Abstract
Porphyromonas gingivalis uses a type IX secretion system (T9SS) to deliver more than 30 proteins to the bacterial surface using a conserved C-terminal domain (CTD) as an outer membrane translocation signal. On the surface, the CTD is cleaved and an anionic lipopolysaccharide (A-PLS) is attached by PorU sortase. Among T9SS cargo proteins are cysteine proteases, gingipains, which are secreted as inactive zymogens requiring removal of an inhibiting N-terminal prodomain (PD) for activation. Here, we have shown that the gingipain proRgpB isolated from the periplasm of a T9SS-deficient P. gingivalis strain was stable and did not undergo autocatalytic activation. Addition of purified, active RgpA or RgpB, but not Lys-specific Kgp, efficiently cleaved the PD of proRgpB but catalytic activity remained inhibited because of inhibition of the catalytic domain in trans by the PD. In contrast, active RgpB was generated from the zymogen, although at a slow rate, by gingipain-null P. gingivalis lysate or intact bacterial cell suspension. This activation was dependent on the presence of the PorU sortase. Interestingly, maturation of proRgpB with the catalytic cysteine residues mutated to Ala expressed in the ΔRgpA mutant strain was indistinguishable from that in the parental strain. Cumulatively, this suggests that PorU not only has sortase activity but is also engaged in activation of gingipain zymogens on the bacterial cell surface.
Keywords: inhibition, limited proteolysis, periodontal disease, posttranslational modification, prodomain, secretion, sortase
1. Introduction
Periodontitis is the one of the most prevalent infection-mediated chronic inflammatory disorder in humans, with severe forms of the disease affecting up to 15% of adults and leading to tooth loss (1-3). The disease is the culmination of a destructive host inflammatory response to the development of a dysbiotic bacterial flora on the tooth surface below the gum line. Porphyromonas gingivalis has been shown to be central to the formation of the dysbiotic microflora in dental plaque (4). The growth of this anaerobic, asaccharolytic Gram-negative bacterium is highly dependent on its proteases, the gingipains, to obtain proteinaceous nutrients, as well as iron and heme (through degradation of hemoglobin) (5-7). The gingipains account for the majority (85%) of the general proteolytic activity produced by this organism and have been implicated as essential virulence factors (8). They have been reported to be indispensable for all stages of the host infection process including: bacterial attachment to host tissues; degradation of intercellular adhesion molecules required for tissue invasion; promotion of inflammation and bleeding through activation and degradation of complement and coagulation factors; dissemination by release of kinins; and dysregulation of the host immune response through degradation of cytokines and their receptors (5, 7, 9).
The gingipains are extracellular proteases unique to the periodontal pathogen P. gingivalis. They belong to the CD clan, family C25 of cysteine peptidases, together with caspases and legumains (10). Like other peptidases of the CD clan, gingipains have strict specificity for a residue in the P1 position of a scissile peptide bond, which can be either arginine (Arg-gingipains or gingipain R, for short, Rgps) or lysine (Lys-gingipain or gingipain K, for short, Kgp) (11,12). There are three members of the gingipain family: RgpA and RgpB hydrolyzing Arg-Xaa bonds, while Kgp cleaves Lys-Xaa peptides bonds (12). They are encoded by three genes: rgpA, rgpB and kgp at separate loci in the P. gingivalis genome and are synthetized as single chain multidomain proteins. The nascent preproRgpB comprises a signal peptide, an N-terminal pro-domain (PD), a catalytic domain (CD), an immunoglobulin superfamily domain (IgSF), and a C-terminal domain (CTD) (Fig. 1). In nascent translation products of the rgpA and kgp genes, additional hemagglutinin/adhesion domains (HA) are inserted between the IgSF and the C-terminal domain (5). Like most proteases, the nascent gingipains are inactive zymogens with their prodomains acting as inhibitors of cognate catalytic domains (13-15). Secreted gingipains are retained on the bacterial cell surface (16). While mature RgpB is a single chain active protease comprising of CD-IgSF, membrane-associated Kgp and RgpA are assembled into very large (>300 kDa), multi-functional, non-covalent complexes composed of CD-IgSF and several HA domains (5). In recent years, significant progress has been made in deciphering the secretion and maturation process of gingipains during their journey to the cell surface (17). With their typical N-terminal signal peptides, they translocate through the inner membrane via the Sec system. At this step the signal peptide is removed by the signal Peptidase I and the newly exposed N-terminal glutamine is enzymatically cyclized to pyroglutamate by a glutamine cyclase localized at the periplasmic face of the inner membrane (18). In the periplasm, gingipains are directed to the newly identified Type IX secretion system (T9SS) by their CTDs, for translocation across the outer membrane (OM) (17, 19). On the extracellular face of the OM, surface-associated sortase PorU, a component of the T9SS, cleaves the CTD domain and conjugates the resulting new-termini to an anionic lipopolysaccharide (A-LPS) to anchor the protein on the bacterial surface (20, 21).
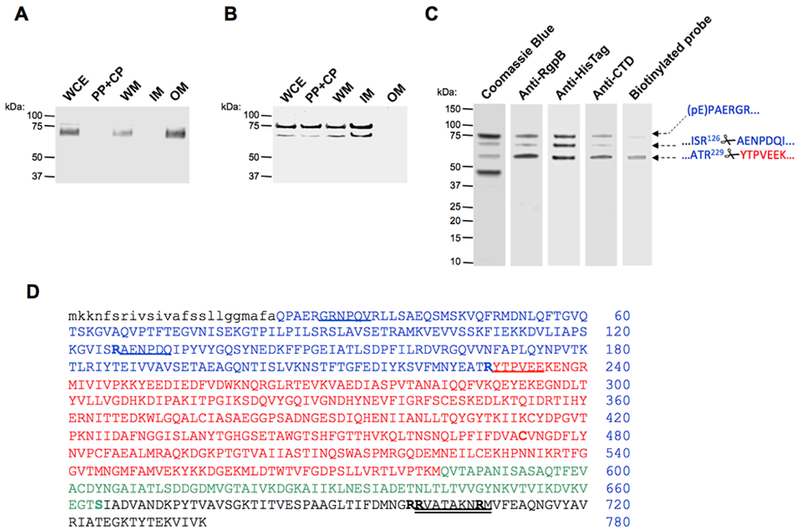
Figure 1. Subcellular location of proRgpB6His produced in wild-type P. gingivalis (panel A) and the T9SS-deficient ΔporN mutant (panel B).
(A & B) Whole cells extracts (WCE) were fractionated into the following fractions: periplasm and cytoplasm (PP + CP), whole membrane (WM), inner-membrane (IM) and outer-membrane (OM). The localization of proRgpB was determined by Western blotting using anti-RgpB polyclonal antibody. (C) ProRgpB6His from the ΔPorN mutant strain was purified by affinity chromatography on Ni-Sepharose and final products were analyzed by SDS-PAGE and Western blotting with anti-RgpB, anti-6His and anti-RgpBCTD antibodies, and by active-site labeling using the specific biotinylated probe, BiRK. N-terminal amino acid sequences of proteins indicated by arrows were determined by automated Edman-degradation and mass spectrometry analysis. (D) The amino acid sequence of preproRgpB with individual domains coded: lowercase: signal sequence; BLUE: N-terminal pro-domain (PD); RED: catalytic domain; GREEN: Ig-like domain; BLACK: C-terminal domain (CTD). R126 and R229 of the pro-domain and sequences identified by the Edman degradation of indicated (arrows) bands are underlined. Double underlined is a RgpBCTD-derived peptide used to obtain rabbit polyclonal anti-RgpBCTD. Residues mutated in the CTD; to Arg (S664) and to Lys (R700, R701 and R708) are highlighted (see Fig 6).
However, the mechanism of progingipain activation remains elusive. Recombinantly expressed proRgpB in Saccharomyces cerevisiae undergoes a quick sequential processing via intermolecular autoproteolysis (22). The first cleavage occurs at the R126-Xaa peptide bond and is followed by removal of remaining part of the prodomain by autocleavage at R229. Finally, the CTD is released and the mature, fully-active gingipain is generated (22). These results contradict the observation that P. gingivalis mutants with defective T9SS accumulate large amounts of proteolytically inactive full-length progingipains in the periplasm (23-26). Furthermore, we found that the complex formed in trans between RgpB and its recombinant pro-fragment is strong (Ki = 6.3 nM) and stable over time (up to 5 days) (13). Moreover, it is now clear that PorU is required for the release of the CTD in vivo (20, 21). To address the apparent discrepancy regarding proRgpB processing and activation, we purified proRgpB zymogen from the periplasm of the T9SS-deficient strain and studied its stability and mechanism for activation.
2. Material and methods
2.1. Material
All material media were sourced from Difco Laboratories (Detroit, MD, USA). The substrates Nα-Benzoyl-L-arginine p-nitroanilide (L-BAPNA) was from Bachem (Torrance, CA, USA). L-Cysteine, the inhibitors leupeptin, tosyl-L-lysine chloromethylketone (TLCK), 4,4’-Dithiodipyridine disulphide, 1, 10-phenanthroline and the dithiothreitol (DTT) were from Sigma (St. Louis, MI, USA). The inhibitors KYT-1 and KYT-38 were from Peptides International, Inc. (Louisville, KY). Ethylenediaminetetraacetic acid (EDTA) from USB Corporation (Cleveland, OH, USA). Ac-biotinyl-Lys-Tyr-6-aminohexanoic-Arg-acyloxymethyl Ketone (BiRK) was synthetized as described previously (22). High molecular weight gingipain R (HRgpA) from P. gingivalis HG66 strain, RgpB with a C-terminal 6His-Tag and Kgp catalytic domain were purified as previously described (19, 27-29).
2.2. Methods
2.2.1. Bacteria cultivation
P. gingivalis wild-type W83 strain and its mutants were cultured in trypticase soy broth (TSB) supplemented with yeast extract 0.5% [w/v], hemin (5 μg/ml) and menadione (1 μg/ml) at 37°C in an anaerobic chamber (Bactron IV; Sheldon Manufacturing Inc., OR) with an atmosphere of 90% nitrogen, 5% carbon dioxide and 5% hydrogen. Erythromycin at 5 μg/ml and/or tetracycline at 1 μg/ml were incorporated into the medium for the growth of P. gingivalis mutants. Escherichia coli strain TOP10 (Invitrogen) and XL-Gold ultracompetent cells (Agilent) were used for plasmids construction and grown in Luria-Bertani (LB) medium and on 1.5% [w/v] agar LB plates. Ampicillin at 100 μg/ml was used for the selection of E. coli clones.
2.2.2. Site-directed mutagenesis
By using a QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) various truncated and site-directed mutagenic mutants of the rgpB gene were constructed. Briefly, the rgpB gene sequence in master plasmid p6HRgpB (19) was modified in mutagenesis reaction using sets of oligonucleotide primers: 664F (GGATGTGAAAGTGGAAGGTACACGTATTGCCGACGTAGCCAATG) /664R: (CATTGGCTACGTCGGCAATACGTGTACCTTCCACTTTCACATCC) for replacing serine (S664) with arginine (S664R), 700F (GACGATCTTCGATATGAACGGCAAGAAGGTAGCTACTGCTAAAAACCGCATG) /700R’ (CATGCGGTTTTTAGCAGTAGCTACCTTCTTGCCGTTCATATCGAAGATCGTC) for replacing arginines (R700/R701) with lysines (RR700/701KK), 708F (CGTGTAGCTACTGCTAAAAACAAGATGGTATTCGAAGCACAAAACGGCGTG) /708R (CACGCCGTTTTGTGCTTCGAATACCATCTTGTTTTTAGCAGTAGCTACACG) for replacing arginine (R708) with lysine (R708K), 700-701-708F (CGTGTAGCTACTGCTAAAAACAAGATGGTATTCGAAGCACAAAACGGCGTG) /700-701-708R (CACGCCGTTTTGTGCTTCGAATACCATCTTGTTTTTAGCAGTAGCTACACG), and Pfu ultra high-fidelity DNA polymerase according to manufacturer’s instructions. Following mutagenesis, the parental template was digested with DpnI and each of mutated plasmids (p6HRgpB/S684R; p6HRgpB/RR700/701KK; p6HRgpB/R708K; p6HRgpB/RRR700/701/708KKK) was transformed into E. coli XL-Gold ultracompetent cells. Clones were then selected on antibiotic selective LB agar and screened for the correct mutation by DNA sequencing of the pertinent region.
Verified p6HRgpB/S684R, p6HRgpB/RR700/701KK, p6HRgpB/R708K and p6HRgpB/RRR700/701/708KKK plasmids were used for generation of isogenic mutants via homologous recombination in P. gingivalis W83 ΔPorN (30) and P. gingivalis W83 RgpB+B strains (19), as previously described (31). Genomic integration by a double crossover event in resulting mutants, all in W83 background (Table 1) was confirmed by PCR and verified by DNA sequencing of the pertinent region of the genome.
Table 1.
P. gingivalis strains and mutants used in this study.
| Strain | Relevant genotype | Source |
|---|---|---|
| W83 | Wild type | Reference strain |
| RgpB+B | rgpB ΔrgpA Emr, Cmr mutant for RgpB complemented | (19) |
| ΔPorU | porU (NCBI: PG_RS00120, old locus PG0026) (Emr) | (30) |
| ΔPorN | porN (NCBI: PG_RS01305, old locus PG0291) (Emr) | (30) |
| ΔK/ΔRAB | kgpΔ598 rgpArgpBΔ410 Tcr Cmr Emr | (32) |
| RgpB+B/RgpBC473A | rgpBp.K736_insHHHHHH/rgpBp.C473A (Tcr) | This study |
| ΔPorN/6HRgpB | porN(Ermr) rgpBp.K736_insHHHHHH (Tcr) | This study |
| ΔPorN/6HRgpB/S664R | porN(Ermr) rgpBp.K736_insHHHHHH/rgpBp.S664R (Tcr) | This study |
| ΔPorN/6HRgpB/RR700/701KK | porN(Ermr) rgpBp.K736_insHHHHHH/rgpBp.RR700/701KK (Tcr) | This study |
| ΔPorN/6HRgpB//R708K | porN(Ermr) rgpBp.K736_insHHHHHH/rgpBp.R708K (Tcr) | This study |
| ΔPorN/6HRgpB/RRR700/701/708KKK | porN(Ermr) rgpBp.K736_insHHHHHH/rgpBp.RRR700/701/708KKK (Tcr) | This study |
| RgpB+B/6HRgpB | ΔrgpA (Cmr) rgpBp.K736_insHHHHHH (Tcr) | This study |
| RgpB+B/6HRgpB/S664R | ΔrgpA(Cmr) rgpBp.K736_insHHHHHH/rgpBp.S664R (Tcr) | This study |
| RgpB+B/6HRgpB/RR700/701KK | ΔrgpA (Cmr) rgpBp.K736_insHHHHHH/rgpBp.RR700/701KK (Tcr) | This study |
| RgpB+B/6HRgpB//R708K | ΔrgpA(Cmr) rgpBp.K736_insHHHHHH/rgpBp.R708K (Tcr) | This study |
| RgpB+B/6HRgpB/RRR700/701/708KKK | ΔrgpA (Cmr) rgpBp.K736_insHHHHHH/rgpBp.RRR700/701/708KKK (Tcr) | This study |
2.2.3. Cell fractionation
Cultures cultivated to early stationary phase were adjusted to OD600=1.5 and 5mL was centrifuged at 5,000 × g for 10 min. Collected cells were washed once with PBS, lysed by ultrasonication in ten 5-seconds pulses (17 W per pulse) in an ice-water bath with 2 seconds of rest between each pulse and designated as whole cell extract (WCE). The membrane debris was pelleted by ultracentrifugation at 150,000 × g for 1 h and the free supernatant was collected as periplasm and cytoplasm extracts (PP + CP). The membranes, washed once with 30 ml PBS prior to being resuspended in 4.5 ml PBS by sonication, were designated as cell extract (CE). 0.5mL of 10% Sarkosyl [w:v] was added to the CE and left to mix gently for 20 min at 4°C to dissolve the inner membrane. The residual Sarkosyl-resistant outer membranes were pelleted by ultracentrifugation at 150,000 × g for 1 h, and the supernatants were designated the inner membrane sample (IM). The pellets were washed once with 30 ml PBS before being resuspended in 5 ml PBS by sonication and designated the outer-membrane sample (OM).
2.2.4. Effect of inhibitor on cell extracts RgpB processing
Inhibitors EDTA (5 mM), TLCK (5 mM), Leupeptin (5 mM) or 4,4’-Dithiodipyridine disulphide (2 mM) were added to bacterial culture adjusted to an OD600=1.5. After centrifugation at 5,000 × g for 10 min, collected cells were washed once, diluted in PBS complemented with the inhibitors and lysed by ultrasonication in ten 5-seconds pulses (17 W per pulse) in an ice-water bath with 2 seconds of rest between each pulse. The presence and forms and RgpB were then analyzed by Western blot.
2.2.5. Purification of proRgpB-6His and mutants
P. gingivalis strain ΔPorU expressing the proRgpB6His were grown for 3 days (early stationary phase) in eTSB media at 37°C and in anaerobic conditions. Cultures were then harvested by centrifugation at 10,000 × g for 25 min. The cells were then washed and resuspended in PBS before lysis by sonication (3 cycles of 10 × 3 s pulses at 17 W). The lysate was clarified by ultracentrifugation at 150,000 × g for 1 hour. Soluble proteins were then dialyzed 2 times against 4 L of Ni-Sepharose binding buffer (20 mM sodium phosphate buffer pH 7.4 supplemented with 500 mM NaCl, 20 mM imidazole and 0.02% NaN3). Proteins of interest were purified by affinity chromatography on Ni-Sepharose™ High Performance (GE Healthcare, Pittsburgh, PA, USA) and eluted in the same buffer + 500 mM imidazole. The protein concentration of the final samples was determined by BCA Assay (Sigma).
Alternatively, the purification was repeated in the same condition but in presence of 4,4’-Dithiodipyridine disulphide. The oxidizing reagent was added in the culture media before centrifugation (at a concentration of 2 mM) and kept in all the buffers throughout the purification process. After the Ni-Sepharose chromatography, the protein of interest was dialyzed against 4 L of Ion exchange buffer (50 mM Bis-Tris buffer, pH 6.5 supplemented with 5 mM CaCl2, 0.02 % NaN3 and 2 mM 4,4’-Dithiodipyridine disulphide). During dialysis, contaminating proteins as well as hemin precipitated out whereas proRgpB6His stayed in solution. Samples were clarified by ultra-centrifugation (100,000 × g, 1 hour). Protein concentration in the final samples was evaluated by a Bradford Assay (BioRad). ProRgpB6His-ΔCTD, proRgpB6His-RR700/701KK, proRgpB6His-R708K, proRgpB6His-RRR700/701/708KKK and proRgpB6His-S664R were all purified in presence of 4,4’-Dithiodipyridine disulphide with the same protocol.
2.2.6. RgpB proteolytic activity
RgpB was incubated at 37°C in activity buffer (200 mM Tris, 5 mM CaCl2, 150 mM NaCl, and 0.02% NaN3, pH 7.6) supplemented with 10 mM L-Cysteine. After 10 min, the chromogenic substrate L-BAPNA was added (0.5 mM) and absorbance at 420 nm was recorded every 15 s over 5 min interval using a spectrophotometer plate-reader Spectramax M5 (Molecular Devices, San Jose, CA).
2.2.7. SDS-PAGE and Western-blot
In order to prevent auto-processing during the preparation of the samples, proteins or cell fractions were incubated for 10 min in presence of 5 mM TLCK before addition of SDS-Sample buffer. The samples were then boiled two times 10 min, first without reducing agent and then with 2% [v:v] 2-mercaptoethanol. The samples were electrophoresed for 1 hour at 150 V on a 4-12% Bis-Tris Gel (Invitrogen) and then stained with SimplyBlue SafeStain (Invitrogen). For Western blot, proteins were electroblotted for 1 hour at 100 V on nitrocellulose membrane and blocked with a 5% skim milk/PBS. The membranes were then incubated with a rabbit polyconal anti-RgpB antibody, a rabbit polyclonal anti-HisTag antibody (GenScript) or a rabbit polyclonal anti-RgpBCTD antibody (custom pAb by Genscript; raised against synthetic peptide RVATAKNRMC) and with the corresponding secondary antibody (anti-rabbit IgG-peroxidase conjugate (Sigma) and anti-mouse IgG-alkaline phosphatase conjugate (Sigma)). Proteins of interest were visualized with AP Conjugate Substrate Kit (BioRad) or with TMB Membrane Peroxidase Substrate (KPL) respectively. Alternatively, proteins of interest were electro-blotted on PVDF membrane for 2 hours at 100 V and subjected to N-terminal sequence analysis.
2.2.8. Active site labeling with activity-based probe
Samples were incubated in activity buffer supplemented with 10 mM L-Cysteine for 15 min at room temperature with the active site biotinylated-probe (10 μM Ac-biotinyl-Lys-Tyr-6-aminohexanoic-Arg-acyloxymethyl ketone (BiRK) (32). The reaction was stopped by addition of TLCK to 10 mM concentration. Samples were treated with reduced sample buffer, denatured at 100°C and resolved on 4-12% Bis-Tris SDS-PAGE gels. After electrotransferred onto nitrocellulose membrane and an incubation step in blocking solution (Sigma), the probe was detected using a streptavidin-AP conjugate (Thermo Fisher Scientific).
2.2.9. Maturation of proRgpB
To assess autoprocessing, native proRgpB or mutant proteins were incubated at 0.4 mg/ml in activity buffer supplemented with or without 10 mM DTT. As a control, the incubation was carried out in the presence of 5 mM TLCK. At various times, aliquots were removed to measure residual RgpB activity using L-BAPNA substrate and to analyze the maturation status by SDS-PAGE, Western-blot and active site probing. To investigate contribution to proRgpB processing and maturation by mature gingipains, the incubation process above was repeated in presence of each of the three active gingipains (RgpB, HRgpA and Kgp) at a molar ratio 1:100 and the processing was followed as described previously. To investigate contribution to proRgpB processing and maturation by other cellular component proteins, cell extracts from wild-type W83, ΔPorU or gingipain-null strain ΔK/ΔRAB (33) were used. The mutant strains were cultivated to early stationary phase and adjusted to OD600=1.5 before being pelleted at 5,000 × g for 10 min. Bacteria were washed and diluted into 5 ml of PBS before sonication (10 times 5-s pulses, 14 W per pulse). ProRgpB (0.4 mg/ml) was then incubated at room temperature in assay buffer supplemented with 10 mM L-Cysteine and in presence of the cell extract at a ratio 1:4 (v/v). Inactivation of gingipains in W83 and ΔPorU was achieved by pre-incubation with 50 μM of inhibitors KYT-1 and KYT-36 for 10 min. To study the potential effect of the outer membrane enzymes on proRpgB maturation, bacteria were washed and diluted in their original volume of RPMI media (Invitrogen). After addition of DTT (10 mM) and proRgpB (0.4 mg/ml) to the bacteria, the processing at room temperature was followed as described previously.
2.2.10. N-terminal sequencing
Samples were resolved on 4–12% NuPAGE gels and then electrotransferred in 10 mM CAPS, 10% methanol, pH 11, onto a PVDF membrane. Protein bands were stained with CBB G-250 staining, excised, and analyzed by automated Edman degradation using a PPSQ / 31B protein sequencer (Shimadzu Biotech) connected to an LC-20AT HPLC equipped with CTO-20A column heater and SPD20A UV detector (Shimadzu Biotech) for on-line PTH analysis.
Statistical analysis
All experiments were performed in at least triplicate, and the results are expressed as the mean ± SEM. Statistical comparisons were performed with Prism 5.0 software (GraphPad), using two-tailed Student t-tests. Differences were considered significant when P < 0.05.
3. Results
3.1. Localization and purification of proRgpB-6His
In order to purify proRgpB, we expressed a recombinant form with a hexahistidine Tag (proRgpB6His) at the C-terminus in the P. gingivalis RgpB6His/ΔPorN double mutant strain to allow accumulation of the proRgpB6His in the periplasm. To determine that the 6His tag was not affecting the proRgpB maturation process and anchorage on the P. gingivalis surface, we first expressed proRgpB6His in the W83 parental strain and analyzed protein localization in subcellular fractions by Western blotting. As expected, the RgpB6His strain showed normal RgpB maturation with the mature protein found associated with the outer membrane fraction as a diffuse band of approximately 70-90 kDa typical for the processed and glycosylated form of the enzyme (Fig 1A). In contrast, in the ΔPorN background, RgpB was found in the intracellular fractions in two forms: one at 75 kDa corresponding to the unprocessed proRgpB6His (missing the signal peptide) and a truncated form of approximately 65 kDa which could correspond to processing at R126 within the pro-domain (Fig 1B). This latter strain was used to purify proRgpB6His by affinity to a Ni-Sepharose column. Analysis of the eluted fractions by SDS-PAGE electrophoresis, Western blot and N-terminal sequencing showed that purified proRgpB6His existed in 3 different forms: the full length proRgpB with a pyroglutamate residue at the N-terminus, indicating that only the signal peptide was processed; and two forms truncated at R126 and R229, respectively, resulting from cleavages inside the PD and at the junction between pro-domain and catalytic domain, respectively (Fig 1C). The band below 50 kDa apparently represents a protein unrelated to RgpB, which co-purified on Ni-Sepharose. Of note, the three forms of proRgpB recognized by anti-Rgp polyclonal antibody were also recognized by the anti-HisTag and anti-RgpBCTD antibodies thus confirming that the protein was not processed at its C-terminus. Labeling of the protein with the activity-based biotinylated probe BiRK that covalently modifies the active site cysteine (32) indicated that the higher molecular mass forms of proRgpB are catalytically inactive and only the loss of the entire PD activates the protease (Fig 1C). Despite the clear presence of an apparent active form of N-terminally-processed RgpB, the chromogenic substrate L-BAPNA failed to be processed by this preparation (data not shown). This result likely is due to the high sensitivity of the active site labeling method compared to an activity assay using a non-optimal substrate.
3.2. In vitro maturation of the proRgpB6His
When purified proRgpB6His was incubated at room temperature in gingipain activity buffer with/without DTT supplementation, we observed a slow autoprocessing of proRgpB under both conditions. The first step of the autoprocessing was cleavage of the entire PD (at Arg-229) which was detected in the reaction mixture as a 20 kDa band by SDS-PAGE. This indicated that no intermediate cleavage occurred inside the pro-fragment at R126 (Fig 2A). In the second step of maturation, the C-terminal domain was truncated as confirmed by the loss of reactivity with the anti-6HisTag antibody (Fig 2B). The molecular mass of the final product of autoprocessing of purified proRgpB was higher than that of the mature RgpB suggesting that only a part of the CTD was removed. This was confirmed by conservation of the reactivity with the anti-RgpBCTD antibody (Fig 2B) developed against a peptide 701RVATAKNRM709 mid-way through the RgpBCTD domain (Fig 1D).
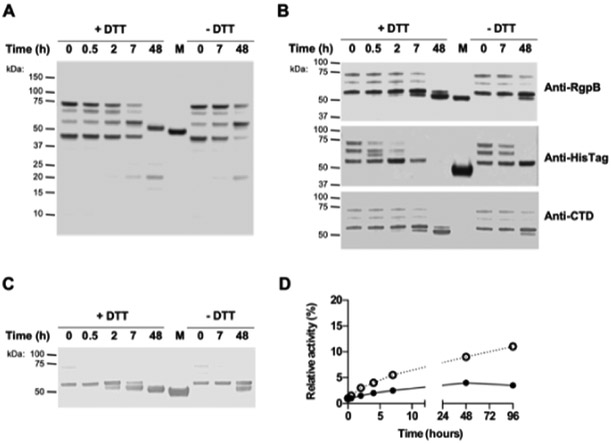
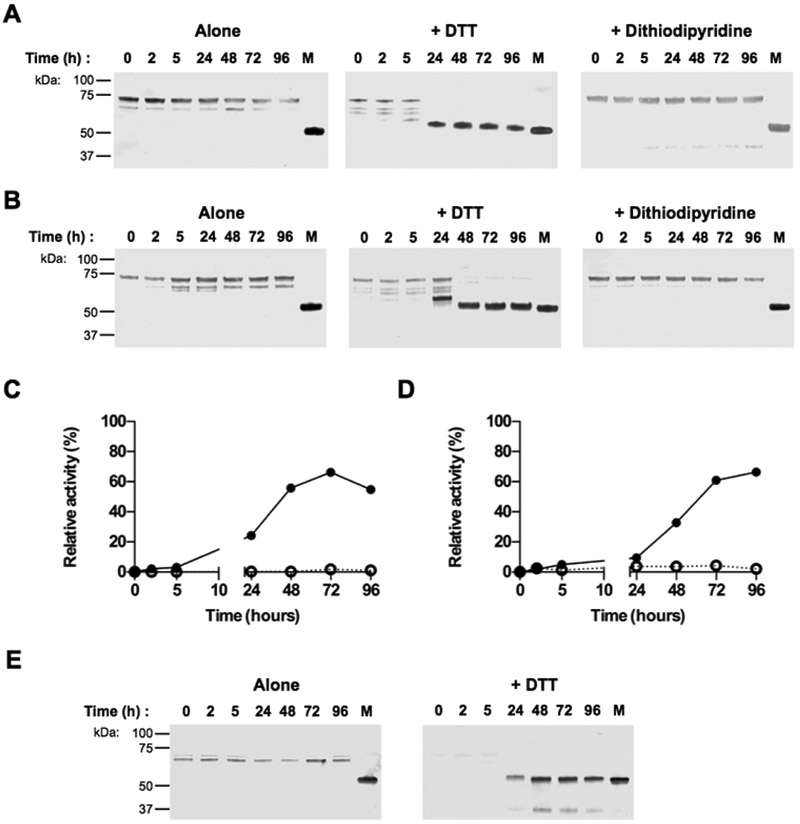
Figure 2. Purified proRgpB6His undergoes partial autoproteolytic processing with limited increase in enzymatic activity.
ProRgpB6His (0.4 mg/ml) was incubated at room temperature in gingipain activity buffer with/without 5 mM DTT supplementation. At various time points, aliquots were removed and subjected to (A) SDS-PAGE electrophoresis, or (B) Western blot analysis with pAb anti-RgpB, mAb anti-6HisTag and pAb anti-CTD domain. (C) Active-site labelling using the biotinylated probe BiRK to detect active forms of partially processed proRgpB. Mature RgpB bearing a C-terminal HisTag (Veillard et al, 2015) was used as a control (lane M). (D) The activity of RgpB at each time point was determined with the chromogenic substrate L-BAPNA. Results are expressed as percentage of activity of the native RgpB incubated in the same condition (n = 3).
Similar to the previous result, the active-site biotinylated probe reacted with RgpB only when the entire PD was processed and CTD truncation did not significantly affect RgpB labeling with the probe (Fig 2C). In the absence of the reducing agent, DTT, during the incubation, proRgpB was processed following the same pattern, but at a slower rate. Cumulatively, these results indicate that the observed rate of processing was dependent on the presence of the active, N-terminally processed RgpB, thus confirming that pro-RgpB maturation can occur via an auto-processing mechanism previously described for the recombinant proRgpB expressed in S. cerevisae (22).
The labeling with the active-site probe correlated well with a time-dependent increase in RgpB activity (Fig 2D). However, even after 96 hours, this activity represented only a fraction (about 10%) of the activity determined for the same amount of the mature active RgpB incubated in parallel. This low level of activity of the in vitro processed proRgpB is apparently due to inhibition of the enzyme by the PD (13,14). This observation indicates that in vitro auto-processing of proRgpB cannot generate the fully active protease and, in addition, it may occur only due to the presence of a small amount of activated RgpB in the preparation of the purified zymogen.
3.3. Purification of non-cleaved full-length proRgpB6His
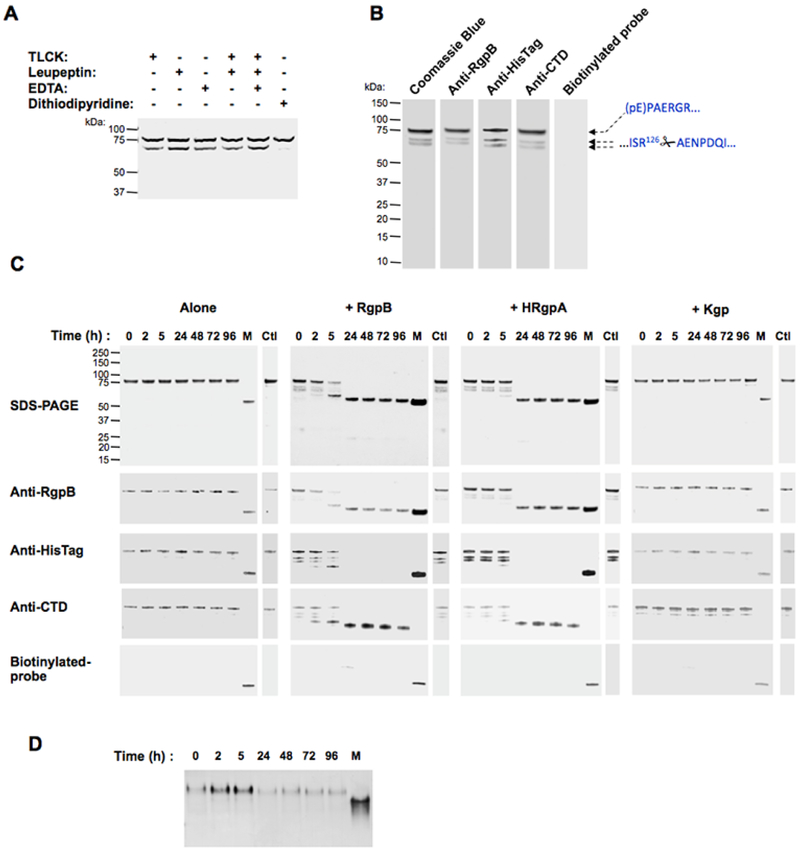
To obtain unprocessed proRgpB, we first tested various inhibitors to stabilize the zymogen during the purification process. We added EDTA (used as a metallo-protease inhibitor), TLCK, leupeptin or dithiodipyridine (a thiol oxidizing agent used as a general and reversible cysteine protease inhibitor) to the culture before collection of bacterial cells as well as to the PBS solution used during the bacterial lysis step. Of these 4 inhibitors, only dithiodipyridine successfully blocked the generation of proRgpB-R126 (Fig 3A), probably due to its ability to penetrate the outer membrane into the periplasm of viable P. gingivalis cells or due to its broad spectrum of protease inhibition. Subsequently, dithiodipyridine was used throughout the proRgpB purification to inhibit autoprocessing. This approach allowed us to isolate a highly pure sample of unprocessed pro-enzyme (Fig 3B). More importantly, despite the fact that excess DTT can reverse the inhibitory activity of dithiodipyridine on RgpB (27), purified proRgpB had no activity towards the L-BAPNA substrate (data not shown) and did not react with the active-site probe even in the presence of high concentrations of DTT.
Figure 3. 4,4’-Dithiodipyridine disulphide protects against autoproteolysis and gingipain-mediated processing of proRgpB6His.
(A) Inhibitors EDTA (5 mM), TLCK (5 mM), Leupeptin (5 mM) or 4,4’-Dithiodipyridine disulphide (2 mM) were added to bacterial cultures adjusted to OD600=1.5. After centrifugation, bacteria were washed once and diluted PBS-containing inhibitors and lysed by ultrasonication. ProRgpB6His present in the intracellular compartments was analyzed by Western blot with anti-RgpB. (B) ProRgpB6His in the T9SS-deficient ΔporN background was purified by affinity chromatography with 2 mM 4,4’-Dithiodipyridine disulphide added at each purification step. Final products were analyzed by SDS-PAGE, by Western blotting and by active-site labeling using the biotinylated probe BiRK. N-terminal sequences were determined by automated Edman-degradation as indicated. (C) ProRgpB6His (0.4 mg/ml) was incubated at room temperature in gingipain activity buffer supplemented with 10 mM DTT. At various times points, aliquots were removed to follow the proteolytic autoprocessing of the enzyme by SDS-PAGE and Western blotting. The active forms of processed proRgpB were identified by active-site labeling using the biotinylated probe BiRK. Mature RgpB bearing a C-terminal HisTag (lane M) was used as positive control. The incubation process was repeated in the presence of each of the three active gingipains (RgpB, HRgpA and Kgp) at a molar ratio 1:100. Incubations for 96 h in presence of TLCK were used as negative control (Ctl). (D) Alternatively, the processing in presence of active RgpB in the reduced conditions was followed by Native-PAGE electrophoresis.
3.4. In vitro processing of full-length proRgpB6His
Compared to proRgpB in a batch containing the partially processed zymogen (Fig 2A), the highly purified full-length proRgpB displayed remarkable stability when incubated under the same conditions (Fig 3C). Accordingly, the active-site probe did not label any form of the enzyme and no activity could be measured using the L-BAPNA substrate (data not shown). This result indicated that proRgpB is not able to process itself, at least by an intra-molecular mechanism. To investigate possible inter-molecular auto-processing, we repeated the incubation in the presence of active, purified RgpB, HRgpA and Kgp. While Kgp had no effect on proRgpB stability, the addition of RgpB and HRgpA led to the processing of the protein following the same pattern observed for proRgpB autoprocessing. Within the first 5 hours of incubation, the entire PD was removed, followed by loss of the C-terminal 6HisTag after 24 h incubation time. However, as observed for proRgpB processing, a part of the CTD domain was still present in the final product as detected by the anti-RgpBCTD antibody. Remarkably, processed RgpB without the PD and truncated at the C-terminus was not labeled by the active site probe and did not display activity towards L-BAPNA despite the presence of DTT. Altogether, these results indicate the inter-molecular processing of proRgpB is not sufficient for activation because the released PD is inhibitory as has already been show using the recombinant PD (13,14). In order to confirm this observation, we analyzed proRgpB processing by native-PAGE electrophoresis. Although proRgpB was processed, it maintained the same apparent mobility on native gel, thus confirming that the PD remains anchored to the catalytic domain after the zymogen is cleaved at R229 (Fig 3D). This observation emphasizes the importance of the cleavage of the PD at R126 by a yet unidentified protease to block the ability of the PD to inhibit mature RgpB.
3.5. The CTD-domain is not involved in inhibition of the catalytic domain
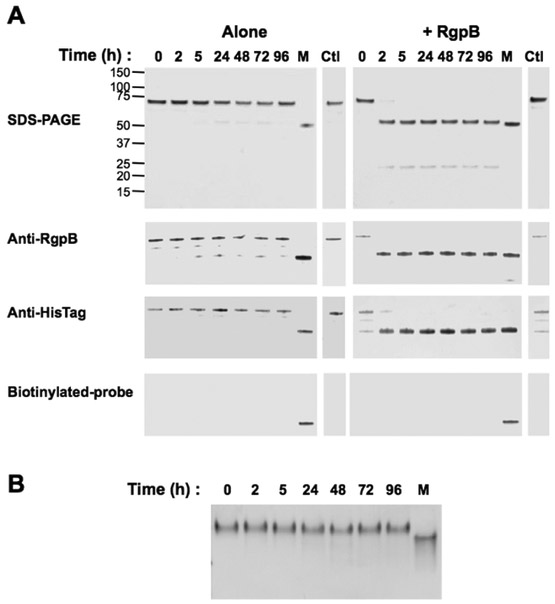
To investigate a potential role of the CTD in processing of proRgpB given the reported maturation process of proRgpB expressed in S. cerevisae (22), we expressed a truncated form of proRgpB: proRgpB6His-ΔCTD in P. gingivalis. Similar to the native enzyme, the truncated form accumulated in the periplasm when expressed in the ΔPorN background. After purification in the presence of dithiopyridine, the proRgpB6His-ΔCTD was stable. However, when incubated with RgpB, the entire PD was quickly processed as observed for the full-length proRgpB6His (Fig 4A). Again, as previously observed, the resulting catalytic domain displayed no activity towards the L-BAPNA substrate (data not shown) nor was it labeled with the active-site biotinylated probe. This lack of activity was confirmed by native-PAGE electrophoresis analysis which confirmed that the PD remained in complex with the catalytic domain (Fig 4B). Taken together, these results indicate that the absence of the CTD did not alter the processing of proRgpB.
Figure 4. The CTD domain does not affect proteolytic processing of proRgpB.
(A) ProRgpB6His-ΔCTD (0.4 mg/ml) was incubated alone or with mature RgpB (molar ratio 1:100) at room temperature in gingipain activity buffer supplemented with 10 mM DTT. At various time points, aliquots were removed to determine proteolytic processing of ProRgpB6His-ΔCTD by SDS-PAGE electrophoresis, Western blot with anti-RgpB or anti-HisTag and active-site labelling with the biotinylated probe BiRK. (B) Time-dependent processing of proRgpB6His-ΔCTD by the mature RgpB was analyzed by 12% native gel electrophoresis. Mature RgpB bearing a C-terminal HisTag (lane M) was used as positive control. Incubations for 96 h in presence of TLCK were used as a negative control (Ctl)
3.6. Release of the CTD
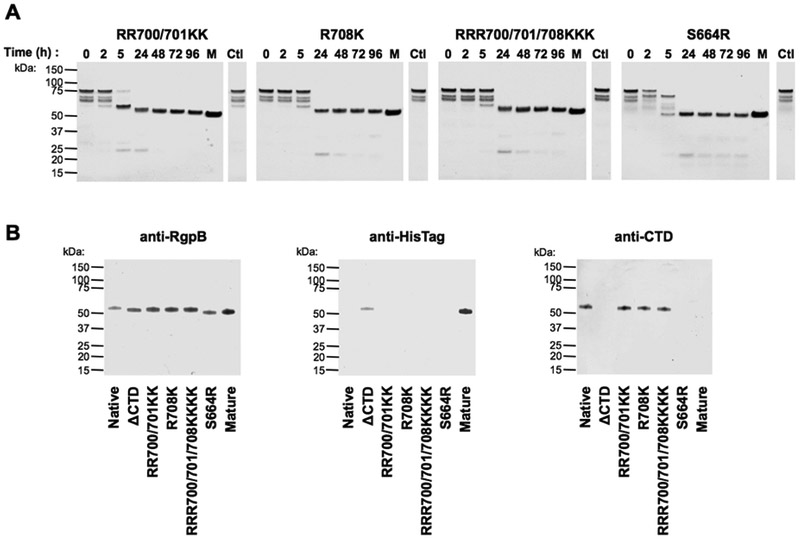
During gingipain secretion via T9SS, the entire CTD is released by PorU sortase (21). In contrast, in vitro processing of proRgpB6His by the mature RgpB results in a different maturation process that involves partial processing of the CTD. Specifically, we observed the loss of the anti-6His region while the anti-RgpBCTD antibody remained able to bind to the C-terminally truncated zymogen. In order to identify the cleavage site responsible of this loss of the 6His tag, we generated, expressed and purified various mutants of proRgpB6His in which the Arg residues of the CTD were substituted for lysines. When incubated with mature RgpB, these mutants, including a triple RRR701/702/708KKK were processed similarly to the native proRgpB6His (Fig 5A). The final product did not display the C-terminal 6His, but was still detected with the anti-RgpBCTD antibody (Fig 5B). This suggests that the cleavage occurred at R721 leaving the intact epitope(s) (701RVATAKNRM709) to be recognized by the anti-RgpBCTD antibody (Fig 1D). Remarkably, only the substitution of S664 at the end of the catalytic domain by an arginine allowed the liberation of the entire CTD domain (Fig 5A and 5B). Thus, these results indicate that RgpB cannot cleave the CTD domain by auto-proteolysis but can remove 15 C-terminal residues by inter-molecular cleavage at R721. This conclusion is consistent with recent studies that PorU is responsible for the processing of all CTD domains of proteins translocated across the outer-membrane by the T9SS in P. gingivalis (20, 21).
Figure 5. Mutations of Arg 700, 701 and 708 residues to Lys do not affect processing of ProRgpB6His at the CTD in contrast to S664R substitution, which facilitates removal of the entire CTD.
(A) ProRgpB6His with mutation(s) in the CTD domain were incubated with mature RgpB (molar ratio 1:100) at room temperature in gingipain activity buffer supplemented with 10 mM DTT. At various times points, aliquots were removed to follow the maturation of the enzyme by SDS-PAGE. (B) The final products obtained after 96 hours of incubation were analysed by Western blotting with pAb anti-RgpB, mAb anti-6His and pAb anti-RgpBCTD domain.
3.7. A protease at the surface of P. gingivalis is essential for proRgpB activation
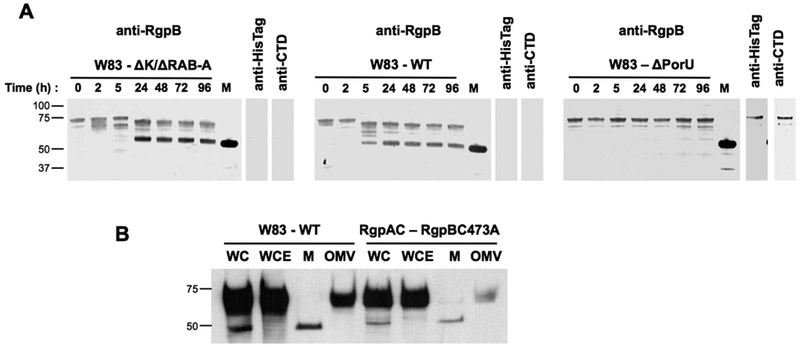
Since proRgpB6His is not able to activate itself, even in the presence of active RgpB, we tested bacterial components to determine their ability to induce zymogen processing and activation. ProRgpB-6His was incubated with cell extracts from P. gingivalis ΔK/ΔRAB-A, a triple mutant strain deficient in gingipain activity. In the absence of DTT, this incubation led to slow processing and accumulation of proRgpB6His truncated at R126 as determined by N-terminal sequencing (Fig 6A). Addition of dithiodipyridine to the reaction media stabilized the proRgpB6His, while treatment with the reducing agent DTT enhanced the cleavage at R229 and the generation of the mature RgpA, lacking both the PD and CTD. These results suggested the presence of a yet unidentified protease in the bacterial extract that can release the PD from the proRgpB6His. The same processing pattern was observed when proRgpB6His was incubated with intact bacterial suspension, indicating that the putative protease is either secreted or localized at the cell surface (Fig 6B). Contrary to processing of proRgpB by active RgpB alone, we observed the generation of the active and functional form of RgpB which had activity towards the L-BAPNA substrate (Fig 6C and 6D). Accordingly, the fully processed 50 kDa RgpB was strongly labeled by the active-site biotinylated probe (Fig 6E). Interestingly, the partially processed proRgpB6His cleaved at R126 (within the PD) that accumulated during the incubation without DTT was also reactive with the active-site probe, indicating that this first cleavage already generated a partially active form of proRgpB.
Figure 6. ProRgpB6His is proteolytically processed by whole cell extracts or intact bacterial cells in gingipain-null P. gingivalis strain.
ProRgpB6His (0.4 mg/ml) was incubated with the (A) whole cell extract (WCE) of P. gingivalis gingipain-deficient strain ΔK/ΔRAB-A or (B) with the whole bacteria diluted in RPMI media (B-RPMI), alone or in the presence of DTT (5 mM) or 4,4’-Dithiodipyridine disulphide (2 mM). At various time points, aliquots were removed to follow the maturation of the zymogen by Western blotting with pAb anti-RgpB. RgpB activity was also recorded during incubation with (black circle) or without DTT (open circle) using the chromogenic substrate L-BAPNA (panel C: WCE; panel D: intact bacterial cells in B-RPMI). Results are expressed as percentage of activity of the mature RgpB incubated under the same condition (n = 3). (E) The active form of RgpB obtained during the incubation with the bacterial suspension was identified by active-site labelling using the biotinylated probe BiRK. Mature RgpB bearing a C-terminal HisTag (lane M) was used as control.
3.8. PorU (PG0026) sortase may function as the proRgpB activating protease
Our results indicated that proRgpB activation was dependent on a secreted or extracellular protease associated with the bacterial outer membrane. PorU (PG0026) was identified as the sortase protease of the T9SS which removes the CTD from secreted proteins (20, 21). Because PorU is a cysteine protease similar to gingipains, we hypothesized this protein is the possible enzyme acting on the PD and CTD domains from proRgpB. To verify this hypothesis, we incubated proRgpB6His with a suspension of wild-type P. gingivalis W83, gingipain-null mutant (ΔK/ΔRAB) and an isogenic mutant in which the porU gene has been deleted. Both wild-type bacteria and the ΔK/ΔRAB gingipain-deficient strain were able to generate mature RgpB (Fig 7A). In contrast, the sortase deficient mutant of P. gingivalis (ΔPorU) failed to process proRgpB6His (Fig 7A). On the other hand proRgpB containing the catalytic Cys473 residue mutated to Ala (proRgpBC473A) showed normal maturation including proteolytic processing and covalent modification with A-LPS when expressed by the ΔRgpA strain (Fig 7B). Collectively, this data strongly suggests that PorU may function not only as the P. gingivalis sortase (removal of the CTD and attachment of A-LPS) but also as the enzyme involved in activation of the proRgpB zymogen through cleavage of the PD. Alternatively, PorU could indirectly mediate cleavage of the PD though an uncharacterized secreted or surface expressed protease.
Figure 7. PorU (PG0026), an extracellular component of T9SS in P. gingivalis, is required for proRgpB processing.
(A) P. gingivalis strains ΔK/ΔRAB, W83 and ΔPorU at early stationary phase were adjusted to OD600=1.5 and centrifuged for 10 min at 5,000 × g. Bacteria were resuspended in RPMI media complemented with 5 mM DTT and 50 μM of gingipain-specific inhibitors KYT-1 and KYT-36. After 15 min, proRgpB6His (0.4 mg/ml) was added and incubated at room temperature. At various time, aliquots were removed to follow the maturation of the enzyme by Western blotting with the pAb anti-RgpB. The final products obtained after 96 hours of incubation were also analysed by Western blotting with mAb anti-6His and pAb anti-RgpBCTD domain. Mature RgpB bearing a C-terminal HisTag (lane M) was used as a control. (B) Comparison of RgpB-WT (expressed in the W83 strain) and RgpBC473A (expressed in the ΔRgpA strain) maturation by Western blot analysis with pAb anti-RgpB (WC, whole culture; WCE, whole cell extract; M, media; OMV, outer-membrane vesicles)
4. Discussion
In contrast to multicellular organisms, control of secreted proteolytic enzymes is often not tightly regulated in the majority of prokaryotes (34). This is mostly due to the release of inactive protease zymogens directly into the extracellular environment where they are rapidly converted into active enzymes by autoproteolytic cleavage of the PD. With several prominent exceptions (35, 36), inhibitory PDs of bacterial proteases often have no structure, and even a short signal peptide can block activation of secreted proteases (37). On the other hand, the gingipains of P. gingivalis, maintain latency using large and structured PDs. In Arg-specific gingipains, the PD is composed of 205 residues and consists of a central 11-stranded β-core divided in two antiparallel β-sandwiches with 3 α-helical elements on the surface (14). Efficient inhibition by the PD is achieved through binding to the catalytic domain with the inhibitory loop positioning Arg126 into the S1 pocket blocking the substrate-binding cleft. The Kgp-PD shares structure similarity and mechanism of inhibition with Rgp-PD and positions Lys129 of the inhibitory loop into this key S1 active site pocket (15). For Kgp activation, cleavage of the PD from the catalytic domain leads to conformational changes in the active site that result in dissociation of the PD. The released Kgp-PD forms dimers, which are no longer inhibitory, resulting in full activation of Kgp. In contrast, the Rgp-PD forms stable inhibitory complexes in trans with processed gingipains (13), suggesting that it needs to be degraded to release the final active Rgps.
Highly purified full length proRgpB and proRgpBΔCTD are stable and do not undergo spontaneous proteolysis for at least 96 h (Fig. 3 and 4), excluding intramolecular autoproteolytic as a mechanism of zymogen activation. On the other hand, intermolecular processing by exogenously added active RgpA or RgpB results in PD removal at Arg229 but the processed enzyme remains inactive. Of note, proRgpB is resistant to proteolysis by Kgp arguing against the involvement of this gingipain in zymogen processing. The cleavage by Rgps was accelerated in the absence of the CTD, but resulted in no activation as detected using the biotinylated activity-based probe (Fig 4A). This lack of proteolytic activation by partial processing of the PD is due to the resulting stable inhibitory complex formed between the cleaved PD and the catalytic domain (Fig 4B).
Gingipains are secreted via the T9SS and are subjected to extensive posttranslational processing including proteolytic cleavage of the PD and the CTD. P. gingivalis mutants deficient in essential components of the T9SS, including PorU, accumulate full length and enzymatically inert progingipains missing only the signal peptide in the periplasm (17). This suggests that proteolytic processing of both the PD and the CTD, as well as processing of hemagglutinin-adhesin domains of RgpA and Kgp, occurs on the bacterial cell surface.
Recently, the structure of the T9SS translocon of Flavobacterium johnsoniae, an extremely large (36-strand) β-barrel transmembrane SprA protein together with 3 accessory proteins, including PorV, has been solved (38). Models derived from this structure, suggest that in P. gingivalis, transported proteins bearing the CTD enter the Sov protein (SprA equivalent) and then exit the translocon laterally onto the OM surface where PorV captures them. PorV, the most abundant component of the T9SS, is a 14-stranded β-barrel protein, which serves as a shuttle for secreted proteins, delivering them to the PorU sortase (also anchored to PorV) (20). PorU functions as a P. gingivalis sortase, which removes the CTD and simultaneously attaches an anionic lipopolysaccharide to the released C-terminal carbonyl group via an isopeptide bond (21). Of note, PorU and gingipain catalytic domains are related and share significant similarity at the primary structure level. Here, we provide strong evidence that beyond sortase activity, PorU also plays a role in proteolytic activation of proRgpB through removal of the PD in a two-step process.
The reason for the relatively slow rate of in vitro activation of proRgpB is not yet clear (Fig. 7). One may speculate that in vivo, during translocation through the T9SS, proRgpB may interact with other T9SS components to adopt a conformation allowing for more efficient processing of the PD. The possibility that proRgpB can take two different conformations is supported by strikingly low zymogenicity of proRgpB expressed in yeast, which is in stark contrast to the stable zymogen isolated form P. gingivalis. The yeast-expressed proRgpB undergoes instant sequential autoproteolytic events, first at Arg126, then at the Arg229, followed by truncation of the CTD. This is reminiscent to the findings for caspases 3 and 7 (belonging to the same CD clan of peptidases as gingipains) in which E. coli expression yields fully active mature caspases 3 and 7 even though their activation in vivo absolutely requires initiator caspases 8 or 9 (39). Although caspases activation in E. coli can be at least partially explained by overexpression, this is unlikely to be the case for recombinant RgpB, which is secreted by yeast in small amounts (27). A possible explanation for subcellular site-specific difference in the zymogen structures could be due to involvement of a chaperone(s) assisting proRgpB folding within the P. gingivalis periplasm but this possibility needs further investigation.
Importantly, we found that proRgpB was processed to full catalytically active form by cell extracts of the gingipain-null mutant ΔK/ΔRAB (Fig 7). This confirms that autoprocessing by gingipains is not sufficient for full protease activation and furthermore, that final processing step to remove the PD requires an additional cell-derived factor. Our results using various knock out cell lines confirmed that purified proRgpB activation was dependent on the presence of PorU (Fig 6 and 7). Unfortunately, all attempts to show direct release of the PD and activation of proRgpB using purified, recombinantly expressed PorU were unsuccessful (data not shown). This lack of processing activity by PorU could be explained by a lack of cofactors (e.g. A-LPS) and interacting proteins (PorV and PorZ) (30) that may be essential for productive presentation of the substrate (proRgpB). Alternatively, PorU may regulate proRgpB activation by indirect action through a protease whose activity or secretion depends on PorU. While further mechanistic insight into proRgpB activation requires further studies, our data suggests that a multi-step activation process likely evolved to protect P. gingivalis from damage or toxicity from premature activation of gingipains.
Acknowledgment
This work was funded by grants from the National Science Center, Poland (UMO-2016/21/B/NZ1/00292 to JP, UMO-2014/15/D/NZ6/02546 to DM, 2016/23/N/NZ1/01513 to ZN) and NIH (DE022597 and DE026280 to JP and R01 EB026332 to MB).
Abbreviation:
- BiRK
Ac-biotinyl-Lys-Tyr-6-aminohexanoic-Arg-acyloxymethyl ketone
- CD
catalytic domain
- CTD
C-terminal domain
- HA
hemagglutinin/adhesion domains
- IgSF
immunoglobulin superfamily domain
- IM
inner membrane
- L-BAPNA
Nα-Benzoyl-L-arginine p-nitroanilide
- NTP
N-terminal pro-domain
- OM
outer membrane
- T9SS
Type IX Secretion System
- TLCK
tosyl-L-lysine chloromethylketone
- TSB
trypticase soy broth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Albandar JM, and Tinoco EM (2002) Global epidemiology of periodontal diseases in children and young persons, Periodontol 2000 29, 153–176. [DOI] [PubMed] [Google Scholar]
- 2.Albandar JM, and Rams TE (2002) Risk factors for periodontitis in children and young persons, Periodontol 2000 29, 207–222. [DOI] [PubMed] [Google Scholar]
- 3.Cobb CM, Williams KB, and Gerkovitch MM (2009) Is the prevalence of periodontitis in the USA in decline?, Periodontol 2000 50, 13–24. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, and Curtis MA (2011) Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement, Cell Host Microbe 10, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, Nguyen KA, and Potempa J (2010) Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins, Periodontol 2000 54, 15–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imamura T (2003) The role of gingipains in the pathogenesis of periodontal disease, J Periodontol 74, 111–118. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien-Simpson NM, Pathirana RD, Walker GD, and Reynolds EC (2009) Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner, Infect Immun 77, 1246–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potempa J, Pike R, and Travis J (1997) Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes, Biol Chem 378, 223–230. [DOI] [PubMed] [Google Scholar]
- 9.Imamura T, Travis J, and Potempa J (2003) The biphasic virulence activities of gingipains: activation and inactivation of host proteins, Curr Protein Pept Sci 4, 443–450. [DOI] [PubMed] [Google Scholar]
- 10.Barrett AJ, and Rawlings ND (2001) Evolutionary lines of cysteine peptidases, Biol Chem 382, 727–733. [DOI] [PubMed] [Google Scholar]
- 11.Schechter I, and Berger A (1967) On the size of the active site in proteases. I. Papain, Biochem Biophys Res Commun 27, 157–162. [DOI] [PubMed] [Google Scholar]
- 12.Veillard F, Potempa B, Poreba M, Drag M, and Potempa J (2012) Gingipain aminopeptidase activities in Porphyromonas gingivalis, Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veillard F, Sztukowska M, Mizgalska D, Ksiazek M, Houston J, Potempa B, Enghild JJ, Thogersen IB, Gomis-Ruth FX, Nguyen KA, and Potempa J (2013) Inhibition of gingipains by their profragments as the mechanism protecting Porphyromonas gingivalis against premature activation of secreted proteases, Biochim Biophys Acta 1830, 4218–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Diego I, Veillard FT, Guevara T, Potempa B, Sztukowska M, Potempa J, and Gomis-Ruth FX (2013) Porphyromonas gingivalis virulence factor gingipain RgpB shows a unique zymogenic mechanism for cysteine peptidases, J Biol Chem 288, 14287–14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomowski A, Uson I, Nowakowska Z, Veillard F, Sztukowska MN, Guevara T, Goulas T, Mizgalska D, Nowak M, Potempa B, Huntington JA, Potempa J, and Gomis-Ruth FX (2017) Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome, J Biol Chem 292, 5724–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potempa J, Pike R, and Travis J (1995) The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain, Infect Immun 63, 1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasica AM, Ksiazek M, Madej M, and Potempa J (2017) The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function, Front Cell Infect Microbiol 7, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochtler M, Mizgalska D, Veillard F, Nowak ML, Houston J, Veith P, Reynolds EC, and Potempa J (2018) The Bacteroidetes Q-Rule: Pyroglutamate in Signal Peptidase I Substrates, Front Microbiol 9, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen KA, Travis J, and Potempa J (2007) Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria?, J Bacteriol 189, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, and Reynolds EC (2012) PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis, J Biol Chem 287, 24605–24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, and Reynolds EC (2015) Porphyromonas gingivalis Type IX Secretion Substrates Are Cleaved and Modified by a Sortase-Like Mechanism, PLoS Pathog 11, e1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikolajczyk J, Boatright KM, Stennicke HR, Nazif T, Potempa J, Bogyo M, and Salvesen GS (2003) Sequential autolytic processing activates the zymogen of Arggingipain, J Biol Chem 278, 10458–10464. [DOI] [PubMed] [Google Scholar]
- 23.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, and Nakayama K (2010) A protein secretion system linked to bacteroidete gliding motility and pathogenesis, Proc Natl Acad Sci U S A 107, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, and Nakayama K (2013) Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system, FEMS Microbiol Lett 338, 68–76. [DOI] [PubMed] [Google Scholar]
- 25.Saiki K, and Konishi K (2010) The role of Sov protein in the secretion of gingipain protease virulence factors of Porphyromonas gingivalis, FEMS Microbiol Lett 302, 166–174. [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro I, Saiki K, and Konishi K (2009) PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system, FEMS Microbiol Lett 292, 261–267. [DOI] [PubMed] [Google Scholar]
- 27.Potempa J, and Nguyen KA (2007) Purification and characterization of gingipains, Curr Protoc Protein Sci Chapter 21, Unit 21 20. [DOI] [PubMed] [Google Scholar]
- 28.Sztukowska M, Veillard F, Potempa B, Bogyo M, Enghild JJ, Thogersen IB, Nguyen KA, and Potempa J (2012) Disruption of gingipain oligomerization into non-covalent cell-surface attached complexes, Biol Chem 393, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veillard F, Potempa B, Guo Y, Ksiazek M, Sztukowska MN, Houston JA, Koneru L, Nguyen KA, and Potempa J (2015) Purification and characterisation of recombinant His-tagged RgpB gingipain from Porphymonas gingivalis, Biol Chem 396, 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thogersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen KA, Potempa J, and Gomis-Ruth FX (2016) Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis, Sci Rep 6, 37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CJ, Parker A, and Rogers MB (1990) Plasmid transformation of Bacteroides spp. by electroporation, Plasmid 24, 100–109. [DOI] [PubMed] [Google Scholar]
- 32.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M (2005) Activity-based probes that target diverse cysteine protease families, Nat Chem Biol 1, 33–38. [DOI] [PubMed] [Google Scholar]
- 33.Rapala-Kozik M, Bras G, Chruscicka B, Karkowska-Kuleta J, Sroka A, Herwald H, Nguyen KA, Eick S, Potempa J, and Kozik A (2011) Adsorption of components of the plasma kinin-forming system on the surface of Porphyromonas gingivalis involves gingipains as the major docking platforms, Infect Immun 79, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kantyka T, Rawlings ND, and Potempa J (2010) Prokaryote-derived protein inhibitors of peptidases: A sketchy occurrence and mostly unknown function, Biochimie 92, 1644–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filipek R, Szczepanowski R, Sabat A, Potempa J, and Bochtler M (2004) Prostaphopain B structure: a comparison of proregion-mediated and staphostatin-mediated protease inhibition, Biochemistry 43, 14306–14315. [DOI] [PubMed] [Google Scholar]
- 36.Mallorqui-Fernandez N, Manandhar SP, Mallorqui-Fernandez G, Uson I, Wawrzonek K, Kantyka T, Sola M, Thogersen IB, Enghild JJ, Potempa J, and Gomis-Ruth FX (2008) A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A, J Biol Chem 283, 2871–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pustelny K, Zdzalik M, Stach N, Stec-Niemczyk J, Cichon P, Czarna A, Popowicz G, Mak P, Drag M, Salvesen GS, Wladyka B, Potempa J, Dubin A, and Dubin G (2014) Staphylococcal SplB serine protease utilizes a novel molecular mechanism of activation, J Biol Chem 289, 15544–15553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauber F, Deme JC, Lea SM, and Berks BC (2018) Type 9 secretion system structures reveal a new protein transport mechanism, Nature 564, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez MLG, and Salvesen GS (2018) A primer on caspase mechanisms, Semin Cell Dev Biol 1397, 17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]