Abstract
Human mesenchymal stem cells (hMSCs) are widely used in clinical research because of their multipotential, immunomodulatory, and reparative properties. Previous studies determined that hMSC spheroids from a three-dimensional (3D) culture possess higher therapeutic efficacy than conventional hMSCs from a monolayer (2D) culture. To date, various 3D culture methods have been developed to form hMSC spheroids but most of them used culture medium containing fetal bovine serum (FBS), which is not suitable for further clinical use. Here, we demonstrate that dissociated single MSCs seeded in induced pluripotent stem medium (MiPS) adhere loosely to the dish and spontaneously migrate to form spheroids during day 3 to day 6. Through component deletion screening and complementation experiments, the knockout serum replacement (KSR) was identified as necessary and sufficient for hMSC spheroid formation. Transcriptome analysis showed that the overall expression profiles were highly similar between 2D culture with FBS and KSR-derived spheroids. Interestingly, genes related to inflammatory response, immune response, and angiogenesis were upregulated in spheroids at day 6 and qPCR results further validated the increased expression level of related genes, including STC1, CCL7, HGF, IL24, and TGFB3. When spheroids were replated in normal FBS medium, cells formed a typical spindle-shaped morphology and FACS results showed that the recovered cells retained MSC-specific surface markers, such as CD73, CD90, and CD105. In summary, we developed a practical and convenient method to generate hMSC spheroids for clinical research and therapy.
1. Introduction
Human mesenchymal stem cells (hMSCs) possess self-renewal and multilineage differentiation potential [1, 2] and are extensively used in clinical studies. hMSCs can be derived from a wide range of tissues, such as the bone marrow, adipose tissue, umbilical cord, placenta, and dental pulp [3], and can be cultured in vitro for several generations. However, clinical data has shown a low survival rate for hMSCs from monolayer two-dimensional (2D) culture when implanted in vivo and optimized approaches for hMSC production are required for clinical application. Recent studies showed that aggregating hMSCs into 3D spheroids increased cell survival [4], stemness [5, 6], anti-inflammatory [7], and proangiogenic [8–10] properties of hMSCs. These data imply that 3D spheroids can be an alternative source for hMSCs in clinical applications.
A variety of in vitro 3D spheroid culture approaches have been developed [11–13], including hanging drop [7, 14–16], precoating of low-adhesive substrates [17], membrane-based aggregation [5, 18, 19], and forced aggregation [20]. However, most of these methods use conditioned medium containing fetal bovine serum (FBS), which contains undefined components and is not recommended for clinical applications [21, 22]. So far, several studies using a serum-free medium have successfully generated characterized hMSC spheroids. The Yloslato group utilized various serum-free and chemically defined xeno-free media, including MSCGM, MesenCult XF, and StemPro XF to generate hMSC spheroids in hanging drops, and found that compact spheroids formed when human serum albumin (HSA) was added into MesenCult XF and StemPro XF medium. Furthermore, they demonstrated that these hMSC spheroids were activated to express higher levels of therapeutic genes, such as TSG6, IL1A, IL1B, and STC1, in StemPro XF medium supplemented with HSA [23]. Meanwhile, Zimmermann and McDevitt utilized the forced aggregation method to form hMSC spheroids in agarose microwells and then cultured them in MesenCult XF medium and they found that hMSC spheroids only grow in size but did not increase the production of immunomodulatory paracrine factor PGE2 and IL-6 and IDO [20]. These studies suggested that hMSC spheroids derived from serum-free medium culture maintained, if not enhanced, the hMSC properties; however, the complicated procedures (hanging drops or gel coating) or instrument (agarose microwells) requirements seriously hinder their potential large-scale applications.
To solve these problems, we developed a novel method to generate hMSC spheroids spontaneously in serum-free condition medium containing knockout serum replacement (KSR). RNA-seq and qPCR results showed that MSC spheroids showed upregulated expression of therapeutic factors, including inflammatory response, immune response, and angiogenesis genes, and spheroid cells retained MSC-specific immunophenotypic markers after replating in FBS culture medium. Overall, our approach provides a convenient and cost-effective method to generate hMSC spheroids with therapeutic potentials.
2. Methods and Materials
2.1. Cell Culture
hMSCs were isolated from umbilical cord tissue and cultured in L-DMEM (Gibco, 11885-084) medium containing 15% FBS (HyClone, sh30084.03) at 37°C in a humidified atmosphere of 5% CO2, and medium was changed every three days. Cells were dissociated with 0.05% Trypsin-EDTA (Invitrogen, 25300062) and passaged at a 1 : 3 ratio when reaching about 80% confluence. Cells from passage 3 to passage 8 were used in this study.
2.2. Formation of Spheroids and Spheroid-Recovered Monolayer MSCs
Cells at passages 3, 5, and 8 were dissociated into single cells with 0.05% Trypsin-EDTA (Invitrogen, 25300062) and seeded at the indicated concentration (1 × 104, 2.5 × 104, 5 × 104, and 1 × 105 per cm2) in human-induced pluripotent stem cell medium (refer to as MiPS), which is DMEM/F12 (Gibco, 11320-033) containing 20% KSR (Gibco, 10828-028), 2 μM L-glutamine (Sigma G8540), 0.1 μM NEAA (Gibco, 11140-050), 0.1 μM 2-mercaptoethanol (Gibco, 21985-023), and 10 ng/ml human bFGF (Invitrogen, PHG0021) or in L-DMEM (Gibco, 11885-084) containing 20% KSR (refer to as L-KSR). Other media containing 20% KSR used in this study include H-DMEM (Life Technologies, 11965-092), DMEM/F12 (Life Technologies, 10565-018), MEM (Gibco, 11095080), and RPMI1640 (Gibco, C22400500BT). Cells were incubated at 37°C in a humidified atmosphere of 5% CO2. To recover monolayer hMSCs from spheroids, spheroids at day 6 in MiPS or L-KSR were collected and washed with PBS and then cultured in L-FBS.
2.3. Image and Video Recording
Cells or spheroids were photographed with a microscope (Zeiss LSM 510) at days 1, 3, and 6, and videos were obtained with a high-contrast instrument (BioTek, Cytation 5) for 72 hours.
2.4. Cell Viability Assay
To measure the cell viability, hMSC spheroids at days 1, 3, and 6 were stained with Calcein-AM (BioLegend, 425201) and PI (Sigma, P4170-25MG) according to the manufacturer's protocol. Briefly, spheroids were incubated in PBS containing Calcein-AM (0.01 μM) and PI (3 μM) at 37°C for 1 h, washed twice with PBS, and then resuspended in MiPS/L-KSR medium.
2.5. Library Construction and RNA Sequencing in a BGISEQ-500 Platform
To perform RNA sequencing, cells were first collected and lysed by TRIzol and total RNA was extracted according to the manufacturer's instructions (Invitrogen, 10296-028). 1 μg of the total RNA sample was purified using poly-T oligo-attached magnetic beads, and the purified mRNA was then fragmented using divalent cations under elevated temperature. First-strand cDNA was generated using reverse transcriptase and random primers, and the second-strand cDNA was synthesized using DNA polymerase I and RNase H. The synthesis product was purified with DNA clean beads, followed by end repair, A-tailing, and subsequent ligation to the adapter. After purification and PCR amplification, the products were subjected to single-strand circularized DNA molecule (ssDNA circle) preparation for final library construction. DNA nanoballs (DNBs) were generated with the ssDNA circle by rolling circle replication (RCR) to intensify the fluorescent signals during the sequencing process. The DNBs were then loaded into the patterned nanoarrays and sequenced on the BGISEQ-500 platform using the paired-end 100 strategy for sequencing and subsequent data analysis [24].
2.6. Initial Processing and Alignment of RNA-seq Data
The FASTQ data of each sample were aligned to the rRNA database (downloaded from NCBI) by SOAPaligner (version 2.21t) to remove rRNAs, and the remaining reads were processed with SOAPnuke (version 1.5.3) [25] to trim adaptors and filter out the low-quality reads. The filtered data were aligned to the hg19 RefSeq transcriptome downloaded from the UCSC Genome Browser database [26] using bowtie2 (version 2.2.5) [27]. Quantification of gene expression levels in raw counts and FPKM for all genes in all samples was performed using RSEM v1.2.4 [28].
2.7. Identification of Differentially Expressed Genes
Differential expression of genes in each group was determined using the R package DESeq2 [29] with default parameters, in which an adjusted p value less than 0.05 and log2 (fold change) > 1 were used to identify significantly differentially expressed genes.
2.8. GO Term and KEGG Enrichment Analysis
Gene ontology and KEGG pathway enrichment were analyzed using DAVID [30], and the BH method was used for multiple testing correction. GO terms with an FDR less than 0.05 were considered as significantly enriched.
2.9. Flow Cytometry
Monolayer MSCs recovered from spheroids at day 6 were harvested and dissociated into single cells by trypsinization and pipetting. To determine cell surface antigen expression, the samples were incubated with the following antibodies: human monoclonal antibodies against CD73 (BioLegend, 344004), CD90 (BioLegend, 328110), and CD105 (BioLegend, 323205). The samples were analyzed using a flow cytometer (BD Biosciences) and gated by forward scatter and side scatter.
2.10. qPCR
Cells were collected and lysed by TRIzol, and total RNA was extracted according to the manufacturer's instructions (Invitrogen, 10296-028). RNA was quantified with a NanoDrop spectrophotometer (Thermo Scientific). 3 μg of total RNA was used for reverse transcription with the PrimeScript First-Strand cDNA Synthesis Kit (Takara, D6110A). Quantitative real-time PCR (qPCR) was performed using the TB Green Premix Ex Taq kit (Takara, RR420A). Thermal cycling was performed with a 7500 real-time PCR desktop machine (Applied Biosystems) by incubating the reactions at 95°C for 20 s followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. The primers for qPCR analyses are as follows: STC1 forward: CACGAGCTGACTTCAACAGGA, reverse: GGATGTGCGTTTGATGTGGG; CCL7 forward: CAGCCAGATGCAATCAATGCC, reverse: TGGAATCCTGAACCCACTTCT; HGF forward: GCTATCGGGGTAAAGACCTACA, reverse: CGTAGCGTACCTCTGGATTGC; IL24 forward: TTGCCTGGGTTTTACCCTGC, reverse: AAGGCTTCCCACAGTTTCTGG; TGFB3 forward: ACTTGCACCACCTTGGACTTC, reverse: GGTCATCACCGTTGGCTCA; and GAPDH forward: GGAGCGAGATCCCTCCAAAAT, reverse: GGCTGTTGTCATACTTCTCATGG.
2.11. Data Analysis
The hMSC spheroid size was measured with the ImageJ software. The mean and standard derivation were calculated with Excel software.
2.12. Ethical Statement
Written informed consent was obtained from donors for all human samples, and all experiments were approved by the BGI ethics committee.
3. Results
3.1. Human Mesenchymal Stem Cells Spontaneously Form Spheroids in Serum-Free Medium Containing KSR
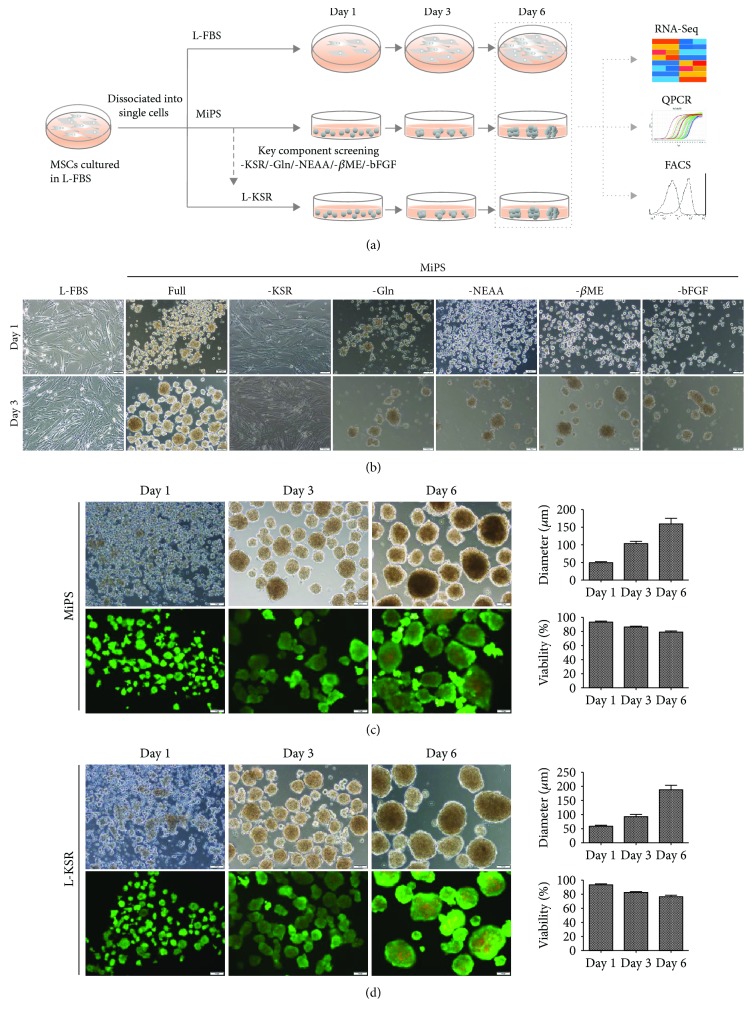
As a substitution of serum, KSR was first used to maintain mouse embryonic stem cells (mESCs). Recently, researchers observed that KSR can promote the proliferation and differentiation of adipose-derived MSCs in monolayer cultures [31, 32] and facilitate the formation of 3D rat testicular culture, indicating that KSR seems to be a suitable substitution of FBS for 2D and 3D cell cultures [33]. To test the effect of KSR-containing medium on hMSC culture, we performed a systematical comparison of different hMSC culture media, including L-FBS, KSR containing MiPS, and optimized L-KSR (Figure 1(a)). Interestingly, when cultured in MiPS medium, a medium originally designed for maintenance and expansion of human embryonic stem cells (hESCs) [34, 35], the dissociated single hMSCs maintained a round cell morphology, attached lightly to the tissue culture dish surface at day 1 and generated spheroids at day 3, while the single hMSCs seeded in L-FBS maintained fibroblast-like morphology (Figure 1(b)). To further determine the key ingredients in MiPS that promote spheroid formation, we conducted a screening that each time, one component was removed from MiPS to establish several incomplete MiPS groups and test the effect of each group on hMSC spheroid formation. We found that most of these incomplete MiPS groups were sufficient to support spontaneously spheroid formation, except for the group without KSR (Figure 1(b)), as they attached to the culture dish, maintained fibroblast-like morphology, and mimicked the cells in L-FBS medium (Figure 1(b)), indicating that KSR is an essential component of MiPS for spheroid formation. To test whether KSR alone was sufficient to generate the hMSC spheroids, we used the basal medium L-DMEM and only added KSR as extra component to culture dissociated hMSCs. Excitingly, we discovered that KSR alone is sufficient for spheroid formation at the concentration as low as 2%, though higher concentration tended to promote a better spheroid formation (). Moreover, the addition of KSR alone into other several basal media, including RPMI1640, DMEM/F12, H-DMEM, and MEM, can also promote the generation of MSC spheroids (), indicating a powerful effect of KSR to substitute serum in this culture medium system for hMSC maintenance. We also tested the effect of initial cell concentration on spheroid formation and found that MSCs could generated spheroids at a concentration as low as 1 × 104/ml in 20% KSR (). To simplify, we used 20% KSR in L-DMEM medium in the subsequent experiments and refer to as L-KSR.
Figure 1.
MSCs at P3 can spontaneously form spheroids in medium containing KSR. (a) Schematic diagram shows the experimental procedure; (b) hMSCs cultured in MiPS and MiPS without KSR/GLn/NEAA/βME/bFGF; (c, d) hMSC spheroids were generated and stained with Calcein-AM/PI in MiPS/KSR at days 1, 3, and 6 on tissue culture dishes. Statistical analysis of the mean diameter and cell viabilities of hMSC spheroids; sizes were measured from captured images of spheroids (n = 12-20); values are mean ± SD (n = 3); % of live cells (Calcein-AM/PI) was determined using flow cytometry; values are mean ± SD (n = 3). Scale bars: 100 μm.
We then analyzed the hMSC spheroid size and cell viability in L-KSR and the MiPS medium system to compare their capability in promoting spheroid formation. In both media, the nearby cells spontaneously migrated and aggregated into small and loose spheroids from day 1 to day 3 and several small spheroids coalesced into large and compact spheroids from day 3 to day 6 (Figures 1(c) and 1(d), supplementary , ), showing no dramatic difference on the formation speed. In addition, the mean diameter of hMSC spheroids increased with the prolongation of culture time in both media. Cell viability identified by Calcein-AM/PI showed that the percentage of dead cells increased in spheroids in both MiPS and L-KSR with cultured time but still maintained a high fraction of viable cells at day 6 (>80%) and no significant difference was observed between the two medium groups (Figures 1(c) and 1(d)), indicating the that effect of L-KSR on hMSC spheroid formation is comparable to MiPS and representing the typical hMSC spheroid phenotype previously reported using a hanging drop protocol [7]. Overall, our data showed that MiPS medium is capable to support MSCs to spontaneously form spheroids and the medium component KSR is necessary and sufficient to generate spheroids.
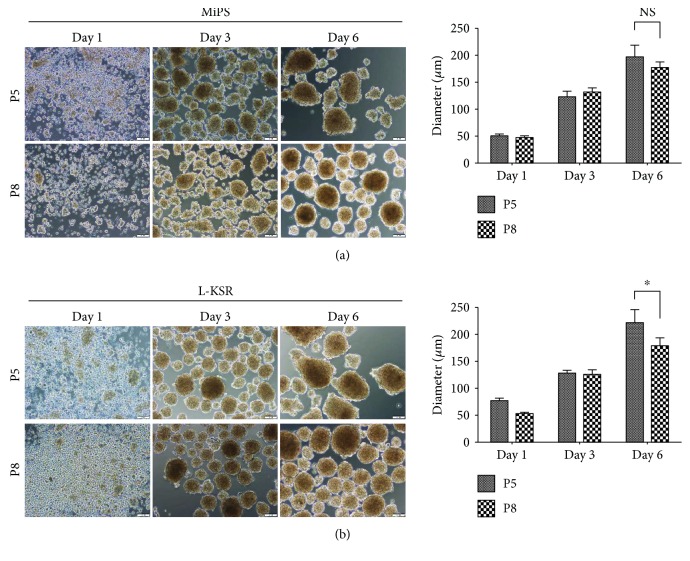
3.2. MSCs at High Passage Retain the Ability to Form Spheroids in KSR Medium
Recent studies have shown that MSCs gradually lose their therapeutic potency due to an increasing senescent cell subset during long-term culture in vitro [36–39]. To test whether hMSCs at high passage could also form spheroids in KSR medium, hMSCs after 5 or 8 passages in FBS containing medium were cultured in MiPS and L-KSR media. Our results showed that cells at P5 and P8 could both form spheroids in MiPS/L-KSR medium (Figures 2(a) and 2(b)), although statistical analysis showed that the spheroid mean diameter from P8 hMSCs slightly decreased when compared to that from P5 (Figures 2(a) and 2(b)). Overall, our data demonstrated that MSCs at high passage still can generate spheroids in KSR medium.
Figure 2.
MSCs at higher passage retain the ability to form spheroids in medium containing KSR. (a) MSCs at P5 and P8 generated spheroids in MiPS at days 1, 3, and 6 on tissue culture dishes. Statistical analysis of the MSC spheroid mean diameter cultured in MiPS; (b) MSCs at P5 and P8 generated spheroids in L-KSR at days 1, 3, and 6. Statistical analysis of the MSC spheroid mean diameter cultured in KSR; sizes were measured from captured images of spheroids (n = 12-20); values are mean ± SD (n = 3). Not significant (NS); P ≥ 0.05 and ∗P < 0.05. Scale bars:100 μm.
3.3. Transcriptomics Analysis Reveals That MSC Spheroids Generated in KSR Medium Obtain Enhanced Expression of Therapeutic Genes
Previous studies demonstrated the expression of anti-inflammatory factors (TNFα, TSG-6, and STC-1), and angiogenic growth factors (ANG, FGF-2, ANGPT-2, and HGF) are significantly increased in MSC spheroid cultures. These results suggested that MSCs cultured in spheroids can enhance cell therapeutic potentials, including anti-inflammation [7] and proangiogenesis [8–10]. To investigate whether the hMSC spheroids generated with our method may acquire similar advantages, we plated P5 and P8 MSCs in L-FBS, MiPS, and L-KSR media, respectively, and collected the cells or hMSC spheroids after 6 days in culturing in those different media for RNA-seq analysis.
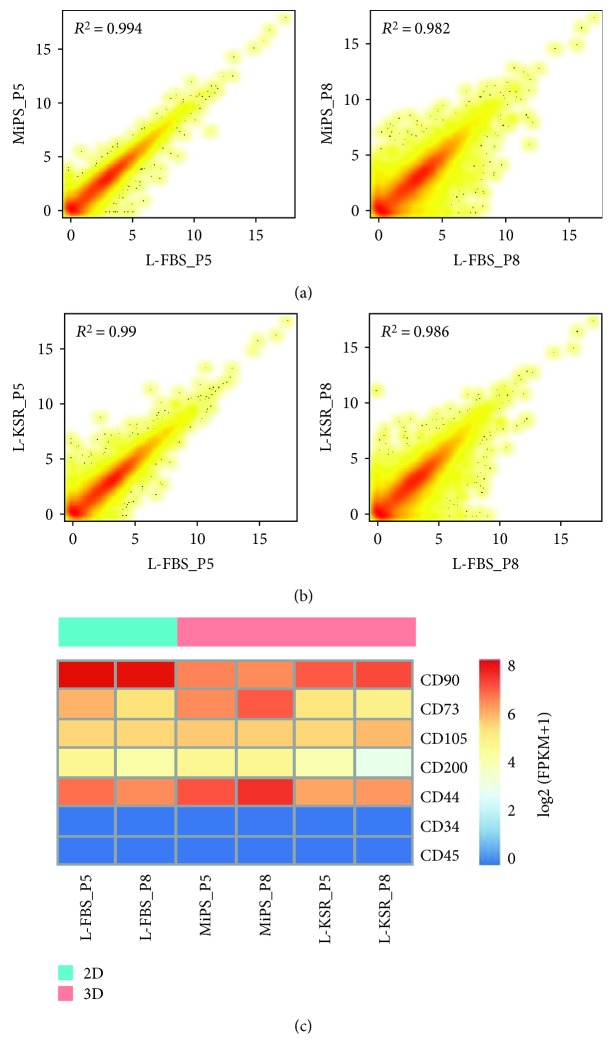
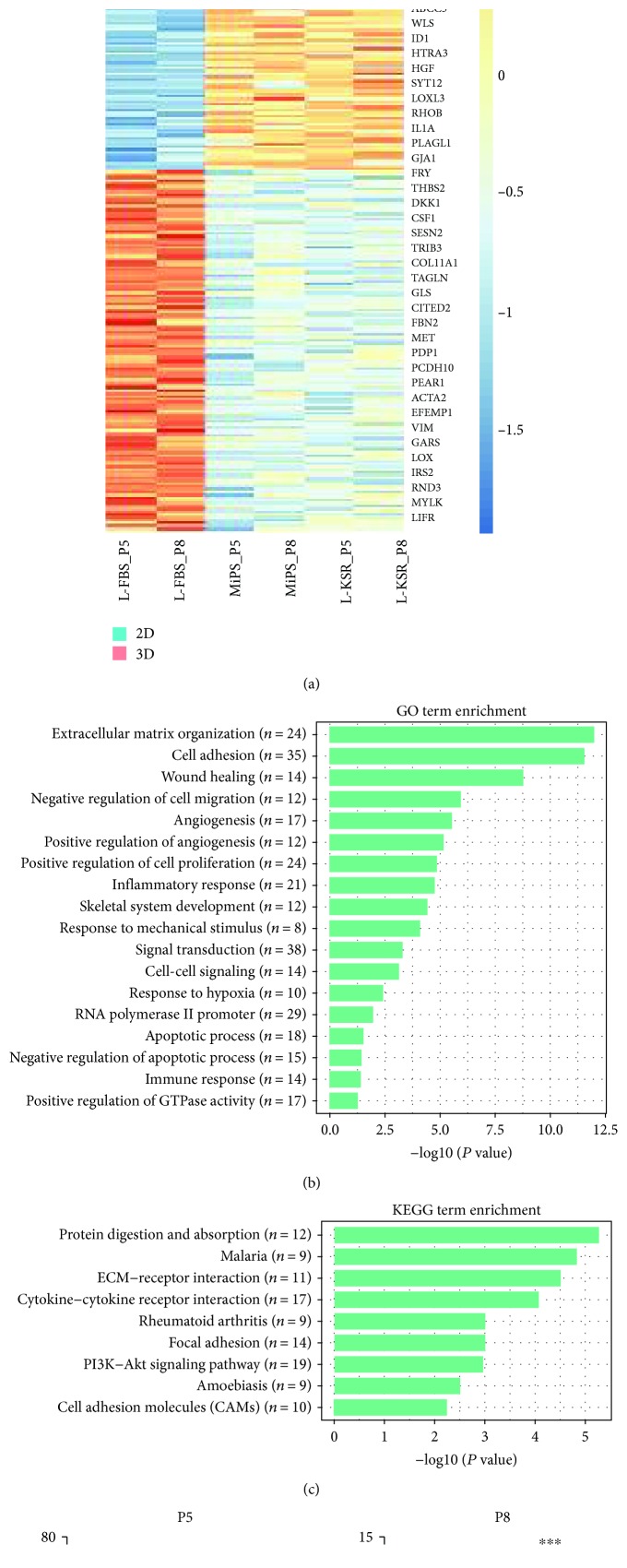
We analyzed the overall transcript expression level in these three groups. Transcriptomic correlation analysis showed that all the samples were highly related (Figure 3(a) and 3(b)) and heatmap results showed that the expression pattern of several important MSC marker genes was similar among the three groups (Figure 3(c)), demonstrating that these samples maintained MSC features. To validate whether genes of therapeutic potential were altered in 3D spheroids, we compared 3D spheroids in MiPS/L-KSR with 2D adherent monolayer MSCs in L-FBS. The heatmap results showed that potentially therapeutic genes, including STC1, CCL7, TNFRSF1B, LIF, TGFB, IL1B, IL1A, and HGF were upregulated in 3D spheroids with MiPS/L-KSR medium, while DKK1 and VIM genes were downregulated (Figure 4(a)). Gene ontology (GO) enrichment analysis showed that these changed genes are associated with extracellular matrix organization, cell adhesion, wounding healing, angiogenesis, inflammatory response, signal transduction, and immune response (Figure 4(b)). KEGG analysis showed that these genes are associated with protein digestion and absorption, ECM-cytokine receptor interaction, focal adhesion, and the P13K-Akt signaling pathway (Figure 4(c)). Our qPCR assay confirmed the upregulated expression of STC1, CCL7, HGF, IL24, and TGFB3 in 3D spheroids with MiPS/L-KSR medium (Figure 4(d)), which are considered as critical genes required for MSCs' function. In summary, the RNA-seq and qPCR results suggested that the expression of potentially therapeutic genes in spheroids can be enhanced in MiPS/L-KSR.
Figure 3.
Transcriptomic correlation analysis of spheroids at day 6 at P5 and P8. (a, b) Comparison of RNA-seq gene expression profiles between spheroids in MiPS/L-KSR at P5 and P8 and their corresponding FBS control. R2 stands for correlation coefficient; (c) the heatmap shows the gene expression level of several specific makers of hMSCs.
Figure 4.
Transcriptomic expression analysis of hMSC spheroids. (a) The heatmap shows scaled expression [log2 (FPKM + 1)] of discriminative genes between 3D spheroids (both in MiPS and in L-KSR) and 2D normal MSCs (in L-FBS medium) at day 6 at P5 and P8. Adjusted P value<0.05. The color scheme is based on z-score distribution from 2 (blue) to 2 (red). (b) Gene ontology (GO) analysis between 3D and 2D; “n” indicates gene numbers. (c) KEGG analysis between 3D and 2D. (d) qPCR result analysis of STC1, CCL7, HGF, IL24, and TGFB3 between 3D and 2D cells at P5 and P8 at day 6. The expression levels of genes of interest to that of GAPDH. Not significant (NS); P ≥ 0.05, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.4. Spheroid-Recovered MSCs Retain Mesenchymal Stem Cell Features
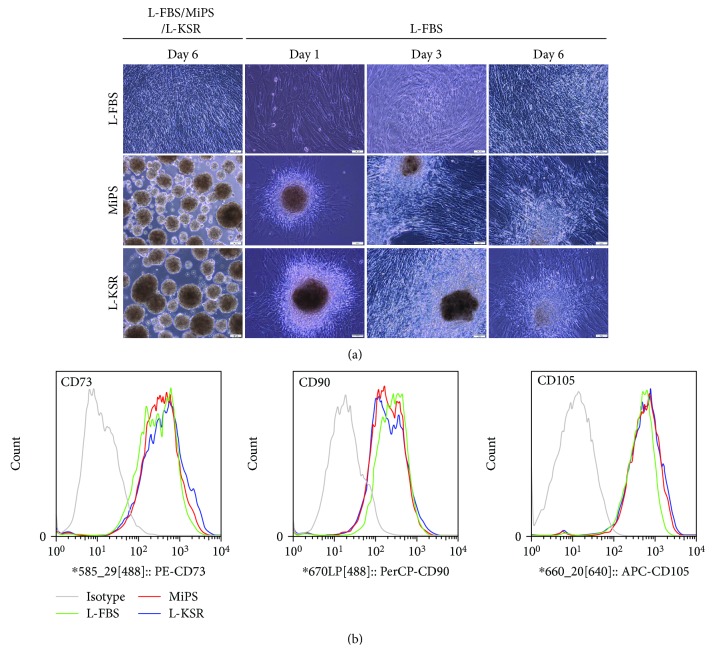
Previous studies showed that the cells in spheroids retained most of the surface epitopes of hMSCs from adherent cultures [4]. To ensure that the MSCs in our KSR culture system retain these MSC properties thus maintaining their value for research and clinical applications, we transferred spheroids derived from P8 MSC at day 6 from MiPS and L-KSR back to standard L-FBS culture medium. Results showed that the MSC spheroids began to attach on to the culture dish surface, and then, from day 1 to day 6, cells in the spheroids migrated out and adhered to culture dish and proliferated (Figure 5(a)). Spheroid-recovered MSCs were collected for FACS analysis. FACS results showed that the expression of MSC markers, including CD73, CD90, and CD105 in MiPS/L-KSR samples were still highly expressed, which is similar to that in normal cultured 2D MSCs (Figure 5(b)).
Figure 5.
Spheroid-recovered hMSCs retain mesenchymal stem cell features. (a) Cells in spheroids at day 6 at passage 8 in MiPS/L-KSR migrated out and adhered to tissue culture dishes when reseeded in L-FBS medium. Cells cultured in FBS as a control; (b) FACS analysis of MSC-positive markers CD73, CD90, and CD105 of spheroid-recovered hMSCs in MiPS/L-KSR. Cells cultured in L-FBS as a control. Scale bars: 100 μm.
4. Discussion
Cells cultured in 3D as spheroids in vitro provide enhanced cell-cell interactions and more closely mimic the natural microenvironment of a tissue. Therefore, it has been widely used in various fields, including tumor biology [40, 41], drug discovery, toxicology screening [42, 43], and organoid research [44]. Previous studies showed that MSCs can form spheroids with a variety of methods. However, most of those methods have to use medium containing FBS or a need for special instruments (agarose microwells) or a complicated procedure (hanging drops or gel coating) [12], hindering their large-scale implication in clinical applications. Here, we developed a convenient method to generate MSC spheroids spontaneously in a novel serum-free formula without any special instrumentation or precoated gels.
First of all, we found that MiPS could prevent cells from adhering to the tissue culture dish and facilitate cells to migrate and aggerate into spheroids (Figure 1(b)). Then, we demonstrated that KSR in MiPS was the only critical active ingredient to promote spheroid formation. In fact, KSR is not only necessary but also sufficient to promote hMSC spheroid formation when added into different basal media (Figures 1(b)–1(d) and ). KSR is a substitute of FBS, and all the components are well defined, consisting of albumin, transferrin, insulin, collagen precursors, amino acids, vitamins, antioxidants, and trace elements [33]. Therefore, KSR is more suitable for clinical-grade MSC production as it eliminates many of the uncertainties encountered when using poorly defined serum supplements. Our study demonstrated that KSR at a concentration as low as 2% could promote hMSC spheroid formation, though a higher concentration tends to generate larger spheroids in a relatively shorter time (Fig. ). It would be interesting to further define the key components in KSR that are pivotal to the spheroid's formation.
Previous studies showed that short-term culture of MSCs in a 3D environment had no significant effect on the level of MSC-specific immunophenotypic marker expression [11]. In this study, we compared the expression pattern of hMSC spheroids derived from MiPS and L-KSR with normal cultured hMSCs through RNA-seq. The overall gene expression pattern, including the MSC marker genes and pluripotency-associated genes, is similar between hMSC spheroids and hMSCs (Figure 3), suggesting that our method did not change the basic properties of hMSCs. Various studies have demonstrated that MSCs in spheroid generated in medium containing FBS have a higher expression level of immunomodulatory-related factors [7, 45]. However, these mediators were not upregulated in spheroids cultured in the chemically defined xeno-free medium [20]. Interestingly, we found that hMSC spheroids generated in KSR medium upregulated the expression of potentially therapeutic genes (Figures 4(a) and 4(d)). The GO term and KEGG analysis of these differentially expressed genes indicated that the signaling pathways enriched in KSR-derived hMSC spheroids were associated with extracellular matrix organization, cell adhesion, wounding healing, angiogenesis, inflammatory response, signal transduction, and immune response, which were considered to be related to the therapeutic function of MSCs (Figures 4(b) and 4(c)). More importantly, when our hMSC spheroids were replated to a tissue culture dish in FBS-contained medium, spindle-shaped cells migrated out and retained MSC properties (Figure 5). These results suggest that the hMSC spheroids may be implanted directly into the body and served as an MSC reservoir to play a sustained therapeutic role, though the real biological functions of the hMSC spheroids generated with our method need further in vivo validation studies.
In conclusion, we developed a practical and convenient method to generate hMSC spheroids in a defined serum-free medium and preliminary studies suggest that it enhanced the therapeutic effect of hMSCs. We anticipate that the hMSC spheroids generated with our method could be widely used for future clinical research and therapy.
Acknowledgments
This research was supported by the P.R. China, MST Special Fund (project name: single cell sequencing based antibody discovery), the Science, Technology and Innovation Commission of Shenzhen municipality (grant number JCYJ20170817145845968), and the Shenzhen Engineering Laboratory for Innovative Molecular Diagnostics (grant number DRC-SZ [2016] 884). We thank Micheal Dean for the help in editing the language and BGI colleagues who helped to produce the high-quality data.
Contributor Information
Weihua Zhao, Email: zwhzyz123@163.com.
Ying Gu, Email: guying@genomics.cn.
Data Availability
The data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb with accession code CNP0000456.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Authors' Contributions
G.D. designed the experiments. G.D., Y.G., Q.C., and Q.D. performed the experiments. S.W. processed the RNA sequencing data. S.W. and G.D. analyzed the data. W.Z. and J.T. collected the umbilical cord. Q.W., Z.S., W.O., J.L., and Y.G. jointly made the discussions. G.D. and S.W. wrote the manuscript. Y.G. and C.L. revised the manuscript. Y.G. supervised the study. All authors reviewed and approved the final manuscript. Guoyi Dong and Shengpeng Wang contributed equally to this work.
Supplementary Materials
Figure S1: MSCs can form spheroids in KSR-contained medium. (A) hMSCs cultured in L-FBS at passage 3; (B) hMSC spheroids at day 6 generated from hMSCs at passage 3 in L-DMEM at the indicated concentration KSR. Statistical analysis of the hMSC spheroid mean diameter cultured in different concentrations of KSR in L-DMEM medium; (C) hMSC spheroids at day 6 generated from hMSCs at passage 3 in various basal media, including RPMI1640, DMEM/F12, H-DMEM, and MEM with 20% KSR. Statistical analysis of the hMSC spheroid mean diameter cultured in various basal media with 20% KSR. Scale bars: 100 μm.
Figure S2: MSCs can form spheroids at different cell concentrations in 20% KSR-containing medium. (A) hMSC spheroids generated from MSCs at passage 3 in L-DMEM with 20% KSR at different cell concentrations; (B) statistical analysis of the hMSC spheroid mean diameter cultured in different concentrations of KSR in L-DMEM medium. Scale bars: 100 μm.
Video 1: MSC cultured in MiPS. The video was obtained by time-lapse photography with a high-contrast instrument for 72 hours.
Video 2: MSC cultured in L-KSR. The video was obtained by time-lapse photography with a high-contrast instrument for 72 hours.
References
- 1.Dominici M., le Blanc K., Mueller I., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger M. F., Mackay A. M., Beck S. C., et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Marquez-Curtis L. A., Janowska-Wieczorek A., McGann L. E., Elliott J. A. W. Mesenchymal stromal cells derived from various tissues: biological, clinical and cryopreservation aspects. Cryobiology. 2015;71(2):181–197. doi: 10.1016/j.cryobiol.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Jiang B., Yan L., Miao Z., Li E., Wong K. H., Xu R. H. Spheroidal formation preserves human stem cells for prolonged time under ambient conditions for facile storage and transportation. Biomaterials. 2017;133:275–286. doi: 10.1016/j.biomaterials.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 5.Cheng N. C., Wang S., Young T. H. The influence of spheroid formation of human adipose-derived stem cells on chitosan films on stemness and differentiation capabilities. Biomaterials. 2012;33(6):1748–1758. doi: 10.1016/j.biomaterials.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 6.Guo L., Zhou Y., Wang S., Wu Y. Epigenetic changes of mesenchymal stem cells in three-dimensional (3D) spheroids. Journal of Cellular and Molecular Medicine. 2014;18(10):2009–2019. doi: 10.1111/jcmm.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartosh T. J., Ylostalo J. H., Mohammadipoor A., et al. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(31):13724–13729. doi: 10.1073/pnas.1008117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhang S. H., Lee S., Shin J. Y., Lee T. J., Kim B. S. Transplantation of cord blood mesenchymal stem cells as spheroids enhances vascularization. Tissue Engineering Part A. 2012;18(19-20):2138–2147. doi: 10.1089/ten.tea.2011.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potapova I. A., Brink P. R., Cohen I. S., Doronin S. V. Culturing of human mesenchymal stem cells as three-dimensional aggregates induces functional expression of CXCR4 that regulates adhesion to endothelial cells. The Journal of Biological Chemistry. 2008;283(19):13100–13107. doi: 10.1074/jbc.M800184200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potapova I. A., Gaudette G. R., Brink P. R., et al. Mesenchymal stem cells support migration, extracellular matrix invasion, proliferation, and survival of endothelial cells in vitro. Stem Cells. 2007;25(7):1761–1768. doi: 10.1634/stemcells.2007-0022. [DOI] [PubMed] [Google Scholar]
- 11.Petrenko Y., Syková E., Kubinová S. The therapeutic potential of three-dimensional multipotent mesenchymal stromal cell spheroids. Stem Cell Research & Therapy. 2017;8(1):p. 94. doi: 10.1186/s13287-017-0558-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cesarz Z., Tamama K. Spheroid culture of mesenchymal stem cells. Stem Cells International. 2016;2016:11. doi: 10.1155/2016/9176357.9176357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends in Biotechnology. 2013;31(2):108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki S., Muneta T., Tsuji K., et al. Properties and usefulness of aggregates of synovial mesenchymal stem cells as a source for cartilage regeneration. Arthritis Research & Therapy. 2012;14(3, article R136) doi: 10.1186/ar3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartosh T. J., Ylostalo J. H. Preparation of anti-inflammatory mesenchymal stem/precursor cells (MSCs) through sphere formation using hanging-drop culture technique. Current Protocols in Stem Cell Biology. 2014;28(1):2B.6.1–2B.6.23. doi: 10.1002/9780470151808.sc02b06s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. Journal of Visualized Experiments. 2011;(51) doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J. H., Han Y. S., Lee S. H. Long-duration three-dimensional spheroid culture promotes angiogenic activities of adipose-derived mesenchymal stem cells. Biomolecules & Therapeutics. 2016;24(3):260–267. doi: 10.4062/biomolther.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh H. Y., Liu B. H., Sieber M., Hsu S. H. Substrate-dependent gene regulation of self-assembled human MSC spheroids on chitosan membranes. BMC Genomics. 2014;15(1):p. 10. doi: 10.1186/1471-2164-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng T. C., Wong C. W., Hsieh F. Y., Hsu S. H. Biomaterial substrate-mediated multicellular spheroid formation and their applications in tissue engineering. Biotechnology Journal. 2017;12(12) doi: 10.1002/biot.201700064. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann J. A., McDevitt T. C. Pre-conditioning mesenchymal stromal cell spheroids for immunomodulatory paracrine factor secretion. Cytotherapy. 2014;16(3):331–345. doi: 10.1016/j.jcyt.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Sharma R. R., Pollock K., Hubel A., McKenna D. Mesenchymal stem or stromal cells: a review of clinical applications and manufacturing practices. Transfusion. 2014;54(5):1418–1437. doi: 10.1111/trf.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spees J. L., Gregory C. A., Singh H., et al. Internalized antigens must be removed to prepare hypoimmunogenic mesenchymal stem cells for cell and gene therapy. Molecular Therapy. 2004;9(5):747–756. doi: 10.1016/j.ymthe.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Ylostalo J. H., Bartosh T. J., Tiblow A., Prockop D. J. Unique characteristics of human mesenchymal stromal/progenitor cells pre-activated in 3-dimensional cultures under different conditions. Cytotherapy. 2014;16(11):1486–1500. doi: 10.1016/j.jcyt.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin S., McPherson J. D., McCombie W. R. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Chen Y., Shi C., et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita P. A., Rhead B., Zweig A. S., et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Research. 2010;39(Database):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalerba P., Kalisky T., Sahoo D., et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nature Biotechnology. 2011;29(12):1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M. I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):p. 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D. W., Sherman B. T., Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund P., Pilgaard L., Duroux M., Fink T., Zachar V. Effect of growth media and serum replacements on the proliferation and differentiation of adipose-derived stem cells. Cytotherapy. 2009;11(2):189–197. doi: 10.1080/14653240902736266. [DOI] [PubMed] [Google Scholar]
- 32.Ho S. T. B., Tanavde V. M., Hui J. H., Lee E. H. Upregulation of adipogenesis and chondrogenesis in MSC serum-free culture. Cell Medicine. 2011;2(1):27–42. doi: 10.3727/215517911X575984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Wang L., Zhang X., et al. The use of knockout serum replacement (KSR) in three dimensional rat testicular cells co-culture model: an improved male reproductive toxicity testing system. Food and Chemical Toxicology. 2017;106(Part A):487–495. doi: 10.1016/j.fct.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Amit M., Carpenter M. K., Inokuma M. S., et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Developmental Biology. 2000;227(2):271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 35.Dakhore S., Nayer B., Hasegawa K. Human pluripotent stem cell culture: current status, challenges, and advancement. Stem Cells International. 2018;2018:17. doi: 10.1155/2018/7396905.7396905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu W. L., Li J., Chen G., Li Q., Tang X., Zhang C. H. Mesenchymal stem cells derived from peripheral blood retain their pluripotency, but undergo senescence during long-term culture. Tissue Engineering Part C: Methods. 2015;21(10):1088–1097. doi: 10.1089/ten.tec.2014.0595. [DOI] [PubMed] [Google Scholar]
- 37.Wagner W., Bork S., Horn P., et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS One. 2009;4(6, article e5846) doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner W., Horn P., Castoldi M., et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3(5, article e2213) doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oja S., Komulainen P., Penttilä A., Nystedt J., Korhonen M. Automated image analysis detects aging in clinical-grade mesenchymal stromal cell cultures. Stem Cell Research & Therapy. 2018;9(1):p. 6. doi: 10.1186/s13287-017-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiswald L. B., Bellet D., Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17(1):1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katt M. E., Placone A. L., Wong A. D., Xu Z. S., Searson P. C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Frontiers in Bioengineering and Biotechnology. 2016;4:p. 12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pampaloni F., Stelzer E. H. K. Three-dimensional cell cultures in toxicology. Biotechnology & Genetic Engineering Reviews. 2009;26(1):117–138. doi: 10.5661/bger-26-117. [DOI] [PubMed] [Google Scholar]
- 43.Thoma C. R., Zimmermann M., Agarkova I., Kelm J. M., Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Advanced Drug Delivery Reviews. 2014;69-70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 45.Bartosh T. J., Ylöstalo J. H., Bazhanov N., Kuhlman J., Prockop D. J. Dynamic compaction of human mesenchymal stem/precursor cells into spheres self-activates caspase-dependent IL1 signaling to enhance secretion of modulators of inflammation and immunity (PGE2, TSG6, and STC1) Stem Cells. 2013;31(11):2443–2456. doi: 10.1002/stem.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: MSCs can form spheroids in KSR-contained medium. (A) hMSCs cultured in L-FBS at passage 3; (B) hMSC spheroids at day 6 generated from hMSCs at passage 3 in L-DMEM at the indicated concentration KSR. Statistical analysis of the hMSC spheroid mean diameter cultured in different concentrations of KSR in L-DMEM medium; (C) hMSC spheroids at day 6 generated from hMSCs at passage 3 in various basal media, including RPMI1640, DMEM/F12, H-DMEM, and MEM with 20% KSR. Statistical analysis of the hMSC spheroid mean diameter cultured in various basal media with 20% KSR. Scale bars: 100 μm.
Figure S2: MSCs can form spheroids at different cell concentrations in 20% KSR-containing medium. (A) hMSC spheroids generated from MSCs at passage 3 in L-DMEM with 20% KSR at different cell concentrations; (B) statistical analysis of the hMSC spheroid mean diameter cultured in different concentrations of KSR in L-DMEM medium. Scale bars: 100 μm.
Video 1: MSC cultured in MiPS. The video was obtained by time-lapse photography with a high-contrast instrument for 72 hours.
Video 2: MSC cultured in L-KSR. The video was obtained by time-lapse photography with a high-contrast instrument for 72 hours.
Data Availability Statement
The data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb with accession code CNP0000456.