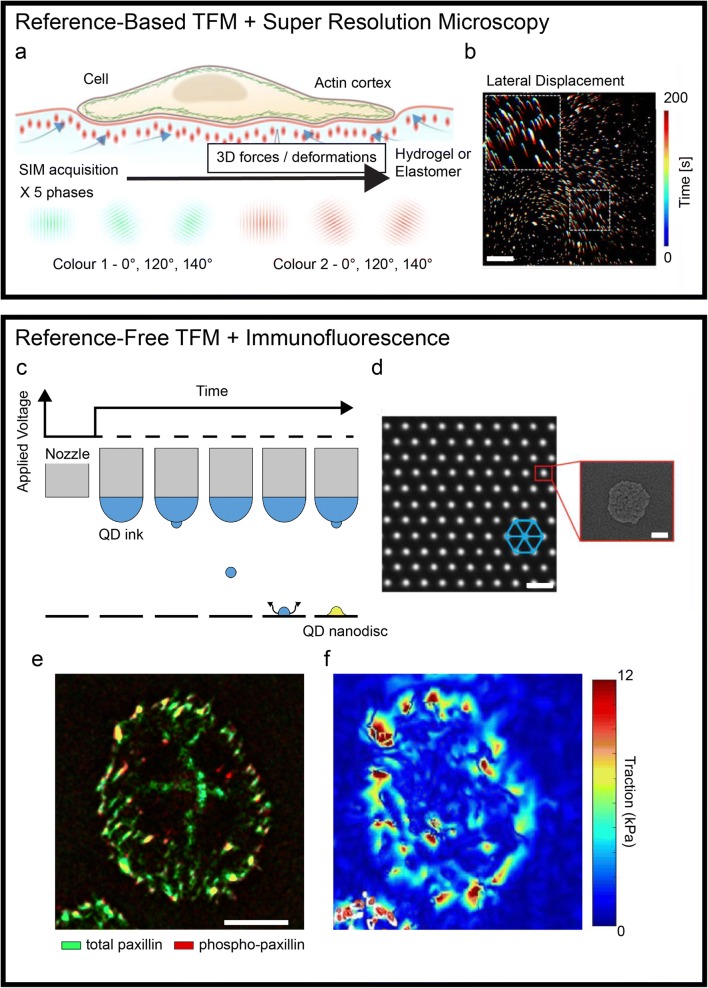
Traction force microscopy (TFM) defines a family of methods for measuring the tractions and stresses generated at the surface by adhering cells. More specifically, these techniques allow the analysis of acto-myosin-mediated cell contractility transmitted to the extracellular environment at the level of integrin adhesion points, the focal adhesions (Kechagia et al. 2019). The resulting forces, typically of the order of a few nNs, are in fact not measured directly but reconstructed from the active deformation imposed by the cell to its surroundings (Roca-Cusachs et al. 2017). To render these deformations, which develop both in-plane and out-of-plane, TFM protocols adopt compliant elastic materials featuring fiducial markers, generally in the form of fluorescent landmarks. The marker’s original placement and their displacement caused by the cell activity are tracked by means of optical microscopy yielding a vectorial field as proxy of the overall deformation. Coupled to an experimental material model of the elastomer (a silicone or a hydrogel) or constitutive model, the displacement field is finally used to derive the actuating cellular forces. With this procedure, TFM enables the dynamic measurement of three dimensional (3D) stress/force fields on the substrate. While TFM methods are proving essential to investigate the mechanical aspects of complex cellular and multicellular activities, technical limitations imposed by the measurement system, the fabrication protocols, or the image post-processing still hinder their applicability to a large range of biological questions. The mechanical activity of cells involves processes occurring at small, submicron scale and with fast, subsecond dynamics, therefore requiring substantial improvement in the temporal and spatial resolution of TFM measurements. Recently, new technologies have addressed some of these outstanding points, yielding improved spatial detail and the precise placement of the optical landmarks, altogether advancing the state of the art in TFM (Fig. 1). Classic TFM methods use elastic substrates (where a continuum behavior is assumed) in which fluorescent nanoparticles are randomly embedded at high density upon curing or gelation. This approach has the potential to yield force maps with high sensitivity, only limited by the spatial resolution at which the information in the displacement field can be sampled. Recent reports have seen the application of super-resolution optical microscopy to disentangle the displacement of dense fluorescent nanoparticles in 3D. The use of super-resolution stimulated emission depletion (STED) microscopy combined with TFM demonstrated a 5-fold improvement in rendering marker displacement yielding a much finer representation of forces compared to standard laser scanning confocal microscopy (Colin-York et al. 2016). This step forward came however at the cost of increasing the acquisition time, which now required a few minutes to gather sufficient information on the marker distribution for each field of view. The tradeoff between high spatial or temporal resolution was recently overcome by a further advancement in the imaging approach. The optimization of 3D super-resolution fluorescent structural illumination microscopy (SIM) for TFM samples achieved a submicron resolution at the speed of few seconds per field of view, yielding unprecedented details in the reconstruction of forces generated by single cells (Fig. 1a, b; (Colin-York et al. 2019)). A random dispersion of fiducial markers precludes the post-processing of TFM samples for the localization of protein activity through immunostaining. Information regarding the load-free position of the fluorescent landmarks is only obtained upon material relaxation, requiring cell deadhesion, and hence the termination of the experiment. To bypass this intrinsic limitation, alternative TFM methods were developed, in which more advanced technologies were applied to the fabrication of substrates featuring the precise placement of optical landmarks. Initially, these substrates consisted of ordered arrays of micron-sized elastomeric pillars generated by soft lithography (Tan et al. 2003). Here, each pillar acts as a spring, bending laterally when a cell adhering to its top applies traction. While achieving a reference-free force reconstruction, the discrete substrates are tainted by a limited spatial resolution (pillars need space to bend) and only provide information on in-plane forces. Early attempts to translate the reference-free concept to continuum TFM were similarly struggling with low spatial resolution and the introduction of topographic structures interfering with cells (Balaban et al. 2001). The development of an innovative printing technology, electrohydrodynamic NanoDrip printing (Galliker et al. 2012), with spatial position accuracy of ~ 20 nm and its optimization to deploy nanodiscs of quantum dots (QDs) with ~ 100 nm of lateral size and thickness below 20 nm allowed to surpass the previous limitations and generate an optimal reference-free continuum TFM platform (Fig. 1c, d; (Bergert et al. 2016)). Confocal TFM (cTFM) substrates comprise QD nanodiscs placed in a crystalline triangular array on a single plane at the surface of a transparent silicone substrate with tunable stiffness with elastic modulus in the 1–100 kPa range. Each of the fluorescent landmarks constitutes a point light source yielding a “bright” and intrinsically confocal signal that is well captured by standard wide-field microscopes, allowing the fast (i.e., hundreds of milliseconds) acquisition of in- and out-of-plane marker displacements. Vectorial maps are generated from a single image (or a Z-stack) without the need of an experimental reference, which is instead computed in silico through an optimization algorithm. Hence, the resulting force patterns can be directly superimposed to the distribution and activity of cellular actuators obtained by immunostaining (Fig. 1d, e). Application of this approach provided multilayered data linking force generation to focal adhesion maturation during the cell cycle (Panagiotakopoulou et al. 2018) and to the activation of mechanotranduction signaling (Bergert et al. 2016). Methods relying on the ordered placement of fiducial markers are, in principle, compatible with on-the-fly force mapping. In this future improvement of TFM, a full representation of the actuating cellular forces shall be rendered seconds after the acquisition of a fluorescent image reporting the landmark displacement in a field of view. Computed at the pace of the biological phenomenon under study, the mechanical activity of cells will then be accessible to educate an ongoing experiment. While this technological development is within reach, it still requires a substantial improvement in the software appliances for the detection, meshing, and reference calculation of fiducial markers. Once available, the on-the-fly analysis will provide the necessary complement to new and upcoming reference-free fabrication protocols (Bergert et al. 2016; Banda et al. 2019) opening to novel applications towards high-resolution and high-throughput TFM investigations.
Fig. 1.
Recent advancements in traction force microscopy. A super-resolution microscopy applied to reference-based TFM substrates (top box). (a) Schematic representation of a structural illumination microscopy (SIM) applied to a compliant TFM substrate. (b) A temporal projection of fluorescent optical landmarks tracked by SIM, rendering the cell-induced displacement upon force generation. Scale bar is 5 μm. Printing technologies for reference-free TFM enabling the combination with immunofluorescence (bottom box). (c) Schematic of electrohydrodynamic NanoDrip printing. Induction of a liquid meniscus and ejection of ink nanodroplets upon application of DC voltage. Once impacted on the underlying substrate, the droplets are vaporized leaving a QD nanodisc (d, inset). The process is reiterated accurately moving the nozzle to obtain a triangular array of fiducial markers (d). Scale bar is 2 μm. (e–d) The resulting ordered placement allows the endpoint staining of cellular components. Maps of focal adhesion markers distribution can be directly overlapped to the corresponding tractions. Scale bar is 10 μm. Adapted from (Colin-York et al. 2019; Galliker et al. 2012; Bergert et al. 2016; Panagiotakopoulou et al. 2018)
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Banda OA, Sabanayagam CR, Slater JH. Reference-free traction force microscopy platform fabricated via two-photon laser scanning lithography enables facile measurement of cell-generated forces. ACS Appl Mater Interfaces. 2019;11:18233–18241. doi: 10.1021/acsami.9b04362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergert M, Lendenmann T, Zundel M, Ehret AE, Panozzo D, Richner P, et al. Confocal reference free traction force microscopy. Nat Commun. 2016;7:12814. doi: 10.1038/ncomms12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin-York H, Shrestha D, Felce JH, Waithe D, Moeendarbary E, Davis SJ, et al. Super-resolved traction force microscopy (STFM) Nano Lett. 2016;16:2633–2638. doi: 10.1021/acs.nanolett.6b00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin-York H, Javanmardi Y, Barbieri L, Li D, Korobchevskaya K, Guo Y, et al. Spatiotemporally super-resolved volumetric traction force microscopy. Nano Lett. 2019;19:4427–4434. doi: 10.1021/acs.nanolett.9b01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliker P, Schneider J, Eghlidi H, Kress S, Sandoghdar V, Poulikakos D. Direct printing of nanostructures by electrostatic autofocussing of ink nanodroplets. Nat Commun. 2012;3:890. doi: 10.1038/ncomms1891. [DOI] [PubMed] [Google Scholar]
- Kechagia JZ, Ivaska J, Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol. 2019;20(8):457–473. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- Panagiotakopoulou M, Lendenmann T, Pramotton FM, Giampietro C, Stefopoulos G, Poulikakos D, et al. Cell cycle-dependent force transmission in cancer cells. Mol Biol Cell. 2018;29:2528–2539. doi: 10.1091/mbc.E17-12-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat Cell Biol. 2017;19:742–751. doi: 10.1038/ncb3564. [DOI] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]