Abstract
Five healthy aging phenotypes were developed in the Long Life Family Study to uncover longevity pathways and determine if healthy aging across multiple systems clustered in a subset of long-lived families. Using blood pressure, memory, pulmonary function, grip strength, and metabolic measures (body mass index, waist circumference and fasting levels of glucose, insulin, triglycerides, lipids, and inflammatory markers), offspring were ranked according to relative health using gender-, age-, and relevant confounder-adjusted z-scores. Based on our prior work, families met a healthy aging phenotype if ≥ 2 and ≥ 50% of their offspring were exceptionally healthy for that respective phenotype. Among 426 families, only two families met criteria for three healthy aging phenotypes and none met criteria for four or more healthy aging phenotypes. Using Spearman correlation, the proportion of offspring within families with exceptionally healthy pulmonary function was correlated with the proportion of offspring within families with exceptional strength (r = 0.19, p = 0.002). The proportion of offspring within families meeting the healthy blood pressure and metabolic phenotypes were also correlated (r = 0.14, p = 0.006), and more families were classified as meeting healthy blood pressure and metabolic phenotypes (Kappa = 0.10, p = 0.02), as well as the healthy pulmonary and blood pressure phenotypes than expected by chance (Kappa = 0.09, p = 0.03). Other phenotypes were weakly correlated (|r| ≤ 0.07) with low pairwise agreement (Kappa ≤ 0.06). Among these families selected for familial longevity, correspondence between healthy aging phenotypes was weak, supporting the heterogeneous nature of longevity and suggesting biological underpinnings of each individual phenotype should be examined separately to determine their shared and unique determinants.
Keywords: Familial longevity, Healthy aging
Introduction
Aging has been characterized as the progressive loss of physiologic integrity, where perturbations in one physiologic system likely initiate or exacerbate dysregulation in other systems, ultimately leading to vulnerabilities to major health outcomes (Cohen 2016; López-Otín et al. 2013; Sanders et al. 2016). The complex multifactorial nature of aging makes studying its biology difficult. Research tends to focus on a single organ system or a single disease; however, this does not capture the interconnectedness of the different physiologic systems (Hu et al. 2016). Shifting the focus to the body as a whole by examining the health of multiple physiologic systems simultaneously may help to identify fundamental processes of aging contributing to health and longevity that can inform common points of intervention to compress morbidity in the growing older adult population.
The Long Life Family Study (LLFS) is a multicenter cohort of two-generation families with clustering of exceptional longevity. In addition to familial longevity, participants in the proband generation from LLFS families had longer health spans; the average age at which 20% of probands had at least one chronic condition was 10 years later than controls without familial longevity from the New England Centenarian Study (Sebastiani et al. 2013). Identifying exceptional health across multiple physiologic domains among individuals with familial longevity may allow for uncovering rare genetic variants responsible for delaying or escaping the onset of common chronic conditions.
For many aspects of health, there are no standard criteria for exceptionally good health. We previously developed five healthy aging phenotypes in the LLFS (Barral et al. 2013; Marron et al. 2018, 2019) that identified families who had a clustering of individuals with exceptional health in memory, strength, pulmonary function, blood pressure, or metabolism. Here, we take a step further by examining the co-occurrence of the five healthy aging phenotypes. Specifically, we sought to determine whether healthy aging phenotypes were correlated among LLFS families, hypothesizing that more families would be classified as exceptionally healthy across multiple phenotypes than expected by chance.
Methods
The Long Life Family Study (LLFS)
The LLFS is a multicenter cohort of two-generation families enriched for longevity. Study centers are based in Boston, Massachusetts; New York, New York; Pittsburgh, Pennsylvania; and Denmark. The overall goal of LLFS is to examine familial and non-familial genetic, environmental, and behavioral determinants of healthy aging and exceptional survival. Families were primarily white (> 99%) and met the following eligibility criteria: (1) one enrolled long-lived participant (proband) aged ≥ 90, (2) at least one enrolled sibling of the proband, (3) at least one enrolled offspring of either the proband or their sibling, and (4) the family’s proband generation had a clustering of members with exceptional survival (Sebastiani et al. 2009). The two generations were labeled as the proband generation consisting of the long-lived individual and their enrolled siblings, and the offspring generation, which included all enrolled offspring of individuals in the proband generation. The LLFS protocol was approved by the Human Research Protection Office of the coordinating center at Washington University, the Regional Scientific Ethical Committees for Southern Denmark, and the Institutional Review Boards at the University of Pittsburgh, Boston University Medical Campus, and Columbia University. All participants provided informed consent. Among 562 families, 136 were excluded from this analysis because they only had one offspring in the study. Thus, the analytic sample size was N = 426 families who had ≥ 2 offspring with information on at least one of the five healthy aging phenotypes. The number of offspring within the 426 families ranged from 2 to 44 (average = 5).
Examination
At the initial study visit (around the time of enrollment, 2006–2009), we collected phenotypic measures and blood samples at the location where the participant was living. Participants self-reported their date of birth, gender, smoking status, and years of education. Immediate and delayed recall was measured using the Logical Memory subtest of the Wechsler Memory Scales-Revised (Wechsler 1987), which assessed ability to recall a short passage immediately and 30 min after. Isometric grip strength was measured twice in the dominant hand (Jamar, Sammons Preston Rolyan, Bolingbrook, IL) and averaged. Forced expiratory volume in 1 s was assessed using a spirometer (EasyOneTM, ndd Medical Technologies, Andover, MA) following American Thoracic Society guidelines. Blood pressure was calculated as the average of three sitting measurements in the right arm with an automated sphygmomanometer (Omron, Kyoto, Japan). Weight was recorded in light clothing using a digital scale (SECA Integra, Hamburg, Germany). Standing height was measured with a portable stadiometer. Waist circumference was measured at the umbilicus with a metal tape. Fasting glucose, insulin, triglycerides, high-density lipoprotein cholesterol, interleukin-6, and high-sensitivity C-reactive protein were measured in blood by the LLFS central laboratory at the University of Minnesota.
Healthy aging phenotypes
Markers used to represent phenotypes
Five healthy aging phenotypes were previously developed in the LLFS for the following domains: memory, strength, pulmonary function, blood pressure, and metabolism (Barral et al. 2013; Marron et al. 2018, 2019). The original aim of developing these healthy aging phenotypes was to quantify health within physiologic domains in order to identify subsets of LLFS families who were exceptionally healthy in a domain to determine genetic traits influencing their exceptional health within that domain. Because very few members of the proband generation remained healthy, the offspring generation was used to define and classify exceptional health within the cohort. Table 1 includes information on phenotype development. The average of immediate and delayed recall was used to develop the healthy memory phenotype, grip strength was used to develop the healthy strength phenotype, forced expiratory volume in 1 s was used to develop the healthy pulmonary phenotype, systolic blood pressure was used to develop the healthy blood pressure phenotype, and the following eight markers were used to develop the healthy metabolic phenotype: body mass index, waist circumference, and fasting levels of glucose, insulin, triglycerides, high-density lipoprotein cholesterol, interleukin-6, and high-sensitivity C-reactive protein.
Table 1.
Description of healthy aging phenotype development and number of families and offspring who met each phenotype
| Healthy aging phenotype | Marker(s) used to develop phenotype | Confounders adjusted for when calculating z-scores | Individual-level healthy aging phenotype criteria | Family-level healthy aging phenotype criteria | Number (%) of offspring who met individual-level phenotypee | Number (%) of families who met family-level phenotypee |
|---|---|---|---|---|---|---|
| Memory | Immediate and delayed recall | Age, sex, education | Adjusted episodic memory z-scorea ≥ 1.0 | ≥ 2 and ≥ 50% of offspring met individual-level healthy memory phenotype |
253 (15%) n = 1683 |
14 (4%) n = 353 |
| Strength | Grip strength | Age, sex, weight, height | Adjusted grip strength z-score ≥ 1.0 | ≥ 2 and ≥ 50% of offspring met individual-level healthy strength phenotype |
293 (14%) n = 2053 |
20 (5%) n = 392 |
| Pulmonary | Forced expiratory volume in one second | Age, sex, weight, height, smoking statusb | Adjusted forced expiratory volume in 1 second z-score ≥ 1.0 | ≥ 2 and ≥ 50% of offspring met individual-level healthy pulmonary phenotype |
305 (15%) n = 1988 |
22 (6%) n = 386 |
| Blood pressure | Systolic blood pressure | Age, sexc | Adjusted systolic blood pressure z-score ≥ − 1.5 and ≤ − 0.5 | ≥ 2 and ≥ 50% of offspring met individual-level healthy blood pressure phenotype |
476 (22%) n = 2211 |
44 (11%) n = 419 |
| Metabolism | Body mass index, waist circumference, glucose, insulin, triglycerides, high-density lipoprotein cholesterol, interleukin-6, high-sensitivity C-reactive protein | Age, sexd | Classification into a subgroup with a healthy profile of all 8 metabolic markers using latent profile analysis | ≥ 2 and ≥ 50% of offspring met individual-level healthy metabolic phenotype |
388 (20%) n = 1987 |
39 (10%) n = 388 |
aEpisodic memory z-score is the average of immediate and delayed recall z-scores
bSmoking classified as ever vs never smokers
cz-scores were calculated excluding participants taking anti-hypertensive medications, these participants were automatically classified as not meeting the healthy blood pressure phenotype
dz-scores for fasting glucose and fasting insulin were calculated excluding participants taking medication for diabetes, these participants were automatically classified as not meeting the healthy metabolic phenotype
eThe number of offspring and families included in each healthy aging phenotype differs because each phenotype was developed using information from as many individuals as possible, where some may have been missing information on one phenotype, but had information on a different phenotype
Calculating z-scores for each phenotype
For each phenotype, we calculated z-scores within the offspring generation using linear regression, while adjusting for age, gender, and specific, critical confounders (Table 1). Insulin, triglycerides and inflammation markers, C-reactive protein and interleukin-6 were log-transformed to normality. Immediate and delayed recall z-scores were calculated separately, while additionally adjusting for education. A single episodic memory z-score was then computed by averaging the immediate and delayed recall z-scores for each individual. The grip strength z-score was additionally adjusted for weight and height and the forced-expiratory volume in 1-s z-score was additionally adjusted for smoking status, weight, and height. When computing systolic blood pressure z-scores, offspring taking anti-hypertensive medication were excluded and automatically classified as not meeting the individual-level healthy blood pressure phenotype. Similarly, when computing fasting glucose z-scores and fasting insulin z-scores for the metabolic phenotype, offspring taking medication for diabetes were excluded and automatically classified as not meeting the healthy metabolic phenotype.
Individual-level phenotype development
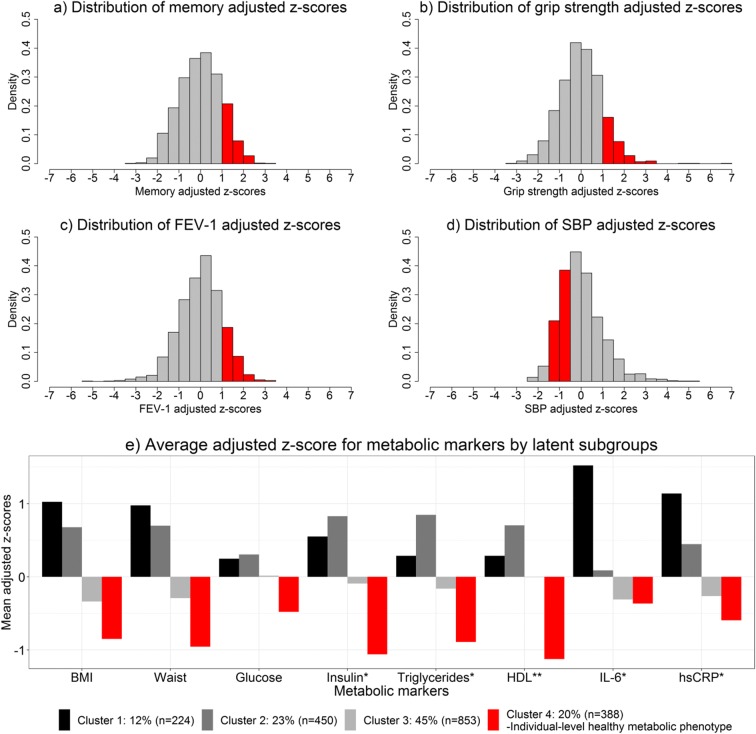
The proportion of offspring classified as meeting the individual-level healthy memory, strength, pulmonary, blood pressure, or metabolic phenotypes is illustrated by the red bars in Fig. 1 a–e. Offspring were classified as meeting the healthy memory, strength, or pulmonary phenotype if their respective adjusted z-score for that phenotype was at least one standard deviation above the overall adjusted mean (Fig. 1 a–c). For example, offspring were classified as having healthy memory if their score was at least one standard deviation better than what you would expect given their age, gender, and education level. The threshold of one standard deviation above the mean was chosen a priori to identify a subset of individuals who had values that were clearly healthy, based on our prior work (Barral 2013; Marron 2018, 2019). Figure 1 d illustrates that an offspring was classified as meeting the healthy blood pressure phenotype if their systolic blood pressure z-score was between − 1.5 and − 0.5. In other words, if they had a systolic blood pressure that was lower than expected for their age and gender, but not too low (Marron et al. 2018).
Fig 1.
Distributions of markers used to develop healthy aging phenotypes, red bars indicate values or cluster classification needed to meet respective healthy metabolic phenotype. a Distribution of memory adjusted z-scores. b Distribution of grip strength adjusted z-scores. c Distribution of FEV-1 adjusted z-scores. d Distribution of SBP adjusted z-scores. e. Average adjusted z-score for metabolic markers by latent subgroups. *Log transformation applied prior to computing z-scores. **HDL z-scores multiplied by − 1
The healthy metabolic phenotype (Marron et al. 2019) was developed by applying a latent profile analysis, using Mclust (version 4, Fraley et al. 2012), to the eight metabolic z-scores among offspring not taking medication for diabetes. All models with more than four latent subgroups had a poor posterior probability of correctly classifying participants into the subgroups (< 80%) or had subgroups that were too sparse (< 5% of participants). Among the well-fitting models with fewer groups, the four-group model was most optimal according to the Bayesian Information Criterion and was used to define offspring as meeting the healthy metabolic phenotype if classified into the latent subgroup that had a healthier average for all eight metabolic markers than expected given age and gender. Figure 1 e illustrates the average values of the eight metabolic markers for the four latent subgroups, of which group four had the healthiest metabolic profile.
Family-level phenotype development
Based on our prior work, families were included if they had ≥ 2 offspring members with available information on the marker that represented the respective phenotype (Barral et al. 2013; Marron et al. 2018, 2019). Families were classified as exceptionally healthy if ≥ 2 and ≥ 50% of their offspring met the respective individual-level healthy aging phenotype (Table 1). For example, a family met the healthy blood pressure phenotype if the majority of their offspring met the individual-level healthy blood pressure phenotype (Marron et al. 2018).
Statistical analysis
The number of families concordant for the healthy aging phenotypes was examined. We also wanted to examine concordance of the healthy aging phenotypes using a continuous measure, so for each family we calculated five proportions, one for each phenotype that described the number of their offspring classified as meeting a specific individual-level healthy aging phenotype out of the total number of offspring in the family. We then determined the correlation of the proportions of offspring within families who met a specific healthy aging phenotype versus the proportion of offspring within families who met a different healthy aging phenotype using Spearman correlation. We applied hierarchical clustering using a centroid component to the proportions of offspring within families meeting each of the healthy aging phenotypes and illustrated the clustering using a dendrogram. Percent positive agreement and Cohen’s kappa statistic were used to quantify pairwise agreement of classifying families as healthy for each pair of healthy aging phenotypes. Positive agreement was calculated as the number of families classified as healthy on two phenotypes divided by the average number of families classified as healthy on either of those phenotypes.
Results
The number of offspring and families who met each of the five healthy aging phenotypes is listed in Table 1. Among the offspring generation, the healthy memory prevalence, healthy strength prevalence, and healthy pulmonary prevalence were all around 15%. Slightly more offspring were classified as meeting the healthy blood pressure phenotype and the healthy metabolic phenotype (22% and 20%, respectively). When examining families, the healthy memory prevalence, healthy strength prevalence, and healthy pulmonary prevalence were all around 5% (Table 1). Similar to the individual-level phenotype, slightly more families met the healthy blood pressure and healthy metabolic phenotypes (11% and 10%, respectively).
The number of healthy aging phenotypes met by each family ranged from zero to three. Although individual offspring may have met criteria for exceptional health, the majority (73%) of families met none of the healthy aging phenotypic criteria, showing a lack of clustering for multiple phenotypes within families. Almost a quarter of families met criteria for one healthy aging phenotype and 5% of families met criteria for two healthy aging phenotypes. Only two (0.5%) families met three healthy aging phenotypic criteria. Among the families who met at least one healthy aging phenotype, almost half met only the healthy blood pressure phenotype or met only the healthy metabolic phenotype. Among the two families who met three healthy aging phenotypes, one family met criteria for the healthy memory, strength, and blood pressure phenotypes, and the other family met criteria for the healthy pulmonary, strength, and blood pressure phenotypes.
Correlations and classification agreement of the five healthy aging phenotypes
For each family, we calculated five proportions that described the number of offspring in the family who met a particular healthy aging phenotype divided by the total number of offspring in that family. Table 2 includes how correlated these proportions are among families. The proportions of offspring within families who met the healthy blood pressure phenotype were correlated with the proportions of offspring within families who met the healthy metabolic phenotypes (r = 0.14, p = 0.006). In addition, the proportions of offspring within families who met the healthy strength and pulmonary phenotypes were correlated (r = 0.19, p = 0.002). All other phenotypes were weakly correlated (|r| ≤ 0.07). Figure 2 illustrates the hierarchical clustering of the proportions of offspring within families who met each of the healthy aging phenotypes, suggesting that the healthy blood pressure and metabolic phenotype and the healthy strength and pulmonary phenotypes were more closely related than others.
Table 2.
Matrix of Spearman correlation coefficients comparing the proportions of offspring in families meeting each phenotype and percent positive agreement and kappa statistics comparing the agreement of classifying families as healthy on two phenotypes among 426 LLFS families, bold entries indicate significant results at p<0.05
| r = Spearman correlation coefficient, p value % positive agreement, к = Kappa statistic, p value n = number of families with information on both phenotypes | Healthy aging phenotypes | ||||
|---|---|---|---|---|---|
| Memory | Strength | Pulmonary | Blood pressure | Metabolic | |
| Memory | ------ | ||||
| Strength | r = 0.04, p = 0.45 | ------ | |||
| 6%, к = 0.02, p = 0.39 | |||||
| n = 342 | |||||
| Pulmonary | r = 0.003, p = 0.96 | r = 0.19,p = 0.002 | ------ | ||
| 0%, к = − 0.05, p = 0.17 | 5%, к = − 0.007, p = 0.45 | ||||
| n = 329 | n = 370 | ||||
| Blood pressure | r = 0.07, p = 0.17 | r = 0.03, p = 0.59 | r = 0.06, p = 0.22 | ------ | |
| 4%, к = − 0.02, p = 0.31 | 13%, к = 0.06, p = 0.08 | 16%,к = 0.09,p = 0.03 | |||
| n = 351 | n = 391 | n = 385 | |||
| Metabolic | r = − 0.02, p = 0.77 | r = 0.07, p = 0.15 | r = 0.04, p = 0.40 | r = 0.14,p = 0.006 | ------ |
| 12%, к = 0.06, p = 0.11 | 3%, к = − 0.04, p = 0.20 | 7%, к = − 0.007, p = 0.45 | 20%,к = 0.10,p = 0.02 | ||
| n = 335 | n = 369 | n = 357 | n = 381 | ||
Fig 2.
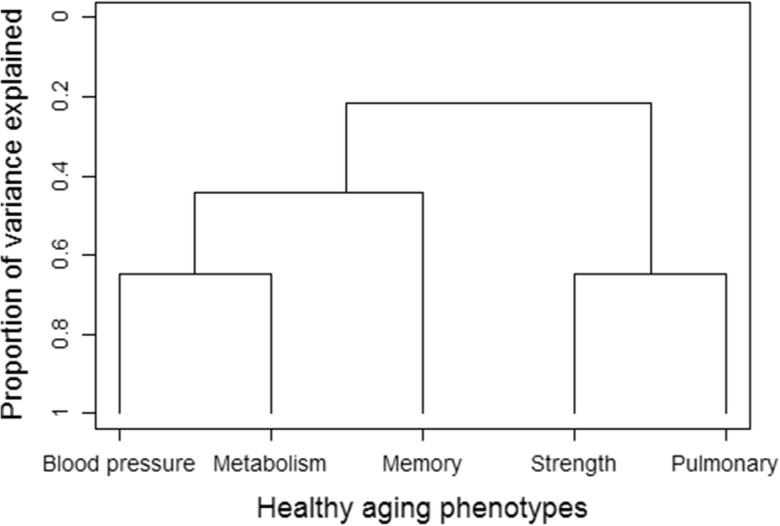
Hierarchical clustering of the proportions of offspring within families meeting each of the five healthy aging phenotypes
When examining pairwise agreement of classifying a family as healthy on a pair of phenotypes, we found significantly more families classified as meeting both the healthy blood pressure and metabolic phenotypes than expected by chance (Table 2). Among the average number of families classified as healthy on either the blood pressure or metabolic phenotype, 20% (8 families) were classified as healthy on both (Kappa = 0.10, p = 0.02). There were also more families who met both the healthy blood pressure and pulmonary phenotypes than expected by chance. Among the average number of families classified as healthy on either the pulmonary or blood pressure phenotype, 16% (5 families) were classified as healthy on both (Kappa = 0.09, p = 0.03). All other phenotypes had low pairwise agreement (Kappa ≤ 0.06).
Discussion
Among families selected for longevity, families who were healthiest in one domain were not likely among the families who were healthiest in other domains. Because aging is a risk factor for aging-related diseases, we expected that individuals who age more slowly would also delay or escape numerous aging-related diseases simultaneously (Andersen et al. 2012). Therefore, we hypothesized that healthy phenotypes would cluster in long-lived families. We did not observe such clustering in the LLFS families and therefore various LLFS families may have different underlying causes for their longevity. There was modest familial correlation between the healthy strength and pulmonary phenotypes and the healthy blood pressure and metabolic phenotypes. Though the correlations were not strong, there was more agreement than expected by chance between families classified as meeting the healthy blood pressure and metabolic phenotypes, as well as between the healthy pulmonary and blood pressure phenotypes.
The associations between healthy pulmonary function with both exceptional strength and healthy blood pressure may reflect an underlying influence of cardio-respiratory fitness. Weak grip strength and hypertension have been associated with higher odds of moderate-to-severe dyspnea among older adults, though hypertension did not remain significant after adjustments (Miner et al. 2016). Adequate respiratory strength is essential to maintaining healthy pulmonary function, and can be assessed by maximal inspiratory and expiratory pressures (Enrıght et al. 1994). Among older adults in the Cardiovascular Health Study, grip strength, but not blood pressure, was an independent correlate of maximal inspiratory and expiratory pressures (Enrıght et al. 1994). Though, blood pressure was marginally associated with maximal inspiratory volume among women, but not men, in the Multi-Ethnic Study of Atherosclerosis (Sachs et al. 2009). Maximal inspiratory pressure has also been associated with future major cardiovascular outcomes among older adults (Van der Palen et al. 2004). In addition, a greater decline in lung function, with values still in a normal range, was associated with a higher risk of incident hypertension over 10 years of follow-up among younger adults (Jacobs et al. 2012). Worse lung function has also been shown to be associated with worse small arterial elasticity (Duprez et al. 2013). In the LLFS, only two families met criteria for three healthy aging phenotypes, of which one met the healthy blood pressure, pulmonary, and strength phenotypes. Health across these three phenotypes may be a consequence of specific protective factors clustering in this family.
The healthy blood pressure and metabolic phenotypes were also significantly associated among the LLFS families. It is well-known that unhealthy metabolic measures and high blood pressure cluster together (Alberti et al. 2009) and together, these, of course, can constitute the metabolic syndrome (Alberti and Zimmet 1998). One hypothesis explaining the association between these two phenotypes involves insulin resistance as a risk factor for both high blood pressure and poor health across multiple metabolic markers, though the pathogenesis of this remains unclear (Alberti et al. 2009). Similarly, a common pathway of insulin sensitivity may be influencing the opposite end of the spectrum, healthy blood pressure and healthy metabolism. Other healthy aging phenotypes were weakly correlated among the LLFS families. Consistent with this finding, components of the Healthy Aging Index, a composite score of multi-system disease burden, were also weakly correlated (Sanders et al. 2014).
We found LLFS families to be heterogeneous across the five healthy aging phenotypes. The low prevalence of LLFS families meeting phenotypic criteria was partly due to our a priori definitions of health for each phenotype. The phenotypes were developed based on health relative to the total sample of LLFS offspring, i.e., a unique sample selected for familial longevity. This approach was deliberately chosen to identify the most extreme families for genetic analysis. Consistent with these findings, linkage signals for phenotypes in LLFS have been shown to be driven by a very small number of families (Barral et al. 2014; Lee et al. 2014). It is quite possible that not all long-lived individuals make it to extreme ages in the same way (Evert et al. 2003; Perls 2006; Ukraintseva et al. 2016). Others have shown that some individuals may make it to extreme ages by delaying the onset of chronic conditions, others by escaping the onset all together. Others survive to extreme ages with chronic conditions, potentially by adapting (Ukraintseva et al. 2010; Ukraintseva et al. 2016).
Different paths to longevity likely reflect different underlying combinations of genetic, environmental, and behavioral factors influencing health over the lifetime (Perls et al. 2006). Even within an individual, aging is heterogeneous, where physiologic systems can have different peaks of health capacity and different rates of decline (Newman and Cauley 2012). It has been hypothesized that a perturbation in one physiologic system can begin or accelerate dysregulation across other systems (Cohen 2016). Because of this complexity, it is reasonable to expect that we would find some LLFS families with exceptional health across multiple physiologic domains. There may still exist robustness in a physiologic system, where functional ability is maintained even in the presence of internal and external disturbances (Kitano 2007). However, others have proposed that robustness in one domain can cause fragility in other domains, a phenomenon described as the evolution of trade-offs (Kriete 2013), further suggesting the low likelihood of maintaining exceptional health across multiple domains.
Several aspects of this analysis are important to keep in mind when considering these findings. First, because we only included families with two or more offspring, we had to exclude 24% of LLFS families with only one offspring. Our predominantly white cohort limits the generalizability of results. Exceptionality was relative to the average of LLFS participants, not the average population. Perhaps, if we had the opportunity to compare findings between LLFS participants and the general population or a cohort representative of people who do not have a familial predisposition for longevity, we might have had more significant and elucidative findings. Strengths of this study include the detailed information on a novel family-based cohort enriched with longevity, available objective measures to develop phenotypes illustrating multiple domains of aging, and in-home visits enabling collection of functional data from participants that may not be healthy enough to leave their home.
Our exceptionally healthy aging phenotypes were heterogeneously distributed among families in this cohort of familial longevity. In other words, the families that were healthiest in one domain were not the healthiest in other domains. This suggests LLFS families may have become enriched with longevity through a diverse set of pathways, where exceptional health across multiple domains may not be necessary to obtaining familial longevity. Thus, to further our understanding of the heterogeneous longevity pathways, it will be important to investigate the genetic and environmental underpinnings of each phenotype separately. Potentially, this will uncover protective factors with pleiotropic effects and unique factors such as rare or even private (unique to a single family) genetic variation.
Funding
This work was supported by the National Institute on Aging at National Institutes of Health cooperative agreements U01-AG023712, U01-AG23744, U01-AG023746, U01-AG023749, and U01-AG023755. Megan Marron is supported on the Epidemiology of Aging training grant at the University of Pittsburgh (National Institute on Aging at National Institutes of Health T32-AG0001810). Dr. Svetlana is additionally supported by the National Institute on Aging of the National Institutes of Health P01 AG043352. Dr. Newman is supported by funding from National Institute of Aging R01 AG023629 Exceptional Survival: Trajectories to Functional Aging (CHS All Stars) and P30 AG024827 Pittsburgh Claude D. Pepper Older Americans Independence Center.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus. Provisional report of a WHO consultation Diabet Med 15(7):539–553. 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S [DOI] [PubMed]
- Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC., Jr Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; national heart, lung, and blood institute; American heart association; world heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012;67(4):395–405. doi: 10.1093/gerona/glr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral S, Cosentino S, Costa R, Andersen SL, Christensen K, Eckfeldt JH, Newman AB, Perls TT, Province MA, Hadley EC, Rossi WK, Mayeux R. Exceptional memory performance in the Long Life Family Study. Neurobiol Aging. 2013;34(11):2445–2448. doi: 10.1016/j.neurobiolaging.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral S, Cosentino S, Christensen K, Newman AB, Perls TT, Province MA, Mayeux R. Common genetic variants on 6q24 associated with exceptional episodic memory performance in the elderly. JAMA Neurol. 2014;71(12):1514–1519. doi: 10.1001/jamaneurol.2014.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA. Complex systems dynamics in aging: new evidence, continuing questions. Biogerontology. 2016;17(1):205–220. doi: 10.1007/s10522-015-9584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez DA, Hearst MO, Lutsey PL, Herrington DM, Ouyang P, Barr RG, Bluemke DA, McAllister D, Carr JJ, Jacobs DR., Jr Associations among lung function, arterial elasticity, and circulating endothelial and inflammation markers: the multiethnic study of atherosclerosis. Hypertension. 2013;61(2):542–548. doi: 10.1161/HYPERTENSIONAHA.111.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrıght PL, Kronmal RA, Manolio TA, Schenker MB, Hyatt RE. Respiratory muscle strength in the elderly. Correlates and reference values. Cardiovascular health study research group. Am J Respir Crit Care Med. 1994;149:430–438. doi: 10.1164/ajrccm.149.2.8306041. [DOI] [PubMed] [Google Scholar]
- Evert J, Lawler E, Bogan H, Perls T. Morbidity profiles of centenarians: survivors, delayers, and escapers. J Gerontol A Biol Sci Med Sci. 2003;58(3):M232–M237. doi: 10.1093/gerona/58.3.M232. [DOI] [PubMed] [Google Scholar]
- Fraley C, Murphy TB, Scrucca L (2012) mclust Version 4 for R: normal mixture modeling for model-based clustering, classification, and density estimation, technical report no. 597. Department of Statistics, University of Washington
- Hu JX, Thomas CE, Brunak S. Network biology concepts in complex disease comorbidities. Nat Rev Genet. 2016;17(10):615–629. doi: 10.1038/nrg.2016.87. [DOI] [PubMed] [Google Scholar]
- Jacobs DR, Jr, Yatsuya H, Hearst MO, Thyagarajan B, Kalhan R, Rosenberg S, Smith LJ, Barr RG, Duprez DA. Rate of decline of forced vital capacity predicts future arterial hypertension: the coronary artery risk development in young adults study. Hypertension. 2012;59(2):219–225. doi: 10.1161/HYPERTENSIONAHA.111.184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Towards a theory of biological robustness. Mol Syst Biol. 2007;3(1):137. doi: 10.1038/msb4100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriete A. Robustness and aging—a systems-level perspective. Biosystems. 2013;112(1):37–48. doi: 10.1016/j.biosystems.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Honig LS, Feitosa M, Kammerer CM, Kang MS, Schupf N, Lin SJ, Sanders JL, Bae HT, Druley T, Perls T, Christensen K, Province M, Mayeux R. Genome wide association and linkage analyses identified three loci—4q25, 17q23. 2, and 10q11. 21—associated with variation in leukocyte telomere length: the Long Life Family Study. Front Genet. 2014;4:310. doi: 10.3389/fgene.2013.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marron MM, Singh J, Boudreau RM, Christensen K, Cosentino S, Feitosa MF, Minster RL, Perls T, Schupf N, Sebastiani P, Ukraintseva S, Wojczynski MK, Newman AB. A novel healthy blood pressure phenotype in the long life family study. J Hypertens. 2018;36(1):43–53. doi: 10.1097/HJH.0000000000001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marron Megan M., Miljkovic Iva, Boudreau Robert M., Christensen Kaare, Feitosa Mary F., Lee Joseph H., Sebastiani Paola, Thyagarajan Bharat, Wojczynski Mary K., Zmuda Joseph M., Newman Anne B. A novel healthy metabolic phenotype developed among a cohort of families enriched for longevity. Metabolism. 2019;94:28–38. doi: 10.1016/j.metabol.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner B, Tinetti ME, Van Ness PH, Han L, Leo-Summers L, Newman AB, Lee PJ, Vaz Fragoso CA. Dyspnea in community-dwelling older persons: a multifactorial geriatric health condition. J Am Geriatr Soc. 2016;64(10):2042–2050. doi: 10.1111/jgs.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AB, Cauley JA. The epidemiology of aging. Dordrecht Heidelberg New York London: Springer; 2012. [Google Scholar]
- Perls Thomas T. The different paths to 100. The American Journal of Clinical Nutrition. 2006;83(2):484S–487S. doi: 10.1093/ajcn/83.2.484S. [DOI] [PubMed] [Google Scholar]
- Sachs MC, Enright PL, Stukovsky KDH, Jiang R, Barr RG. Performance of maximum inspiratory pressure tests and maximum inspiratory pressure reference equations for 4 race/ethnic groups. Respir Care. 2009;54(10):1321–1328. [PMC free article] [PubMed] [Google Scholar]
- Sanders JL, Minster RL, Barmada MM, Matteini AM, Boudreau RM, Christensen K, Mayeux R, Borecki IB, Zhang Q, Perls T, Newman AB. Heritability of and mortality prediction with a longevity phenotype: the healthy aging index. J Gerontol A Biol Sci Med Sci. 2014;69(4):479–485. doi: 10.1093/gerona/glt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JL, Boudreau RM, Newman AB. Understanding the aging process using epidemiologic approaches. In: Newman AB, Cauley JA, editors. The epidemiology of aging. Dordrecht Heidelberg New York London: Springer; 2016. pp. 187–214. [Google Scholar]
- Sebastiani P, Hadley EC, Province M, Christensen K, Rossi W, Perls TT, Ash AS. A family longevity selection score: ranking sibships by their longevity, size, and availability for study. Am J Epidemiol. 2009;170(12):1555–1562. doi: 10.1093/aje/kwp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Sun FX, Andersen SL, Lee JH, Wojczynski MK, Sanders JL, Yashin A, Newman AB, Perls TT. Families enriched for exceptional longevity also have increased health-span: findings from the long life family study. Front Public Health. 2013;1:38. doi: 10.3389/fpubh.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukraintseva SV, Arbeev KG, Akushevich I, Kulminski A, Arbeeva L, Culminskaya I, Akushevich L, Yashin AI. Trade-offs between cancer and other diseases: do they exist and influence longevity? Rejuvenation Res. 2010;13(4):387–396. doi: 10.1089/rej.2009.0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukraintseva S, Yashin A, Arbeev K, Kulminski A, Akushevich I, Wu D, Joshi G, Land KC, Stallard E. Puzzling role of genetic risk factors in human longevity: “risk alleles” as pro-longevity variants. Biogerontology. 2016;17(1):109–127. doi: 10.1007/s10522-015-9600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Palen J, Rea T, Manolio T, Lumley T, Newman AB, Tracy RP, Enright PL, Psaty BM. Respiratory muscle strength and the risk of incident cardiovascular events. Thorax. 2004;59(12):1063–1067. doi: 10.1136/thx.2004.021915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. WMS-R: Wechsler memory scale-revised: manual. New York: Psychological Corporation; 1987. [Google Scholar]