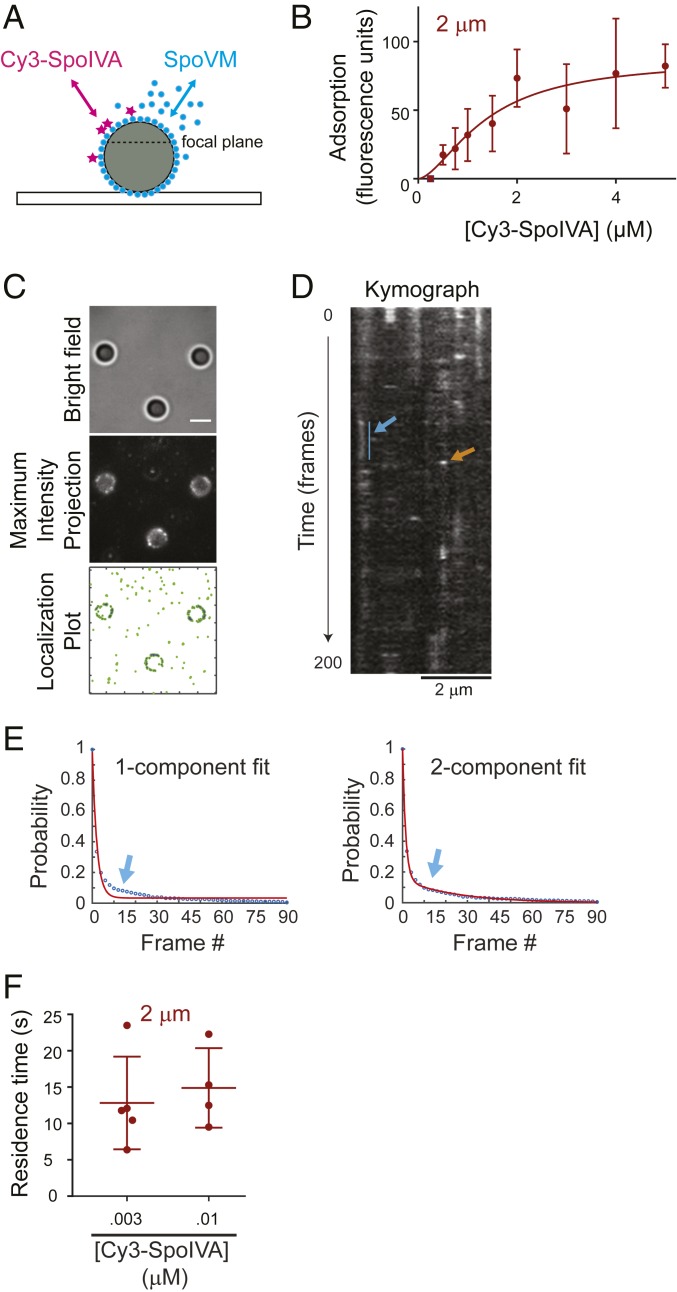
We next examined the dissociation kinetics of single molecules of SpoVM-Cy3 from differently curved membranes in vitro. To this end, we placed either 2- or 8-μm SSLBs on glass coverslips and incubated them with defined concentrations of SpoVM-Cy3. To enable single-molecule imaging at different SpoVM concentrations, we added increasing amounts of unlabeled SpoVM while maintaining a constant concentration (3 nM for 2-μm SSLBs; 10 nM for 8-μm SSLBs) of SpoVM-Cy3. The appearance and disappearance of fluorescent molecules on individual 2-μm (Movie S1) or 8-μm (Movie S2) SSLBs were imaged every 500 ms for 200 frames using highly inclined and laminated optical sheet microscopy (HiLo) illumination (Fig. 2A), wherein the excitation source illuminates the sample at an angle to reduce background fluorescence (24). The aggregate number of localization events from the movies were compiled to produce a maximum-intensity projection (Fig. 2 B, Center) that permitted isolation of individual localization events that occurred on the surface of SSLBs (Fig. 2 B, Right, indicated in blue) to examine their appearance and disappearance over time. We noticed 2 different localization phenomena (shown in a time-lapse series of images of a single representative SSLB in Fig. 2C and of 3 representative SSLBs in the kymograph in Fig. 2D). The first was the appearance of fluorescent puncta that appeared for only one frame (≤500 ms; Fig. 2 C and D, orange arrow). Control experiments in which biotinylated Cy3 was tethered onto neutravidin-coated coverslips revealed a half-life of Cy3 photobleaching of ∼28 s (SI Appendix, Fig. S1A); since the photobleaching time was far longer than the residence time of this population, we concluded that the disappearance of these short-lived localizations likely indicates the adsorbance, followed by rapid desorption, of a SpoVM-Cy3 molecule. The second observed pattern was the persistent localization of fluorescent puncta that lasted for multiple frames (Fig. 2 C and D, blue arrow). Fitting all of the residence time data to a single-component exponential decay revealed a relatively poor fit (R2 = 0.9899; Fig. 2E), whereas a 2-component exponential decay model resulted in an improved fit (R2 = 0.9995; Fig. 2F). This suggested that SpoVM-Cy3 adsorption to SSLBs could be explained by 2 separate populations of molecules: A short-lived population that we interpret as nonspecific interactions with the membrane and a long-lived population that specifically interacted with the membrane surface. For each dataset, we computed the survival probability of either SpoVM (Fig. 2G) or SpoVMP9A (Fig. 2H) bound to 2-μm SSLBs or 8-μm SSLBs. While SpoVM had a similar probability of being bound for ≤4.5 s on both 2- and 8-μm SSLBs, the protein had a higher probability of staying bound for >4.5 s on 2-μm SSLBs (Fig. 2G). In contrast, SpoVMP9A displayed similar probabilities of being bound for any length of time on either SSLB (Fig. 2H).
Fig. 2.
SpoVM-Cy3 displays longer residence times on more convex membranes in vitro. (A) Schematic of single-molecule fluorescence microscopy experimental setup: excitation light (green) illuminates the SSLB (circle) on a glass coverslip at an angle to indicate HiLo. The dashed line represents approximate focal plane. (B, Left) SSLBs visualized using bright-field illumination. (B, Center) Maximum-intensity projection of total binding events during one acquisition, aggregated from every fluorescence image of SpoVM-Cy3 molecules binding to SSLBs taken through time and projected into a 2-dimensional image. (B, Right) Localization plot of total SpoVM-Cy3 localization events (green) or those on the surface of SSLBs (blue). (Scale bar: 2 μm.) (C) Frames from a movie of SpoVM-Cy3 binding on a 2-μm SSLB; acquisition time: 500 ms. SpoVM-Cy3 molecules persisted for multiple frames (blue) or just one frame (orange). (Scale bar: 1 μm.) (D) Kymograph of SpoVM-Cy3 binding on three 2-μm SSLBs observed for 200 frames. Each binding space is individually plotted along the x axis in its corresponding temporal location throughout the acquisition. Brightness indicates a SpoVM-Cy3 molecule is bound. Short-lived (orange) and long-lived (blue) interactions are indicated. (Scale bar: 1 μm.) (E and F) Cumulative distribution function of SpoVM duration on SSLBs fit with either 1-component (E) or 2-component (F) exponential decay model. The arrow indicates parameter space of data better fit using a 2-component model. (G and H) Survival probability of SpoVM (G) or SpoVMP9A (H) bound to 2-μm (maroon) or 8-μm (blue) SSLBs. (I and J) Residence times of SpoVM-Cy3 (I) or SpoVMP9A-Cy3 (J) bound to 2-μm (maroon) or 8-μm (blue) SSLBs at indicated protein concentrations. Data points represent one region of interest (ROI) containing 1 to 28 SSLBs; each ROI imaged with an exposure time of 500 ms for 200 total frames; 7 to 10 ROIs analyzed for each protein concentration. (I) SpoVM-Cy3 binding on 2-μm (n = 53 ROI) or 8-μm (n = 49 ROI) SSLBs. (J) SpoVMP9A-Cy3 binding on 2-μm (n = 58 ROI) or 8-μm (n = 35 ROI) SSLBs (SI Appendix, Table S2). To maintain single-molecule imaging conditions, protein concentrations above 3 nM (2-μm SSLBs) or 10 nM (8-μm SSLBs) were achieved by adding unlabeled protein. (K and L) Mean residence times of SpoVM-Cy3 (K) or SpoVMP9A-Cy3 (L) of short- and long-lived species on 2-μm (maroon) or 8-μm (blue) SSLBs. P values: ** < 0.01; *** < 0.005; **** < 0.001; otherwise, >0.05.
We next analyzed residence times of the long-lived SpoVM-Cy3 species on either 2- or 8-μm SSLBs (Fig. 2I). The average residence time of SpoVM-Cy3 was 7.5 (± 1.5) s on 2-μm SSLBs, and 3.0 (± 0.4) s on 8-μm SSLBs and was largely similar at different protein concentrations, indicating that SpoVM-Cy3 specifically resides ∼2.5-fold longer on more highly curved membranes (Fig. 2K). In contrast, SpoVMP9A-Cy3 displayed an average residence time of 7.3 (± 2.0) s on 2-μm SSLBs and 4.8 (± 1.6) s on 8-μm SSLBs (Fig. 2J), which reflected only a ∼1.5-fold increase in residence time on the more convex membranes (Fig. 2L). Since off-rate (koff) is the inverse of residence time, this corresponds to a slower off-rate for SpoVM-Cy3 of 0.13 s−1 on the 2-μm beads compared to 0.33 s−1 on the 8-μm beads. Using the measured Khalf value of 0.64 µM for SpoVM-Cy3 binding onto 2-μm beads (Fig. 1C and SI Appendix, Table S1), and the measured off-rate of 0.13 s−1, we calculated an on-rate of 0.20 µM−1 s−1 for SpoVM onto 2-μm beads; similarly, using Khalf = 1.1 µM and an off-rate of 0.33 s−1 for SpoVM-Cy3 binding to 8-µM beads, this corresponds to an on-rate of 0.30 µM−1 s−1.
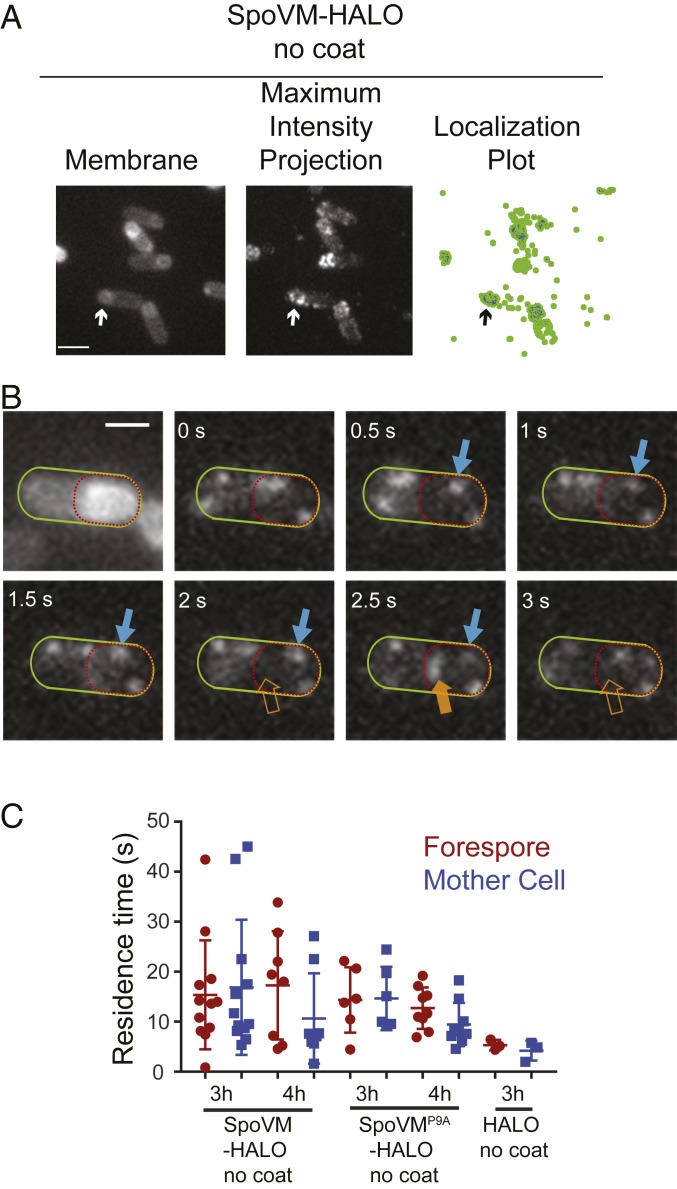
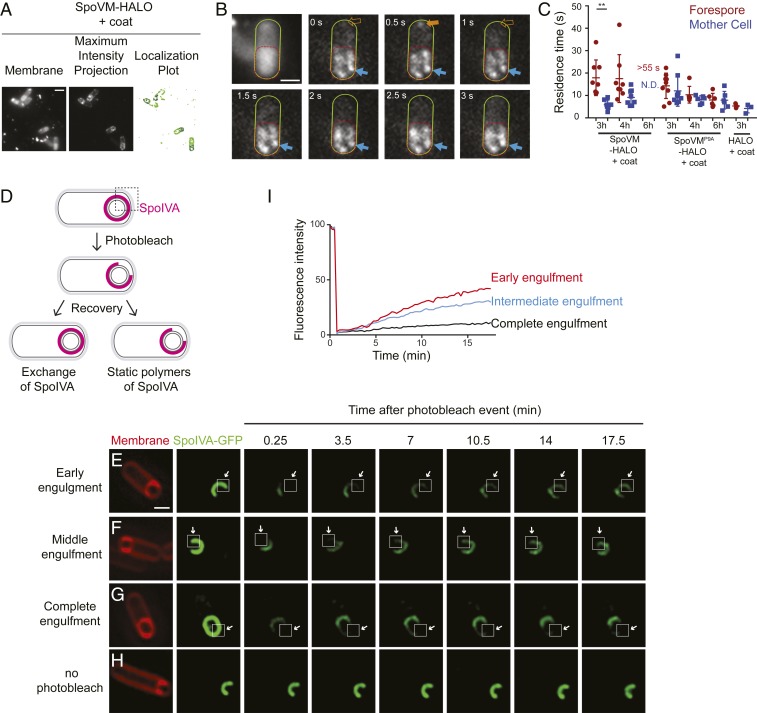
We next measured the residence times of single molecules of SpoVM in vivo on the convex forespore membranes and the concave cytosolic surface of the mother cell. To visualize single molecules of SpoVM in vivo, we engineered a B. subtilis strain that produced, at a single chromosomal locus under control of the native spoVM promoter, SpoVM fused to the HaloTag protein, which binds to the membrane-permeable dye JF549 that can be titrated to label only a small fraction of SpoVM-HALO molecules (25). In the presence of excess dye (50 nM), SpoVM-HALOJF549 localized primarily to the forespore surface (SI Appendix, Fig. S2 A–C), indicating that the HaloTag did not interfere with SpoVM localization; localization of HaloTag fusions to other SpoVM variants also resembled the localization patterns of GFP fusions to those variants (SI Appendix, Fig. S2 D–F). Finally, immunoblotting cell extracts revealed that the HaloTag fusions remained intact in vivo (SI Appendix, Fig. S2G). To mimic the in vitro setup (Fig. 2) in which additional coat proteins were not added, we measured residence times of SpoVM-HALOJF549 in cells harboring a deletion of the spoIVA gene, which prevents subsequent coat assembly (17). In the presence of limiting amounts of JF549 dye (200 pM), we observed the appearance and disappearance of single molecules of SpoVM-HALOJF549 on various membranes (Movie S3 and Fig. 3A). As with the localization of SpoVM-Cy3 on SSLBs, we noticed that some SpoVM-HALOJF549 molecules persisted on a membrane for multiple frames (Fig. 3B, blue arrows), whereas others were present for just one frame (≤500 ms; Fig. 3B, orange arrow). Since we measured a photobleaching rate of JF549 of ∼55 s (SI Appendix, Fig. S1B), we conclude that these short-lived species likely represented adsorbance, followed rapidly by desorption of single SpoVM-HALOJF549 molecules.
Fig. 3.
SpoVM-HALO displays longer residence times on the forespore in vivo. (A, Left) Fluorescence micrograph of sporulating B. subtilis (EAP26) harboring SpoVM-HALO in the absence of coat assembly imaged 3 h after induction of sporulation using MitoTracker Green FM to visualize membranes. (A, Center) Maximum-intensity projection of aggregate SpoVM-HALOJF549 binding events over time. (A, Right) Localization plot, forespore-localized SpoVM-HALOJF549 (blue); all measured localizations (green). Arrow indicates a forespore. (B) Frames from a movie of SpoVM-HALO binding events, visualized using 200 pM JF549, in a representative sporangium. Mother cell is outlined in green, forespore in red. Long-lived (blue) or short-lived (orange) SpoVM-HALOJF549 species are indicated. (C) Residence times of SpoVM-HALOJF549, SpoVMP9A-HALOJF549, or HALOJF549 binding to forespore (maroon) or mother cell (blue) membranes. Data points represent one region of interest, each containing 1 to 8 cells (SI Appendix, Table S3). P values for all datasets: >0.05. (Scale bars: 2 μm.)
Three hours after the onset of sporulation, the average residence time of SpoVM-HALOJF549 on the forespore membrane was 15.4 (± 10.4) s, similar to its residence time on the mother cell membrane: 16.9 (± 12.9) s (Fig. 3C). Four hours after sporulation induction, the average residence time at the forespore membrane remained at 17.3 (± 10.1) s, whereas the residence time at the mother cell membrane was just 10.7 (± 8.4) s (Fig. 3C), suggesting that, in the absence of coat assembly, slight preferential localization of SpoVM in vivo only occurs at a relatively later stage in sporulation and that a significant population of SpoVM, presumably at equilibrium, mislocalizes to the mother cell membrane (Fig. 1B). The residence times of SpoVMP9A-HALOJF549 at the forespore was 12.7 (± 3.9) s and 9.5 (± 4.1) s at the mother cell after 4 h. In summary, the in vivo and in vitro data suggest that SpoVM displays a relatively higher residence time (and therefore a slower off-rate) on more convex surfaces such as that found on the forespore. In vitro, in a defined environment, this increase in residence time occurs relatively quickly, but in vivo, where multiple membranes of different curvatures are present, in the absence of coat assembly, the increase in residence time is more subtle and takes longer to manifest.
Subsequent Coat Assembly Drives Stable Association of SpoVM on the Membrane.
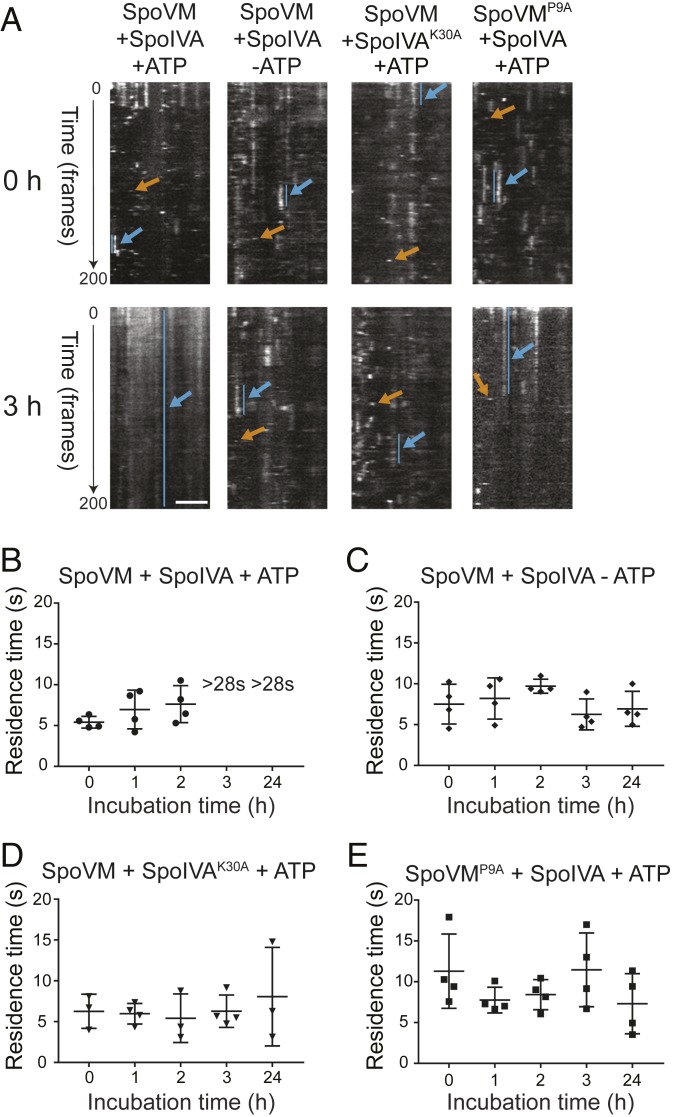
SpoVM directly recruits and tethers SpoIVA, the major coat morphogenetic protein that hydrolyzes ATP to irreversibly polymerize into a platform that recruits other coat proteins (16, 18, 19). Although SpoVM displayed a lower off-rate on more positively curved membranes in vitro, in the absence of coat assembly in vivo, SpoVM only displayed a modest difference in off-rates between the forespore surface and the mother cell membrane, which manifested at a later stage of sporulation. Previously, we had reported that SpoVM mislocalizes in vivo in the absence of SpoIVA, suggesting that both proteins are reciprocally required for the proper localization of each other (ref. 16 and Fig. 1B). To test the influence of SpoIVA polymerization on the binding kinetics of SpoVM, we measured the residence times of SpoVM-Cy3 on 2-µm SSLBs in vitro in the presence of purified SpoIVA. In the presence of ATP, we observed short- and long-lived binding events of SpoVM-Cy3 at the onset of our measurement (Fig. 4A, orange and blue arrows, respectively; Movie S4). After 3 h, though, our measurements were dominated by long-lived SpoVM-Cy3 binding to SSLBs that ultimately photobleached (Fig. 4A and Movie S5). Examination of intermediate time points revealed that, in the presence of SpoIVA and ATP, SpoVM-Cy3 initially exhibited residence times of 5.4 (± 0.6) s, 7.0 (± 2.1) s, and 7.6 (± 2.0) s at 0, 1, and 2 h, respectively (Fig. 4B): an average residence time of 6.7 s that corresponds to a koff value of 0.15 s−1. At 3 h, single-molecule resolution could not be achieved due to high-density packing of SpoVM-Cy3 molecules, which eventually photobleached, suggesting that SpoVM did not exhibit a measurable off-rate when SpoIVA polymerizes. In the absence of ATP, we continued to observe short- and long-lived binding events of SpoVM-Cy3 even at 3 h (Fig. 4A), displaying residence times of 7.5 (± 2.1), 8.2 (± 2.2), and 9.7 (± 0.8) s at 0, 1, and 2 h, respectively (Fig. 4C). Unlike in the presence of ATP (Fig. 4B), at 3 h and even 24 h, we continued to measure residence times (and therefore calculate off-rates) for SpoVM-Cy3 of 6.3 (± 1.6) and 7.0 (±1.9) s, respectively (Fig. 4C). Likewise, incubating SpoVM-Cy3 in the presence of ATP, but with a SpoIVA variant that does not bind ATP (SpoIVAK30A) (18), or incubating SpoVMP9A-Cy3 (which does not interact with SpoIVA; ref. 26) with ATP and SpoIVA, did not result in an increase in residence times and permitted us to measure residence times even at 24 h (Fig. 4 A, D, and E). The data suggest that polymerization of SpoIVA atop membrane-bound SpoVM molecules could ultimately prevent the dissociation of SpoVM molecules from the membrane. Similar residence times were obtained when we repeated these experiments using 8-µm SSLBs (SI Appendix, Fig. S3), indicating that, if SpoVM properly inserts onto a membrane and can recruit SpoIVA, SpoIVA polymerization can nonetheless occur in vitro and prevent membrane dissociation of SpoVM.
Fig. 4.
SpoIVA polymerization drives stable association of SpoVM on the membrane. (A) Kymographs of SpoVM-Cy3 binding on 2-μm SSLBs in the presence of SpoIVA and ATP, presence of SpoIVA alone, presence of SpoIVAK30A and ATP, or SpoVMP9A in the presence of SpoIVA and ATP, taken immediately after (0 h) (Top) or 3 h after (Bottom) initiating binding reaction. (Scale bar: 2 μm.) Long-lived species, blue; short-lived species, orange. (B–D) Residence times of SpoVM-Cy3 measured 0, 1, 2, 3, and 24 h after initiating binding reaction in the presence of SpoIVA and ATP (B), SpoIVA alone (C), or SpoIVAK30A and ATP (D). “>28 s” indicates that single-molecule imaging conditions could not be attained and that residence times likely exceeded the timescale for photobleaching. (E) Residence times of SpoVMP9A in the presence of SpoIVA and ATP (SI Appendix, Table S4).
Next, we measured SpoVM-HALO binding kinetics in vivo in the presence of SpoIVA. In the presence of limiting amounts of JF549 dye, we observed the appearance and disappearance of single molecules of SpoVM-HALOJF549 on the forespore and mother cell membranes (Movie S6 and Fig. 5A) and observed both short- and long-lived binding events (Fig. 5B). Three hours into sporulation, SpoVM displayed mean residence times of 17.9 (± 7.4) s on the forespore and 5.9 (± 1.9) s on the mother cell, suggesting that it distinguished between the 2 membranes (Fig. 5C). Six hours after sporulation induction, we were unable to measure an off-rate for SpoVM-HALOJF549, indicating its stable association with the forespore. In contrast, SpoVMP9A displayed similar residence times of 14.3 (± 5.3) s on the forespore and 12.1 (± 6.5) s on the mother cell 3 h after the onset of sporulation, a pattern that largely persisted 6 h into sporulation (Fig. 5C). Taken together, the in vitro and in vivo results indicate that the ATP-dependent polymerization of SpoIVA atop SpoVM drives the irreversible association of SpoVM in the forespore membrane, thereby reinforcing the preferential localization of SpoVM around the forespore.
Fig. 5.
SpoIVA polymerization drives longer SpoVM-HALO residence times in vivo. (A, Left) Fluorescence micrograph of sporulating wild-type (WT) B. subtilis (EAP5) harboring SpoVM-HALO in a WT background imaged 3 h after sporulation induction using MitoTracker Green FM to visualize membranes. (A, Center) Maximum-intensity projection of aggregate SpoVM-HALO binding events over time. (A, Right) Localization plot of forespore-localized SpoVM-HALOJF549 (blue) and all measured localizations (green). (B) Frames from a movie of SpoVM-Cy3 binding events visualized using 200 pM JF549. Mother cell outlined in green; forespore outlined in red; long-lived (blue) or short-lived (orange) SpoVM-HALOJF549. (C) Residence times of SpoVM-HALOJF549, SpoVMP9A-HALOJF549, or HALOJF549 binding to forespore (maroon) or mother cell (blue) membranes. Data points are one region of interest (ROI), each containing 2 to 14 cells (SI Appendix, Table S5). “>55 s”: residence time was greater than photobleaching rate of 55 s; “N.D.”: binding events “not detected.” (D) Schematic of sporangium harboring GFP-SpoIVA (magenta) photobleached where indicated (dashed box). Fluorescence recovery indicates exchange of labeled SpoIVA (Left); no recovery (Right) indicates formation of SpoIVA polymers. (E–H) Fluorescence recovery of GFP-SpoIVA in sporulating cells (KR160) after photobleaching an area of the engulfing membrane (arrow) during early (E), intermediate (F), or after completion of (G), engulfment, or no photobleaching (H). Square indicates estimated region that received maximum photobleaching. First column: membranes visualized using FM4-64; subsequent columns: fluorescence from GFP-SpoIVA. (I) Quantification of fluorescence recovery during early (n = 22 cells) and middle (n = 23 cells) engulfment and after completion of engulfment (n = 24 cells). Acquisitions of 5 unique ROIs were analyzed. P values: ** < 0.01; otherwise, >0.05. (Scale bars: 2 μm.)
To define the stage of sporulation at which SpoIVA polymerizes in vivo, we photobleached an area of the engulfing membrane of strains producing SpoIVA-GFP and monitored fluorescence recovery after photobleaching (FRAP) (Fig. 5D). Rapid recovery of fluorescence would indicate incorporation of SpoIVA or the exchange of unpolymerized SpoIVA at that site, whereas the inability to recover fluorescence would indicate formation of static SpoIVA polymers. One possible limitation of these measurements is that we can only measure bulk polymerization of SpoIVA in a relatively large area at the forespore surface and cannot distinguish individual SpoIVA polymerization events that may occur in a relatively small area. At early or intermediate stages of engulfment (determined by measuring fluorescence intensity of the engulfing membrane labeled with FM4-64; ref. 27), ∼50% of prephotobleaching levels of SpoIVA-GFP fluorescence recovered by ∼17.5 min (Fig. 5 E, F, and I). In contrast, upon completing engulfment, we did not measure appreciable SpoIVA-GFP fluorescence recovery (Fig. 5G), suggesting little new SpoIVA incorporation or exchange out of the coat basement layer. These data indicate that the majority of SpoIVA molecules are in a polymerized state on the surface of the forespore at a time that the forespore becomes fully engulfed, but the size of the SpoIVA polymers and the extent of polymerization is not known.
Simulation of SpoVM and SpoIVA Kinetics Reveals Dynamic Coat-Assembly Process.
Our model of spore coat assembly initiation predicts an on- and off-rate for SpoVM, and that the subsequent polymerization of recruited SpoIVA molecules could effectively eliminate the off-rate of SpoVM from the membrane. Thus far, we had experimentally determined the off-rate of SpoVM in the presence of SpoIVA and could calculate the apparent on-rate. To add to the complexity of the model, we next sought to measure the adsorption kinetics of labeled SpoIVA onto membranes saturated with bound SpoVM (Fig. 6A). We therefore first incubated 2-µm SSLBs with 3 µM SpoVM in the presence of increasing concentrations of purified Cy3-SpoIVA (in the absence of ATP to prevent polymerization) to generate a saturation binding curve (Fig. 6B), which revealed that SpoIVA bound to SpoVM-covered membranes with Khalf value of 1.4 ± 0.9 µM. Next, we determined residence times of purified Cy3-SpoIVA incubated with SSLBs saturated with SpoVM using HiLo microscopy (Fig. 6C) and observed that, as with SpoVM, SpoIVA displayed both short-lived and long-lived binding behavior (Fig. 6D). Consistent with the existence of 2 populations, the distribution of observed residence times fit a 2-component exponential decay (Fig. 6 E and F). Cy3-SpoIVA displayed an average residence time on SpoVM-saturated SSLBs of 13.8 (± 5.4) s (Fig. 6F and SI Appendix, Table S6), which corresponds to an off-rate of 0.073 s−1. Using the Khalf value of 1.4 µM, we calculated an on-rate for SpoIVA onto SpoVM-coated SSLBs to be 0.05 µM−1s−1.
Fig. 6.
Kinetics of SpoIVA recruitment onto SpoVM-saturated SSLBs. (A) Schematic of an SSLB (gray) saturated with unlabeled SpoVM (blue) incubated with limiting amounts of Cy3-SpoIVA (magenta). (B) Adsorption of increasing concentrations of Cy3-SpoIVA onto SpoVM-saturated 2-µm SSLBs in vitro. Fluorescence intensity on each SSLB measured using flow cytometry. Data fit is as in Fig. 1. Data points are mean values; error bars: SD (n = ∼20,000). (C) SpoVM-saturated 2-μm SSLBs incubated with Cy3-SpoIVA visualized using bright field (Top); maximum-intensity projection of aggregate Cy3-SpoIVA molecule binding events over time (Middle); localization plot of total Cy3-SpoIVA localization events (green) or those that occur on SSLBs (blue) (Bottom). (D) Kymograph of Cy3-SpoIVA molecules binding on a SpoVM-saturated 2-μm SSLB observed for 200 frames. Short-lived (orange) and long-lived (blue) interactions are indicated. (E) Cumulative distribution function of SpoIVA duration on SpoVM-saturated SSLBs fit with 1-component (Left) or 2-component (Right) exponential decay model. Arrow: parameter space of data that was better fit with using a 2-component model. (F) Residence times of long-lived Cy3-SpoIVA bound to SpoVM-saturated 2-μm SSLBs at Cy3-SpoIVA concentrations indicated. Data points are one region of interest containing 3 to 14 SSLBs. (Scale bars: 2 μm.)
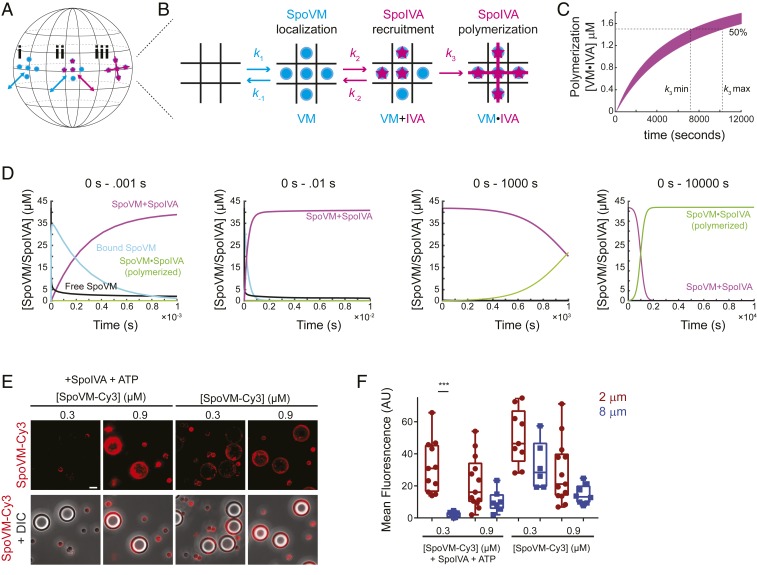
To construct and test a quantitative model that described the initiation of coat assembly, we first modeled the spherical surface of the forespore as a periodic lattice in which each SpoVM binding site had a maximum of four neighboring sites (Fig. 7 A and B). SpoVM binding to each of these sites used the on-rate (k1) and off-rate (k-1) that were determined in the presence of SpoIVA. Next, we modeled the recruitment of SpoIVA, displaying the calculated on-rate (k2), as being dependent upon the binding of 5 SpoVM molecules. We modeled this as a reversible step that used the measured off-rate (k-2) when 2 or less SpoIVA concurrent binding events occur. The model then required 2 more inputs: 1) a rate of polymerization of SpoIVA (k3), which would be irreversible and, as an assumption, would require at least 3 adjacent bound SpoIVA molecules to induce polymerization of SpoIVA; and 2) concentrations of SpoVM and SpoIVA.
Fig. 7.
A quantitative model for coat assembly initiation incorporating dynamic, but preferential, localization of SpoVM coupled to localized polymerization of recruited SpoIVA. (A and B) Schematic of the spherical forespore surface (A) modeled as a 2-dimensional lattice containing 3 coat assembly intermediates (B): 1) bound SpoVM (blue), 2) SpoIVA (magenta) recruited to bound SpoVM, and 3) polymerized SpoIVA atop membrane-bound SpoVM. (k1 and k−1) and (k2 and k−2): association and dissociation rates of SpoVM on and from the membrane or SpoIVA on and from membrane-bound SpoVM, respectively. (k3): polymerization rate of 3 or more adjacently recruited SpoIVA molecules. (C) Range of values for k3 determined by running multiple simulations with experimentally determined (k1 and k−1) and (k2 and k−2) that resulted in 50% SpoIVA polymerization between 2 to 3 h. (D) Simulation, using cellular concentrations of SpoVM and SpoIVA, that tracks abundance of assembly intermediates over time. First graph: first millisecond of the reaction, SpoVM binds to membrane (blue) and recruits SpoIVA to produce membranes bound to both SpoVM and SpoIVA (magenta). Black: free SpoVM; green: membranes harboring polymerized SpoIVA atop SpoVM. Second and third graphs: first 10 milliseconds and 1,000 s of the simulation. Polymerized SpoIVA atop membranes shown in green. Fourth graph: first 10,000 s of the simulation. Polymerization of SpoIVA drives depletion of membranes simply bound by SpoVM and SpoIVA (magenta). (E) Fluorescence microscopy of SpoVM-Cy3 (0.3 or 0.9 µM) binding to a mixed population of 2 and 8 µm SSLBs either in the presence (Left) or absence (Right) of SpoIVA and ATP. (E) Cy3 fluorescence (top row); merge, Cy3 and DIC (bottom row). (Scale bar: 4 μm.) (F) Quantification of SpoVM-Cy3 fluorescence bound to 2-µm (maroon) or 8-µm (blue) SSLBs. P values: *** < 0.005; otherwise, >0.05.
Since we could not directly measure k3, we determined a range for this constant based on observation that the irreversible binding of SpoVM to SSLBs in the presence of SpoIVA and ATP occurred between 2 and 3 h after initiation of the reaction in vitro (Fig. 4B). We therefore fixed the values for k1, k-1, k2, and k-2, and varied values for k3 in the presence of 3 µM of each protein that resulted in 50% polymerized SpoIVA between 2 and 3 h (Fig. 7C). We estimated the number of binding sites atop 2-μm SSLBs saturated with SpoVM using flow cytometry by measuring the fluorescence intensity of bound molecules of SpoIVA labeled with AlexaFluor488 and calculating the number of molecules by comparison with beads labeled with a known quantity of fluorescent molecules that produced equivalent fluorescence intensity. Our simulations identified a relatively narrow region of parameter space for k3 between 1.2 × 10−4 and 1.5 × 10−4. We therefore used an average value of 1.4 × 10−4 for k3.
Finally, we determined the in vivo concentrations of SpoVM and SpoIVA by immunoblotting extracts of sporulating B. subtilis producing SpoVM-GFP at a single ectopic locus from its native promoter 2, 3, and 4 h after induction of sporulation using antisera raised against purified SpoIVA or GFP and comparing the intensity of the band to that of immunoblots of purified SpoIVA or SpoVM-GFP at known concentrations (SI Appendix, Fig. S4). The number of spores in each culture was then determined by enumerating heat resistant colonies produced 24 h after the initiation of sporulation. This analysis yielded 45,000 ± 31,000 molecules of SpoIVA and 55,000 ± 23,000 molecules of SpoVM-GFP per sporangium at 2 h (SI Appendix, Fig. S4 and Table S7). Assuming a mother cell volume of 2 fL (1 µm × 1 µm × 2 µm), this equates to a nearly equimolar intracellular concentration of 38 µM for SpoIVA and 46 µM for SpoVM. For simplicity, we ran our simulations using 42 µM SpoVM and SpoIVA.
Running the simulation for 10,000 s revealed four trackable populations: free SpoVM, SpoVM bound to the forespore, bound SpoVM with recruited SpoIVA, and polymerized SpoIVA atop forespore-bound SpoVM. In the first 10−3 s, free SpoVM was rapidly depleted (Fig. 7D, first panel, black) as SpoVM bound to the forespore surface increased (Fig. 7D, cyan). However, the concentration of binding sites bound by SpoVM did not reach 100%, as SpoIVA was recruited to forespore-bound SpoVM (Fig. 7D, magenta). This product, consisting of forespores bound to SpoVM and recruited, but unpolymerized, SpoIVA remained stable for the next ∼10 min (Fig. 7D, third panel). However, within ∼15 min, 50% of the population consisted of forespores covered with polymerized SpoIVA (Fig. 7D, green), and by ∼30 min, 100% of the population consisted of forespores covered in a polymerized SpoIVA (Fig. 7D, fourth panel). Taken together, the incorporation of in vitro-derived kinetic constants into a simulation of coat assembly revealed a complex interplay between SpoVM and SpoIVA in which recruitment of SpoVM to the forespore surface is followed rapidly by recruitment and polymerization of SpoIVA. The polymerization of SpoIVA ultimately prevents the dissociation of both proteins from the membrane surface and results in the rapid accumulation of a stable proteinaceous structure that covers the forespore surface.
Discussion
In this report, we have investigated how bacteria initiate de novo assembly of the surface of endospores. Whereas surfaces of cells resulting from symmetrical cell division assemble in a templated manner that requires preexisting cell surface material, the morphogenesis of the endospore surface is unique in several ways. In B. subtilis endospores, the outermost surface (the “coat”) is entirely proteinaceous, and not composed of lipids (like mammalian cells and gram-negative bacteria) or cell wall material (like plants, fungi, archaea, or gram-positive bacteria) and is assembled in a fashion that does not require preexisting spore coat material. Additionally, coat assembly occurs not in an oxidizing extracytoplasmic environment but in the reducing environment of the progenitor cell’s cytoplasm. Assembly of the coat therefore represents a model for understanding of how the localized intracellular assembly of a supramolecular static structures can initiate.
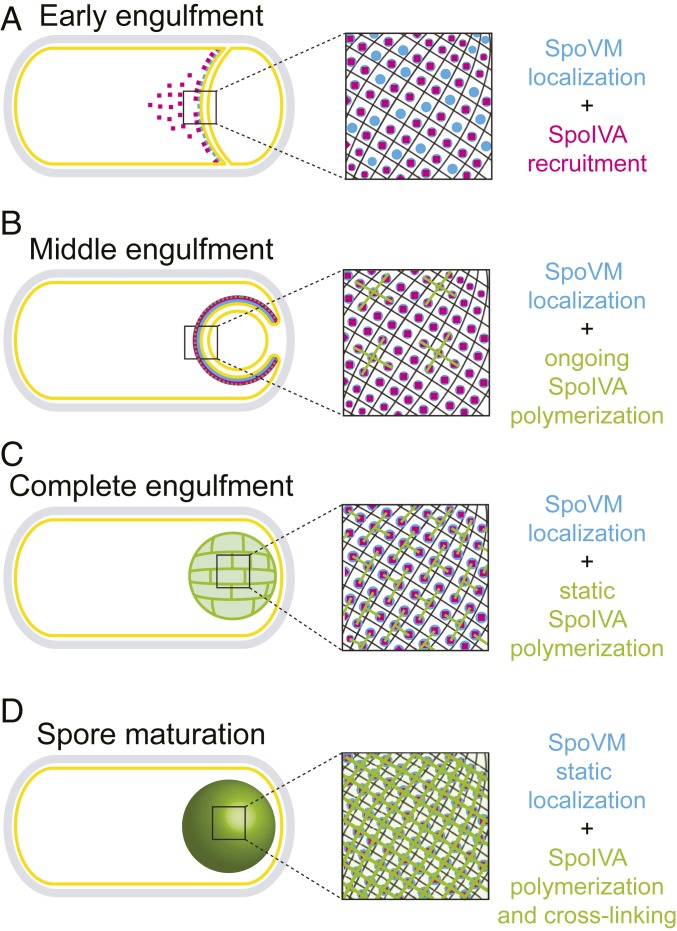
Coat assembly initiates with 2 proteins: a small structural protein, SpoVM, that starts the reaction by marking the forespore surface as the proper site for coat assembly and anchoring the assembling coat onto the outer forespore membrane; and SpoIVA, a structural ATPase recruited by SpoVM that polymerizes in an ATP hydrolysis-dependent manner to form a stable platform for subsequent recruitment of coat proteins (4). Previously, we have mechanistically studied both proteins largely in isolation to demonstrate the membrane curvature-dependent localization of SpoVM (12, 13, 15) and the ATP-dependent polymerization of SpoIVA in solution (18, 19). Here, we propose a 2-dimensional ratchet model that provides a unified picture of the role of both proteins in the concerted initiation of coat assembly (Fig. 8) (28). In our model, SpoVM, an amphipathic α-helical peripheral membrane protein (29), inserts spontaneously into lipid bilayers and can bind and unbind membranes. Previously, we measured the kinetics of bulk adsorption of SpoVM onto differently curved membranes and proposed a quantitative model (based on holding the off-rate constant for computational simplicity) in which a faster on-rate could explain the higher affinity of SpoVM toward more positively curved membranes (13). Here, we tested this hypothesis directly and discovered that the increased intrinsic affinity is actually largely defined by a difference in off-rate, as SpoVM displays a longer residence time (reduced off-rate) on membranes of preferred curvature as it samples all available membranes. The longer residence time of SpoVM on the forespore surface makes it more likely to recruit SpoIVA to that site. Recruitment of a threshold number of SpoIVA molecules that are adjacent to one another atop membrane-bound SpoVM results in localized SpoIVA polymerization that prevents SpoVM from coming off the forespore surface and effectively halts further sampling of other membranes. Polymerization of SpoIVA drives the irreversible association of SpoVM molecules at the membrane surface, and ultimately the assembly of a static SpoVM-SpoIVA structure atop the forespore. In the absence of SpoIVA polymerization, the ratcheting mechanism fails and SpoVM continues to sample other membranes in the developing sporangium.
Fig. 8.
Model for spore coat basement-layer assembly via a ratcheting mechanism. (A) At the onset of engulfment, SpoVM (blue) preferentially embeds onto the convex engulfing membrane and recruits SpoIVA (magenta). (B) As engulfment proceeds, SpoIVA polymerizes on the forespore surface. (C) At the end of engulfment, the bulk of SpoIVA molecules are in a polymer, but the lack of extensive cross-linking still permits some escape of SpoVM. (D) As the forespore matures, cross-linking of SpoIVA polymers progresses, which traps the underlying SpoVM.
Several lines of evidence are consistent with this model. In an in vitro system utilizing membranes of different defined convex curvatures, SpoVM-Cy3 molecules displayed longer residence times on more convex surfaces but nonetheless sampled differently curved membranes. However, the addition of purified SpoIVA in the presence of ATP resulted in the irreversible association of SpoVM on the membrane. Since this trapping of SpoVM on the membrane was not observed in the absence of ATP, we propose that this event coincides with SpoIVA polymerization (Fig. 4B). Stable association of SpoVM-HALO was also observed in vivo in the presence of SpoIVA (Fig. 5C). Further, consistent with the model, in a competition binding experiment using a low concentration of SpoVM that did not permit discrimination between differently curved membranes, the addition of SpoIVA and ATP drove the preferential adsorption of SpoVM onto the more convex membrane surface (Fig. 7 E and F). This observation could explain the in vivo codependence of SpoVM and SpoIVA for each other’s proper localization (Fig. 1B and ref. 16), wherein SpoVM is required to initially recruit and anchor SpoIVA to the forespore surface and subsequent SpoIVA polymerization prevents the further dissociation of SpoVM from the membrane. To test the feasibility of our model, we measured or calculated the on- and off- rates for SpoVM onto the membrane and the on- and off-rates of SpoIVA onto SpoVM-covered membranes and estimated the rate constant for SpoIVA polymerization based on the time scales we observed in vitro. Using these parameters and in vivo concentrations of SpoVM and SpoIVA present in the cell 2 h into sporulation (SI Appendix, Fig. S4), our simulation indicated that SpoVM would attach to the membrane and recruit SpoIVA in the first millisecond. This state would persist for the next ∼10 min, followed by polymerization of SpoIVA over the next ∼20 min, which led to an extremely stable state in which SpoVM and polymerized SpoIVA remained associated on the forespore. Interestingly, this timescale corresponds well to the in vivo observation using FRAP that SpoIVA polymerization is only appreciable after the completion of engulfment (Fig. 5 E–H), which occurs ∼30 min after the buildup of SpoIVA and SpoVM in the mother cell after asymmetric division. Why, then, would individual molecules of SpoVM continue to exhibit an off-rate for an additional ∼3 to 4 h in vitro and in vivo before becoming trapped on the forespore? In solution, SpoIVA polymerizes into filaments, but it was also shown to display significant lateral interactions (18). We propose that, by the end of engulfment, most SpoIVA molecules have polymerized, but that most of these polymers are relatively short or do not display extensive cross-linking. We predict that this small extent of polymerization is enough to prevent free exchange of SpoIVA molecules (detected using FRAP) but that the extent of cross-linking between SpoIVA polymers at this stage is not enough to prevent the off-rate for underlying SpoVM molecules. As sporulation proceeds, we predict that SpoIVA cross-linking becomes more extensive and eventually prevents SpoVM molecules from coming off the membrane (Fig. 8). Our model therefore invokes 3 separate events: dynamic localization of SpoVM, recruitment of SpoIVA, and polymerization of SpoIVA, which work in concert to localize and assemble a supramolecular structure atop a 2-dimensional membrane surface.
A major remaining challenge is to understand the structural and ultrastructural basis for how SpoIVA polymers physically cover the forespore surface to form a platform for subsequent coat recruitment. Structurally, SpoIVA undergoes a massive conformational change in response to ATP hydrolysis, which we have proposed results in a polymerization-competent form of the protein (19). Solving the atomic structure of SpoIVA may therefore reveal how nucleotide hydrolysis can set up SpoIVA to polymerize. On a larger scale, ultrastructural analysis of the basement layer of the coat may reveal how SpoIVA polymers arrange themselves atop the forespore. In vitro, SpoIVA assembles into filaments (18), but it is unclear how this 1-dimensional polymer can coat a 2-dimensional surface. Nuclear lamins in eukaryotes are intermediate filaments that help maintain the shape of the nucleus. Although lamins do not require a nucleotide to assemble, they, like SpoIVA, form static polymers can arrange themselves into a mesh-like pattern in vitro that may provide a mechanism for covering a membrane surface (30). Indeed, atomic force micrographs of spore surfaces have revealed filament-like features near the basement layer of the coat, but it is unclear if these features are built with SpoIVA (31). It will be interesting to see if a ratchet-like mechanism, like the one we have proposed for the initiation of bacterial spore coat assembly, can be invoked for other scenarios where static cytoskeletal proteins assemble on a membrane surface.
Materials and Methods
Please see SI Appendix for detailed methods.
Strain Construction and General Methods.
All B. subtilis strains are derivatives of PY79 (32). Strain genotypes and general methods described in SI Appendix. Significance tests were done using unpaired t tests. P values: * < 0.05, ** < 0.01, *** < 0.005, and **** < 0.001.
Epifluorescence Microscopy.
B. subtilis strains were induced to sporulate by resuspension in Sterlini-Mandelstam medium supplemented as required with 80 µg/mL threonine if the thr locus was disrupted. Cells were imaged as previously described (33).
SSLB Preparation.
SSLBs were made as described previously (12, 13) using 67% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 23.2% 1,2-dioleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DOPG), and 9.8% cardiolipin (Avanti).
Flow Cytometry.
Flow cytometry was performed as described previously (13). Briefly, SpoVM-Cy3 or SpoVMP9A-Cy3 were incubated with 2- or 8-µm SSLBs in buffer A: 50 mM tris(hydroxymethyl)aminomethane at pH 7.5, 400 mM NaCl, and 10 mM MgCl2. To ensure equal surface areas, each reaction contained either 1 µL (10 mg/mL) of 2-µm SSLBs or 4 µL (10 mg/mL) of 8-µm SSLBs for a total volume of 40 µL.
Single Molecule Fluorescence Microscopy.
Single-molecule imaging was conducted on a custom-built Olympus IX 71 microscope coupled with a 150× oil-immersion objective lens (numerical aperture = 1.45; Olympus), a multiband dichroic (405/488/561/633 BrightLine quad-band bandpass filter; Semrock) and a piezo z-stage (Applied Scientific Instrumentation).
Simulation.
Beads were represented as periodic 2-dimensional square lattices such that each binding site has n = 4 nearest neighbors (P: SpoVM protein; B: SSLB; A: SpoIVA protein). k1, k−1indicate association and dissociations rates of SpoVM binding to bead. SpoIVA binding occurred only after SpoVM binding. k2 and k−2 indicate association and dissociation rates of SpoIVA interaction with SpoVM-bound beads. After more than 60% of the square lattices had been occupied, further SpoIVA binding would induce polymerization with irreversible association rate of k3. k1, k−1, k2, and k−2 were experimentally determined. The range for k3 was determined indirectly based on the polymerization time observed for SpoIVA in vitro and initial protein concentrations of SpoVM and SpoIVA were based on in vivo concentrations of SpoVM and SpoIVA. Kinetics of the process is described by the ordinary differential equations (SI Appendix, Eqs. 1–4) solved using Runge–Kutta method in Matlab (ode23 function).
Supplementary Material
Acknowledgments
We thank K. Wolcott of the National Cancer Institute (NCI) Flow Cytometry Core Facility for technical assistance and training and S. Gottesman, S. Wickner, G. Storz, A. Khare, M. Maurizi, H. Vishwasrao, and members of the K.S.R. laboratory for discussions. This work was funded by the Intramural Research Program of the NIH Advanced Imaging and Microscopy Resource (J.C. and H.S.), the National Institute of Biomedical Imaging and Bioengineering (H.S.), and the NCI Center for Cancer Research (K.S.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907397116/-/DCSupplemental.
References
- 1.Hovorakova M., Lesot H., Peterka M., Peterkova R., Early development of the human dentition revisited. J. Anat. 233, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan S., Scholey J. M., Assembly, functions and evolution of archaella, flagella and cilia. Curr. Biol. 28, R278–R292 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Waring G. L., Morphogenesis of the eggshell in Drosophila. Int. Rev. Cytol. 198, 67–108 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Driks A., Eichenberger P., The spore coat. Microbiol. Spectr. 4, (2016). [DOI] [PubMed] [Google Scholar]
- 5.Setlow P., Spore resistance properties. Microbiol. Spectr. 2, (2014). [DOI] [PubMed] [Google Scholar]
- 6.Henriques A. O., Moran C. P. Jr, Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61, 555–588 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Tan I. S., Ramamurthi K. S., Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 6, 212–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins D., Dworkin J., Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36, 131–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenney P. T., Driks A., Eichenberger P., The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11, 33–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin P. A., et al. , An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 9, 761–771 (1993). [DOI] [PubMed] [Google Scholar]
- 11.van Ooij C., Losick R., Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 185, 1391–1398 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramamurthi K. S., Lecuyer S., Stone H. A., Losick R., Geometric cue for protein localization in a bacterium. Science 323, 1354–1357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E. Y., Tyndall E. R., Huang K. C., Tian F., Ramamurthi K. S., Dash-and-Recruit mechanism drives membrane curvature recognition by the small bacterial protein SpoVM. Cell Syst. 5, 518–526.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prajapati R. S., Ogura T., Cutting S. M., Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim. Biophys. Acta 1475, 353–359 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Gill R. L., Jr, et al. , Structural basis for the geometry-driven localization of a small protein. Proc. Natl. Acad. Sci. U.S.A. 112, E1908–E1915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramamurthi K. S., Clapham K. R., Losick R., Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 62, 1547–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Roels S., Driks A., Losick R., Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174, 575–585 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramamurthi K. S., Losick R., ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol. Cell 31, 406–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaing J. P., Nagy A., Anantharaman V., Aravind L., Ramamurthi K. S., ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc. Natl. Acad. Sci. U.S.A. 110, E151–E160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castaing J. P., et al. , An autoinhibitory conformation of the Bacillus subtilis spore coat protein SpoIVA prevents its premature ATP-independent aggregation. FEMS Microbiol. Lett. 358, 145–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price K. D., Losick R., A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181, 781–790 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayerl T. M., Bloom M., Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys. J. 58, 357–362 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan G., Rouiller I., Colman D. R., Lennox R. B., Supported bilayers formed from different phospholipids on spherical silica substrates. Langmuir 25, 5455–5458 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Santos S., et al. , Optically sectioned fluorescence endomicroscopy with hybrid-illumination imaging through a flexible fiber bundle. J. Biomed. Opt. 14, 030502 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Grimm J. B., et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebmeier S. E., Tan I. S., Clapham K. R., Ramamurthi K. S., Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol. Microbiol. 84, 682–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley E. P., et al. , Spatiotemporally regulated proteolysis to dissect the role of vegetative proteins during Bacillus subtilis sporulation: Cell-specific requirement of σH and σA. Mol. Microbiol. 108, 45–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ait-Haddou R., Herzog W., Brownian ratchet models of molecular motors. Cell Biochem. Biophys. 38, 191–214 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Giménez-Andrés M., Čopič A., Antonny B., The many faces of amphipathic helices. Biomolecules 8, E45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funkhouser C. M., et al. , Mechanical model of blebbing in nuclear lamin meshworks. Proc. Natl. Acad. Sci. U.S.A. 110, 3248–3253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plomp M., Carroll A. M., Setlow P., Malkin A. J., Architecture and assembly of the Bacillus subtilis spore coat. PLoS One 9, e108560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youngman P., Perkins J. B., Losick R., Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12, 1–9 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Eswaramoorthy P., et al. , Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet. 10, e1004526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.