Abstract
Pomegranate (Punica granatum L.) peel is a potential source of polyphenols known for their activity against foodborne pathogen bacteria. In this study, the effects of pomegranate peel extraction time (10–60 min), agitation speed (120–180 rpm), and solvent/solid ratio (10–30) on phytochemical content and antibacterial activity were determined. Response surface methodology (RSM) and artificial neural network (ANN) methods were used, respectively, for multiresponse optimization and predictive modelling. Compared with the original conditions, the total phenolic content (TPC), the total flavonoid content (TFC), and the total anthocyanin content (TAC) increased by 56.22, 63.47, and 64.6%, respectively. Defined by minimal inhibitory concentration (MIC), the maximum of antibacterial activity was higher than that from preoptimized conditions. With an extraction time of 11 min, an agitation speed 125 rpm, and a solvent/solid ratio of 12, anti-S. aureus activity remarkably decreased from 1.56 to 0.171 mg/mL. Model comparisons through the coefficient of determination (R2) and mean square error (MSE) showed that ANN models were better than the RSM model in predicting the photochemical content and antibacterial activity. To explore the mode of action of the pomegranate peel extract (PPE) at optimal conditions against S. aureus and S. enterica, Chapman and Xylose Lysine Deoxycholate broth media were artificially contaminated at 104 CFU/mL. By using statistical approach, linear (ANOVA), and general (ANCOVA) models, PPE was demonstrated to control the two dominant foodborne pathogens by suppressing bacterial growth.
1. Introduction
Biological wastes, annually generated in important amounts from the agro-food industries, arouse a significant disposal burden when they are directly disposed to soil or landfill under poor management [1, 2]. In this line, the fruit processing industry, such as pomegranate, implies the raw material transformation to increase the added-value products [3–5]. Therefore, a huge amount of solid waste was generated. For example, for the pomegranate processing, approximately 500 g/kg of the pomegranate fruit composed from inedible portion, i.e., the peel and the remaining half has about 400 g/kg of juice and 100 g/kg of seeds [6]. Obviously, they still have notable content of polyphenols such as flavonoids, anthocyanins, hydroxybenzoic acids, hydroxycinnamic acids, and tannins [7–10] with beneficial effects on human health. Reported data demonstrate that phenolic compounds from pomegranate peel have diverse beneficial properties to health including anticancer activity [11–14], antidiarrheal activity [15], higher free radical scavenging activities [16], and strong antimicrobial activity [13, 17, 18]. In this context, several studies have reported the efficacy of pomegranate peel extracts (PPEs) to inhibit the growth of Gram-positive and Gram-negative bacteria, which are foodborne pathogens, spoilage bacteria, and human pathogens [19–23].
The extraction of pomegranate peels was primarily conducted for separating the phenolic compounds from the plant tissues. In this regard, the polarity of the solvent, extraction temperature, solid–solvent ratio, and particle size are influential parameters for extraction process [24–26]. Extraction of phenolic compounds from pomegranate peel has been reported [27–31], and in the most of these works, organic solvents such as methanol and acetone came up as suitable extraction solvents to reach good yields. However, environmentally benign and nontoxic food-grade organic solvents, such as ethanol, are recommended by the US Food and Drug Administration for extraction purposes [26, 32, 33].
As a result of the inability of conventional single variable optimization in the assessment of the individual and combined interactions between the process parameters, using statistical modelling techniques is better and increasingly common way. One such approach is the response surface methodology (RSM), a collection of statistical and mathematical techniques useful in many engineering applications, was demonstrated to be able to optimize a process with a minimal amount of experimental data. As a statistical tool, RSM can model the impact of various process factors, both individually and through their cumulative interactions, providing an indication of best combination of parameters and response prediction [34]. In recent years, artificial neural network (ANN) is finding increasing use as a predictive tool in an extensive range of disciplines because of its ability to employ learning algorithms. ANN discerns also input–output relationships for complex and nonlinear multifactor systems using the generic structure and ability to learn from historical data [35, 36]. Recently, RSM and ANN methods have been used jointly for both modelling and optimizing natural product extraction processes and biological activities, and the obtained models showed strong correlation with experimental results [35, 36].
On the other hand, despite the intensive research activities carried out in the past decades, it was estimated that less than 10% of the known plant species in the world has been studied for antimicrobial activities, and data are lacking regarding their compositions and detailed antimicrobial mechanisms [37]. In this line, studies on the understanding of the mechanism of natural antimicrobial action, such as PPE, by the development of predictive mathematical models are still scarce. Furthermore, predictive inactivation models have been developed in liquid laboratory media that can mimic the microbial environment [38, 39].
As part of further research work pertaining to the exploration of the bioactive compounds and biological control against foodborne bacterial pathogens, in this study, PPE was optimized using RSM and ANN techniques to enhance simultaneously the total phenolic content (TPC), total flavonoid content (TFC), total anthocyanin content (TAC), and anti-Staphylococcus aureus and anti-Salmonella enterica subsp. enterica serovar Typhimurium activities. Equally, by skillfully using statistical approach (linear (ANOVA) and general (ANCOVA) models), results from this study provide insight into PPE effectiveness against S. aureus and S. enterica.
To the best of our knowledge, data on (i) the comparison of RSM and ANN techniques for pomegranate peel optimization for bioactive compounds and biological control and (ii) intensive investigation of the inactivation mode against two dominant foodborne pathogens are not available in the literature.
2. Materials and Methods
2.1. Plant Material
Pomegranate fruits (Punica granatum L.) were obtained during commercial harvest in November 2017 from local farms of Sfax (N: 34.4426°, E: 10.4537°), a central coastal region in Tunisia. Peels were cleaned, washed with running water, air-dried under ambient conditions, and reduced to a fine powder using an electric grinder (Moulinex, France) to get 40-mesh size powder.
2.2. Extraction Conditions
Pomegranate peel powders were subjected to extraction through the maceration technique with ethanol (Novachim, Bucharest, Romania) as the solvent. Using a shaker (Heidolph, Unimax 2010; Elektro GmbH, Kelheim, Germany), extractions were performed in Erlenmeyer flasks (Schott Duran, Voor't labo, Eeklo, Belgium) containing 2.0 g of the pomegranate peel powder sample. All extractions were performed at 20°C (cold extraction) for different extraction time (X1: 10–60 min), agitation speed (X2: 120–180 rpm) and solvent/solid ratio (X3: 10–30). Extraction time and agitation speed were controlled from the panel of the instruments. After extraction, each mixture was centrifuged (Sorvall Biofuge Stratos, ThermoScientific, Hanau, Germany) at 13,000×g for 10 min at 4°C. For removal of peel particles, the supernatant was filtered through cotton wool and Whatman paper No. 1 (Whatman Ltd, England) and then collected. Each extraction mixture was evaporated in a rotary evaporator (Laborota 4000, Heidolph, Milan, Italy) at 40°C obtaining a yellow–brown residue from peels that were immediately analyzed. PPEs were weighed to calculate the recovery extraction (%). The extraction procedure was carried out in triplicates.
2.3. Phytochemical Study of PPE
2.3.1. Quantification of the TPC
TPCs from PPE were quantified by using the Folin–Ciocalteu method [40] adapted for a 96-well plate assay. PPEs were evaluated at the final concentration of 100 μg/mL. 100 μL of 0.2 N Folin–Ciocalteu reagent (Sigma-Aldrich GmbH, Steinheim, Germany) was added to a 96-well plate (SPL Life Sciences, Pocheon, Gyeonggi, South Korea) containing 20 μL of each PPE with 80% ethanol and kept for 5 min in darkness at room temperature. 80 μL of sodium carbonate at 75 g/L (Scharlau, Barcelona, Spain) was added to each well, and the plate was then incubated for 30 min at room temperature with slightly shaking in the darkness. Absorbance at 725 nm was measured in the spectrophotometer (PG Instrument Ltd. Model T60, United Kingdom). Gallic acid at 0–200 μg/L (Sigma-Aldrich GmbH, Steinheim, Germany) was applied as standard. Results were expressed as mgGA equivalents/g dry sample (mg GAE/g) using the linear equation based on the calibration curve. Each assay was carried out in triplicates.
2.3.2. Quantification of the TFC
TFCs in PPE samples were determined using the AlCl3 colorimetric method [41] adapted to 96-well plate. In brief, 100 μL of 2% AlCl3 (Scharlau, Barcelona, Spain) was added to 100 μL of each PPE in ethanol, and the plate was then incubated for 15 min at room temperature in the dark. The absorbance was determined at 430 nm. PPEs were evaluated at the final concentration of 100 μg/mL. Quercetin at 0–50 μg/mL (Sigma-Aldrich GmbH, Steinheim, Germany) was used as the standard. The total flavonoid contents were expressed as milligrams of quercetin equivalents mg QE/g of PPE. All the experiments were carried out in triplicates.
2.3.3. Quantification of the TAC
TAC was evaluated by the pH differential method using two buffer systems: 25 mM potassium chloride (Sigma-Aldrich GmbH, Steinheim, Germany) solution (pH 1.0) and 0.4 M sodium acetate (Loba Chemie Pvt. Ltd., Mumbai, India) buffer (pH 4.5). PPEs were mixed with the corresponding buffers, and absorbances were determined simultaneously at 510 and 700 nm, respectively, after 15 min of incubation at 23°C [42].
| (1) |
TAC, expressed as mg of cyanidin-3-glucoside equivalents per 100 g of PPE (mg cy-3-glu/100 g), had been calculated as follows:
| (2) |
where A: absorbance; MW: molecular weight (449.2 g/mol); and ε: molar absorptivity of cyanidin-3-glucoside (26,900 M−1·cm−1).
2.4. Antibacterial Assays
2.4.1. Bacterial Strains and Culture Conditions
Target bacteria strains were obtained from international culture collections (ATCC). They included Gram-positive bacterium: Staphylococcus aureus ATCC 6538 and a Gram-negative bacterium: Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028. S. aureus ATCC 6538 was grown on Chapman medium (Oxoid, Hampshire, United Kingdom), and S. enterica ATCC 14028 was cultivated on Xylose Lysine Deoxycholate (XLD, Oxoid CM0469, Basingstoke, United Kingdom) at 37°C for 24 h. For antagonist tests, the final inoculum concentration used for each indicator bacterium was 106 CFU/ml [43].
2.4.2. Determination of Minimum Inhibitory Concentrations (MICs)
The MIC of each PPE was determined against S. aureus ATCC 6538 and S. enterica ATCC 14028. The test was performed in sterile 96-well microplates with a final volume in each microplate well of 100 μl. A stock solution of each extract, ranged between 0.0487 and 6.24 mg/mL, was prepared. To each test well, 10 μl of cell suspension was added to final inoculum concentration of 106 CFU/ml of bacterium. Positive growth control well consisted of S. aureus ATCC 6538 and S. enterica ATCC 14028, respectively, growth in Chapman and XLD media. Dimethyl sulfoxide (DMSO) (Loba Chemie Pvt. Ltd., Mumbai, India)/water (1/9) was used as the negative control. The plates were then covered with sterile plate covers and incubated for 24 h [44]. MIC was defined as the lowest concentration of each extract in which the microorganism did not demonstrate visible growth after incubation. As an indicator of microorganism growth, 25 μl of Thiazolyl Blue Tetrazolium Bromide (MTT) (Sigma-Aldrich, Taufkirchen, Germany), indicator solution (0.5 mg/mL) dissolved in sterile water was added to the wells and incubated at 30 min. The colourless tetrazolium salt acts as an electron acceptor and was reduced to a red-coloured formazan product by biologically active organisms. Where microbial growth was inhibited, the solution in the well remained clear after incubation with MTT. The determinations of MIC values were done in triplicates.
2.5. Box–Behnken Design and Optimization by RSM
The extraction methodology was developed following the Box–Behnken design, and RSM was used to analyze the relationship between the measured responses and the individual and combined effects. Significant variables were optimized for enhanced phytochemical contents (TPC, TFC, and TAC), and anti-S. aureus and anti-S. enterica activities employing the Box–Behnken design [45].
The extraction time, agitation speed, and solvent/solid ratio were analyzed at three levels low, medium, and high coded as (−1), (0), and (+1), respectively, in fifteen runs. These variables: extraction time (X1: −1 : 10; 0 : 35; +1 : 60 min), agitation speed (X2: −1 : 120; 0 : 150; +1 : 180 rpm), and solvent/solid ratio (X3: −1 : 10; 0 : 20; +1 : 30) were selected for studying the effect and significance on TPC, TFC, TAC and the antibacterial activity.
The dummy variables were used to calculate the standard error. Each run was carried out in three replicates. The behavior of the system was explained by a second-order polynomial equation:
| (3) |
where Y is the predicted response, β0 is the offset term, βi is the linear effect, βii is the squared effect, βij is the interaction effect, and Xi is the coded value of independent variables under study. This design was used to evaluate the main effects, interaction effects, and quadratic effects. It is also used to optimize the levels of parameters for enhancing the five responses. The statistical software Minitab 15.0 version was used for the experimental design and data analysis. Three-dimensional response surface plots were drawn to illustrate the relationship between the responses and the experimental levels of each independent variable. Optima levels of the variables for the maximum TPC, TFC, TAC, and the antibacterial activity was determined by the response optimizer tool of the software.
2.6. ANN-Based Modelling
To forecast nonlinear processes via neural networks, the network type Multilayered Perceptron (MLP) was used [46, 47]. These analyses can be used to generate a system that would support forecast procedures. Modelling of the MLP network was achieved via tanh.
In this study, in order to guarantee the simplest network design, the networks were constructed with one hidden layer: the input layer consists of three neurons (X1, X2, and X3), while the output layer had five neurons (TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activities). To measure the performance of ANN models for approximating the desired output of the five responses, the coefficient of determination (R2) was calculated. In fact, R2 can be defined as a standard criterion for the evaluation of statistical performance and employed to verify the accuracy of the predictive ability of the assembled models [48]. The training data set was applied to train the ANN to locate the overall comprehensive model between its inputs and outputs. The test data were used to validate and corroborate the predictive value of the expanded networks.
Neural Network module of STATISTICA 8.0 software was used in modelling the ANN. The data were categorized into two parts: training (80%) and testing (20%). In the network, there were three inputs and one output, corresponding to the TPC, TFC, TAC, and the antibacterial activity. The hidden neurons were optimized by building various MLP with hidden nodes from 1 to 10. Networks with hidden nodes greater than 10 were not developed because of the predictive capabilities decreasing with the decrease in the number of intermediate units.
2.7. Time-Kill Assay of PPE on Viable Counts of S. aureus ATCC 6538 and S. enterica ATCC 14028
The PPE effect on the inhibition of S. aureus ATCC 6538 and S. enterica ATCC 14028 was assessed by sequential sampling and counting viable bacteria, respectively, in the Chapman and the XLD broth. Firstly, S. aureus ATCC 6538 and S. enterica ATCC 14028 growth reached the beginning of the exponential phase (∼104 CFU/ml). After three hours of incubation time, PPEs were added separately in four different concentrations (0.171, 0.342, 0.684, and 1.368 mg/mL for S. aureus and 0.555, 1.11, 2.22, and 4.44 for S. enterica) then incubated 37°C for 24 hours. At various points of incubation time: 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 16, 18, 20, 22, 24, and 26 hours, the numbers of CFU were determined by plating the samples on Chapman agar for S. aureus ATCC 6538 and XLD agar for S. enterica ATCC 14028 and then counting of the colonies that appeared. Controls were prepared under the same experimental conditions as mentioned above but without PPE addition. Each test was performed in triplicates.
2.8. Statistical Analysis
Measurements were carried out in triplicates and repeated three times. A one-way analysis of variance (ANOVA) and Tukey's post-hoc test were performed to determine significant differences between the responses using the Statistical Package for the Social Sciences (SPSS) software (SPSS Ltd. Woking, United Kingdom). Means and standard errors were calculated. Differences among the mean values of the various responses were determined by the least significant difference test. A probability level of p < 0.05 was used in testing the statistical significance of all the experimental data.
Plate count data were converted to logarithms prior to their statistical treatment. Linear mixed models assuming the error to compare CFU values among treatments with different time periods were used. Mixed models were fitted using SPSS 19 and followed by post hoc contrasts through the origin. The interpretation of the statistical output of a mixed model requires an understanding of how to explain the relationships among the fixed and random effects.
3. Results and Discussion
The effect of extraction time, agitation speed, and solvent/solid ratio on the recovery extraction of PPE was shown in Table 1. Recovery extraction of different experimental runs ranged between 14.4 ± 0.51 and 19.98 ± 0.95%. Among the fifteen runs, the highest (p < 0.05) PPE recovery extraction was recorded in run 2 with a value of 19.98 ± 0.95%. A similar trend has been observed by Malviya et al. [49] studying the effect of using different solvents on the extraction yield from pomegranate peels. In fact, these authors reported that recovery extraction was about 15%.
Table 1.
Box–Behnken design and ANN models of variables (in coded levels), with phytochemical contents (TPC, TFC, and TAC) and anti-S. aureus and S.enterica activity responses.
| Run order | (X1) | (X2) | (X3) | Recovery extraction (%) | Phytochemical content | Antibacterial activity | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPC (mg GAE/g) | TFC (mg QE/g) | TAC (mg cy-3-glu/100 g) | S. aureus (MIC mg/ml) | S. enterica (MIC mg/ml) | |||||||||||||||
| Exp. | Pred. RSM | Pred. ANN | Exp. | Pred. RSM | Pred. ANN | Exp. | Pred. RSM | Pred. ANN | Exp. | Pred. RSM | Pred. ANN | Exp. | Pred. RSM | Pred. ANN | |||||
| 1 | 10 (−1) | 150 (0) | 10 (−1) | 18.45 ± 0.92c | 123.8 ± 6.19cd | 127.485 | 128.671 | 37.11 ± 1.85f | 35.249 | 37.06 | 4.78 ± 0.22c | 4.607 | 4.817 | 0.39 ± 0.00b | 0.34 | 0.395 | 0.195 ± 0.00c | 0.188 | 0.193 |
| 2 | 10 (−1) | 180 (+1) | 20 (0) | 19.98 ± 0.95d | 118.02 ± 5.91c | 115.312 | 118.15 | 30.02 ± 1.45d | 28.892 | 30.006 | 4.64 ± 0.21c | 4.884 | 4.828 | 0.39 ± 0.00b | 0.492 | 0.382 | 0.195 ± 0.00c | 0.171 | 0.195 |
| 3 | 60 (+1) | 150 (0) | 10 (−1) | 17.3 ± 0.86b | 109.6 ± 4.48bc | 106.637 | 109.849 | 37.09 ± 1.84f | 35.535 | 36.606 | 11.61 ± 0.52h | 10.085 | 11.582 | 0.097 ± 0.00a | 0.224 | 0.128 | 0.195 ± 0.00c | 0.185 | 0.194 |
| 4 | 35 (0) | 150 (0) | 20 (0) | 17 ± 0.85b | 70.02 ± 3.49a | 70.00 | 71.156 | 22.00 ± 1.22a | 23.037 | 23.019 | 4.64 ± 0.21c | 4.633 | 4.586 | 1.56 ± 0.00d | 1.56 | 1.509 | 0.781 ± 0.00b | 0.780 | 0.772 |
| 5 | 10 (−1) | 120 (−1) | 20 (0) | 15.6 ± 0.7ab | 130 ± 6.47d | 126.162 | 129.789 | 33.35 ± 1.62de | 34.749 | 33.507 | 3.71 ± 0.17b | 2.520 | 3.81 | 0.39 ± 0.00b | 0.464 | 0.386 | 0.39 ± 0.00a | 0.325 | 0.393 |
| 6 | 35 (0) | 180 (+1) | 10 (−1) | 15 ± 0.75a | 85.00 ± 4.23ab | 84.125 | 84.729 | 25.00 ± 1.22b | 27.862 | 23.019 | 9.75 ± 0.46g | 9.385 | 9.808 | 0.781 ± 0.00c | 0.547 | 0.794 | 0.39 ± 0.00a | 0.424 | 0.388 |
| 7 | 60 (+1) | 120 (−1) | 20 (0) | 15.15 ± 0.6a | 99.91 ± 4.92b | 102.712 | 100.108 | 29.00 ± 1.44cd | 30.108 | 28.917 | 3.94 ± 0.12b | 3.406 | 4.033 | 0.39 ± 0.00b | 0.347 | 0.411 | 0.195 ± 0.00c | 0.197 | 0.198 |
| 8 | 35 (0) | 120 (−1) | 10 (−1) | 16.7 ± 0.45b | 112.5 ± 5.61bc | 112.65 | 112.16 | 30.01 ± 1.5d | 30.357 | 29.897 | 7.01 ± 0.33f | 8.766 | 6.982 | 0.195 ± 0.00e | 0.170 | 0.206 | 0.195 ± 0.00c | 0.196 | 0.197 |
| 9 | 35 (0) | 150 (0) | 20 (0) | 14.4 ± 0.51a | 70.00 ± 3.41a | 70.00 | 71.156 | 22.01 ± 1.02a | 23.037 | 23.019 | 4.61 ± 0.21c | 4.633 | 4.586 | 1.56 ± 0.00d | 1.56 | 1.509 | 0.781 ± 0.00b | 0.780 | 0.772 |
| 10 | 35 (0) | 150 (0) | 20 (0) | 17.85 ± 0.82bc | 70.01 ± 3.33a | 70.00 | 71.156 | 22.00 ± 1.09a | 23.037 | 23.111 | 4.64 ± 0.22c | 4.633 | 4.586 | 1.56 ± 0.00d | 1.56 | 1.509 | 0.781 ± 0.00b | 0.780 | 0.772 |
| 11 | 10 (−1) | 150 (0) | 30 (+1) | 17.05 ± 0.79b | 106.5 ± 5.25bc | 109.462 | 107.043 | 35.50 ± 1.75e | 36.965 | 35.706 | 5.32 ± 0.20d | 5.547 | 4.112 | 0.39 ± 0.00b | 0.263 | 0.411 | 0.195 ± 0.00c | 0.193 | 0.193 |
| 12 | 60 (+1) | 150 (0) | 30 (+1) | 15 ± 0.66a | 95.8 ± 4.77b | 92.112 | 95.755 | 28.00 ± 1.39c | 29.751 | 28.151 | 6.38 ± 0.30e | 6.554 | 6.384 | 0.195 ± 0.00e | 0.245 | 0.206 | 0.39 ± 0.00a | 0.364 | 0.369 |
| 13 | 35 (0) | 120 (−1) | 30 (+1) | 17.27 ± 0.84b | 73.6 ± 3.66a | 74.475 | 73.895 | 33.36 ± 1.66de | 30.508 | 33.071 | 3.02 ± 0.14a | 3.068 | 3.341 | 0.39 ± 0.00b | 0.442 | 0.391 | 0.39 ± 0.00a | 0.414 | 0.415 |
| 14 | 35 (0) | 180 (+1) | 30 (+1) | 17.35 ± 0.36b | 89.9 ± 4.44ab | 89.75 | 89.397 | 24.00 ± 1.15b | 23.643 | 24.154 | 14.86 ± 0.71i | 12.792 | 14.434 | 0.195 ± 0.00e | 0.219 | 0.197 | 0.39 ± 0.00a | 0.381 | 0.382 |
| 15 | 60 (+1) | 180 (+1) | 20 (0) | 17 ± 0.49b | 96.6 ± 4.81b | 100.437 | 96.482 | 28.06 ± 1.32c | 26.605 | 28.151 | 7.89 ± 0.37f | 9.484 | 7.961 | 0.781 ± 0.00c | 0.475 | 0.797 | 0.39 ± 0.00a | 0.363 | 0.374 |
Variables: extraction time (X1), agitation speed (X2), and solvent/solid ratio (X3). ±: Standard deviation of three replicates; Averages with different letters (a–i) in the same column, for each parameter, are different (p < 0.05).
To develop an empirical model, an experimental design has been formulated by examining the interaction of different associated parameters X1 (time of extraction), X2 (agitation speed), and X3 (solvent/solid ratio). The design arrangement and experimental results of the extraction are shown in Table 1. In order to evaluate the fitness of response function, a total of 15 designed experiments were conducted for optimizing X1, X2, and X3 in a multivariable system. According to equation (3), YTPC, YTFC, YTAC, Yanti-S. aureus, and Yanti-S. enterica were calculated into the generalized model. Equally, the linear and quadratic effects of independent variables (X1, X2, and X3) of the multiple regression coefficients were calculated, and their interactions were also analyzed for regression coefficients in the RSM study.
3.1. Model Fitting and Analysis of Response Surfaces
The adequacy and fitness of the models were judged by the lack-of-fit significance and R2. For all responses, the statistical analysis indicated that the proposed model was adequate, possessing no significant lack-of-fit, and with very satisfactory values of R2. The latter was estimated to be 98.39, 89.26, 86.41, 97.98, and 97.98% for the TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activities, respectively. As shown in Table 2, with no lack-of-fit, p values of the total model of all responses were less than 0.05. Besides, F values from all regression models (F[TPC] = 33.87, p=0.001; F[TFC] = 4.62, p=0.043; F[TAC] = 3.53, p=0.049; F[anti-S. aureus] = 26.88, p=0.001 and F[anti-S. enterica] = 26.13, p=0.001) were significant. Second-order polynomial equations were used to study the relation between the input process variables (X1, X2, and X3) and their five respective responses. So, the second-order polynomial coefficient for each term of the equation was determined through multiple regression analysis using RSM.
Table 2.
ANOVA results of process variables against TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activity responses.
| Responses | Regression | Sum of square | F value | p value | R 2 | R adjusted 2 |
|---|---|---|---|---|---|---|
| TPC | Linear | 1347.15 | 24.51 | 0.002∗ | 98.39 | 95.48 |
| Quadratic | 3735.17 | 67.96 | 0.000∗∗ | |||
| Interaction | 501.60 | 9.13 | 0.018∗ | |||
| Total model | 5583.92 | 33.87 | 0.001∗∗ | |||
|
| ||||||
| TFC | Linear | 76.072 | 3.24 | 0.079 | 89.26 | 72.92 |
| Quadratic | 228.990 | 9.75 | 0.016∗ | |||
| Interaction | 20.225 | 0.86 | 0.119 | |||
| Total model | 325.287 | 4.62 | 0.043∗ | |||
|
| ||||||
| TAC | Linear | 69.208 | 4.25 | 0.048∗ | 86.41 | 71.95 |
| Quadratic | 37.531 | 2.94 | 0.078 | |||
| Interaction | 28.690 | 5.42 | 0.031∗ | |||
| Total model | 135.429 | 3.53 | 0.049∗ | |||
|
| ||||||
| S. aureus | Linear | 0.022 | 2.18 | 0.153 | 97.98 | 94.33 |
| Quadratic | 3.561 | 78.07 | 0.000∗∗ | |||
| Interaction | 0.094 | 2.08 | 0.222 | |||
| Total model | 3.678 | 26.88 | 0.001∗∗ | |||
|
| ||||||
| S. enterica | Linear | 0.001 | 1.77 | 0.215 | 97.92 | 94.17 |
| Quadratic | 0.718 | 71.08 | 0.000∗∗ | |||
| Interaction | 0.072 | 7.17 | 0.029∗ | |||
| Total model | 0.809 | 26.13 | 0.001∗∗ | |||
∗ p=0.05; ∗∗p=0.001; ∗∗∗p < 0.001.
3.1.1. Effect of Extraction Variables on Phytochemical Contents
(1) TPC. As shown in Table 1, the TPCs of different experimental runs ranged between 70.2 and 123.8 mg GAE/g. The data obtained from the experiments were used to estimate the coefficients by regression analysis. p values were used to evaluate the significance of different coefficients, which provides the information required to understand the interaction patterns among the experimental variables. p-values less than 0.05 are always accepted as the values for statistical significance with a confidence level greater than 95%, and smaller p values refer to a larger significance of the respective coefficient. As presented in Table 2, the linear (p=0.002), the quadratic (p ≤ 0.001), and interaction (p=0.018) coefficients were significant. The response of activity can be expressed in the following regression equation:
| (4) |
The quadratic term coefficients (X12, X22, and X32) and the interaction coefficients (X1X2, X1X3, and X2X3) have a positive effect while X1, X2, and X3 exhibit a negative effect on the TPC. A longer extraction time presents a negative effect on the TPC. Meanwhile, no influence of this parameter was found, indicating that there is no benefit in using contact times greater than 15 min. Similar effects of time (10–30 min) on TPC pomegranate peel extracted by ultrasonic procedure was observed [33]. On the same line, when the solvent/solid ratio increased from 10 to 30, the extraction efficiency of the TPC did not vary significantly. Dailey et al. in 2015 and Medouni-Adrar et al. in 2015 found that the extraction time and sample-to-solvent ratios have respectively a negative effect on the Macadamia (Macadamia tetraphylla) skin waste and the Algerian skin grape TPCs [50, 51]. The 3-D response surface plot of the regression equation (4) was presented in Figure 1(a). From the graphical representation, it can be concluded that the TPC does not depend on the agitation speed and extraction time. In this regard, the TPC increased with the increase of the solvent/solid ratio from 12 to 18, and then slightly decreased from 19 to 23. With a minimal value of extraction time, the maximum of TPC in the PPE was observed. Likewise, Dailey and Vuong observed that TPC did not change significantly by extraction time (10–30 min) of Macadamia tetraphylla [50]. However, Pinelo et al. showed that the TPC of grape extracts decrease with the increase of the extraction time (30–90 min) [52].
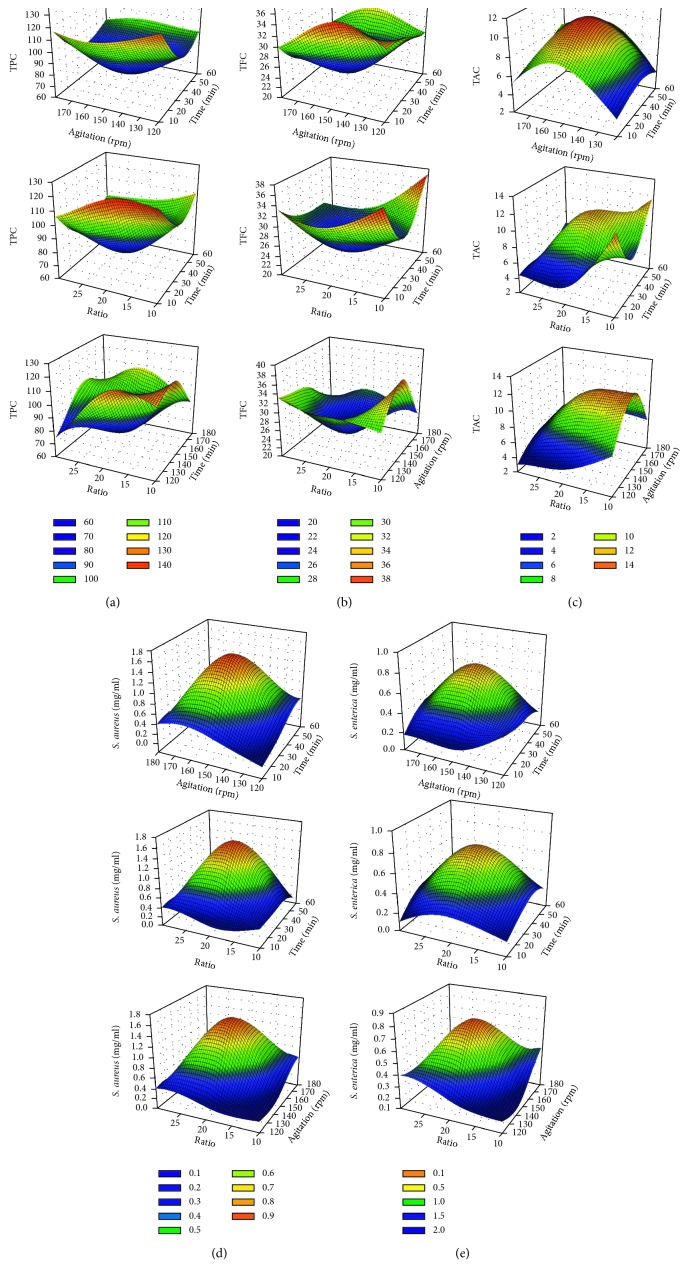
Figure 1.
Three-dimensional presentation of the developed response surface model for the TPC (a), TFC (b), TAC (c), anti-S. aureus (d), and anti-S. enterica (e) activities from pomegranate peel with respect to extraction time, agitation speed, and solvent/solid ratio.
Based on numerical optimization, the predicted models with the maximum TPC was 124.5 mg GAE/g, when the optimal values of test factors were: time of extraction (10 min), agitation speed (135 rpm) and solvent/solid ratio (15) (Table 1, Figure 2(a)). In fact, an increase of 56.224% was shown between the original (70 mg GAE/g) and the optimized TPC (124.5 mg GAE/g).
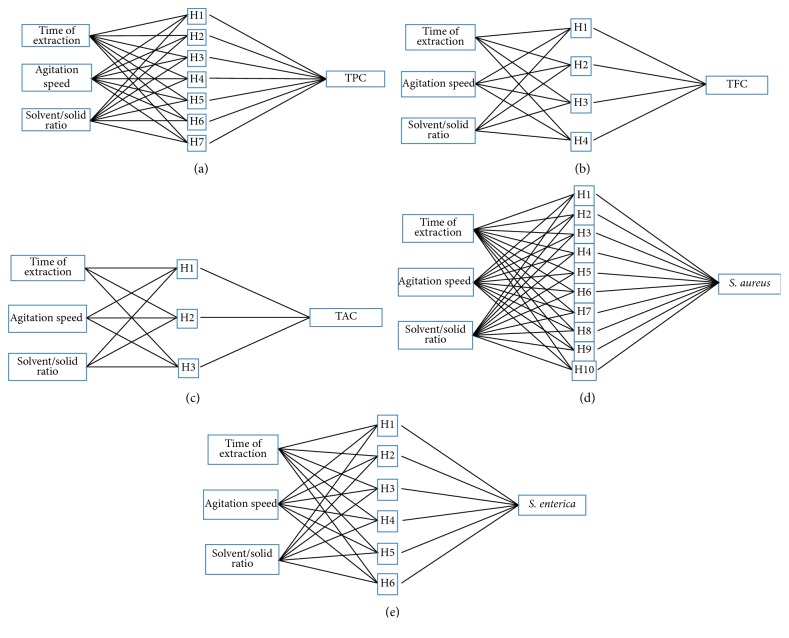
Figure 2.
Graphical representation of the MLP neural network of the TPC (a), TFC (b), TAC (c), anti-S. aureus (d), and anti-S. enterica (e) activities with respect to the extraction time, agitation speed, and solvent/solid ratio.
(2) TFC. The maximum and minimum TFCs were 37.11 and 22.00 mg QE/g, respectively (Table 1). The quadratic models (X12, X22, and X32) have a positive effect on TFC while linear and interaction models of time of extraction and solvent/solid ratio have negative effects. The generalized second-order polynomial equation proposed for TFC is as follows:
| (5) |
Figure 1(b) shows the influence of the three studied parameters on the TFC. In this regard, the increase of agitation speed from 140 rpm to 150 rpm increased the TFC. The increase of extraction time and solvent/solid ratio has a negative effect on the TFC (Figure 1(b)). Dailey and Vuong has demonstrated the negative interaction between the time and ratio on TFC [50]. On the contrary, Sood and Gupta observed that the interaction ((ethanol/pomegranate powder ratio) and (ethanol/extraction time)) has a positive effect on the TFC [34].
Moreover, greater extraction time (X1), having a negative impact on the TPC and TFC, increase the chances of free radicals formation which can be scavenged by phenolic compounds [53, 54]. Equally, plant cells contain enzymes, in particular polyphenol oxidase, capable of altering the phenolic compounds and contribute to enzymatic browning reactions [55]. Furthermore, excessive time is not useful to extract more phenolic compounds [56, 57]. The optimization allowed attaining a maximum TFC of 36.292 mg QE/g with a 63.47% of increase compared with standard conditions. To get this TFC, the time of extraction was set at 8.5 min, agitation speed at 132 rpm, and solvent/solid ratio at 11.25.
(3) TAC. As shown in Table 2, the interaction effect was the most significant (F-value of 5.42, p=0.031), followed by the linear effects (F values of 4.25, p=0.048).
The regression equation showing the mathematical relationship of process variables for the TAC is as follows:
| (6) |
Figure 1(c) showed that the increment in agitation speed had both positive and negative (or dual) effects on the TAC. More precisely, enhancement of the intensity level of the agitation speed up to 130 rpm led to a negligible increase in the TAC. By a further increment of the intensity level >160 rpm, the TAC was decreased. In a fixed extraction time, the TAC can also be increased by the increasing the solvent/solid ratio up to 15.5 (Figure 1(c)).
The optimization allowed as well a maximum TAC of 7.182 mg cy-3-glu/100 g under the following conditions: time of extraction of 35 min, agitation speed of 155 rpm, and solvent/solid ratio of 17. These conditions contributed to a 64.6% increase of TAC in comparison with standard conditions.
3.1.2. Effect of Extraction Variables on the Antibacterial Activity
(1) Anti-S. aureus activity. The anti-S. aureus activity (MIC, Yanti-S. aureus) of PPE, varied from 0.097 to 1.56 mg/ml, was significantly (p < 0.05) affected by the quadratic effects (F values of 78.07, p ≤ 0.001) (Tables 1 and 2). The response of anti-S. aureus activity can be expressed by the following regression equation:
| (7) |
Equation (7) showed that X2 and the interaction coefficients (X1X2 and X1X3) have a positive effect, while, quadratic term coefficients and the single effect of X1 and X3 showed a negative effect on the MICanti-S. aureus activity. The 3D response surfaces based on equation (7) was presented in Figure 1(d): the dark violet color regions in each response surface plot represent the regions where maximum anti–S. aureus activity was observed. At a high agitation speed (180 rpm), and by increasing the extraction time, MIC anti-S. aureus activity was decreased with a maximum at a level ranged between 15 and 20 min. The interaction effect X1X3 showed that the anti-S. aureus activity goes to a maximum activity at 0.2 mg/ml at a high value of extraction time (60 min) and at a minimum level of ratio. While, minimum anti-S. aureus activity was obtained at the middle levels of extraction time (30–40 min), agitation speed (145–155 rpm), and solvent/solid ratio (17–22). At optimized conditions extraction time at 11 min, agitation speed 125 rpm and solvent/solid ratio 12, anti-S. aureus activity remarkably decreased from 1.56 to 0.171 mg/mL.
PPE is widely recognized for its effectiveness against a broad spectrum of bacterial pathogens. Our results are in the same line with the obtained results reported by Dahham et al. and Ismail et al. who studied the antimicrobial effect of PPE against S. aureus [58, 59].
(2) Anti-S. enterica activity. Regression analysis data such as estimated coefficients and p-value were shown in Table 2, and the response of anti-S. enterica activity was predicted by the following regression equation:
| (8) |
All linear and interactions (X1X2 and X1X3) exhibit a positive effect on anti-S. enterica activity, while quadratic term coefficients (X12, X22, and X32) and interaction X2X3 present a negative effect. At a constant ratio of solvent to solid (X3), the increasing of extraction time (X1) to 60 min and a minimal speed agitation (X2) to 120 rpm increases the anti-S. enterica activity (Figure 1(e)). The best conditions to produce the minimal MIC against S. enterica, at 0.555 mg/ml, were resulted in the following responses: time of extraction, 12 min; agitation speed, 129.675 rpm, and solvent/solid ratio; 22.5. These conditions contributed to an increase of 71.19 % in MICanti-S. enterica activity in comparison with standard conditions.
Wafa et al. reported that MICs values of pomegranate ethanolic extract against Salmonella strains ranged between 10.75 and 12.5 mg/mL [19]. Results reported by Choi et al. showed that the MIC values of PPE against Salmonella strains ranged from 0.0625 to 1 mg/mL [60]. Among the tested Salmonella strains, Salmonella typhimurium and Salmonella anatum were found to be more sensitive with an MIC value of 0.25 mg/mL [61]. Different inhibition profile of PPE against microorganisms could be in part due to the different extraction solvents, the plant variety, variations in the geographical, and the climatic conditions [60].
3.2. Artificial Neural Network Prediction on Phytochemical Contents and Antibacterial Activity
ANN has been widely used as a cutting-edge tool for the simulation and optimization of bioactive compounds extraction from different matrices [62, 63]. In this study, ANN was used in combination with an experimental design in order to obtain maximum phytochemical contents with the highest antibacterial activity. Figure 2 showed the structure of the multilayer perceptron ANN model with a back-propagation learning algorithm, designed for the relevant process. The first step of ANN modelling was the optimization of a neural network with the aim of obtaining an ANN model with minimal dimension and minimal errors in training and testing. The design of experiments and their respective experimental yields were used for training the network. The input layer represents vectors constituted of noncoded variables: time of extraction, agitation speed, and solvent/solid ratio, and the output layer represents TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activities. In the ANN structure, the unit number in the hidden layer was set as 7, 4, 3, 10, and 6 for the node number [64, 65]. For TPC, the most suitable model is an MLP trained with 3 independent variables for the input parameters (3-7-1): 3 input neurons, 7 hidden neurons, and 1 output (Figure 2(a)). The results of the trained model were created with high R2 (R2 = 99.88% for test). The topology of the ANN architecture illustrated in Figures 2(b)–2(c), showed a neural structure of (3-4-1) and (3-3-1) respectively for TFC and TAC with R2 = 99.58 and 99.21%, respectively. For anti-S. aureus and S. enterica activities, the robustness network architecture with (3-10-1) and (3-6-1), respectively, were represented in Figures 2(d)–2(e). Furthermore, given R2 were 99.13 and 99.67%, which is totally satisfactory.
The predicted data by ANN model were given along with the RSM predicted and experimental values (Table 1, Figure 3). In fact, for predicted ANN, the relative percent deviation between the phytochemical contents and the antibacterial activities obtained experimentally for the same extraction conditions were respectively: 1.005, 1.003, 1.004, 1.031, and 1.005% for the TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activities.
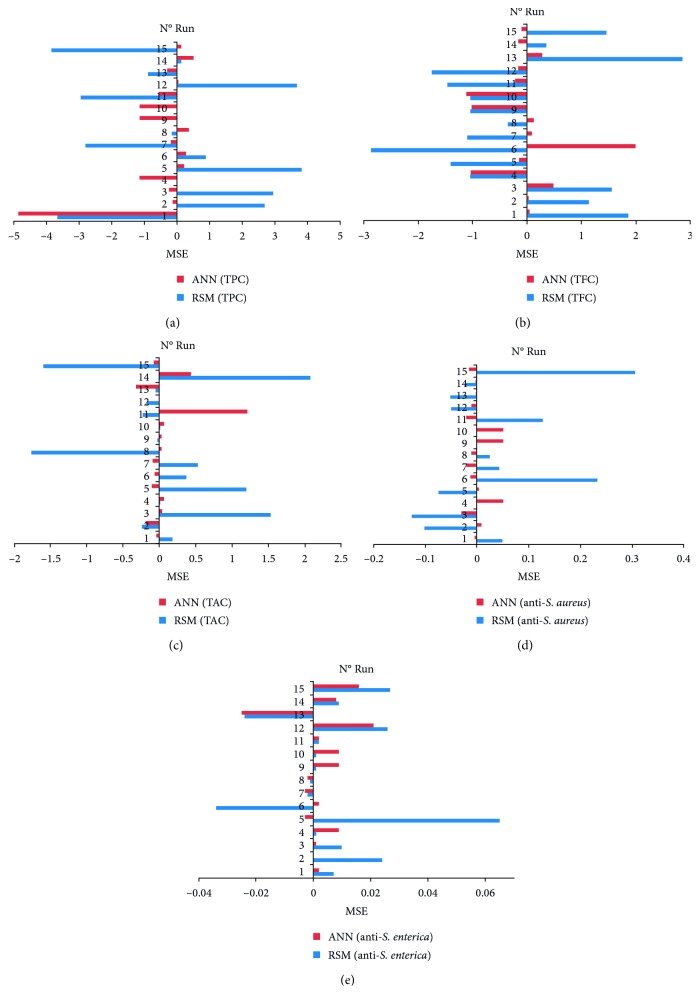
Figure 3.
Comparison of the predictive capacity of RSM and ANN for five model outputs.
3.3. Comparison of RSM and ANN Models
For a comparison of both well trained ANN and RSM models, R2 and mean square error (MSE) were used as statistical indicators (Table 1, Figure 3).
| (9) |
Hence, for phytochemical contents and antibacterial activities, obtained results from RSM models indicated that the measured R2 between the observed and the predicted data were acceptable with R2 > 70%. Furthermore, ANN models showed higher accuracies when compared with RSM models. The comparison between results with predicted outputs from ANN and RSM was shown in Figure 3. ANN model improved the adjustments in comparison with the RSM model. ANN model presents an improvement of 4.4, 26.66, 28, 4.8 and 5.5% in terms of R2 concerning the TPC, TFC, TAC, anti-S. aureus, and anti-S. enterica activities (Figure 3). To illustrate the difference between the mentioned variables, enlarged versions of the simulation output based on the ANN and RSM models are presented in Figure 3. The predictive superiority of the ANN model over the RSM model for data fitting and the estimation capability has already been demonstrated in previous studies [64–68].
3.4. Prediction of the Antibacterial Activity of PPE
To elucidate a better understanding of the PPE as an antibacterial agent, the linear (ANOVA) and general (ANCOVA) models were mathematically used to model the growth rate of S. aureus ATCC 6538 and S. enterica subsp. enterica serovar Typhimurium ATCC 14028. Challenge experiments were conducted to evaluate PPE effects on the growth S. aureus and S. enteric, respectively, in Chapman and XLD media. Therefore, S. aureus and S. enterica kinetics were performed using different concentrations of PPE (MIC, 2 × MIC, 4 × MIC, and 8 × MIC). To investigate the effects of PPE, S. aureus and S. enterica growth were followed during 26 h and evaluated with respect to the control (culture without PPE addition). Moreover, it should be noted that the PPE addition to bacterial cells was realized after 3 hours of incubation when growth reached the beginning of the exponential phase (Cell∼104 CFU/ml).
Viable cell counts log10 (CFU/ml) in absence Control (0MIC) and in presence of 1 × MIC, 2 × MIC, 4 × MI and 8 × MIC of pomegranate peel extracts. The time of extracts addition was 3 hours. ±: Standard deviation of three replicates; Values with a different letter (a–e) within a row of the same time are significantly different (p < 0.05); Values with a different letter (A–L) within a column of the same concentration are significantly different (p < 0.05).
3.4.1. Influence of the PPE on In Vitro S. aureus and S. enterica Inhibition Using Linear Model (ANOVA)
A rapid killing action occurred at 1 hour after addition of all tested MICs (incubation time of 4 hours). For S. aureus, these population numbers were 0.74, 1.02, 1.32, and 1.85 log10 CFU/mL lower (p < 0.05) than the control numbers. Meanwhile, for S. enterica, 1 h post PPE addition, at all tested MICs, PPE showed a reduction (p < 0.05) of 0.24, 0.4, 0.71, and 1.15 log10 CFU/mL compared with control samples (Table 3). At 4 × MIC, no viable cells were observed after 13 and 11h respectively for S. aureus and S. entreica. Interestingly, at 8 × CMI, the early logarithmic growth phase of S. aureus ATCC 6538 resulted in a rapid decrease during 5 hours, while 7 hours were enough to inhibit S. enterica ATCC 14028 (Table 3). These results clearly demonstrated that PPE has bactericidal activity against the Gram-positive (S. aureus ATCC 6538) and the Gram-negative (S. enterica ATCC 14028) bacteria.
Table 3.
Influence of the dose of PPE addition on the growth of Staphylococcus aureus ATCC 6538 and Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028 in vitro using linear model (ANOVA).
| Staphylococcus aureus ATCC 6538 | Salmonella enterica subsp. enterica serovar typhimurium ATCC 14028 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (h) | 0 × MIC | 1 × MIC | 2 × MIC | 4 × MIC | 8 × MIC | Time (h) | 0 × MIC | 1 × MIC | 2 × MIC | 4 × MIC | 8 × MIC |
| 0 | 4.00 ± 0.18aAB | 4.00 ± 0.19aF | 4.00 ± 0.15aH | 4.00 ± 0.17aI | 4.00 ± 0.18aF | 0 | 4.00 ± 0.16aA | 4.00 ± 0.20aI | 4.00 ± 0.16aI | 4.00 ± 0.18aH | 4.00 ± 0.19aG |
| 1 | 4.35 ± 0.21aA | 4.35 ± 0.20aGH | 4.35 ± 0.19aHI | 4.35 ± 0.20aJ | 4.35 ± 0.21aGH | 1 | 4.25 ± 0.17aAB | 4.25 ± 0.18aJ | 4.25 ± 0.20aJ | 4.25 ± 0.17aI | 4.25 ± 0.20aH |
| 2 | 4.80 ± 0.22aAB | 4.80 ± 0.24aHI | 4.80 ± 0.22aJ | 4.80 ± 0.19aK | 4.80 ± 0.18aHI | 2 | 4.50 ± 0.21aB | 4.50 ± 0.22aK | 4.50 ± 0.19aK | 4.50 ± 0.21aK | 4.50 ± 0.22aI |
| 3 | 5.02 ± 0.24aAB | 5.02 ± 0.22aI | 5.02 ± 0.25aK | 5.02 ± 0.20aL | 5.02 ± 0.23aI | 3 | 4.93 ± 0.24aC | 4.93 ± 0.23aL | 4.93 ± 0.24aM | 4.93 ± 0.21aL | 4.93 ± 0.24aJ |
| 4 | 5.34 ± 0.25eAB | 4.63 ± 0.22dH | 4.35 ± 0.19cI | 4.05 ± 0.19bI | 3.52 ± 0.16aE | 4 | 5.15 ± 0.24dC | 4.91 ± 0.22cdL | 4.75 ± 0.22cL | 4.44 ± 0.21bK | 4.00 ± 0.19aG |
| 5 | 6.57 ± 0.32eB | 4.60 ± 0.22dH | 4.20 ± 0.20cHI | 3.81 ± 0.180bH | 3.01 ± 0.14aD | 5 | 5.80 ± 0.27dD | 4.80 ± 0.23cL | 4.54 ± 0.21bcK | 4.21 ± 0.00bI | 3.22 ± 0.00aF |
| 6 | 7.09 ± 0.34eBC | 4.50 ± 0.21dH | 4.15 ± 0.19cH | 3.70 ± 0.17bH | 2.51 ± 0.12aC | 6 | 6.02 ± 0.28dE | 4.50 ± 0.21cK | 4.22 ± 0.20bcIJ | 4.00 ± 0.18bH | 2.55 ± 0.12aE |
| 7 | 8.25 ± 0.39eBC | 4.24 ± 0.18dG | 3.60 ± 0.17cG | 3.22 ± 0.15bG | 1.89 ± 0.09aB | 7 | 6.15 ± 0.29dEF | 4.30 ± 0.19cJ | 3.81 ± 0.18bH | 3.58 ± 0.17bG | 2.00 ± 0.08aD |
| 8 | 8.52 ± 0.16eBC | 3.91 ± 0.18dF | 3.44 ± 0.16cF | 3.00 ± 0.14bF | 1.00 ± 0.00aA | 8 | 6.25 ± 0.29eF | 4.00 ± 0.19dI | 3.45 ± 0.16cG | 3.00 ± 0.12bF | 1.75 ± 0.07aC |
| 9 | 9.02 ± 0.43eC | 3.65 ± 0.17dE | 3.00 ± 0.13cE | 2.61 ± 0.12bE | 1.00 ± 0.00aA | 9 | 6.45 ± 0.30dFG | 3.51 ± 0.16cH | 3.33 ± 0.15cG | 2.54 ± 0.12bE | 1.33 ± 0.06aB |
| 10 | 9.32 ± 0.44dCD | 3.11 ± 0.14cD | 2.74 ± 0.13bD | 2.42 ± 0.11bD | 1.00 ± 0.00aA | 10 | 6.55 ± 0.31dG | 3.26 ± 0.15cG | 3.00 ± 0.14cF | 2.11 ± 0.10bD | 1.00 ± 0.00aA |
| 11 | 9.56 ± 0.45eCD | 2.77 ± 0.13dC | 2.41 ± 0.11cC | 2.00 ± 0.09bC | 1.00 ± 0.00aA | 11 | 6.74 ± 0.32eGH | 3.04 ± 0.15dF | 2.71 ± 0.13cE | 1.75 ± 0.08bC | 1.00 ± 0.00aA |
| 12 | 9.80 ± 0.47eCD | 2.40 ± 0.11dBC | 1.90 ± 0.09cB | 1.55 ± 0.07bB | 1.00 ± 0.00aA | 12 | 6.85 ± 0.33eH | 2.75 ± 0.14dE | 2.42 ± 0.11cD | 1.44 ± 0.06bB | 1.00 ± 0.00aA |
| 14 | 10.52 ± 0.50dD | 2.35 ± 0.11cBC | 1.44 ± 0.07bAB | 1.22 ± 0.05abAB | 1.00 ± 0.00aA | 14 | 6.98 ± 0.33dH | 2.22 ± 0.10cD | 1.90 ± 0.09bC | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 16 | 10.77 ± 0.51dCD | 2.25 ± 0.10cB | 1.20 ± 0.05bAB | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 16 | 7.02 ± 0.30dH | 1.75 ± 0.07cC | 1.51 ± 0.07bB | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 18 | 11.02 ± 0.52cCD | 2.14 ± 0.10bAB | 1.10 ± 0.03aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 18 | 7.12 ± 0.29dHI | 1.66 ± 0.07cBC | 1.25 ± 0.06bAB | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 20 | 11.25 ± 0.50cD | 2.00 ± 0.07bAB | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 20 | 7.25 ± 0.33cHI | 1.44 ± 0.06bB | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 22 | 11.36 ± 0.54cD | 1.74 ± 0.08bAB | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 22 | 7.35 ± 0.33dJ | 1.25 ± 0.05bAB | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 24 | 11.50 ± 0.39cD | 1.25 ± 0.07bA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 24 | 7.53 ± 0.34bJ | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
| 26 | 11.55 ± 0.55cD | 1.00 ± 0.06bA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 26 | 7.55 ± 0.36bJ | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA | 1.00 ± 0.00aA |
Table 4 shows the overall differences between 5 trials for each bacterium at all tested MICs (1 × MIC, 2 × MIC, 4 × MIC and 8 × MIC) and 20 different times. For one fixed time, significant differences (p < 0.001) were observed between treatments trials and times (Trial × Time). The five treatment groups significantly increased the anti-S. aureus (p < 0.001) and anti-S. enterica (p < 0.001) activities. Indeed, this result confirmed the previous results showed in Table 3 where PPE exerts dose-dependent bactericidal effects.
Table 4.
Statistics test of main and interaction effects of univariate mixed analyses of variance for all dependent variables and effects (Trial and Time) with 95% confidence intervals.
| Outcome | Effect | Level number | Parameter number | F | dfnum/dfden | p value |
|---|---|---|---|---|---|---|
| Fixed trial | ||||||
| Anti-S. aureus activity | Intercept | 1 | 1 | 206.094∗ | 1/60 | <0.001 |
| Time | 20 | 19 | 0.657 | 19/60 | 0.845 | |
| Trial | 1 | 1 | 66.187∗ | 1/60 | <0.001 | |
| Time × trial | 20 | 19 | 1.291 | 19/60 | 0.224 | |
| Anti-S. enterica activity | Intercept | 1 | 1 | 66.171 | 1/60 | 1.000 |
| Time | 20 | 19 | 0.240 | 19/60 | 0.412 | |
| Trial | 1 | 1 | 84.122∗ | 1/60 | <0.001 | |
| Time × trial | 20 | 19 | 1.540 | 19/60 | 0.104 | |
|
| ||||||
| Fixed time | ||||||
| Anti-S. aureus activity | Intercept | 1 | 1 | 1226.229∗ | 1/90 | <0.001 |
| Trial | 1 | 1 | 46.654∗ | 1/90 | <0.001 | |
| Time | 5 | 4 | 3.616∗ | 4/90 | 0.009 | |
| Trial × time | 5 | 4 | 95.230∗ | 4/90 | <0.001 | |
| Anti-S. enterica activity | Intercept | 1 | 1 | 387.703∗ | 1/90 | <0.001 |
| Trial | 1 | 1 | 21.093∗ | 1/90 | <0.001 | |
| Time | 5 | 4 | 1.228 | 4/90 | 0.305 | |
| Trial × time | 5 | 4 | 15.225∗ | 4/90 | <0.001 | |
dfnum/dfden = degrees of freedom of numerator/denominator.
3.4.2. Influence of the PPE on In Vitro S. aureus and S. enterica Inhibition Using General Linear Model (ANCOVA)
A descriptive statistics using a mixed model of the time-related survival of S. aureus and S. enterica following treatment with various PPE concentrations was presented in Table 5. p-value < 0.05 was always accepted value for statistical significance with confidence level >95% and smaller p values refer to a larger significance of the respective coefficient [69, 70]. As shown in Table 5, for S. aureus, a significant effect (p < 0.05) was noticed at 0, 1, 2, 3, 4, and 5 hours. In fact, in the beginning of experimentation, the first 2 hours after PPE addition were primordial for S. aureus inhibition. Concerning S. enterica inhibition, 3 hours of post PPE addition was effective. In contrast, no significant differences (p > 0.05) were found between groups at any time point 2 and 3 hours post PPE addition, respectively, on S. aureus and S. enterica inhibition (Table 5).
Table 5.
Analyses of covariance for all dependent variables and effect of S. aureus ATCC 6538 and S. enterica ATCC 14028 activities.
| Parameter | Staphylococcus aureus ATCC 6538 | Salmonella enterica subsp. enterica serovar typhimurium ATCC 14028 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | df | t | Sig. | LB | UB | Estimate | SE | df | t | Sig. | LB | UB | |
| Intercept | 9.44 | 2.432 | 60 | 3.88 | 0.000∗∗∗ | 4.573 | 14.306 | 6320 | 3203.043 | 0 | 1.973 | 1.000 | −79955.35 | 92595.35 |
| Hour 0 | −5.44 | 3.44 | 60 | −1.581 | 0.119 | −12.321 | 1.441 | −6316 | 4529.787 | 0 | −1.394 | 1.000 | −128327.77 | 115695.77 |
| Hour 1 | −5.089 | 3.44 | 60 | −1.479 | 0.144 | −11.97 | 1.792 | −2070 | 4529.787 | 0 | −0.457 | 1.000 | −124081.77 | 119941.77 |
| Hour 2 | −4.64 | 3.44 | 60 | −1.349 | 0.182 | −11.521 | 2.241 | −1820 | 4529.787 | 0 | −0.402 | 1.000 | −123831.77 | 120191.77 |
| Hour 3 | −4.42 | 3.44 | 60 | −1.285 | 0.204 | −11.301 | 2.461 | −1420 | 4529.787 | 0 | −0.313 | 1.000 | −123431.77 | 120591.77 |
| Hour 4 | −3.859 | 3.44 | 60 | −1.122 | 0.266 | −10.74 | 3.022 | −844 | 4529.787 | 0 | −0.186 | 1.000 | −122855.77 | 121167.77 |
| Hour 5 | −2.623 | 3.44 | 60 | −0.763 | 0.449 | −9.504 | 4.258 | −89 | 4529.787 | 0 | −0.020 | 1.000 | −122100.77 | 121922.77 |
| Hour 6 | −2.079 | 3.44 | 60 | −0.605 | 0.548 | −8.961 | 4.801 | 166 | 4529.787 | 0 | 0.037 | 1.000 | −121845.77 | 122177.77 |
| Hour 7 | −1.248 | 3.44 | 60 | −0.363 | 0.718 | −8.129 | 5.633 | 352 | 4529.787 | 0 | 0.078 | 1.000 | −121659.77 | 122363.77 |
| Hour 8 | −1.018 | 3.44 | 60 | −0.296 | 0.768 | −7.9 | 5.863 | 348 | 4529.787 | 0 | 0.077 | 1.000 | −121663.77 | 122359.77 |
| Hour 9 | −0.672 | 3.44 | 60 | −0.196 | 0.846 | −7.554 | 6.208 | 422.8 | 4529.787 | 0 | 0.093 | 1.000 | −121588.97 | 122434.57 |
| Hour 10 | −0.51 | 3.44 | 60 | −0.148 | 0.882 | −7.392 | 6.37 | 500 | 4529.787 | 0 | 0.110 | 1.000 | −121511.77 | 122511.77 |
| Hour 11 | −0.712 | 3.44 | 60 | −0.207 | 0.837 | −7.593 | 6.169 | 491 | 4529.787 | 0 | 0.108 | 1.000 | −121520.77 | 122502.77 |
| Hour 12 | −0.665 | 3.44 | 60 | −0.193 | 0.847 | −7.546 | 6.216 | 511 | 4529.787 | 0 | 0.113 | 1.000 | −121500.77 | 122522.77 |
| Hour 14 | −0.181 | 3.44 | 60 | −0.053 | 0.958 | −7.062 | 6.7 | 304 | 4529.787 | 0 | 0.067 | 1.000 | −121707.77 | 122315.77 |
| Hour 16 | −0.029 | 3.44 | 60 | −0.008 | 0.993 | −6.91 | 6.852 | 11 | 4529.787 | 0 | 0.002 | 1.000 | −122000.77 | 122022.77 |
| Hour 18 | 0.096 | 3.44 | 60 | 0.028 | 0.978 | −6.785 | 6.977 | −44 | 4529.787 | 0 | −0.010 | 1.000 | −122055.77 | 121967.77 |
| Hour 20 | 0.205 | 3.44 | 60 | 0.060 | 0.953 | −6.676 | 7.086 | −70 | 4529.787 | 0 | −0.015 | 1.000 | −122081.77 | 121941.77 |
| Hour 22 | 0.183 | 3.44 | 60 | 0.053 | 0.958 | −6.698 | 7.064 | −190 | 4529.787 | 0 | −0.042 | 1.000 | −122201.77 | 121821.77 |
| Hour 24 | 0.087 | 3.44 | 60 | 0.025 | 0.980 | −6.794 | 6.968 | −120 | 4529.787 | 0 | −0.026 | 1.000 | −122131.77 | 121891.77 |
| Hour 26 | 0a | 0 | 60 | — | — | — | 0a | 0 | — | — | — | — | — | |
| Trial | −2.11 | 0.733 | 60 | −2.877 | 0.006∗∗ | −3.577 | −0.642 | −1330 | 422.878 | 60 | −3.145 | 0.003∗∗ | −2175.882 | −484.117 |
| Hour 0 × trial | 2.11 | 1.037 | 60 | 2.034 | 0.016∗ | 0.035 | 4.184 | 1330 | 598.04 | 60 | 2.224 | 0.009∗ | 133.741 | 2526.258 |
| Hour 1 × trial | 2.11 | 1.037 | 60 | 2.034 | 0.022∗ | 0.035 | 4.184 | 1330 | 598.04 | 60 | 2.224 | 0.011∗ | 133.741 | 2526.25 |
| Hour 2 × trial | 2.11 | 1.037 | 60 | 2.034 | 0.029∗ | 0.035 | 4.184 | 1330 | 598.04 | 60 | 2.224 | 0.015∗ | 133.741 | 2526.258 |
| Hour 3 × trial | 2.11 | 1.037 | 60 | 2.034 | 0.034∗ | 0.035 | 4.184 | 1330 | 598.04 | 60 | 2.224 | 0.030∗ | 133.741 | 2526.258 |
| Hour 4 × trial | 1.691 | 1.037 | 60 | 1.631 | 0.044 | −0.3835 | 3.766 | 1054 | 598.04 | 60 | 1.762 | 0.039 | −142.258 | 2250.258 |
| Hour 5 × trial | 1.326 | 1.037 | 60 | 1.279 | 0.046 | −0.7486 | 3.401 | 755 | 598.04 | 60 | 1.262 | 0.044 | −441.258 | 1951.258 |
| Hour 6 × trial | 1.13 | 1.037 | 60 | 1.090 | 0.206 | −0.9447 | 3.205 | 586 | 598.04 | 60 | 0.980 | 0..049 | −610.258 | 1782.258 |
| Hour 7 × trial | 0.775 | 1.037 | 60 | 0.748 | 0.208 | −1.299 | 2.85 | 428 | 598.04 | 60 | 0.716 | 0.077 | −768.258 | 1624.258 |
| Hour 8 × trial | 0.574 | 1.037 | 60 | 0.554 | 0.481 | −1.5 | 2.649 | 352 | 598.04 | 60 | 0.589 | 0.258 | −844.258 | 1548.258 |
| Hour 9 × trial | 0.476 | 1.037 | 60 | 0.459 | 0.548 | −1.598 | 2.551 | 256.6 | 598.04 | 60 | 0.429 | 0.369 | −939.658 | 1452.858 |
| Hour 10 × trial | 0.376 | 1.037 | 60 | 0.363 | 0.718 | −1.698 | 2.451 | 144 | 598.04 | 60 | 0.241 | 0.511 | −1052.258 | 1340.258 |
| Hour 11 × trial | 0.358 | 1.037 | 60 | 0.345 | 0.731 | −1.716 | 2.432 | 115 | 598.04 | 60 | 0.192 | 0.748 | −1081.258 | 1311.258 |
| Hour 12 × trial | 0.275 | 1.037 | 60 | 0.265 | 0.792 | −1.799 | 2.349 | 29 | 598.04 | 60 | 0.048 | 0.861 | −1167.258 | 1225.258 |
| Hour 14 × trial | 0.111 | 1.037 | 60 | 0.107 | 0.915 | −1.96 | 2.185 | 14 | 598.04 | 60 | 0.023 | 0.981 | −1182.258 | 1210.2584 |
| Hour 16 × trial | 0.045 | 1.037 | 60 | 0.043 | 0.966 | −2.029 | 2.119 | 51 | 598.04 | 60 | 0.085 | 0.932 | −1145.258 | 1247.258 |
| Hour 18 × trial | 0.006 | 1.037 | 60 | 0.006 | 0.995 | −2.068 | 2.08 | 40 | 598.04 | 60 | 0.067 | 0.947 | −1156.258 | 1236.258 |
| Hour 20 × trial | −0.029 | 1.037 | 60 | −0.028 | 0.978 | −2.103 | 2.045 | 30 | 598.04 | 60 | 0.050 | 0.960 | −1166.258 | 1226.258 |
| Hour 22 × trial | −0.029 | 1.037 | 60 | −0.028 | 0.978 | −2.103 | 2.045 | 50 | 598.04 | 60 | 0.084 | 0.934 | −1146.258 | 1246.258 |
| Hour 24 × trial | −0.015 | 1.037 | 60 | −0.015 | 0.988 | 0–2.09 | 2.059 | 30 | 598.04 | 60 | 0.050 | 0.960 | −1166.258 | 1226.258 |
| Hour 26 × trial | 0a | 0 | — | — | — | — | — | 0a | 0 | — | — | — | — | — |
SE : Standard Error; df: Degrees of freedom; t: the Student t-statistic; Sig.: the p value (associated with the correlation); LB : Lower bound. UB : Upper bound; Trial 1: negative control; Trial 2 : 1 × MIC. Trial 3 : 2 × MIC; Trial 4 : 4 × MIC and Trial 5 : 8 × MIC; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, aThis parameter is set to zero because it is redundant;b Dependent variable: CFU.
The effects of treatments, time, and their interaction on the inhibition of S. aureus and S. enterica were shown in Table 6. A significant interaction (p < 0.05) between all treatments samples and bacterial growth time was remarked. With the lowest p value, 8 × MICs (Trial 5), and incubation was found highly significant (p < 0.01) effect regarding inhibition of S. aureus and S. enterica (Table 6). It is obvious that high concentrations of 1.368 mg/mL against S. aureus) and (4.44 mg/mL against S. enterica) coupled with time have an active action on inhibition of studied bacteria.
Table 6.
S. aureus ATCC 6538 and S. enterica ATCC 14028 behavior estimates of incubation time (hour) fixed effects with 95% confidence intervalsb.
| Parameter | Staphylococcus aureus ATCC 6538 | Salmonella enterica subsp. enterica serovar typhimurium ATCC 14028 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | df | t | Sig. | LB | UB | Estimate | SE | df | t | Sig. | LB | UB | |
| Intercept | 3.632 | 0.289 | 90 | 12.565 | 0.000∗∗∗ | 3.058 | 4.206 | 3133.002 | 452.338 | 90 | 6.926 | 0.000∗∗∗ | 2234.353 | 4031.651 |
| Time | −0.144 | 0.021 | 90 | −6.68 | 0.000∗∗∗ | −0.187 | −0.101 | 1077.153 | 639.702 | 90 | 1.684 | 0.000∗∗∗ | −193.727 | 2348.035 |
| Trial 1 | 1.455 | 0.408 | 90 | 3.56 | 0.807 | 0.643 | 2.267 | 1263.938 | 639.702 | 90 | 1.976 | 0.639 | −6.942 | 2534.819 |
| Trial 2 | 1.121 | 0.408 | 90 | 2.743 | 0.306 | 0.309 | 1.933 | 1091.692 | 639.702 | 90 | 1.707 | 0.632 | −179.188 | 2362.573 |
| Trial 3 | 1.084 | 0.408 | 90 | 2.652 | 0.430 | 0.272 | 1.896 | 818.011 | 639.702 | 90 | 1.279 | 0.595 | −452.869 | 2088.892 |
| Trial 4 | 0.8132 | 0.408 | 90 | 1.989 | — | 0.000 | 1.625 | 0a | 0 | — | — | — | — | — |
| Trial 5 | 0a | 0 | — | — | — | — | −110.793 | 33.894 | 90 | −3.269 | 0.002∗∗ | −178.131 | −43.456 | |
| Trial 1 × time | 0.456 | 0.03 | 90 | 14.885 | 0.049 | 0.395 | 0.516 | 277.036 | 47.934 | 90 | 5.780 | 0.006 | 181.806 | 372.266 |
| Trial 2 × time | −0.007 | 0.03 | 90 | −0.245 | 0.009∗ | −0.068 | 0.053 | −22.531 | 47.934 | 90 | −0.470 | 0.050 | −117.76 | 72.698 |
| Trial 3 × time | −0.031 | 0.03 | 90 | −1.03 | 0.007∗∗ | −0.092 | 0.029 | −23.058 | 47.934 | 90 | −0.481 | 0.041 | −118.288 | 72.17 |
| Trial 4 × time | −0.0242 | 0.03 | 90 | −0.793 | 0.004∗ | −0.085 | 0.036 | −25.565 | 47.934 | 90 | −0.533 | 0.031 | −120.795 | 69.664 |
| Trial 5 × time | 0a | 0 | — | — | 0.001 | — | — | 0a | 0 | — | — | 0.010 | — | — |
SE : Standard error; df: degrees of freedom; t: the student t-statistic; Sig.: the p value (associated with the correlation); LB : Lower bound. UB : Upper bound; Trial 1: negative control; Trial 2 : 1 × MIC. Trial 3 : 2 × MIC; Trial 4 : 4 × MIC and Trial 5 : 8 × MIC; ∗p < 0.05; ∗p < 0.01; ∗∗∗p < 0.001, aThis parameter is set to zero because it is redundant; bDependent variable: CFU.
The covariance parameters are presented in Table 7(A). Intercept variances are estimated as 5.379 and 1.788 for S. aureus and S. enterica, respectively. The null hypothesis for this parameter is a variance of zero, which would indicate that a random effect is not needed, and the statistical test is called a Wald Z statistic [71]. For S. aureus and S. enterica, the two hypotheses Wald Z = 0.000 where p=0.015; for anti-S. aureus activity and p=0.018; for anti-S. enterica activity was accepted (Table 7(A)). Equally, as illustrated in Table 7(B) for fixed time, the two hypotheses Wald Z = 0.815, p=0.038 and Wald Z = 0.000, p=0.031 as well as Wald Z = 0.000, p=0.029 were accepted. Furthermore, results from Table 7 showed that interactions of the trial type and incubation time have an important role in inhibition of S. aureus and S. enterica.
Table 7.
Estimates of covariance parameters in S. aureus ATCC 6538 and S. enterica ATCC 14028 behavior estimates of incubation time (A) and trial (B) fixed effecta.
| Staphylococcus aureus ATCC 6538 | Salmonella enterica subsp. enterica serovar typhimurium ATCC 14028 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | Wald Z | Sig. | LB | UB | Estimate | SE | Wald Z | Sig. | LB | UB | ||
| A trial fixed | Residual | 5.379 | 0.982239 | 5.477 | 0.000 | 3.761580 | 7.694 | 1.788 | 0.326 | 5.477 | 0.000 | 1.25 | 2.557 |
| Time [subject = id] variance | 0.000a | 0.000 | 0.000 | 0.015 | — | — | 0.000a | 0.000 | 0.000 | 0.018 | — | — | |
|
| |||||||||||||
| B time fixed | Residual | 0.278 | 0.082 | 3.354 | 0.001 | 0.155110 | 0.499 | 0.278 | 0.082955 | 3.354 | 0.001 | 0.155 | 0.499 |
| Trial [subject = id] variance | 0.278a | 0.000 | 0.815 | 0.038 | — | — | 0.2780a | 0.000000 | 0.000 | 0.029 | — | — | |
aDependent variable: CFU.
4. Conclusion
Recent years have witnessed an indisputable interest in plants for exploring and promoting their bioactive compounds in the health field. In this study, the use of pomegranate peel as a natural alternative source to produce biological compounds, phytochemical contents (TPC, TFC, and TAC), and anti-S. aureus and anti-S. enterica activities, has been evaluated and optimized. Both RSM and ANN have been used to generate the model on all five responses from PPE. The extraction time, agitation speed, and solvent/solid ratio of pomegranate peel markedly influenced phytochemical contents and antibacterial activity. Under optimal conditions, TPC (124.5 mg GAE/g), TFC (36.292 mg QE/g), TAC (7.182 mg cy-3-glu/100 g), MICanti-S. aureus (0.171 mg/mL), and MICanti-S. enterica (0.555 mg/mL) were maximal. Equally, this work highlights the great anti-S. aureus and anti-S. enterica effects of PPE at optimized conditions. The mode of action study confirms our results and indicates that PPE exerts a dose-dependent bactericidal effect against these foodborne bacterial pathogens. Therefore, PPE can be considered as a strong and promising tool for future application as a safe method for the preservation of food products.
Acknowledgments
This research was funded by the Tunisian Ministry of Higher Education and Scientific Research (Program contract 2015–2018 of the Laboratory of Microorganisms and Biomolecules (LR15CBS05) of the Center of Biotechnology of Sfax, Tunisia).
Data Availability
All data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors have declared that there are no conflicts of interest.
References
- 1.Ferronato N., Rada E. C., Gorritty Portillo M. A., Cioca L. I., Ragazzi M., Torretta V. Introduction of the circular economy within developing regions: a comparative analysis of advantages and opportunities for waste valorization. Journal of Environmental Management. 2019;230:366–378. doi: 10.1016/j.jenvman.2018.09.095. [DOI] [PubMed] [Google Scholar]
- 2.Padeyanda Y., Jang Y.-C., Ko Y., Yi S. Evaluation of environmental impacts of food waste management by material flow analysis (MFA) and life cycle assessment (LCA) Journal of Material Cycles and Waste Management. 2016;18(3):493–508. doi: 10.1007/s10163-016-0510-3. [DOI] [Google Scholar]
- 3.Ververi M., Goula A. M. Pomegranate peel and orange juice by-product as new biosorbents of phenolic compounds from olive mill wastewaters. Chemical Engineering and Processing - Process Intensification. 2019;138:86–96. doi: 10.1016/j.cep.2019.03.010. [DOI] [Google Scholar]
- 4.Turrini F., Donno D., Boggia R., et al. An innovative green extraction and re-use strategy to valorize food supplement by-products: Castanea sativa bud preparations as case study. Food Research International. 2019;115:276–282. doi: 10.1016/j.foodres.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Caballero E., Soto C. Biorefinery. Cham, Switzerland: Springer; 2019. Valorization of agro-industrial waste into bioactive compounds: techno-economic considerations; pp. 235–252. [DOI] [Google Scholar]
- 6.Nag S., Sit N. Optimization of ultrasound assisted enzymatic extraction of polyphenols from pomegranate peels based on phytochemical content and antioxidant property. Journal of Food Measurement and Characterization. 2018;12(3):1734–1743. doi: 10.1007/s11694-018-9788-2. [DOI] [Google Scholar]
- 7.Lima M. C., Paiva de Sousa C., Fernandez-Prada C., Harel J., Dubreuil J. D., de Souza E. L. A review of the current evidence of fruit phenolic compounds as potential antimicrobials against pathogenic bacteria. Microbial Pathogenesis. 2019;130:259–270. doi: 10.1016/j.micpath.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 8.El‐Hadary A. E., Ramadan M. F. Phenolic profiles, antihyperglycemic, antihyperlipidemic, and antioxidant properties of pomegranate (Punica granatum) peel extract. Journal of Food Biochemistry. 2019;43 doi: 10.1111/jfbc.12803.e12803 [DOI] [PubMed] [Google Scholar]
- 9.Russo M., Cacciola F., Arena K., et al. Characterization of the polyphenolic fraction of pomegranate samples by comprehensive two-dimensional liquid chromatography coupled to mass spectrometry detection. Natural Product Research. 2019 doi: 10.1080/14786419.2018.1561690. In press. [DOI] [PubMed] [Google Scholar]
- 10.Russo M., Fanali C., Tripodo G., et al. Analysis of phenolic compounds in different parts of pomegranate (Punica granatum) fruit by HPLC-PDA-ESI/MS and evaluation of their antioxidant activity: application to different Italian varieties. Analytical and Bioanalytical Chemistry. 410:3507–3520. doi: 10.1007/s00216-018-0854-8. [DOI] [PubMed] [Google Scholar]
- 11.Sadeghipour A., Eidi M., Ilchizadeh Kavgani A., Ghahramani R., Shahabzadeh S., Anissian A. Lipid lowering effect of Punica granatum L. peel in high lipid diet fed male rats. Evidence-Based Complementary and Alternative Medicine. 2014;2014:5. doi: 10.1155/2014/432650.432650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forouzanfar F., Afkhami Goli A., Asadpour E., Ghorbani A., Sadeghnia H. R. Protective effect of Punica granatum L. against serum/glucose deprivation-induced PC12 cells injury. Evidence-Based Complementary and Alternative Medicine. 2013;2013:p. 9. doi: 10.1155/2013/716730.716730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh B., Singh J. P., Kaur A., Singh N. Phenolic compounds as beneficial phytochemicals in pomegranate (Punica granatum L.) peel: a review. Food Chemistry. 2018;261:75–86. doi: 10.1016/j.foodchem.2018.04.039. [DOI] [PubMed] [Google Scholar]
- 14.Mastrogiovanni F., Bernini R., Basiricò L., et al. Antioxidant and anti-inflammatory effects of pomegranate peel extracts on bovine mammary epithelial cells BME-UV1. Natural Product Research. 2019:1–5. doi: 10.1080/14786419.2018.1508149. In press. [DOI] [PubMed] [Google Scholar]
- 15.Zhao S.-S., Ma D.-X., Zhu Y., et al. Antidiarrheal effect of bioactivity-guided fractions and bioactive components of pomegranate (Punica granatum L.) peels. Neurogastroenterology and Motility. 2018;30(7) doi: 10.1111/nmo.13364.e13364 [DOI] [PubMed] [Google Scholar]
- 16.Di Stefano V., Pitonzo R., Novara M. E., et al. Antioxidant activity and phenolic composition in pomegranate (Punica granatum L.) genotypes from south Italy by UHPLC-Orbitrap-MS approach. Journal of the Science of Food and Agriculture. 2019;99(3):1038–1045. doi: 10.1002/jsfa.9270. [DOI] [PubMed] [Google Scholar]
- 17.Alexandre E. M. C., Silva S., Santos S. A. O., et al. Antimicrobial activity of pomegranate peel extracts performed by high pressure and enzymatic assisted extraction. Food Research International. 2019;115:167–176. doi: 10.1016/j.foodres.2018.08.044. [DOI] [PubMed] [Google Scholar]
- 18.Indurkar S. J., Rathod V. K. Aqueous two-phase extraction of punicalagin (α+β) from pomegranate peel by response surface methodology. Separation Science and Technology. 2019;54(1):51–58. doi: 10.1080/01496395.2018.1488866. [DOI] [Google Scholar]
- 19.Wafa B. A., Makni M., Ammar S., et al. Antimicrobial effect of the Tunisian Nana variety Punica granatum L. extracts against Salmonella enterica (serovars Kentucky and Enteritidis) isolated from chicken meat and phenolic composition of its peel extract. International Journal of Food Microbiology. 2017;241:123–131. doi: 10.1016/j.ijfoodmicro.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Singh B., Singh J. P., Kaur A., Singh N. Antimicrobial potential of pomegranate peel: a review. International Journal of Food Science and Technology. 2019;54(4):959–965. doi: 10.1111/ijfs.13964. [DOI] [Google Scholar]
- 21.Bouarab Chibane L., Degraeve P., Ferhout H., Bouajila J., Oulahal N. Plant antimicrobial polyphenols as potential natural food preservatives. Journal of the Science of Food and Agriculture. 2019;99(4):1457–1474. doi: 10.1002/jsfa.9357. [DOI] [PubMed] [Google Scholar]
- 22.Al-Zoreky N. S. Antimicrobial activity of pomegranate (Punica granatum L.) fruit peels. International Journal of Food Microbiology. 2019;134:244–248. doi: 10.1016/j.ijfoodmicro.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Smaoui S., Hlima H. B., Mtibaa A. C., et al. Pomegranate peel as phenolic compounds source: advanced analytical strategies and practical use in meat products. Meat Science. 2019;158:107914–107933. doi: 10.1016/j.meatsci.2019.107914. [DOI] [PubMed] [Google Scholar]
- 24.Trigo J. P., Alexandre E. M., Saraiva J. A., Pintado M. E. High value-added compounds from fruit and vegetable by-products–characterization, bioactivities, and application in the development of novel food products. Critical Reviews of Food Science and Nutrition. 2019 doi: 10.1080/10408398.2019.1572588. In press. [DOI] [PubMed] [Google Scholar]
- 25.Skenderidis P., Mitsagga C., Giavasis I., et al. The in vitro antimicrobial activity assessment of ultrasound assisted Lycium barbarum fruit extracts and pomegranate fruit peels. Journal of Food Measurement and Characterization. 2019;13(3):2017–2031. doi: 10.1007/s11694-019-00123-6. [DOI] [Google Scholar]
- 26.Venkataramanamma D., Aruna P., Singh R. P. Standardization of the conditions for extraction of polyphenols from pomegranate peel. Journal of Food Science and Technology. 2016;53(5):2497–2503. doi: 10.1007/s13197-016-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R. P., Chidambara Murthy K. N., Jayaprakasha G. K. Studies on the antioxidant activity of pomegranate (punicagranatum) peel and seed extracts using in vitro models. Journal of Agricultural and Food Chemistry. 2002;50(1):81–86. doi: 10.1021/jf010865b. [DOI] [PubMed] [Google Scholar]
- 28.Negi P. S., Jayaprakasha G. K., Jena B. S. Antioxidant and antimutagenic activities of pomegranate peel extracts. Food Chemistry. 2003;80(3):393–397. doi: 10.1016/s0308-8146(02)00279-0. [DOI] [Google Scholar]
- 29.Yasoubi P., Barzegar M., Sahari M. A., Azizi M. H. Total phenolic contents and antioxidant activity of pomegranate (Punica granatum L.) peel extracts. Journal of Agricultural Science and Technology. 2016;9:35–42. [Google Scholar]
- 30.Viuda-Martos M., Ruiz-Navajas Y., Fernández-López J., Sendra E., Sayas-Barberá E., Pérez-Álvarez J. A. Antioxidant properties of pomegranate (Punica granatum L.) bagasses obtained as co-product in the juice extraction. Food Research International. 2011;44(5):1217–1223. doi: 10.1016/j.foodres.2010.10.057. [DOI] [Google Scholar]
- 31.Pan Z., Qu W., Ma H., Atungulu G. G., McHugh T. H. Continuous and pulsed ultrasound-assisted extractions of antioxidants from pomegranate peel. Ultrasonics Sonochemistry. 2012;19(2):365–372. doi: 10.1016/j.ultsonch.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Bartnik D. D., Mohler C. M., Houlihan M. Methods for the production of food grade extracts. 2006. US Patent 20060088627.
- 33.Tabaraki R., Heidarizadi E., Benvidi A. Optimization of ultrasonic-assisted extraction of pomegranate (Punica granatum L.) peel antioxidants by response surface methodology. Separation and Purification Technology. 2012;98:16–23. doi: 10.1016/j.seppur.2012.06.038. [DOI] [Google Scholar]
- 34.Sood A., Gupta M. Extraction process optimization for bioactive compounds in pomegranate peel. Food Bioscience. 2015;12:100–106. doi: 10.1016/j.fbio.2015.09.004. [DOI] [Google Scholar]
- 35.Amdoun R., Benyoussef E.-H., Benamghar A., Khelifi L. Prediction of hyoscyamine content in Datura stramonium L. hairy roots using different modeling approaches: response Surface Methodology (RSM), Artificial Neural Network (ANN) and Kriging. Biochemical Engineering Journal. 2019;144:8–17. doi: 10.1016/j.bej.2019.01.002. [DOI] [Google Scholar]
- 36.Alara O. R., Abdurahman N. H., Afolabi H. K., Olalere O. A. Efficient extraction of antioxidants from Vernonia cinerea leaves: comparing response surface methodology and artificial neural network. Beni-Suef University Journal of Basic and Applied Sciences. 2018;7(3):276–285. doi: 10.1016/j.bjbas.2018.03.007. [DOI] [Google Scholar]
- 37.Yuan W., Yuk H.-G. Antimicrobial efficacy of Syzygium antisepticum plant extract against Staphylococcus aureus and methicillin-resistant S. aureus and its application potential with cooked chicken. Food Microbiology. 2018;72:176–184. doi: 10.1016/j.fm.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Garre A., Egea J. A., Esnoz A., Palop A., Fernandez P. S. Tail or artefact? Illustration of the impact that uncertainty of the serial dilution and cell enumeration methods has on microbial inactivation. Food Research International. 2019;119:76–83. doi: 10.1016/j.foodres.2019.01.059. [DOI] [PubMed] [Google Scholar]
- 39.Possas A., Pérez-Rodríguez F., Valero A., Rincón F., García-Gimeno R. M. Mathematical approach for the Listeria monocytogenes inactivation during high hydrostatic pressure processing of a simulated meat medium. Innovative Food Science and Emerging Technologies. 2018;47:271–278. doi: 10.1016/j.ifset.2018.03.012. [DOI] [Google Scholar]
- 40.Singleton V. L., Rossi J. A. Colorimetry of total phenolics with phosphomolybdic phosphotungstic acid reagents. American Journal of Enology and Viticulture. 1965;16:144–158. [Google Scholar]
- 41.Quettier-Deleu C., Gressier B., Vasseur J., et al. Phenolic compounds and antioxidant activities of buckwheat (Fagopyrum esculentum Moench) hulls and flour. Journal of Ethnopharmacology. 2000;72(1-2):35–42. doi: 10.1016/s0378-8741(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 42.Çam M., Hışıl Y., Durmaz G. Classification of eight pomegranate juices based on antioxidant capacity measured by four methods. Food Chemistry. 2019;112:721–726. [Google Scholar]
- 43.Smaoui S., Hsouna A. B., Lahmar A., et al. Bio-preservative effect of the essential oil of the endemic Mentha piperita used alone and in combination with BacTN635 in stored minced beef meat. Meat Science. 2016;117:196–204. doi: 10.1016/j.meatsci.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Gulluce M., Sahin F., Sokmen M., et al. Antimicrobial and antioxidant properties of the essential oils and methanol extract from Mentha longifolia L. ssp. Longifolia. Food Chemistry. 2007;103(4):1449–1456. doi: 10.1016/j.foodchem.2006.10.061. [DOI] [Google Scholar]
- 45.Box G. E. P., Behnken D. W. Some new three level designs for the study of quantitative variables. Technometrics. 1960;2(4):455–475. doi: 10.1080/00401706.1960.10489912. [DOI] [Google Scholar]
- 46.Senthilkumar T., Chattopadhyay S. K., Miranda L. R. Optimization of activated carbon preparation from pomegranate peel (punica granatumPeel) using RSM. Chemical Engineering Communications. 2017;204(2):238–248. doi: 10.1080/00986445.2016.1262358. [DOI] [Google Scholar]
- 47.Simić V. M., Rajković K. M., Stojičević S. S., Veličković D. T., Nikolić N. Č., Lazić I. T. Optimization of microwave-assisted extraction of total polyphenolic compounds from chokeberries by response surface methodology and artificial neural network. Separation Science and Technology. 2016;160:89–97. [Google Scholar]
- 48.Kline R. B. Principles and Practice of Structural Equation Modeling. New York, NY, USA: Guilford publications; 2015. [Google Scholar]
- 49.Malviya S., Arvind A., Jha A., Hettiarachchy N. Antioxidant and antibacterial potential of pomegranate peel extracts. Journal of Food Science and Technology. 2014;51(12):4132–4137. doi: 10.1007/s13197-013-0956-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dailey A., Vuong Q. Optimization of aqueous extraction conditions for recovery of phenolic content and antioxidant properties from Macadamia (Macadamia tetraphylla) skin waste. Antioxidants. 2015;4(4):699–718. doi: 10.3390/antiox4040699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medouni-Adrar S., Boulekbache-Makhlouf L., Cadot Y., et al. Optimization of the recovery of phenolic compounds from Algerian grape by-products. Industrial Crops and Products. 2015;77:123–132. doi: 10.1016/j.indcrop.2015.08.039. [DOI] [Google Scholar]
- 52.Pinelo M., Rubilar M., Jerez M., Sineiro J., Núñez M. J. Effect of solvent, temperature, and solvent-to-solid ratio on the total phenolic content and antiradical activity of extracts from different components of grape pomace. Journal of Agricultural and Food Chemistry. 2005;53(6):2111–2117. doi: 10.1021/jf0488110. [DOI] [PubMed] [Google Scholar]
- 53.Naczk M., Shahidi F. Extraction and analysis of phenolics in food. Journal of Chromatography A. 2004;1054(1-2):95–111. doi: 10.1016/s0021-9673(04)01409-8. [DOI] [PubMed] [Google Scholar]
- 54.Irakli M., Chatzopoulou P., Ekateriniadou L. Optimization of ultrasound-assisted extraction of phenolic compounds: oleuropein, phenolic acids, phenolic alcohols and flavonoids from olive leaves and evaluation of its antioxidant activities. Industrial Crops and Products. 2018;124:382–388. doi: 10.1016/j.indcrop.2018.07.070. [DOI] [Google Scholar]
- 55.Singh B., Suri K., Shevkani K., Kaur A., Kaur A., Singh N. Enzymatic browning of fruit and vegetables: a Review. Enzymes in Food Technology. 2018;4:63–78. doi: 10.1007/978-981-13-1933-4_4. [DOI] [Google Scholar]
- 56.Silva E., Rogez H., Larondelle Y. Optimization of extraction of phenolics from Inga edulis leaves using response surface methodology. Separation and Purification Technology. 2007;55(3):381–387. doi: 10.1016/j.seppur.2007.01.008. [DOI] [Google Scholar]
- 57.Wissam Z., Ghada B., Wassim A., Warid K. Effective extraction of polyphenols and proanthocyanidins from pomegranate’s peel. International Journal of Pharmacy and Pharmaceutical Sciences. 2012;4:675–682. [Google Scholar]
- 58.Dahham S. S., Ali M. N., Tabassum H., Khan M. Studies on antibacterial and antifungal activity of pomegranate (Punica granatum L.) American-eurasian Journal Of Agricultural and Environmental Sciences. 2010;9:273–281. [Google Scholar]
- 59.Ismail T., Akhtar S., Sestili P., Riaz M., Ismail A., Labbe R. G. Antioxidant, antimicrobial and urease inhibitory activities of phenolics-rich pomegranate peel hydro-alcoholic extracts. Journal of Food Biochemistry. 2016;40(4):550–558. doi: 10.1111/jfbc.12250. [DOI] [Google Scholar]
- 60.Choi J.-G., Kang O.-H., Lee Y.-S., et al. In vitro and in vivo antibacterial activity of Punica granatum Peel ethanol extract against Salmonella. Evidence-Based Complementary and Alternative Medicine. 2011;2011:1–8. doi: 10.1093/ecam/nep105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayouni E. A., Miled K., Boubaker S., et al. Hydroalcoholic extract based-ointment from Punica granatum L. peels with enhanced in vivo healing potential on dermal wounds. Phytomedicine. 2011;18(11):976–984. doi: 10.1016/j.phymed.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 62.Uysal S., Cvetanović A., Zengin G., Zeković Z., Mahomoodally M. F., Bera O. Optimization of maceration conditions for improving the extraction of phenolic compounds and antioxidant effects of Momordica Charantia L. leaves through response surface methodology (RSM) and artificial neural networks (ANNs) Analytical Letters. 2019;52(13):2150–2163. doi: 10.1080/00032719.2019.1599007. [DOI] [Google Scholar]
- 63.Anastácio A., Silva R., Carvalho I. S. Phenolics extraction from sweet potato peels: modelling and optimization by response surface modelling and artificial neural network. Journal of Food Science and Technology. 2016;53(12):4117–4125. doi: 10.1007/s13197-016-2354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smaoui S., Ennouri K., Chakchouk-Mtibaa A., et al. Modeling-based optimization approaches for the development of Anti- Agrobacterium tumefaciens activity using Streptomyces sp TN71. Microbial Pathogenesis. 2018;119:19–27. doi: 10.1016/j.micpath.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Smaoui S., Ennouri K., Chakchouk-Mtibaa A., et al. Statistical versus artificial intelligence -based modeling for the optimization of antifungal activity against Fusarium oxysporum using Streptomyces sp. strain TN71. Journal de Mycologie Médicale. 2018;28(3):551–560. doi: 10.1016/j.mycmed.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 66.Ennouri K., Ayed R. B., Mazzarello M., Ottaviani E., Hertelli F., Azzouz H. Classical and bayesian predictions applied to Bacillus toxin production. 3 Biotech. 2016;6:206–212. doi: 10.1007/s13205-016-0527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iglesias-Carres L., Mas-Capdevila A., Bravo F. I., Aragonès G., Muguerza B., Arola-Arnal A. Optimization of a polyphenol extraction method for sweet orange pulp (Citrus sinensis L.) to identify phenolic compounds consumed from sweet oranges. PLoS One. 2019;14(1):p. e0211267. doi: 10.1371/journal.pone.0211267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Şahin S., Samli R., Tan A. S. B., et al. Solvent-free microwave-assisted extraction of polyphenols from olive tree leaves: antioxidant and antimicrobial properties. Molecules. 2017;22:1056–1068. doi: 10.3390/molecules22071056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braïek O. B., Smaoui S., Ennouri K., Morandi S., Cremonesi P., Hani K. RAPD-PCR characterisation of two Enterococcus lactis strains and their potential on Listeria monocytogenes growth behaviour in stored chicken breast meats: generalised linear mixed-effects approaches. LWT- Food Science and Technology. 2019;99:244–253. [Google Scholar]
- 70.Mhalla D., Bouaziz A., Ennouri K., et al. Antimicrobial activity and bioguided fractionation of Rumex tingitanus extracts for meat preservation. Meat Science. 2017;125:22–29. doi: 10.1016/j.meatsci.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 71.Meyers L. S., Gamst G., Guarina A. J. Applied Multivariate Research: Design and Interpretation. Thousand Oaks, CA, USA: Sage; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used to support the findings of this study are included within the article.