Abstract
Kinetochores connect mitotic-spindle microtubules with chromosomes, allowing microtubule depolymerization to pull chromosomes apart during anaphase while resisting detachment as the microtubule shortens. The DASH/Dam1 heterodecamer (DASH/Dam1c), an essential component of yeast kinetochores, assembles into a microtubule-encircling ring. The ring associates with rod-like Ndc80 complexes to organize the kinetochore-microtubule interface. We report the cryo-EM structure (at ~4.5 Å resolution) of a DASH/Dam1c ring and a molecular model of its ordered components, validated by evolutionary direct-coupling analysis. Integrating this structure with that of the Ndc80 complex and with published interaction data yields a molecular picture of kinetochore-microtubule attachment, including how flexible, C-terminal extensions of DASH/Dam1c subunits project and contact widely separated sites on the Ndc80c rod and how phosphorylation at previously identified sites might regulate kinetochore assembly.
One Sentence Summary:
A cryo-EM derived structure of the DASH/Dam1 complex suggests how microtubule depolymerization drives chromosome separation.
Kinetochore linkages between chromosomes and spindle microtubules (MTs) persist despite rapid gain and loss of tubulin subunits during cell division (1). The capacity to follow cycles of polymerization and depolymerization without dissociation is an important and puzzling property of the kinetochore-MT interface. The puzzle is particularly evident for “point-centromere” kinetochores of budding yeast, each of which connects a single MT with a small (~125–200 bp) and well-defined centromere (2, 3). Although motor proteins participate during an initial, side-on attachment to MTs (4), the energy for chromosome movement during anaphase comes from the GTPase activity of tubulin and the polymerization-depolymerization events it governs (5–7).
Stable association with the plus end of a MT while the latter lengthens and shortens depends most directly on two essential molecular components of the budding-yeast kinetochore -- the heterotetrameric Ndc80 complex (Ndc80c) and the heterodecameric DASH/Dam1 complex (DASH/Dam1c). Ndc80c is an end-to-end, rod-like assembly of two heterodimers (Ndc80:Nuf2 and Spc24:Spc25), with a ~580 Å long, coiled-coil shaft and small, globular domains at either end (8, 9). It establishes the principal kinetochore axis, by associating with the MT at its Ndc80:Nuf2 end and with chromosome-proximal substructures at its Spc24:Spc25 end. A single kinetochore contains multiple copies of Ndc80c; the most recent estimates from quantitative fluorescence microscopy and biochemistry are in the range 6 to 10 (10–12).
DASH/Dam1c is a more compact structure that assembles into a ring encircling the MT, organizing the Ndc80c rods and potentially other kinetochore components (13, 14). Ring formation under physiological conditions in vitro by recombinant DASH/Dam1c requires MTs, and at high enough concentrations, DASH/Dam1c rings (or helical spirals) decorate the entire MT length. The abundance of the ten subunits in yeast cells indicates that kinetochores in vivo have no more than one or two DASH/Dam1c rings (Table S1) (15). DASH/Dam1c and Ndc80c interact with each other in addition to their separate interactions with MTs (16). Specific phosphorylation regulates most of these contacts, suggesting that a DASH/Dam1c:Ndc80c assembly controls both the timing of end-on attachment and the mechanical coupling of chromosome separation to MT shortening (17, 18). DASH/Dam1c is not present in most metazoans, which instead have the three-protein Ska complex (19). The two complexes appear to have analogous functions (20–22).
Multiple sequence alignments and secondary-structure predictions (23) allowed us to identify likely α-helical regions in the ten DASH/Dam1c subunits and to make corresponding truncations in all of them (Table S2). We prepared multi-gene expression vectors containing these truncated subunits, individually and in various combinations (see Materials and Methods, Fig. S1) and analyzed integrity of the expressed complexes by affinity purification and gel electrophoresis (Fig. S2A, B). We did so for nine fungal species in addition to S. cerevisiae (Table S3, Fig. S2C). A 115 kDa Chaetomium thermophilum DASH/Dam1c construct, comprising the ten subunits, crystallized as hexagonal rods, but Bragg peaks extended only to very low resolution (60–40 Å) (Fig. S2D–J). The apparent unit cell dimensions (Table S4) suggested that the crystallized DASH/Dam1c had formed rings or helical spirals, like those observed when it has assembled around MTs (13, 24, 25). Dynamic light scattering (DLS) and negative-stain EM showed that C. thermophilum DASH/Dam1c assembled into tubes upon incubation with increasing concentrations of monovalent salts (e.g. sodium formate or ammonium formate), consistent with the high-salt conditions that gave the crystals.
For cryo-EM, we locked assembled DASH/Dam1c with bifunctional crosslinkers in high-salt buffer and purified the tubes by size exclusion chromatography in low salt (Fig. S3). The tubes are one-dimensional crystals formed by alternate stacking of DASH/Dam1c rings with 17 protomers (C17 symmetry, Fig. S4). Poor long-range order limited the resolution of an initial helical reconstruction (Fig. S5). We therefore carried out a symmetric reconstruction at 6.5 Å resolution of individual DASH/Dam1c double rings, by masking images and reference volumes (Fig. S6). At this stage, we validated our reconstruction by tilt-pair analysis, which also established the correct enantiomer (Fig. S7). We then independently refined sets of four adjacent protomers in a ring (“asymmetric refinement” in FREALIGN), improving the nominal resolution to 4.5 Å (Fig. S6). Three-dimensional classification did not yield better maps, presumably because selecting particles from stacks had eliminated bad ones and because the rings in a stack have a continuum of small structural distortions or variations.
We built a de novo DASH/Dam1c molecular model into the asymmetric reconstruction. We could obtain a reliable model because the structure consists mostly of α-helices, which are well resolved at 4.5 Å resolution and for which more than one large side chain can establish correct sequence register for the rest of the helix (Fig. S8). After an initial manual build, we used RosettaCM (26) to find low energy conformations that fit the density map and then refined the structure with PHENIX (27). We validated the model by predicting molecular contacts within DASH/Dam1c by direct coupling analysis (DCA) of residue co-evolution (28). For this purpose, we identified and annotated all DASH/Dam1c genes from about 1300 fungal genomes as input for the DCA (see Materials and Methods). Fig. S9 shows residue pairs with the highest coupling scores mapped onto our molecular model. The analysis confirmed the subunit assignment and sequence register. DCA has successfully predicted folded protein structure (29); its application to validating residue assignments is likely to be a powerful tool for interpreting cryo-EM maps, when a suitably large and diverse set of sequences is available. The final model contains amino-acid residues for Ask1 (13–78), Dad1 (18–76), Dad2 (25–95, 109–116), Dad3 (18–82), Dad4 (3–70), Dam1 (53–107), Duo1 (49–121), Hsk3 (22–77), Spc19 (7–112) and Spc34 (3–48, 112–199). Data collection and model statistics are in Table S5.
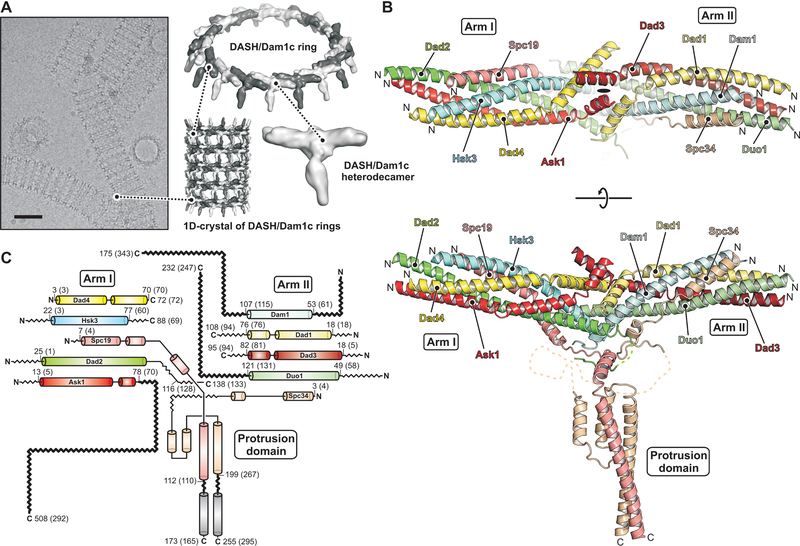
DASH/Dam1c is a 160 Å long, rod-shaped complex, with two coiled-coil arms that merge in the middle, where a protruding domain extends roughly perpendicular to the arms (Fig. 1). The overall shape agrees well with a published, low-resolution envelope of the S. cerevisiae complex (24). Both coiled-coil domains are parallel five-helix bundles, with the N termini of the subunits at the distal ends of the arms, generally consistent with previous assignments (30). Arm I contains Ask1, Dad2, Dad4, Hsk3 and the N-proximal segment of Spc19; arm II, Dad3, Duo1, Dad1, Dam1 and the N-proximal segment of Spc34. The subunits of the two five-helix bundles interdigitate in the middle of the complex, forming what was called at low resolution a “central domain”. The C-terminal halves of Spc19 and Spc34 form the base and extension of the protrusion domain. Part of the base, formed by Spc34, is not α-helical, less conserved, and likely flexible. We did not attempt to model it (Fig. S10A).
Fig. 1. Cryo-EM structure of the DASH/Dam1c heterodecamer.
(A) Electron cryo-micrograph of DASH/Dam1c helical tubes (1D-crystals) in vitreous ice recorded at 1.9 μm defocus. The scale bar corresponds to 50 nm. 1D-crystals formed by sequential stacking of DASH/Dam1c rings, composed of 17 heterodecamers. (B) Ribbon diagram of the DASH/Dam1c structure with subunits colored: Ask1 (red), Dad2 (green), Dad4 (yellow), Hsk3 (cyan), Spc19 (salmon), Dad3 (firebrick), Duo1 (palegreen), Dad1 (paleyellow), Dam1 (palecyan) and Spc34 (wheat). On top, the view of the complex is along the approximate two-fold symmetry axis. (C) Secondary structure diagram. The same color scheme is used as in (B). α helices are shown as cylinders. First and last residue numbers included in the model, are shown (numbers in parenthesis are the corresponding residues in Saccharomyces cerevisiae). C-terminal residue numbers of all subunits are shown. α helices not modeled in the structure, but inferred from secondary structure prediction, are shown in gray. Zig-zag lines show flexible loops, N- and C-terminal extensions that were not included in the expression construct or observed in the cryo-EM map (functionally important termini of Dam1, Duo1, Ask1 and Spc19:Spc34 are in bold). The description of a previous low-resolution structure called Arm I the “top rod” and Arm II, the “bottom rod” (24).
A layer of buried hydrophobic amino-acid side chains lines the core of the central domain (Fig. S10B), and hydrophobic residues also line the interiors of the two arms. The structure shows that removal of any of the ten subunits would expose hydrophobic patches and probably induce degradation of DASH/Dam1c in the cell. Indeed, omitting Hsk3 from co-expression disrupts arm I, leading to dissociation of Ask1, Dad2 and Dad4, and leaving a hexameric complex of the remaining subunits. Similarly, omitting Dam1 disrupts arm II so that Duo1, Dad1 and Dad3 no longer associate with the complex (31).
Apart from the protrusion domain, the DASH/Dam1c heterodecamer has approximate internal two-fold symmetry relating the left and right arms (Fig. 1B). Each subunit in one arm has a corresponding subunit in the other arm, with similar organization and three-dimensional conformation. Sequence analysis has shown homology between some DASH/Dam1c subunits, (20). Our structure shows that each subunit has a structural paralog, and we can assign the five pairs as Ask1:Dad3, Dad2:Duo1, Dad4:Dad1, Hsk3:Dam1 and Spc19:Spc34. Contemporary DASH/Dam1c may thus have evolved from one or more gene duplication events.
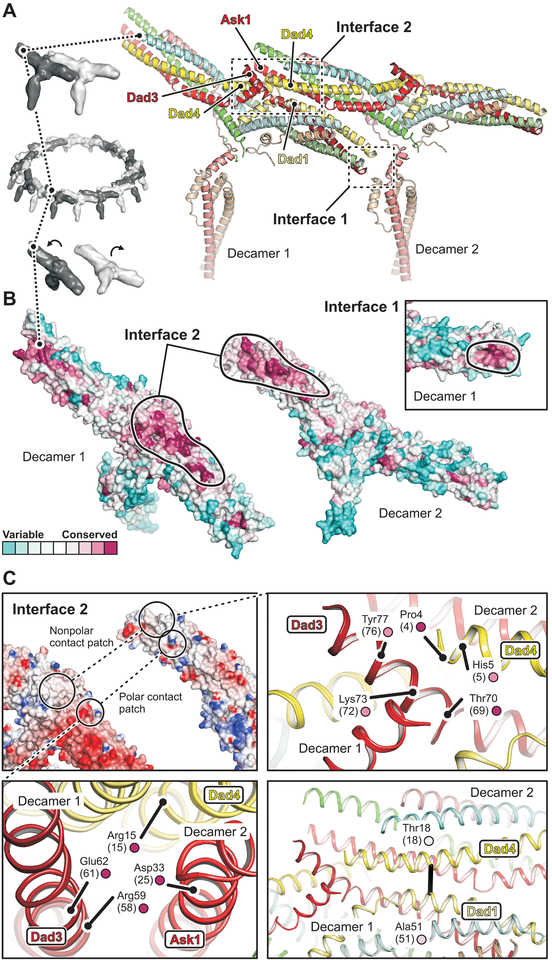
The stacking interactions between rings in our in vitro assembled tubes are probably adventitious contacts resulting from the high protein and high salt concentrations that allow tube formation and crystallization. There is good evidence, however, that the lateral interactions between protomers within a ring, is the physiological mechanism of DASH/Dam1c oligomerization. (i) The same relative heterodecamer orientation (Fig. 2A) is present in a low-resolution reconstruction of a S. cerevisiae DASH/Dam1c dimer (24). (ii) The diameter of the ring is very similar to that of the ring formed by S. cerevisiae DASH/Dam1c (25) and consistent with the requirement that it surround a MT. (iii) Surface amino-acid residues involved in the lateral inter-complex interactions are conserved (Fig. 2B). (iv) We detect coevolution of residues in the inter-complex interface (Fig. 2C, bottom right).
Fig. 2. Structural basis of DASH/Dam1c oligomerization.
(A) Close-up view of two adjacent protomers (labeled decamer 1 and 2) within the DASH/Dam1c ring. Interfaces 1 and 2 are boxed. The subunits are colored as in Fig. 1. (B) Surface residue conservation of DASH/Dam1c in the fungal kingdom. Conservation scores were calculated with ConSurf (46) and mapped onto our structure. The protomers are flipped open, exposing interface 2. Black lines surround the inter-complex binding footprints. The inset shows a close-up view of interface 1 for decamer 1 only. (C) Interface 2 has patches of polar and hydrophobic contacts. Top left, surface representation of the two protomers colored according to the electrostatic potential, calculated with DelPhi (47). Bottom left, close-up view of the polar contact patch. Relevant amino-acids are labeled with residue numbers (S. cerevisiae numbers in parenthesis) and conservation score (small circle, color as in B). Top right, close-up view of the hydrophobic patch. Bottom right, interface molecular contact inferred from DCA of residue coevolution shown as a black line.
There are two interfaces between DASH/Dam1c heterodecamers within the lateral, interprotomer contacts. At interface 2, arm I of one complex (decamer 2) binds the central domain and proximal parts of arm II of an adjacent complex (decamer 1). At interface 1, the base of the protrusion domain of decamer 2 engages the arm II tip of an adjacent decamer 1 (Fig. 2A). From our multiple amino-acid sequence alignments, we calculated DASH/Dam1c residue conservation scores within the fungal kingdom. Fig. 2B shows that the highest conservation scores of surface residues overlap with the footprints of the oligomerization interfaces. At interface 2 (Fig. 2C, top left), we find a tight cluster of charged residues where Ask1 (Asp33) and Dad4 (Arg15) of decamer 2 face Dad3 (Arg59, Glu62) of decamer 1 (Fig. 2C, bottom left); these four residues have the highest conservation scores. Moreover, conserved Pro4 at the tip of the Dad4 helix (decamer 2) contacts a patch of hydrophobic residues (Thr70, Lys73, Ala74, Tyr77) at the end of the Dad3 helix (decamer 1) (Fig. 2C, top right). Because we could not model part of the base of the protrusion domain, folded from Spc34 sequence, we could not analyze specific interactions at interface 1.
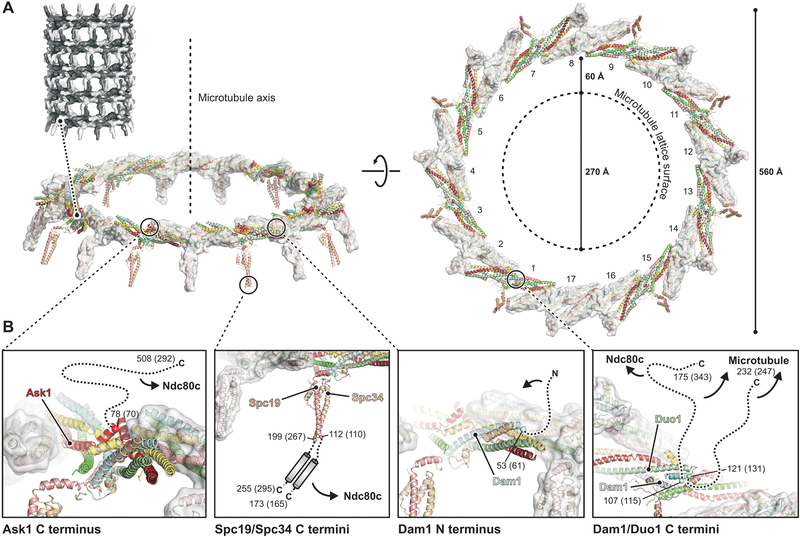
Under our experimental conditions, 17 C. thermophilum DASH/Dam1c protomers polymerized into a ring with outer diameter of 560 Å, about twice that of a MT (Fig. 3A). The ring has one protomer more than the 16-membered rings reported for the S. cerevisiae complex (25). Insertion of an additional DASH/Dam1 complex does not substantially change the diameter of the ring (Fig. S11A), which could vary among species. Even within a species, there may be some variation from ring to ring in the number of protomers. When encircling a MT, the gap between the MT lattice surface and DASH/Dam1c is about 60 Å. Oligomerization interfaces 1 and 2 determine the relative orientation of adjacent protomers; ring closure defines their orientation with respect to the MT.
Fig. 3. The DASH/Dam1c ring with flexibly projecting extensions.
(A) Structure of the 17-membered ring. Heterodecamers are show as ribbons and colored as in Fig. 1. Protomers are alternately shown with transparent surface. The MT axis (side view, left) and lattice surface (top view, right) are shown as dashed lines. (B) Close-up views of the anchor points on the ring, where flexibly projecting extension are attached. Cα atoms of terminal amino acids modeled in the cryo-EM map are shown as spheres with residue numbers (S. cerevisiae numbers in parenthesis). Flexible extensions are drawn schematically as dashed lines, except for Spc19/Spc34 (center left panel), for which we indicate a C-terminal heterodimeric coiled coil based on secondary structure prediction and molecular contacts inferred from DCA of residue coevolution.
Near the N terminus of Dam1, phosphorylation of Ser20 (in S. cerevisiae) by Ipl1/AuroraB inhibits cooperative binding of DASH/Dam1c to MTs (17, 32). The location of the Dam1 N- terminus, on the outside of the ring (Fig. 3B, center right), suggests that Ser20 phosphorylation could modulate DASH/Dam1c oligomerization either by destabilizing an interaction with a neighboring protomer or by stabilizing an autoinhibitory interaction within the same protomer. The phosphorylation site is conserved among budding yeast (Data S1), and the length of the Dam1 N-terminal extension could accommodate either option.
Flexible polypeptide extensions that link partner protein complexes are a general feature of kinetochore architecture (33, 34). Apart from ring formation, in which DASH/Dam1c cores interact directly, contacts with both MT and Ndc80c are through subunit extensions predicted to be conformationally variable (Fig. 1C, Data S1). Although these extensions were either omitted from our construct or undefined (owing to flexibility) in the cryo-EM map, the structure of the DASH/Dam1c core and its orientation with respect to the MT together determine the directions in which the flexible extensions project and hence their likely contacts.
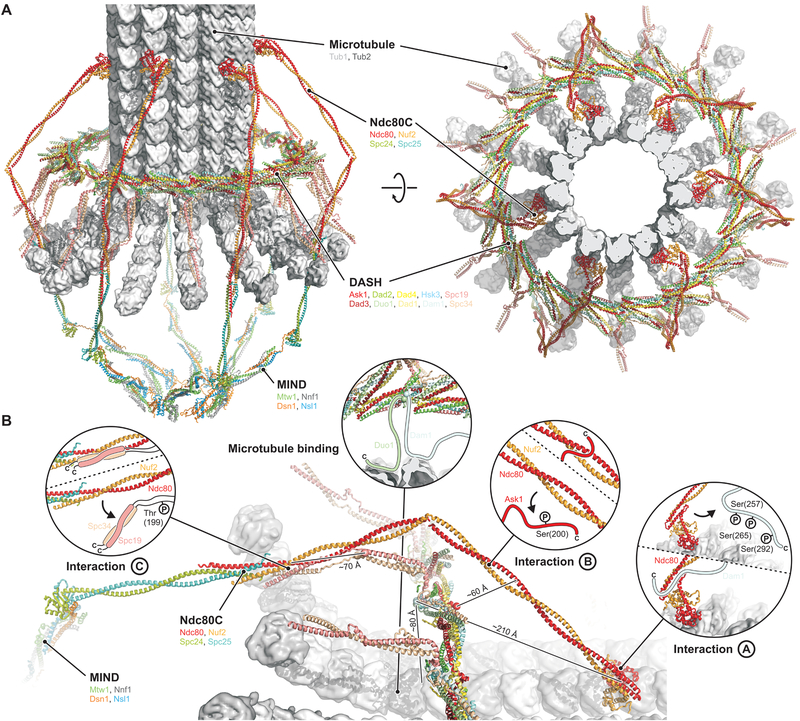
The structure of the DASH/Dam1c ring also suggests how to interpret three sets of contact points, detected by chemical crosslinking of Ndc80c with Dam1, Ask1 and Spc34, respectively (35), leading to the interactions illustrated by arrows in Fig. 3B and to the model for the MT-proximal components of the yeast kinetochore shown in Fig 4. The lengths of the C-terminal extensions of Dam1, Ask1, and Spc34 and the directions in which they project might permit all three Ndc80c contacts to be established with a single DASH/Dam1c ring (Fig. 4B). The molecular organization of this assembly is also consistent with the assumption that a DASH/Dam1c ring encircles a MT plus-end with curled protofilaments, as observed in tomographic reconstructions of kinetochore MTs (36).
Fig. 4. Model of the yeast kinetochore-MT interface.
(A) Overview from the side (left) and top (right). (B) Close-up view and interactions of DASH/Dam1c with the MT and Ndc80c. Approximate distances between the attachments of the flexible extensions on the DASH/Dam1c ring and their target sites, defined by crosslinking, are shown. Interactions A, B and C are between Ndc80c and DASH/Dam1 subunits Dam1, Ask1 and Spc34:Spc19, respectively. S. cerevisiae residues known to be phosphorylated by Ipl1/Aurora B are shown in the insets, and arrows indicate release of Ndc80c binding upon phosphorylation.
The C-terminal halves of Dam1 and Duo1 bridge the gap between the DASH/Dam1c ring and the MT and contact the MT surface (31, 37). The points from which the Dam1 and Duo1 extensions emanate are next to each other on the inside of the ring (Fig. 3B, right). Their proximity explains extensive crosslinking of Dam1 and Duo C-terminal extensions with each other (37, 38). Both also crosslink to tubulin at positions on the outer surface of a MT (37). Low-resolution density, likely from Dam1 and Duo1, is present in the reconstruction of a MT-encircling, DASH/Dam1c ring from S. cerevisiae (Fig. S11B) (24, 25). The long Dam1 C-terminal projection in S. cerevisiae crosslinks to Ndc80 (35) and hence has a dual role (Fig. 3B, right). The site of crosslinking, a hairpin at the junction between the Ndc80 globular head and the coiled-coiled shaft, is also the site of a deletion that affects chromosome segregation (18). When Ndc80c attaches to a MT, this loop is close to the MT surface (39). Because of the high symmetry of the DASH/Dam1c ring, however, the same Dam1 subunit need not participate in both MT and Ndc80 contacts at the same time. In some fungal species, including C. thermophilum, the C-terminal extension of Dam1 may not be long enough to extend from a DASH/Dam1c ring to the Ndc80 globular head (Data S1).
The Ask1 C-terminal extension projects from the outside of the ring (Fig. 3B, left). Its contact on the Ndc80:Nuf2 shaft (35), possibly also involving residues of the loop (16), is well within reach of the Ask1 arm, which would emerge from just beneath the Ndc80c shaft as positioned in Fig. 4. The Spc19 and Spc34 C-terminal extensions project from the tip of the protrusion domain. Secondary structure prediction (Data S1) and the second highest peak in our Spc19:Spc34 molecular contact map from DCA suggest that a heterodimeric Spc19:Spc34 coiled coil is flexibly tethered to the structured protrusion domain by two short linkers (Fig. 3B, center left). The observed crosslink toward the C terminus of Nuf2 (35), near the junction with Spc24:Spc25, and binding of S. cerevisiae Spc19 residues 128–165 and Nuf2 residues 399–429 (40), then sets the polarity of the DASH/Dam1c ring with respect to the MT, with the protrusion pointing toward the plus end. As the C-terminal Spc19:Spc34 binding module is anchored to the structured core of the DASH/Dam1c ring through a relatively short linker, the Ndc80:Nuf2 coiled-coil shaft probably bends around the DASH/Dam1c ring, perhaps near the position of the “Ndc80 loop”, bringing the Spc24:Spc25 tip of Ndc80c closer to the kinetochore central axis where it binds the MIND adaptor complex (Fig. 4A).
DASH/Dam1c and Ndc80c are both essential for kinetochore-MT attachments that are stable enough to withstand the forces generated by MT depolymerization (41, 42). A rigid DASH/Dam1c ring that interacts through flexible extensions with the MT and Ndc80c has several potential functional advantages. (i) Junctions created by a flexible peptide extension that fits into a specific docking site can adapt to different relative orientations of the partner complexes, as required by variable stoichiometry and unmatched symmetry in the kinetochore (e.g. a 13-protofilament MT surrounded by a 16- or 17-protomer DASH/Dam1c ring and 6–8 Ndc80c complexes). (ii) These junctions allow distinct interactions at different stages of mitosis: budding-yeast kinetochores first attach to the lateral surface of a MT, along which they migrate toward the spindle pole. DASH/Dam1c is dispensable at this stage, but it is necessary for the transition to stable end-on attachments (4). Adaptability of the DASH/Dam1c contacts with Ndc80c is probably a functional requirement for this process. (iii) Peptide segments facilitate regulation of the assembly state and interaction affinities by post-translational modifications in DASH/Dam1c (17, 43). Unfolded and extended polypeptides are good substrates for kinases and other modifying enzymes, as they fit readily into the enzymatic active sites. Ipl1/Aurora B phosphorylation of DASH/Dam1c subunits switches off all three sets interactions with Ndc80c (Fig. 4B) (35) in order to reset MT attachments during activation of the spindle checkpoint. (iv) Docked-peptide interactions are generally of modest strength, so that individual contacts dissociate and rebind rapidly, allowing a disassembling agent, such as the Ipl1 kinase, to access the DASH/Dam1c peptide segments for modification and to release the kinetochore from the MT. (v) Multiple weak contacts, flexible distance constraints, and incommensurate symmetries enable the organized assembly of Ndc80 and DASH/Dam1c to track the plus end of a depolymerizing MT by stochastic, “hand-over-hand” biased diffusion. The role of DASH/Dam1c ring as a structural organizer of Ndc80c is central to this process. (vi) Coordinated structural changes in the cage-like structure of the kinetochore, made possible by flexibility of the DASH/Dam1c:Ndc80c interactions (and of linkages in other parts of the assembly), may be part of the tension-sensing mechanism implicated in setting and releasing the mitotic spindle assembly checkpoint. Compliance of the overall structure can reposition components in response to tension and convert force into changes in molecular proximity.
The molecularly detailed model of the kinetochore-MT connection in budding yeast (Fig. 4), derived from the DASH/Dam1c and Ndc80c structures and from the constraints of published biochemical and genetic data, unifies two proposals for how the energy released by MT depolymerization drives chromosome separation at anaphase (44, 45). One model invokes biased diffusion, in which rapid, asynchronous association and dissociation of multiple, flexible contacts allows any individual contact to break and form again further along the shortening MT. The other invokes a conformational wave (e.g., protofilament curling) that actively pushes the kinetochore in the direction of shortening. Protofilment curling against a closed ring probably exerts force directly, but MT contacts, from the Dam1:Duo1 extensions and from the organized set of Ndc80 complexes, must break and reform to allow MT shortening to progress. Thus, docking and undocking of flexible connections between defined subunits can allow a kinetochore to track the end of the MT through cycles of growth and shrinkage.
Supplementary Material
Acknowledgments
We thank Chen Xu (University of Massachusetts Medical School) for instructing S.J. in cryo-EM; Chuan Hong, Zhiheng Yu (Janelia Research Campus), Melissa Chambers and Zongli Li (Harvard Medical School) for help with data collection; Stephen Hinshaw (Harvard Medical School) for valuable comments on the manuscript; Annie Bellemare and Adrian Tsang (Concordia University) for providing C. thermophilum cDNA; Alexis Rohou, Timothy Grant and Nikolaus Grigorieff (Janelia Research Campus) for helpful discussions and a pre-release version of FrealignX; the Northeastern Collaborative Access Team (NE-CAT) beamline staff for help with x-ray data collection. MycoCosm sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov in collaboration with the user community.
Funding: Some of the research was conducted at the Advanced Photon Source (APS), on the NE-CAT beamlines, funded by NIH grant P41 GM103403; the Pilatus 6M detector on beam line 24-ID-C is funded by NIH-ORIP HEI grant S10 RR029205; APS is operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. S.J. was a HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG-2004–09). S.C.H. is Investigator in the Howard Hughes Medical Institute.
Footnotes
Competing interests: No competing interests.
Data and materials availability: The cryo-EM symmetric and asymmetric reconstructions are deposited in the Electron Microscopy Data Bank (EMDB accession numbers EMD-7446, EMD-7469). The DASH/Dam1c structure is deposited in the Protein Data Bank (PDB accession number 6CFZ).
References and Notes
- 1.Biggins S, The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194, 817–846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuyama S, Biggins S, Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U. S. A 104, 14706–14711 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winey M et al. , Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129, 1601–1615 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU, Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178, 269–281 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grishchuk EL, McIntosh JR, Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25, 4888–4896 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koshland DE, Mitchison TJ, Kirschner MW, Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 331, 499–504 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Tytell JD, Sorger PK, Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172, 861–874 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei RR, Sorger PK, Harrison SC, Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. U. S. A 102, 5363–5367 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciferri C et al. , Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem 280, 29088–29095 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Aravamudhan P, Felzer-Kim I, Joglekar AP, The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr. Biol 23, 770–774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP, Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr. Biol 24, 1437–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekgoz Altunkaya G et al. , CCAN Assembly Configures Composite Binding Interfaces to Promote Cross-Linking of Ndc80 Complexes at the Kinetochore. Curr. Biol 26, 2370–2378 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Miranda JJ, De Wulf P, Sorger PK, Harrison SC, The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol 12, 138–143 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Westermann S et al. , Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17, 277–290 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S et al. , Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Maure JF et al. , The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr. Biol 21, 207–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheeseman IM et al. , Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S, Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 200, 21–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeyaprakash AA et al. , Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell 46, 274–286 (2012). [DOI] [PubMed] [Google Scholar]
- 20.van Hooff JJ, Snel B, Kops GJ, Unique phylogenetic distributions of the Ska and Dam1 complexes support functional analogy and suggest multiple parallel displacements of Ska by Dam1. Genome Biol. Evol 9, 1295–1303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janczyk PL et al. , Mechanism of Ska Recruitment by Ndc80 Complexes to Kinetochores. Dev. Cell 41, 438–449 e434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheerambathur DK et al. , Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex. Dev. Cell 41, 424–437 e424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yachdav G et al. , PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HW et al. , Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol 14, 721–726 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Ramey VH et al. , The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol. Biol. Cell 22, 457–466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y et al. , High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morcos F et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U. S. A 108, E1293–1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopf TA et al. , Three-dimensional structures of membrane proteins from genomic sequencing. Cell 149, 1607–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramey VH et al. , Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol. Biol. Cell 22, 4335–4342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miranda JJ, King DS, Harrison SC, Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol. Biol. Cell 18, 2503–2510 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gestaut DR et al. , Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10, 407–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC, Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell 167, 1014–1027 e1012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenni S, Dimitrova YN, Valverde R, Hinshaw SM, Harrison SC, Molecular structures of yeast kinetochore subcomplexes and their roles in chromosome segregation. Cold Spring Harb. Symp. Quant. Biol 82, (2017). [DOI] [PubMed] [Google Scholar]
- 35.Kim JO et al. , The Ndc80 complex bridges two Dam1 complex rings. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIntosh JR et al. , Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 200, 459–474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legal T, Zou J, Sochaj A, Rappsilber J, Welburn JP, Molecular architecture of the Dam1 complex-microtubule interaction. Open Biol. 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zelter A et al. , The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat. Commun 6, 8673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alushin GM et al. , The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467, 805–810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y et al. , Coiled-coil networking shapes cell molecular machinery. Mol. Biol. Cell 23, 3911–3922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tien JF et al. , Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189, 713–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lampert F, Hornung P, Westermann S, The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189, 641–649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latham JA, Chosed RJ, Wang S, Dent SY, Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 146, 709–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asbury CL, Tien JF, Davis TN, Kinetochores’ gripping feat: conformational wave or biased diffusion? Trends Cell Biol. 21, 38–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asbury CL, Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles. Biology (Basel) 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashkenazy H et al. , ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L et al. , DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 5, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benson DA et al. , GenBank. Nucleic Acids Res. 41, D36–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC, MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice P, Longden I, Bleasby A, EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Gibson DG et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Deneke J, Ziegelin G, Lurz R, Lanka E, The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. U. S. A 97, 7721–7726 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfahl M, Effect of DNA denaturants on the lac repressor-operator interaction. Biochim. Biophys. Acta 520, 285–290 (1978). [DOI] [PubMed] [Google Scholar]
- 55.Pfahl M, Tight-binding repressors of the lac operon: selection system and in vitro analysis. J. Bacteriol 137, 137–145 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfahl M, Mapping of I Gene mutations which lead to repressors with increased affinity for lac operator. J. Mol. Biol 147, 175–178 (1981). [DOI] [PubMed] [Google Scholar]
- 57.Daber R, Stayrook S, Rosenberg A, Lewis M, Structural analysis of lac repressor bound to allosteric effectors. J. Mol. Biol 370, 609–619 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalodimos CG et al. , Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain. EMBO J. 21, 2866–2876 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W, XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bell JM, Chen M, Baldwin PR, Ludtke SJ, High resolution single particle refinement in EMAN2.1. Methods 100, 25–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant T, Grigorieff N, Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mastronarde DN, Held SR, Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grant T, Grigorieff N, Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol 192, 204–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang K, Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grigorieff N, Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Methods Enzymol. 579, 191–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosenthal PB, Henderson R, Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Briegel A, Pilhofer M, Mastronarde DN, Jensen GJ, The challenge of determining handedness in electron tomography and the use of DNA origami gold nanoparticle helices as molecular standards. J. Struct. Biol 183, 95–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G, Tomato bushy stunt virus at 2.9 A resolution. Nature 276, 368–373 (1978). [DOI] [PubMed] [Google Scholar]
- 72.Jones TA, Zou JY, Cowan SW, Kjeldgaard M, Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991). [DOI] [PubMed] [Google Scholar]
- 73.Ikeuchi A, Sasaki Y, Kawarasaki Y, Yamane T, Exhaustive identification of interaction domains using a high-throughput method based on two-hybrid screening and PCR-convergence: molecular dissection of a kinetochore subunit Spc34p. Nucleic Acids Res. 31, 6953–6962 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ikeuchi A, Nakano H, Kamiya T, Yamane T, Kawarasaki Y, A method for reverse interactome analysis: High-resolution mapping of interdomain interaction network in Dam1 complex and its specific disorganization based on the interaction domain expression. Biotechnol. Prog 26, 945–953 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Wang RY et al. , De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat. Methods 12, 335–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grigoriev IV et al. , MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Notredame C, Higgins DG, Heringa J, T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Cock PJ et al. , Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stanke M, Steinkamp R, Waack S, Morgenstern B, AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32, W309–312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C et al. , BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Misawa K, Kuma K, Miyata T, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T, trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roure B, Rodriguez-Ezpeleta N, Philippe H, SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol. Biol 7 Suppl 1, S2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ekeberg M, Lovkvist C, Lan Y, Weigt M, Aurell E, Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 87, 012707 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Ekeberg M, Hartonen T, Aurell E, Fast pseudolikelihood maximization for direct-coupling analysis of protein structure from many homologous amino-acid sequences. J. Comput. Phys 276, 341–356 (2014). [Google Scholar]
- 87.Zhang R, Alushin GM, Brown A, Nogales E, Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 162, 849–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valverde R, Ingram J, Harrison SC, Conserved Tetramer Junction in the Kinetochore Ndc80 Complex. Cell Rep. 17, 1915–1922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang HW et al. , Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol 383, 894–903 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huis In ‘t Veld PJ et al. , Molecular basis of outer kinetochore assembly on CENP-T. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maiolica A et al. , Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Desfosses A, Ciuffa R, Gutsche I, Sachse C, SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol 185, 15–26 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Shaikh TR et al. , SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc 3, 1941–1974 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Egelman EH, A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 95.Behrmann E et al. , Real-space processing of helical filaments in SPARX. J. Struct. Biol 177, 302–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hohn M et al. , SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol 157, 47–55 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Waterman DG et al. , Diffraction-geometry refinement in the DIALS framework. Acta Crystallogr. D Struct. Biol 72, 558–575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kucukelbir A, Sigworth FJ, Tagare HD, Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones DT, Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.