Abstract
Hypotheses of the origins of RNA and DNA are generally centered on the prebiotic synthesis of a pristine system (pre-RNA or RNA), which gives rise to its descendent. However, a lack of specificity in the synthesis of genetic polymers would likely result in chimeric sequences; the roles and fate of such sequences are unknown. Here we show that chimeras, exemplified by mixed TNA-RNA and RNA-DNA oligonucleotides, preferentially bind to, and act as templates for, homogeneous TNA-, RNA- and DNA-ligands. The chimeric-templates can act as a catalyst, mediating the ligation of oligomers to give homogenous-backbone sequences, and the regeneration of the chimeric templates potentiates a scenario for possible cross-catalytic cycle with amplification. This process provides a proof-of-principle demonstration of a heterogeneity-to-homogeneity scenario while giving credence to the idea that DNA could appear concurrently with RNA instead of being its later descendent.
Graphical Abstract

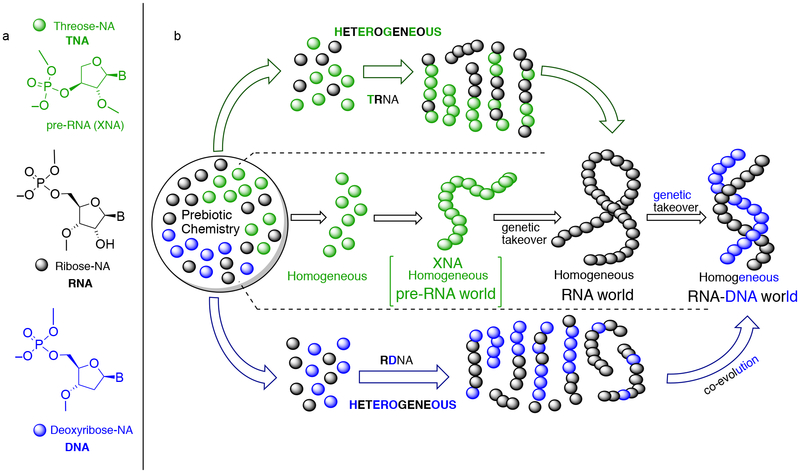
The RNA World hypothesis proposes the emergence of self-replicating and catalytic RNA giving rise later to proteins and DNA (Fig. 1b, middle).1,2 Models posit the existence of a genetic polymer – whether RNA or its precursor – with a homogeneous backbone that transitions to its homogenous-backbone successor1,3–10 This transition is proposed to occur despite the difficulties2,11–14 associated with generation of the pristine oligomers using prebiotic chemistry,15,16 and the challenge of replacing one genetic polymer with another2,17–21 in the absence of any sophisticated discrimination mechanism during the transition in a pre-biological world.13,22 However, there is a growing realization23–25 that most prebiotic pathways26,27 would lead to nucleic acid oligomers consisting of mixed backbone units.14,17,19,28 In this context, RNA containing a mixture of 2′,5′-and 3′,5′-linkages,29,18,19 and chimeric RNA-DNA systems,17,21 have been investigated (and it has been shown that these types of backbone-heterogeneity compromise aptamer function17–19), and we have shown that RNA-DNA chimeras consistently form weaker duplexes.14 Though chimeric RNA-DNA genomes are known in extant biology30 and such chimeras containing nonheritable-backbone-heterogeneity have been postulated to be useful in the emergence of functional nucleic acids,17,19 questions have been raised about their role as enhanced-templates for replication17,31 generating polymers with homogeneous-backbones14. For pre-RNA to RNA transitions, Orgel has speculated two extreme possibilities using TNA (Fig. 1a)32 as an example: (a) an all TNA-organism converting to all RNA-organism, and (b) a gradual replacement of TNA residues by RNA residues within the oligomeric system.33 The second scenario leads to a continuous-pathway from TNA to RNA, via chimeric sequences.33 We have proposed a heterogeneity-to-homogeneity scenario34 for the emergence of RNA and DNA13,14, and argued that based on certain criteria such as the stability and functional advantages inherent to homogeneous-backbone-polymers, their emergence would be a natural consequence even when starting from a mixture of its constituent building blocks (Fig. 1b, top and bottom).13 A demonstration that chimeric TNA-RNA sequences (TRNA, Fig. 1b, top) or RNA-DNA sequences (RDNA, Fig. 1b, bottom) can enable the non-enzymatic emergence of homogeneous backbone oligonucleotide (RNA or DNA) starting from mixtures of chimeric sequences would provide support to the heterogeneity-to-homogeneity scenario.13
Figure 1. The prebiotic clutter generated heterogeneity-to-homogeneity scenario versus the biology inspired paradigm of replacing one homogeneous genetic system with its homogeneous genetic successor.
(a) Constitutional formula representation of the three oligonucleotide building blocks investigated in this study. (b) Three possible scenarios for the emergence of RNA and DNA from prebiotic chemistry. Middle: the classical RNA world concept where the formation of a pristine and homogeneous RNA (or pre-RNA) leads to its homogeneous-backbone successor DNA (or RNA). Top: a heterogeneous mixture of TNA (pre-RNA) and RNA forming chimeric TRNA sequences that transition to homogenous RNA, which then gives rise to DNA. Bottom: a heterogeneous RNA-DNA mixture progressing/co-evolving via chimeric RDNA sequences directly to homogeneous RNA and DNA simultaneously.
Results
TNA-RNA chimeric sequences function as templates for RNA ligands.
We selected TNA32 – a Watson-Crick base-pairing system able to cross-pair with RNA32,35– as a model pre-RNA polymer,13 based on the prebiotic availability of the sugars27,36–39 (Fig. 1a). We investigated TNA-RNA chimeric sequences (TRNA) which exhibited peculiar base-pairing properties even though TNA formed strong and stable duplexes with complementary RNA strands (Supplementary Table 1 & 2).32 First, in general TRNA formed weaker duplexes compared to the unmodified strands. Second, based on which sugar (threose or ribose) unit contained a purine (A) or pyrimidine (T), TRNA demonstrated unpredictable duplex stabilities (Fig. 2a). Unexpectedly, TRNA non-self-complementary strands which showed weak affinity for each other (Fig. 2a, entry 7), formed stronger duplexes with the corresponding complementary RNA (or TNA) sequences (Fig. 2a, entries 6 and 8), a behavior which was general for sequences containing all four nucleobases (Supplementary Table 3 and Figs. 7–13).
Figure 2. The preferential association with, and ligation of homogeneous ligands by, chimeric TRNA template over chimeric ligands.
(a) Thermal stability of TRNA chimeric duplexes in 1 M NaCl, 10 mM Na2HPO4, 100 μM EDTA, pH 7.2; a = [5μM], b = [2μM] duplex concentration; c = Entry 7, no clear sigmoidal transition in UV-thermal melt was observed. (b) Comparison of the rate of ligation reaction at 4°C of homogeneous-RNA (RL1+RL2) and heterogeneous TNA-RNA ligands (CL1+CL2) on heterogeneous TNA-RNA template, CT1. (c) EDC-mediated ligation reaction at 4 °C of mixture of homogeneous-RNA (RL1, RL2) and chimeric TNA-RNA ligands (CL1, CL2) using TRNA chimeric sequence (CT1) as template. (d) comparison of the amounts of products RP1 and CP1 produced in the reaction mixture in 2c; see supplementary Fig. 24, for conditions. A, T = RNA; T = DNA; a, t = TNA. Line in graph (2b) is drawn as guide to indicate the trend and is not a mathematical curve fitting. % yields are calculated with respect to the template CT1. Experiments were run in triplicate and the error range is less than ± 5%; error bars represent standard deviation.
The preferential association of chimeric TRNA sequences with homogeneous RNA (or TNA) sequences (Fig. 2a, entries 6, 8) implied that chimeric-sequences could act selectively as templates for the non-enzymatic ligation of homogeneous-sugar-backbone ligands, and thereby facilitate the emergence of homogeneous-backbone-oligomer (e.g. RNA), starting from a mixture of oligonucleotides. To test this proof-of-concept, we employed the widely used water soluble 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) mediated ligation conditions40 of homogeneous-RNA ligands templated by TRNA-chimeric- and RNA-templates, and compared it with ligation of the chimeric TRNA ligands (Fig. 2b). The 3′-NH2 modified TNA-ligand41 and 3′-NH2-deoxynucleotide (TNH2) terminated RNA-ligand42 was used to conduct the ligation-reaction within reasonable time-frame, since the corresponding TNA-3′-OH and RNA-3′-OH residues react very slowly (Supplementary Figs. 14–17). The single phosphoramidate linkage was shown to have no special effect on duplex stability (Supplementary Fig. 12). The reactions were monitored by anion-exchange chromatography (AEC), and products were confirmed by comparison with standards and matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Supplementary Figs. 18–28). As expected from a previous study41, the efficiency and the rate of ligation reactions paralleled the affinity (and thermal stability) of the templates for ligands, in the following order: RNA template with TNA ligands ≈ RNA template with RNA ligands ≥ TRNA chimeric template with RNA ligands >> RNA template with TRNA chimeric ligands >>>> TRNA chimeric template with TRNA chimeric ligands (Supplementary Figs. 18–23). Control reactions lacking the template(s) showed no product formation (Supplementary Figs. 25–28). We then examined the ligation behavior of the mixture of all four ligands in the presence of the chimeric TRNA template (Fig. 2c) and observed only the formation and growth of the RNA-product from homogeneous RNA-ligands, with no discernible chimeric-TRNA product from heterogeneous TRNA ligands by AEC (Fig. 2d; Supplementary Fig. 24). However, MALDI-TOF analysis of the reaction of chimeric TRNA ligands with chimeric TRNA template at 24 h did show traces of the chimeric TRNA-product (Supplementary Fig. 18). We have not investigated intensively a parallel scenario for the emergence of homogeneous TNA sequences43 (due to the investment in synthesizing the various TNA 3′-NH2-phosphoramidites), though we expect a similar propensity32 based on the observation that homogeneous-TNA ligands also were preferentially ligated by the chimeric TRNA template (Supplementary Fig. 20).
RNA-DNA chimeric templates ligate complementary RNA and DNA ligands.
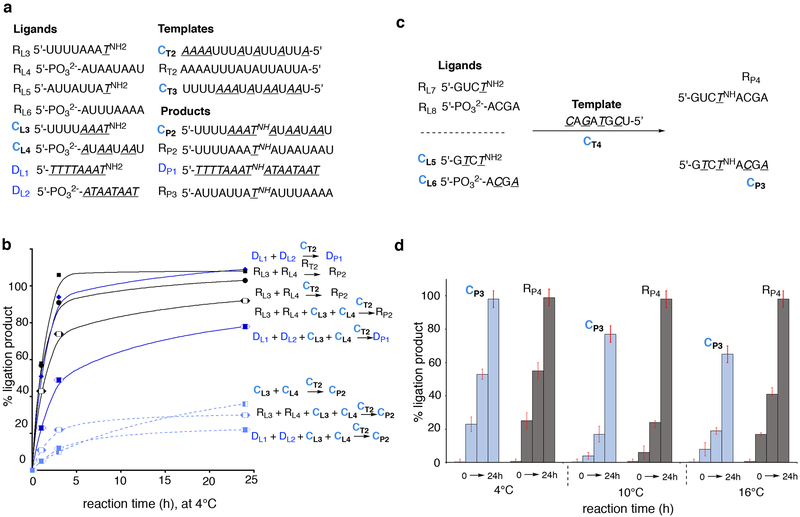
The above results inspired us to investigate mixed DNA and RNA chimeric sequences based on (a) our previous studies of RNA-DNA (RDNA) chimeras14 and the plausible coexistence and coevolution of RNA and DNA in prebiotic scenarios17,21,28,44, and (b) the ease of commercial/synthetic availability of diverse RDNA chimeric sequences. We studied a series of RDNA chimeric sequences, (Supplementary Table 4), which, again, formed stronger duplexes with complementary homogeneous RNA over corresponding complementary chimeric RDNA (Supplementary Table 5 and Supplementary Figs. 29–35). To test whether the preferential association of RDNA with RNA would also translate to selective ligation of RNA ligands (as seen in the TRNA system), we investigated the ligation behavior of a hexadecamer chimeric RDNA template (CT2, Fig. 3a) with RNA and RDNA ligands containing 3′-NH2 deoxynucleotide units. Ligation of RNA sequences (RL3 and RL4) on the chimeric RDNA template (CT2) was not only faster than the corresponding ligation of chimeric RDNA ligands (CL3 and CL4 on CT2; Fig. 3b), but was almost equal to the efficiency of RNA ligands RL3 and RL4 (or chimeric CL3 and CL4 ligands; Supplementary Fig. 39) on an RNA template, RT2 (Supplementary Figs. 36–46).
Figure 3. Chimeric RDNA templates preferentially associate and ligate homogeneous RNA and DNA ligands over chimeric ligands.
(a) The list of homogeneous and chimeric sequences used in this study. (b) Comparison of ligation efficiency by hexadecameric (AU)-RDNA template CT2, with RNA (RL3, RL4), DNA (DL1, DL2) and chimeric RDNA (CL3, CL4) ligands, showing the consistent preferential formation of homogeneous ligation products, RP2 and DP1 over chimeric ligation products CP2. (c and d) Comparison of ligation efficiency by octameric (A, U/T, G, C)-RDNA template CT4, with RNA (RL7, RL8), and chimeric RDNA (CL5, CL6) ligands, showing the influence of temperature on the preferential formation of homogeneous ligation products, RP4 over chimeric ligation products CP3. See supplementary figs. 50–52 for EDC-ligation reaction conditions. A, U, G, C = RNA; A, T, G, C = DNA. Lines in graph (3a) are drawn as guide indicating the trend and are not mathematical curve fittings. % yields were calculated with respect to the template CT2 or RT2 or CT4 respectively. Experiments were run in triplicate and the error range is less than ± 5%; error bars represent standard deviation.
The duplex formation in octameric homogeneous and chimeric sequences containing all five canonical nucleosides again showed preferential association of homogeneous-backbone sequences with complementary chimeric templates (Supplementary Table 5). Based on this, we investigated the ligation reaction mediated by the chimeric template (CT4) with RNA and chimeric ligands shown in Fig. 3c. The results revealed a temperature-dependent ligation behavior that was not observed in the hexadecameric-AU system (Supplementary Figs. 50–52). While at lower temperatures (4°C) there was little difference between the rate of ligation between the two systems, the rate of ligation of chimeric ligands and the amounts of products formed at higher temperature (10 and 16°C) differed considerably with preference for the ligation product from homogeneous-ligands on the chimeric template (Fig. 3d and Supplementary Fig. 52). This indicates that temperature could also control and modulate the overall dynamics and distribution of the end-products.
The trend of preferential association correlating with ligation capacity of CT2 also extended to DNA ligands (DL1, DL2), in place of RNA ligands, giving rise to the homogeneous DNA product DP1 (Fig. 3b), and was valid even when starting from a pool of mixed RL3+RL4+CL3+CL4 ligands or DL1+DL2+CL3+CL4 ligands (Fig. 3b, Supplementary Figs. 47–49). When all ligands (RL3, RL4, DL1, DL2, CL3 and CL4) were added to chimeric RDNA template CT2 in a single pot, three major ligation products, RP2 (38%), DP1 (20%) and an RNA-DNA cross ligation product (RDP1, 75%) were formed at 24h; no chimeric product from CL3+CL4 was detected (Supplementary Figs. 53–54). The nature of cross-ligation product was confirmed with appropriate control experiments and shown to be the result of DL1-RL4 ligation (Supplementary Figs. 55–59). Replacing the chimeric template with RNA template, under otherwise identical conditions, gave RP2 (65%), DP1 (12%) and 62% of RDP1 and RDP2 (RL3-DL2), indicating that RNA template also gives rise to significant cross-ligation products (Supplementary Figs. 60–61). Changing the ratios of RNA ligand (RL3+RL4) to DNA ligands (DL1+DL2) affected the product distribution (Supplementary Fig. 59) implying that generation of chimeric oligomers (along with homogeneous backbone oligomeric products) have to be reckoned with; these chimeric oligomer products should, in turn, help in the formation of homogeneous RNA and DNA ligation products. While this hypothesis is reinforced by the results in Fig. 3, it was demonstrated to be so by isolating RDP1 and using it as a template with RNA ligands producing RP3 efficiently in 108% yield (Supplementary Fig. 62). The above results show that from a mixed system with two different oligonucleotides (e.g. RDNA) there is indeed the possibility of the simultaneous emergence of the two respective homogeneous-nucleotide polymers (e.g. RNA and DNA).
RNA-DNA chimeric-templates are better in overcoming template-product inhibition.
The above observations suggest that chimeric-templates could provide a solution to the problem of product inhibition (Fig. 4a), where the continuous production of the product is curtailed due to the strong association of the initially formed template-product complex.45–48 For instance, RNA-ligands RL3 and RL4 in the presence of the RP2-RP3 RNA duplex under the EDC-activation conditions showed no production of RP2 even after 24 hours, indicative of classic product inhibition behavior; but the addition of chimeric template CT2 led to the formation of more RP2 within a matter of few hours (Supplementary Figs. 67–68). As outlined in Fig. 4b, if there was the adventitious presence/formation of a complementary RNA partner (RP3, from its corresponding ligands RL5 and RL6) in the mixture containing the chimeric duplex (RP2-CT2), it would induce the formation of the stronger RNA (RP2-RP3) duplex. This should release the original chimeric RDNA template for another round of ligation of RL3 and RL4 forming more of RP2, and result in a continuous accumulation of duplex RP2-RP3, with the chimeric template CT2 taking the role of a catalyst producing more of RP2 from its respective ligands. To test this scenario, we first conducted a step-wise addition of RNA ligands RL3 and RL4 to RDNA chimeric template CT2, leading to the formation of the product RP2 (97% in 20 h, Fig. 4c). Then, ligands RL5 and RL6 were added to this mixture. The formation of the second ligation product RP3 (21% in 1 h increasing to 77% in 24 h, Fig. 4c), indicated that the in situ generated first ligation product RP2 was indeed acting as a template (Supplementary Figs. 64–66). More encouragingly, with higher ligand ratios of RL3 and RL4, an increased amount of the first ligation product RP2 (251% with respect to CT2) and of the second ligation product RP3 (204%) was observed after 24 hours (Fig. 4c, Supplementary Fig. 65 and Table). This indicates that the chimeric template CT2 was indeed being released to take part in a turn-over, which in turn leads to formation of more RP2. Pertinent control experiments confirmed the need for all components to be present for this system to operate; importantly, CT2 itself did not serve as a template to ligate RL5 and RL6 and did not produce RP3 (Supplementary Figs. 45, 66). Encouraged by these results, we set up a one-pot experiment where all components, CT2+RL3+RL4+RL5+RL6, were mixed from the beginning and observed the concomitant production of the two RNA ligation products RP2 and RP3 (as efficiently as the step-wise addition experiment) (Fig. 4c). The presence of chimeric template CT2 in a mixed-one-pot-system not only initiated the ligation process, but also acted as a turn-over intermediary down-stream, potentially enabling continuous production of RP2 and RP3 by mitigating the inhibition by template-product complex. This process was mainly driven by the preference of a thermodynamically stable homogeneous-backbone duplex RP2:RP3. Control reactions for RL3+RL4 or RL5+RL6 ligands without CT2 template showed no observable background ligation reactions. However, when all four ligands (absent CT2) were mixed together, 33% RP2, 20% RP3 and 13% cross-ligation products (probably from RL3+RL6 and/or RL5+RL4) were formed but more slowly at 24 h (Supplementary Fig. 70), as opposed to 259% of RP2 and 191% of RP3 with no cross-ligation products in the presence of chimeric template CT2 (Supplementary Fig. 69). The background ligation-reactions were eliminated when ligand concentrations were lowered from 200 μM to 20 μM each; and only in the presence of 10 μM chimeric template CT2, the formation RP2 (83%) and RP3 (18%) in 24 h was observed (Supplementary Figs. 73–76). Furthermore, we tested whether the presence of the complementary ligands (CL3+CL4, Fig. 3a) leading to the CT2-CP2 duplex would prevent further copying of the first two RNA ligands RL4+RL3 and also impact the next round of copying when all four RNA ligands RL4+RL3+RL5+RL6 are present. In both cases, in 24 h at 4°C, corresponding RNA products formed in good yields; 92% of RP2 (with 30% of CP2) for the first experiment and in the second scenario, 83% RP2 and 16% RP3, with no discernible peak for CP2 in the chromatogram trace (Supplementary Figs. 47 and 77).
Figure 4. The beneficial role for chimeric RDNA template in overcoming the template-product inhibition based on thermodynamic stability of duplexes.
(a) The expected difference between chimeric RDNA-RNA duplex and the homogeneous RNA-RNA duplex in being able to overcome the template-product inhibition. (b) Schematic representation of the proposal that hexadecameric (AU)-RDNA template CT2 with RNA ligands RL3+RL4 produces RP2, which in the presence of RL5, RL6 is expected to lead to RP3, based on the greater thermodynamic stability of the RP2:RP3 duplex over the RP2:CT2 duplex, and release the CT2 for another round of ligation reaction. (c) Time course of the EDC-mediated-ligation experiments documenting the effect of change in ratio of ligands, and the sequential-addition of ligands RL5+RL6 (0 h) followed by RL5+RL6 (at 20 h) versus all-in-one-pot reaction on the production of RP2 and RP3. (d) Comparison of the amount of RP3 formed by the homogeneous RNA template RT2 versus chimeric RDNA template CT2 (at 48 h) demonstrating the higher efficiency of CT2 in mediating the formation of RP3 by overcoming the template-product inhibition. See supplementary figs. 63–78 for EDC-ligation conditions. A, U = RNA; A, T = DNA. Lines in graph (4c) are drawn as guide indicating the trend and are not mathematical curve fittings. % yields were calculated with respect to the template CT2 or RT2 respectively. Experiments were run in triplicate and the error range is less than ± 5%; error bars represent standard deviation.
In order to assess the efficiency of chimeric template CT2 versus that of the corresponding homogeneous backbone RNA counterpart RT2, the all-in-one-pot reaction was repeated but with RNA template RT2 in place of CT2. In this case, as expected, the production of RP2 at 48 h was comparable (99% for RT2 versus 109% for CT2); however, RP3 formation dropped by almost half to 18% (for RT2) when compared to 30% (for CT2) indicating template-product inhibition by the stronger RT2:RP2 complex meant that RP2 was less available for ligating RL5+RL6 (Fig. 4d). The advantage of CT2 over RT2 was more apparent when the ratio of ligands was changed to 5(RL3+RL4):2(RL5+RL6) with CT2 producing 178% of RP2 and 77% of RP3 when compared to 119% of RP2 and 43% of RP3 with RNA template, RT2 (Fig. 4d). This strongly suggests that CT2 is better able to dissociate from the CT2:RP2 template-product complex while the RNA template RT2 is limited by the classic RT2:RP2 template-product inhibition and is, therefore, unable to recycle to produce more RP2 and RP3. In fact, CT2 consistently outperformed RT2 in the production of RP3 for all other combinations of ligand ratios (Fig 4d, Supplementary Fig. 78), indicative of the beneficial role played by chimeric templates in moving towards the emergence of homogeneous-backbone sequences. But for this to be possible, this phenomenon must hold good for other strands in terms of length and sequence diversity. Given the limitations imposed by the EDC-ligation-chemistries and analysis of the chimeric sequences involved, we set up a proof-of-principle experiment as in Fig. 4b but with octameric AUGC containing chimeric template CT4 (Supplementary Fig. 79), since it also showed a preference for the complementary homogeneous ligands over the chimeric counterparts as seen in Fig. 3d. As expected, the chimeric template CT4 was efficient in producing the homogeneous product RP4 and RP5 (Supplementary Fig. 80) overcoming the template-product inhibition even in the presence of all four ligands (RL7+RL8+RL9+RL10), paralleling the observations for the AU-based system. Thus, the ability of the chimeric template to give rise to homogeneous backbones (the heterogeneity-to-homogeneity paradigm) seems to be still operative in this RDNA-chimeric system even when shortening the length of the template and expanding the sequence diversity.
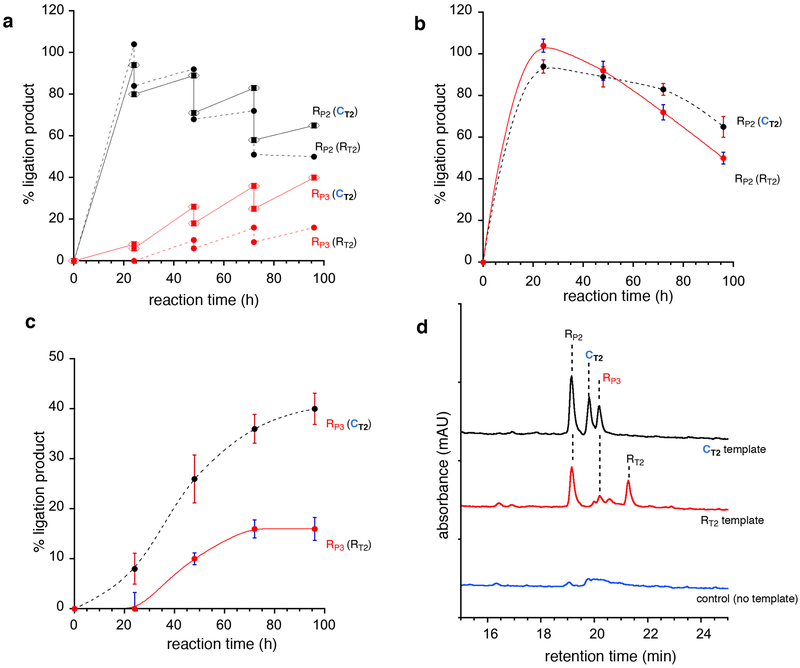
We then examined the effect of step-wise dilution (as a selection pressure) on the efficiency of the templates in overcoming the template-product inhibition, asking the question – which of the templates, chimeric-RDNA or the homogeneous-RNA would produce the ligation products more efficiently as the step-wise dilution was continued? Using the AU-system outlined in Fig. 4a we conducted a step-wise dilution experiment in parallel with templates CT2 and RT2 containing the complementary RNA-ligands (RL3, RL4, RL5 and RL6) where, every 24 h a portion of the reaction mixture was removed and fresh ligands and EDC were added, such that the concentrations of the ligands remained constant, but the template concentration decreased with each dilution-step (Supplementary Figs. 91–93). As seen from Fig. 5a, as the step-wise dilution was implemented at 24 h intervals, the formation of RP2 and RP3 was observed in both cases; while there was a concomitant drop in the product concentration (by 2 μM) at each dilution step, the amount of RP2 and RP3 increased to level greater than the previous value with progress of time. The amount of the first ligation product RP2 was almost the same between the chimeric-(CT2) and homogeneous-(RT2) template containing vials over the first two-steps (48 h) of dilution, with CT2 performing slightly better than RT2 as the dilution steps were continued (72–96 h, Fig. 5b). However, there was a remarkable difference in the production of the second ligation product RP3 with increasing step-wise dilutions; the chimeric template CT2 outperformed the homogeneous template RT2 in producing RP3 by ca 250% (Fig. 5c), even as the concentration of the templates were going down with each step of dilution. A comparison of the chromatogram traces at 96 h (Fig. 5d) shows the dramatic difference and highlights the ability of CT2 to be a superior template17 for the production of homogeneous product RP3, demonstrating the ability of chimeric template CT2 to better bypass the template-product inhibition and turn-over even under dilute conditions when compared to RT2. Appropriate controls without the template showed no product formation (Fig. 5d).
Figure 5. Comparison of the efficiency between chimeric RDNA (CT2) and RNA (RT2) templates in producing the final ligation product RP3 under step-wise dilution conditions, demonstrating the superior ability of CT2 to act as a template for ligation with turn over.
(a) Production of the ligation products RP2 and RP3 in the stepwise-dilution (in 24 h intervals) experiment with templates CT2 and RT2, over a period of 96 h, containing all four ligands RL3, RL4, RL5 and RL6; the drops at 24, 48, and 72 h indicate the dilution step. (b) Time course contrast between the templates CT2 and RT2 for the production of the first ligation product RP2 formed from RL3 and RL4. (c) Comparison of the efficiency of production of the second ligation product RP3 (from RL5 and RL6) between the templates CT2 and RT2. (d) Chromatogram traces at 96 h after three stepwise-dilution juxtaposing the three parallel experiments in the presence of CT2 (top trace), RT2 (middle trace) and containing no template (bottom trace). See supplementary figs. 91–93 for EDC-ligation conditions (at 4 °C). For CT2, RT2, RP2, RP3, RL3, RL4, RL5 and RL6 see Fig. 4a. Lines in graphs (5a-c) are drawn as guide indicating the trend and are not mathematical curve fittings. % yields were calculated with respect to the template CT2. Experiments were run in triplicate and the error range is less than ± 5%; error bars represent standard deviation.
RNA-DNA chimeric templates harbor the potential for cross-catalytic self-replication.
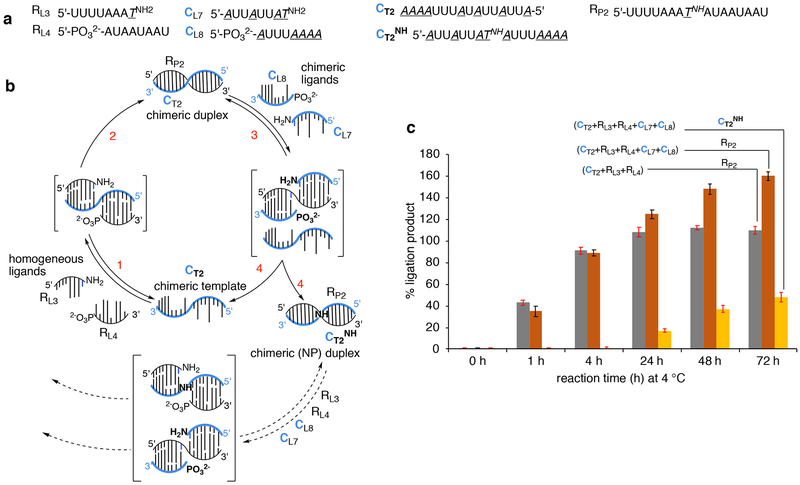
The promise of turnover of RNA ligation (Fig. 4b) when coupled with the observation that RDNA (CP2) chimeric products can also be formed on the RNA template (Supplementary Fig. 39) suggested that the catalytic chimeric template (CT2) could also be regenerated in the same reaction mixture if the corresponding chimeric ligands (CL7+CL8) are present (Fig. 6). If this would be possible, then the regeneration of the catalytic template CT2 could allow for a cross-catalytic cycle to be operative, which would be expected to lead to the amplification of the homogeneous RNA product RP2 (Fig. 6b). To test this possibility, we set up a one-pot EDC-ligation reaction with RNA ligands RL3+RL4 along with chimeric ligands CL7+CL8 in the presence of chimeric template CT2 (Fig. 6). We observed within 1–4 h the formation of expected product RP2 (90%), which now can act as the template for the chimeric ligands CL7+CL8. Indeed, by 24 h, formation of the phosphoramide-linked equivalent of CT2 (CT2NH, 16%) was clearly observed, and kept increasing with time to 36% in 48 h and to 48% in 72 h. And, in parallel, the amount of RP2 increased accordingly to 125% in 24 h, to 148% in 48 h and to 160% in 72h (Supplementary Fig. 94). This is well above the levels of RP2 produced in the ligation reaction mediated by CT2 in the presence of only RL4+RL3 and lacking the chimeric ligands (Fig. 6c), where the amount of RP2 leveled at around 108% by 72 h. Thus, the chimeric template mediated ligation process shows potential for cross-catalytic self-replicating systems that can result in amplification of the down-stream product. Further systematic investigations are ongoing to understand the scope and limitation of this system. In all of the experiments described in this work no discernible degradation of the homogeneous or chimeric templates or products was observed (confirmed by comparing with an external standard of oligonucleotide dT24 added to the samples just before analysis).
Figure 6. Experiment for testing the possibility of cross-catalytic amplification in oligonucleotide replication via regeneration of the chimeric RDNA (CT2) template.
(a) The sequences of oligonucleotides used in this investigation; CT2NH is the same as chimeric template CT2 but with a single phosphoramidate (NP) link at the ligation junction. (b) Schematic representation of the hypothesis that the presence of chimeric ligands CL7 and CL8 (complementary to RP2) could induce the regeneration of the chimeric template CT2NH leading to further production of RP2. The concomitant release of CT2 also creates the potential for another round of ligation reaction. (c) Comparison of the amount of RP2 produced from the combination of CT2+RL3+RL4 (1:5:5) versus the combination of CT2+RL3+RL4+CL7+CL8 (1:5:5:2:2), demonstrating the regeneration of chimeric template CT2NH along with higher and increasing production of RP2 in the latter combination. See supplementary Fig. 94 for experimental conditions. % yields were calculated with respect to the template CT2. Experiments were run in duplicate and the error range is less than ± 5%; error bars represent standard deviation.
Discussion
The results described in this work have confirmed experimentally the beneficial roles of chimeric sequences (backbone-heterogeneity) in nucleic acid replication, augmenting the evolution of functionality in mosaic nucleic acids17; and suggest that the nucleobase sequence-information encoded in heterogeneous-backbones can indeed be heritable for chemical evolution (similar to homogeneous-backbone systems). In these chimeric systems, there is the added advantage of (a) by-passing the template-product inhibition problem commonly encountered in the non-enzymatic replication of nucleic acids (unlike the homogeneous-backbone systems), and (b) moving towards (cross-catalytic) self-replication of the chimeric-templates, that eventually are able to assist in the transition from heterogeneity-to-homogeneity in nucleic acid systems13,14. Whether the preference for homogeneous-backbone ligands by chimeric templates (dictated by the thermodynamic stability of duplex formation) could be a general phenomenon for oligonucleotides composed of other different sugar-backbones/nucleobases that are able to cross-pair needs further examples (such as chimeras of 2′,5′-RNA with 3′,5′-RNA)19,49,50 to validate its scope and limitations.
For the work described here, however, there are some issues still to be addressed: firstly, the use of EDC-mediated-ligation combined with 3′-NH2 modified deoxynucleotide in this proof-of-principle study is not considered to be plausibly prebiotic. To this end, we are exploring the use of other prebiotically plausible phosphorylation-activation combined with oligomerization and ligation/recombination chemistries that may be compatible with the replication conditions.51–54 We briefly explored the use of enzymes (T4 DNA ligase and T4 RNA ligase 2) with canonical RNA, DNA and RDNA chimeric sequences, to check if ligases could be used to overcome the limitations of (a) the side reactions with chemical (EDC) activation55 and (b) the need for synthesizing sequences with the 3′-deoxy-NH2 modification – so that we may be able to push towards many rounds of replication and sequence analysis within a shorter time span, but have had limited success (Supplementary Figs. 95–103). We are exploring other ligases to expand the sequence-space and length parameters to overcome restrictions imposed by the EDC chemical-ligation methods.55, 56
Secondly, longer homogeneous products formed in the scenario described above are unlikely to work as continuous templates and may not provide the solution when moving towards sustained replication of longer homogeneous strands relying on thermodynamic-driven effects alone. One possible solution (alluded to in this work) is that chimeric-templates can facilitate indirect replication by catalysing the accumulation of homogeneous strands. The product homogenous strands can act as information storage, but cannot be directly replicated. Therefore, other mechanisms need to be invoked to allow the transfer of information stored in the homogeneous strands.56 One straightforward pathway consistent with the above heterogeneity-to-homogeneity scenario would be for the homogeneous RNA strands to give rise to functional ribozymes (ligase or polymerase) with the capability to take over the replication the homogeneous strands.57 Other pathways could involve the beneficial effects provided by different classes of molecules that have not been considered in this study. For example, two other components, primordial (depsi)peptides58 and protocells59 should be invoked, since they would have been an important part of any prebiotic scenario; they are as elementary as, if not more than, the nucleotide building blocks.53,60 Including them would be the next logical step to test the idea whether they could have not only aided in the transition from heterogeneity-to-homogeneity,34 but could also play a role in enabling the replication of information stored in the longer homogeneous RNA and DNA strands by overcoming the slower kinetics of strand exchange in replication of homogeneous RNA and DNA strands as strand lengths increase.61,62
Finally, in a prebiotic context, the possibility of oligomerizing on chimeric templates starting with monomeric building blocks has to be considered along-side the ligation chemistry demonstrated in this study.8 In our work, we were influenced by the duplex stabilities and reasoned that (a) the selectivity expressed at the ligand-template level may not translate to the level of weaker monomer-template associations and (b) based on earlier studies,8,63 oligomerization of monomers would be biased towards G and C containing sequences (due to their stronger association) over A and U residues. Also, as argued by others,64,65 the presence of dimers and trimers along with monomers in a prebiotic clutter may lead to selective incorporation of the higher order oligomers (dimers and trimers) over the monomers and, therefore, the ligation process may have an advantage over the oligomerization process. It would be necessary to test the limits of oligomerization with monomers in a chimeric scenario to observe what the preference is, both in terms of effect of the sugar and base residue (based on the nearest-neighboring nucleotide).49,50
The results reported in this study have two-fold implications for the emergence of homogeneous-backbone nucleic acids. First, starting from a mixture of binary-chimeric-systems e.g. RDNA, (a possibility that is strengthened by the recent report44 by Sutherland and co-workers on the plausibly prebiotic conversion of RNA nucleotides to DNA nucleos(t)ides), there is the potential for the simultaneous emergence of the two respective homogeneous-polymeric and communicating informational systems (RNA and DNA). This is opposed to the often-suggested sequential –RNA as the forerunner and DNA being the successor–paradigm. The successive replication cycles42,66,67 are expected to lead, simultaneously, to the two respective strands containing the homogeneous sugar-backbone (RNA and DNA), as indicated by the results in Figs. 3–6. Therefore, if RNA and DNA could have appeared together, then there is no need for genetic takeover by the new informational system (DNA) from an older system (RNA), a suggestion that has been made implicitly and explicitly by others,14,17,23–25,28,68,69 since there is neither a predecessor nor a successor in this scenario. This is also true for the supposed pre-RNA to RNA transition33; for example, there is no need for RNA being the descendent of TNA, when TRNA can simultaneously give rise to TNA and RNA. Second, the generality of this phenomenon–exemplified by RDNA and TRNA chimera systems–lends experimental credence to a point that is implied in Fig. 1, and one that has been discussed before;13,21,28 namely, a clean and directed prebiotic synthesis of a nucleotide building block of a particular oligonucleotide (e.g. TNA or RNA or DNA) is not an absolute requisite for a homogeneous-backbone nucleic acid like RNA to emerge. In other words, as is suggested in Fig. 1, the appearance of system with homogeneous nucleotide-backbone repeat units can be achieved at the emergent level of a replicating polymer.34 Therefore, a mixture of diverse nucleotides can, via the formation of mixtures of oligonucleotides and the ensuing emergent property of template-mediated ligation, tend towards homogeneous-nucleotide backbone systems.13 This process can include alternative linker units and alternative nucleobases,10,17,19,70 and chirality of the building blocks71. Which means, the appearance of a homogeneous-backbone homochiral polymer with a set of uniform building blocks from a prebiotic mixture is a natural outcome of chemical evolution,14 without the need for invoking the predecessor-successor models of extant biology.34,68,72
Supplementary Material
Acknowledgements
The work was supported by a grant from NASA (NNX14AP59G) and the Simons Foundation to R. K. (327124). S. B. thanks NASA Astrobiology Postdoctoral Program for a fellowship. We thank S. F. Dowdy laboratory for the use of their instrument for MALDI-ToF analysis. We are grateful to Jack Szostak, Irene Chen, Dieter Braun, Ulrich Muller, Luke Leman and our lab members for helpful discussions. Correspondence and requests for materials should be addressed to R.K. (rkrishna@scripps.edu).
Footnotes
Data Availability Statement
Full experimental details and data are provided in the Supplementary Information. The raw data that support the findings of this study are available from the corresponding author upon reasonable request. Supplementary information and chemical compound information are available in the online version of the paper. Reprints and permissions information is available online at www.nature.com/reprints.
Competing interests
The authors declare no competing interests.
Supplementary Information
Supplementary Methods, Supplementary Figures, Supplementary Tables and Supplementary References.
REFERENCES
- 1.Gilbert W Origin of Life - the RNA World. Nature 319, 618–618 (1986). [Google Scholar]
- 2.Joyce GF & Orgel LE Progress toward understanding the origin of the RNA world. Cold Spring Harbor Monograph Series 43, 23–56 (2006). [Google Scholar]
- 3.Gesteland RF, Cech TR & Atkins JF The RNA world: the nature of modern RNA suggests a prebiotic RNA. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor,1999). [Google Scholar]
- 4.Schwartz A Origins of the RNA world In Brack A (Ed.), The Molecular Origins of Life: Assembling Pieces of the Puzzle. pp. 237–254 (Cambridge: Cambridge University Press, 1998). [Google Scholar]
- 5.Orgel LE Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol 39, 99–123 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Anastasi C et al. RNA: prebiotic product, or biotic invention? Chem. Biodivers 4, 721–739 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Robertson MP & Joyce GF The origins of the RNA world. Cold Spring Harbor Perspectives in Biology 2012; 4:a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szostak J The eightfold path to non-enzymatic RNA replication. J. Sys. Chem 3, 2 (2012). [Google Scholar]
- 9.Higgs PG & Lehman N The RNA world: molecular cooperation at the origin of life. Nat. Rev. Genet 16, 7–17 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Hud NV, Cafferty BJ, Krishnamurthy R & Williams LD The origin of RNA and “my grandfather’s axe”. Chem. Biol 20, 466–474 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Joyce GF The antiquity of RNA-based evolution. Nature 418, 214–221 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Islam S & Powner MW Prebiotic systems chemistry: complexity overcoming clutter. Chem. 2, 470–501 (2017). [Google Scholar]
- 13.Krishnamurthy R On the emergence of RNA. Isr. J. Chem 55, 837–850 (2015). [Google Scholar]
- 14.Gavette JV, Stoop M, Hud NV & Krishnamurthy R RNA–DNA chimeras in the context of an RNA world transition to an RNA/DNA world. Angew. Chem. Int. Ed 55, 13204–13209 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Joyce GF, Schwartz AW, Miller SL & Orgel LE The case for an ancestral genetic system involving simple analogs of the nucleotides. Proc. Natl. Acad. Sci. U.S.A 84, 4398–4402 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazcano A & Miller SL The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell 85, 793–798 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Trevino SG, Zhang N, Elenko MP, Luptak A & Szostak JW Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl. Acad. Sci. U.S.A 108, 13492–13497, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng J et al. Structural insights into the effects of 2′−5′ linkages on the RNA duplex. Proc. Natl. Acad. Sci. U.S.A 111, 3050–3055, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelhart AE, Powner MW & Szostak JW Functional RNAs exhibit tolerance for non-heritable 2′–5′ versus 3′–5′ backbone heterogeneity. Nat. Chem 5, 390–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eschenmoser A The TNA-Family of nucleic acid systems: properties and prospects. Orig. Life Evol. Biosph 34, 277–306 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Powner MW, Zheng S-L & Szostak JW Multicomponent assembly of proposed DNA precursors in water. J. Am. Chem. Soc 134, 13889–13895, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szostak JW An optimal degree of physical and chemical heterogeneity for the origin of life? Phil. Trans. R. Soc. B: Biol. Sci 366, 2894–2901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orgel LE & Lohrmann R Prebiotic chemistry and nucleic acid replication. Acc. Chem. Res 7, 368–377 (1974). [Google Scholar]
- 24.Woese CR The Genetic Code: The Molecular Basis for Genetic Expression. New York: Harper and Row, 1967. [Google Scholar]
- 25.Oró J & Stephen-Sherwood E The Prebiotic synthesis of oligonucleotides In: Oró J, Miller SL, Ponnamperuma C, Young RS (eds) Cosmochemical Evolution and the Origins of Life. (Springer, Dordrecht, 1974). [PubMed] [Google Scholar]
- 26.Becker S et al. Wet-dry cycles enable the parallel origin of canonical and non-canonical nucleosides by continuous synthesis. Nat. Commun 9, 163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam S et al. Detection of potential TNA and RNA nucleoside precursors in a prebiotic mixture by pure shift diffusion-ordered NMR spectroscopy. Chem. Eur. J 19, 4586–4595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland JD & Whitfield JN Prebiotic chemistry: a bioorganic perspective. Tetrahedron 53, 11493–11527 (1997). [Google Scholar]
- 29.Usher D Early chemical evolution of nucleic acids: a theoretical model. Science 196, 311–313 (1977). [DOI] [PubMed] [Google Scholar]
- 30.Mehta AP et al. Bacterial genome containing chimeric DNA–RNA sequences. J. Am. Chem. Soc 140, 11464–11473 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Song X-P, Maiti M & Herdewijn P Enzymatic synthesis of DNA employing pyrophosphate-linked dinucleotide substrates. J. Sys. Chem 2, 3 (2011). [Google Scholar]
- 32.Schoning K-U et al. Chemistry of alpha-aminonitriles. 37. The alpha-L-threofuranosyl-(3′->2′)-oligonucleotide system (“TNA”): Synthesis and pairing properties. Helv. Chim. Acta 85, 4111–4153 (2002). [Google Scholar]
- 33.Orgel LE Perspectives: Origin of life: A simpler nucleic acid. Science 290, 1306–1307 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Krishnamurthy R Giving rise to life: transition from prebiotic chemistry to protobiology. Acc. Chem. Res 50, 455–459 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Pallan PS et al. Why does TNA cross-pair more strongly with RNA than with DNA? An answer from X-ray analysis. Angew. Chem. Int. Ed 42, 5893–5895 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Butlerov A Formation synthetique d’une substance sucree. Acad. Sci 53, 145–147 (1861). [Google Scholar]
- 37.Kim H-J et al. Synthesis of carbohydrates in mineral-guided prebiotic cycles. J. Am. Chem. Soc 133, 9457–9468 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Kim H-J & Benner SA Prebiotic stereoselective synthesis of purine and noncanonical pyrimidine nucleotide from nucleobases and phosphorylated carbohydrates. Proc. Natl. Acad. Sci. U.S.A 114, 11315–11320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber AL & Pizzarello S The peptide-catalyzed stereospecific synthesis of tetroses: a possible model for prebiotic molecular evolution. Proc. Natl. Acad. Sci. U.S.A 103, 12713–12717 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Kiedrowski G A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed 25, 932–935 (1986). [Google Scholar]
- 41.Wu X, Delgado G, Krishnamurthy R & Eschenmoser A 2,6-Diaminopurine in TNA: effect on duplex stabilities and on the efficiency of template-controlled ligations. Org. Lett 4, 1283–1286 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Derr J et al. Prebiotically plausible mechanisms increase compositional diversity of nucleic acid sequences. Nucleic Acids Res. 40, 4711–4722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu HY, Zhang S & Chaput JC Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem 4, 183–187 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Green N, Gibard C, Krishnamurthy R & Sutherland J Prebiotic phosphorylation of 2-thiouridine provides either nucleotides or DNA building blocks via photoreduction. Nat. Chem 11, 457–462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernando C, Von Kiedrowski G & Szathmáry E A Stochastic model of nonenzymatic nucleic acid replication: “elongators” sequester replicators. J. Mol. Evol 64, 572–585, (2007). [DOI] [PubMed] [Google Scholar]
- 46.Grossmann TN, Strohbach A & Seitz O Achieving turnover in DNA-templated reactions. ChemBioChem 9, 2185–2192 (2008). [DOI] [PubMed] [Google Scholar]
- 47.He C, Gállego I, Laughlin B, Grover MA & Hud NV A viscous solvent enables information transfer from gene-length nucleic acids in a model prebiotic replication cycle. Nat. Chem 9, 318–324 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Duim H & Otto S Towards open-ended evolution in self-replicating molecular systems. Beil. J. Org. Chem 13, 1189–1203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ertem G & Ferris JP Synthesis of RNA oligomers on heterogeneous templates. Nature 379, 238–240 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Prakash TP, Roberts C & Switzer C Activity of 2′,5′-linked RNA in the template-directed oligomerization of mononucleotides. Angew. Chem. Int. Ed 36, 1522–1523 (1997). [Google Scholar]
- 51.Mutschler H et al. Random-sequence genetic oligomer pools display an innate potential for ligation and recombination. eLife 7, e43022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lutay AV, Chernolovskaya EL, Zenkova MA & Vlassov VV The nonenzymatic template-directed ligation of oligonucleotides. Biogeosciences 3, 243–249 (2006). [Google Scholar]
- 53.Gibard C, Bhowmik S, Karki M, Kim E-K & Krishnamurthy R Phosphorylation, oligomerization and self-assembly in water under potential prebiotic conditions. Nat. Chem 10, 212–217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaidya N et al. Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Taran O, Thoennessen O, Achilles K & von Kiedrowski G Synthesis of information-carrying polymers of mixed sequences from double stranded short deoxynucleotides. J. Sys. Chem 1, 9 (2010). [Google Scholar]
- 56.Edeleva E et al. Continuous nonenzymatic cross-replication of DNA strands with in situ activated DNA oligonucleotides. Chem. Sci 10, 5807–5814 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mutschler H, Wochner A & Holliger P Freeze-thaw cycles as drivers of complex ribozyme assembly. Nat. Chem 7, 502–508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forsythe JG et al. Ester-mediated amide bond formation driven by wet–dry cycles: a possible path to polypeptides on the prebiotic earth. Angew. Chem. Int. Ed 127, 10009–10013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joyce GF & Szostak JW Protocells and RNA self-replication. Cold Spring Harbor Perspectives in Biology 2018; 10:a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruiz-Mirazo K, Briones C & de la Escosura A Prebiotic systems chemistry: new perspectives for the origins of life. Chem. Rev 114, 285–366 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Kamat NP, Tobe S, Hill IT & Szostak JW Electrostatic localization of RNA to protocell membranes by cationic hydrophobic peptides. Angew. Chem. Int. Ed 54, 11735–11739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen IA & Walde P From self-assembled vesicles to protocells. Cold Spring Harbor Perspectives in Biology 2, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce GF, Inoue T & Orgel LE Non-enzymic template-directed synthesis on RNA random copolymers. Poly(C, U) templates. J. Mol. Biol 176, 279–306 (1984). [DOI] [PubMed] [Google Scholar]
- 64.Prywes N, Blain JC, Del Frate F & Szostak JW Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife 5:e17756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeffer D, Sosson M & Richert C Enzyme-free ligation of dimers and trimers to RNA primers. Nucl. Acids Res 47, 3836–3845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leu K, Obermayer B, Rajamani S, Gerland U & Chen IA The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucl. Acids Res 39, 8135–8147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tupper A, Shi K & Higgs P The role of templating in the emergence of RNA from the prebiotic chemical mixture. Life 7, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dworkin JP, Lazcano A & Miller SL The roads to and from the RNA world. J. Theor. Biol 222, 127–134 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Brewin N Catalytic Role for RNA in DNA Replication. Nat. New Biol 236, 101 (1972). [DOI] [PubMed] [Google Scholar]
- 70.Krishnamurthy R RNA as an emergent entity: An understanding gained through studying its nonfunctional alternatives. Synlett 25, 1511–1517 (2014). [Google Scholar]
- 71.Ribó JM, Hochberg D, Crusats J, El-Hachemi Z & Moyano A Spontaneous mirror symmetry breaking and origin of biological homochirality. J. R. Soc. Interface 14: 20170699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnamurthy R Life’s biological chemistry: A destiny or destination starting from prebiotic chemistry? Chem. Eur. J 24, 16708–16715 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.