Abstract
Fusobacterium nucleatum (F. nucleatum) plays key roles in the initiation and progression of periodontitis. However, the pathogenic effect of F. nucleatum on human oral tissues and cells has not been fully evaluated. In this study, we aimed to analyze the pathogenic effects of F. nucleatum on human gingival fibroblasts (GFs) and clarify the potential mechanisms. RNA-sequencing analysis confirmed that F. nucleatum significantly altered the gene expression of GF as the stimulation time increased. Cell counting and EdU-labeling assays indicated that F. nucleatum inhibited GF proliferation and promoted cell apoptosis in a time- and dose-dependent manner. In addition, cell apoptosis, intracellular reactive oxygen species (ROS) generation, and proinflammatory cytokine production were dramatically elevated after F. nucleatum stimulation. Furthermore, we found that the AKT/MAPK and NF-κB signaling pathways were significantly activated by F. nucleatum infection and that a large number of genes related to cellular proliferation, apoptosis, ROS, and inflammatory cytokine production downstream of AKT/MAPK and NF-κB signaling pathways were significantly altered in F. nucleatum-stimulated GFs. These findings suggest that F. nucleatum inhibits GF proliferation and promotes cell apoptosis, ROS generation, and inflammatory cytokine production partly by activating the AKT/MAPK and NF-κB signaling pathways. Our study opens a new window for understanding the pathogenic effects of periodontal pathogens on the host oral system.
1. Introduction
Periodontal diseases are prevalent globally, with approximately 60% of the adult population suffering from mild, moderate, or aggressive periodontitis [1, 2]. As a chronic inflammatory disease, periodontitis is characterized by the destruction of the gingiva, periodontal ligament, cementum, and alveolar bone and is initiated by the invasion of specific oral pathogens that colonize dental plaque biofilms on the tooth surface [3–5]. The pathogenesis of periodontitis is complex, and excessive tissue destruction occurs as a result of interactions between pathogenic bacteria and the host immune inflammatory response [3]. Previous studies have confirmed that periodontitis is commonly caused by gram-negative bacterial infection such as Porphyromonas gingivalis (P. gingivalis), Tannerella forsythia, and Aggregatibacter actinomycetemcomitans [6–9]. However, Fusobacterium nucleatum (F. nucleatum), which is also a fundamental periodontal pathogen, has not been extensively studied in the field of periodontics, and the pathogenicity of F. nucleatum has not been extensively studied in oral tissues and cells.
F. nucleatum, as a bridging bacterium, transfers critical periodontal pathogens to periodontal infectious sites and recruits and activates local immune cells, which results in tooth-supporting tissue destruction [10, 11]. F. nucleatum has been identified as a high-frequency pathogen in periodontal disease [12] and many other infectious diseases, such as ventriculitis and brain abscesses [13], liver abscesses [14, 15], lung abscesses [16], septicemia-related infections [17], pelvic inflammatory disease [18], and intrauterine infections [19–21]. F. nucleatum attacks host tissues and obstructs the healing of damaged oral tissues by secreting large amounts of ammonia and butyrate [22, 23] and accelerates the initiation and progression of colorectal cancer by promoting tumor cell proliferation [24]. However, the effects of F. nucleatum on gingival fibroblast (GF) proliferation and apoptosis have not been reported.
The host response to pathogenic invasion is the determining factor of human health. As the first line of physical and chemical defense against infection, gingival epithelial cells release antimicrobial peptides such as human β-defensins to repress plaque biofilm formation, and activate innate and adaptive immune responses to protect the host from injury [25, 26]. Pathogens penetrate the connective tissue, directly damage the periodontal supporting tissues, and then induce the destruction of the integrity of the gingival epithelium [5, 27]. Human GFs, the predominant cell type of the periodontal connective tissue, are activated and recruited to sites of infection to regulate the occurrence and development of diseases [28–30]. It has been reported that GFs directly act on oral pathogens to defend against the progression of the infection by secreting various types of cytokines and chemokines, such as interleukin- (IL-) 1β, IL-6, IL-8, and tumor necrosis factor- (TNF-) α [31, 32]. Cytokine production is a crucial component for host defense against pathogenic invasion [31, 32]. A previous study confirmed that GFs showed no tolerance to bacterial stimulation and could continuously respond to exogenous stimuli and produce high levels of inflammatory cytokines [33]. However, the understanding of the pathogenic effects of F. nucleatum on inflammatory cytokine production by GFs and the potential mechanism has not been completely elucidated.
It has been reported that bacterial infection induces the mitochondrial electron transport for aerobic respiration and then dramatically elevates the intracellular reactive oxygen species (ROS) levels, which play critical roles in regulating cellular proliferation, apoptosis, and inflammatory response [34–36]. F. nucleatum has been shown to induce bladder cancer cell apoptosis through mediating ROS generation and mitochondrial dysfunction [34]. F. nucleatum enhances proinflammatory cytokine production by causing the impairment of autophagic flux in Caco-2 cells [35]. However, the effects of F. nucleatum on GF ROS generation remain unclear.
In this study, we aimed to explore the comprehensive gene expression profile of GFs over an F. nucleatum stimulation time course. We validated the effects of F. nucleatum on GF proliferation, apoptosis, intracellular ROS generation, and inflammatory cytokine production by biological experiments. Furthermore, we investigated the potential mechanism by which F. nucleatum regulated the biological properties of GFs according to the significantly enriched signaling pathways.
2. Materials and Methods
2.1. Human Subjects and Ethical Statements
This study was approved by the Medical Ethical Committee of the Stomatology School, Shandong University (Protocol Number: 20170101). Five healthy individuals aged 30-35 who underwent impacted tooth extraction at the Department of Oral and Maxillofacial Surgery, Stomatology Hospital of Shandong Province, were recruited. All individuals were informed about the research project and signed the informed consent form according to the Helsinki Declaration of 1975.
2.2. Cell Isolation and Culture
The excised gingival tissues were immediately immersed in Dulbecco's modified Eagle's medium (DMEM, HyClone, Logan, UT, USA) with 5% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin, Sigma-Aldrich, St Louis, MO, USA) and quickly transferred to the laboratory. Then, the free gingival tissue samples were washed, and the epithelium was removed. The samples were minced into small fragments of approximately 1-3 mm2 and digested for 2 h at 37°C by 3 mg/ml collagenase I (Sigma Aldrich) and 4 mg/ml Dispase II (Invitrogen, Carlsbad, CA, USA). The primary GFs were cultured with DMEM containing 20% fetal bovine serum (FBS, Biological Industries, Kibbutz, Israel) at 37°C in a humidified atmosphere of 5% CO2. Cells were fed fresh medium every three days until the cell monolayer reached 80-90% confluence. GFs were trypsinized and passaged at a dilution ratio of 1 : 3 to expand the culture in 10% FBS medium. The fourth passage cells were used for the following experiments, and cells were cultured in 10% FBS DMEM in all experiments.
2.3. RNA-Sequencing (RNA-Seq) Analysis
A total of 54 samples from 5 individuals were sequenced to analyze the gene expression at the whole genome level at BGI (Beijing Genomics Institute, Shenzhen, China) by RNA-seq (each individual sample included 1 sample at time point 0 h; 5 control samples at 2 h, 6 h, 12 h, 24 h, and 48 h with no F. nucleatum treatment; and 5 experimental samples with F. nucleatum stimulation at multiplicity of infection (MOI) of 100 at the same time points; sufficient RNA could not be extracted from one treated sample at the 12 h time point). Total RNA was isolated, evaluated for quality, reverse-transcribed to cDNA, and sequenced on the BGISEQ-500 platform. After a quality control (QC) step (Agilent 2100 Bioanalyzer, Santa Clara, California, USA), the clean reads were mapped to the reference genome (GRCh38) via hierarchical indexing for spliced alignment of transcripts (HISAT) (v2.0.4) [37]. Bowtie 2 (v2.2.5) [38] was used to map the clean reads to the reference transcripts, and the gene expression level for each sample was calculated by RSEM [39]. The correlation among all samples was detected by principal component analysis (PCA). Based on the gene expression level, DESeq2 and PoissonDis algorithms were used to detect the differentially expressed genes (DEGs) between the control groups and the F. nucleatum-treated groups by an absolute of log base 2 of fold change in DEGs ≥ 1 and an adjusted P value (%) < 5%. Gene ontology (GO) was used for screening and annotating DEGs. WEGO software [40] was used to generate the GO functional classification file. Pathway enrichment analysis of DEGs was performed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [41]. A Venn diagram was used to show the overlapping genes among the DEGs of each group. The protein-protein interaction network was generated by the STRING [42] website with default parameters. The Pathview [43] website was used to analyze the signaling pathway activation after F. nucleatum stimulation. In addition, Venn diagrams and heat maps were used to present the expression signatures of the DEGs involved in the GO biological processes, including cell proliferation (GO: 0008283), apoptotic process (GO: 0006915), response to reactive oxygen species (GO: 0000302), and defense response (GO: 0006952). The RNA sequence data have been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE118691.
2.4. Cell Proliferation Assay
GFs were seeded in 6-well (1 × 105 cells/well) plates in growth medium with 10% FBS. Cells were left untreated or incubated with F. nucleatum at multiplicities of infection (MOIs) (F. nucleatum : cell of 10 : 1, 50 : 1, 100 : 1, 200 : 1, and 400 : 1). Cells were counted every other day using a hemocytometer (Corning, Corning, NY, USA) or an automated cell counter (Countstar, Shanghai, China). To assess the cell proliferation rate, cells were inoculated in 24-well plates (5 × 104 cells/well) and treated with the indicated concentration of F. nucleatum. The 5-ethynyl-2′-deoxyuridine labeling assay was used to evaluate the cell proliferation rate according to the instructions of an EdU Apollo DNA in vitro kit (RiboBio, Guangzhou, China). The experiment was performed in sextuplicate and repeated three times.
2.5. Cell Apoptosis and Viability Analysis
Cell apoptosis was analyzed according to the instructions of an Annexin V-FITC/PI kit (Dojindo, Kumamoto, Japan). GFs were seeded in 6-well plates (2 × 105cells/well) and stimulated with different densities of F. nucleatum for 2, 6, 12, 24, 36, and 48 h. Afterward, cells were trypsinized, washed with phosphate-buffered saline (PBS), stained with a PI-conjugated anti-annexin V antibody for 15 min at room temperature in the dark, and then subjected to flow cytometry; the data were analyzed by Accuri C6 Plus software (Becton Dickinson, Franklin Lakes, NJ, USA). Cell viability was detected by trypan blue (Solarbio) staining. GFs treated with 4% formaldehyde were used as a positive control, and untreated cells were used as a negative control. Trypan blue was added to the monolayers at a 1 : 10 dilution and incubated for 10 min. The cells were observed immediately under the microscope and photographed (Olympus, Tokyo, Japan). The experiment was performed in triplicate and repeated three times.
2.6. Measurement of Intracellular ROS
Intracellular ROS levels were detected by a 2′,7′-dichlorofluorescein diacetate (DCFH-DA) assay. GFs were seeded in 6-well plates and treated with F. nucleatum at the above-mentioned MOIs for 2, 6, 12, 24, 36, and 48 h. Cells were incubated with DCFH-DA (1 : 1000) for 20 min at 37°C in a cell incubator in the dark. Flow cytometry (Becton Dickinson) was used to determine the intracellular ROS production. The flow cytometry data were analyzed by Accuri C6 Plus software. Each experiment was performed in triplicate and repeated three times.
2.7. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
GFs were seeded in 6-well plates and cultured with F. nucleatum. GFs were collected at 6 different time points (0 h, 2 h, 6 h, 12 h, 24 h, and 48 h), and total RNA was extracted with TRIzol® (CWBIO, Beijing, China). The mRNA concentration was determined using an ultramicro spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). One microgram of mRNA was reverse-transcribed to cDNA using a HiFiScript cDNA Synthesis kit (CWBIO). qRT-PCR was performed with an UltraSYBR Mixture (CWBIO) on a LightCycler 96 Real-Time PCR System (Roche, Basel, Switzerland) in triplicate. Briefly, the hot start enzyme was activated (95°C for 10 min), and the cDNA was then amplified for 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s and extension at 72°C for 32 s. Data were analyzed using the 2(-ΔΔCt) method. The sequences of the primers for amplification are shown in Supplementary . The experiment was performed in triplicate and repeated three times.
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
GFs were seeded in 6-well plates and treated with F. nucleatum at an MOI of 100 : 1. Cell culture supernatants were collected and centrifuged at 12,000 rpm for 5 min at 4°C. The levels of secreted IL-6, IL-8, IL-1β, and TNF-α protein were measured by ELISA (BioLegend, San Diego, CA, USA) according to the manufacturer's instructions. The optical density values were measured by a microplate reader at 450 nm and 570 nm, and the 570 nm values were subtracted from the absorbance at 450 nm in the subsequent data analysis. The experiment was performed in triplicate and repeated three times.
2.9. Western Blot Assay
GFs with or without F. nucleatum stimulation were harvested with a RIPA lysis buffer containing 1% protease inhibitors and 1% phosphatase inhibitors (Solarbio, Beijing, China). Protein concentrations were measured according to bicinchoninic acid (BCA) assays, and proteins (20 μg/lane) were separated to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% nonfat milk for 1 h, incubated with primary antibodies overnight at 4°C, and then incubated with horseradish peroxidase-conjugated secondary antibodies (1 : 10 000; Proteintech, Chicago, IN, USA) for 1 h at room temperature. The protein bands were visualized with enhanced chemiluminescence reagents (Millipore) and scanned using an extra-sensitive imager (Amersham Imager 600; GE Healthcare Life Sciences, Pittsburgh, PA, USA). ImageJ 1.44 software (NIH, Bethesda, Maryland, USA) was used to quantify the protein expression levels. The primary antibodies and dilution ratio were as follows: rabbit anti-NF-κB p65 (1 : 10000; Abcam, Cambridge, UK), rabbit anti-phospho-NF-κB p65 (1 : 1000; Abcam), rabbit anti-IκBα (1 : 5000; Abcam), rabbit anti-phospho-IκBα (1 : 1000;Abcam), rabbit anti-p38 (1 : 1000; Cell Signaling Technology, Danvers, MA, USA), rabbit anti-phospho-p38 (1 : 1000; Cell Signaling Technology), rabbit anti-JNK (1 : 1000; Cell Signaling Technology), rabbit anti-phospho-JNK (1 : 1000; Cell Signaling Technology), rabbit anti-ERK1/2 (1 : 1000; Cell Signaling Technology), rabbit anti-phospho-ERK1/2 (1 : 1000; Cell Signaling Technology), rabbit anti-AKT (1 : 1000; Cell Signaling Technology), rabbit anti-phospho-AKT (1 : 1000; Cell Signaling Technology), rabbit anti-p53 (1 : 1000; Abcam), and rabbit anti-phospho-p53 (1 : 1000;Abcam). The experiment was performed in triplicate and repeated three times.
2.10. Cell Immunocytochemistry Assay
To detect F. nucleatum-induced nuclear translocation of NF-κB p65 and p-p65, GFs were seeded in coverslip containing 24-well plates at a density of 2 × l04 cells/well and treated with or without F. nucleatum for 5 min to 120 min. Cells were fixed with 4% paraformaldehyde and blocked with 10% normal goat serum in PBS for 1 h. Then, cells were incubated with primary antibodies against p65 and p-p65 overnight at 4°C. The next day, the cells were incubated with an Alexa Fluor 594-conjugated goat anti-rabbit IgG secondary antibody (1 : 500) in the dark for 1 h, and the nuclei were visualized using 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) for 5 min. The images were captured by fluorescence microscopy (Olympus BX53, Tokyo, Japan). The experiment was performed in triplicate and repeated three times.
2.11. Statistical Analysis
All data were expressed as the mean ± standard deviation (SD). Tests were analyzed using GraphPad Prism software (version 6, Software MacKiev, Boston, MA, USA), and differences among more than two groups were analyzed by one-way or two-way ANOVA followed by Tukey's honestly significant difference (HSD) comparison test. Variance between two groups was compared by a multiple t-test. P < 0.05 was considered indicative of statistical significance.
3. Results
3.1. RNA-Seq Analysis of F. nucleatum-Stimulated GFs over Time
To better understand the global responses of GFs to F. nucleatum infection over time, we performed a genome-wide transcriptome analysis by RNA-seq to determine the global changes in gene expression. A total of 100 Gb of sequence data was generated from 54 samples, and 23.90 million reads for each sample were generated on average (Supplementary ). After QC, the clean reads were mapped to the reference genomes and transcripts with mapping percentages of 94.40% and 87.67%, respectively (Supplementary and Supplementary ). A total of 18691 genes were detected, and the expression levels were calculated with RSEM (data can be obtained through GSE118691). The correlation analysis of the PCA on the whole-genome gene expression levels showed that the normal cells were relatively stable, while the F. nucleatum-stimulated GFs gradually deviated from normal conditions as the stimulus time increased (Supplementary ). These results suggest that the effect of F. nucleatum stimulation on GFs may accumulate over time.
To characterize the DEGs influenced by F. nucleatum, the gene expression profiles of normal GFs versus infected cells at 2, 6, 12, 24, and 48 h were compared at each time point. Approximately 228, 374, 616, 1208, and 1334 genes were found to be upregulated, and 28, 147, 448, 1302, and 2145 genes were downregulated at 2, 6, 12, 24, and 48 h, respectively (Figure 1(a)). The overlap analysis of the DEGs among the 5 time points identified that 62 genes were differentially expressed between nonstimulated and F. nucleatum-stimulated GFs across the entire time course, and the number of DEGs continuously increased as the stimulation time increased (Figure 1(b)). These results suggest that the response of GFs to F. nucleatum infection increases over time, and a longer duration F. nucleatum stimulation might induce more side-effects than a short duration infection.
Figure 1.
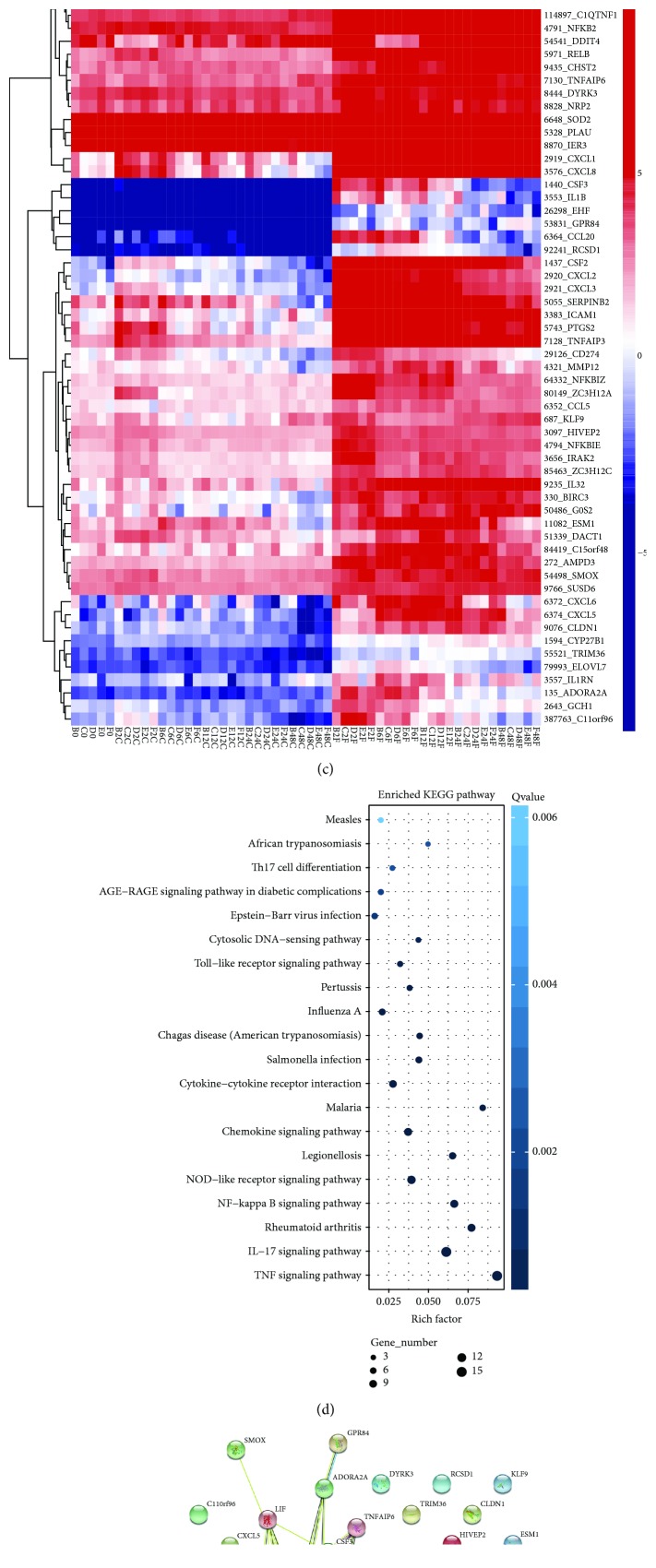
RNA-sequencing analysis of GFs stimulated with F. nucleatum (MOI of 100). (a) The number of DEGs in 54 samples after F. nucleatum stimulation at 2 h, 6 h, 12 h, 24 h, and 48 h. (b) Venn diagram summarizing the overlapping DEGs among the five time points. (c) Heat map of the 62 overlapping DEGs. (d) KEGG enrichment analysis of the 62 overlapping DEGs. (e) Network of 62 overlapping DEGs after F. nucleatum stimulation at 2 h, 6 h, 12 h, 24 h, and 48 h.
The heat map showed that the gene expression levels of 62 DEGs were upregulated and that some of these genes, such as IL-6, IL-1β, CCL2, CXCL2, CXCL8, and CSF3, are closely associated with host defense responses to bacterial infection (Figure 1(c)). The KEGG annotation showed that the top 5 enriched pathways of these DEGs are immune-associated, namely, the TNF-signaling pathway, IL-17 signaling pathway, rheumatoid arthritis, NF-κB signaling pathway, and NOD-like receptor signaling pathway (Figure 1(d)). The protein-protein interaction network of 62 DEGs showed the potentially complex interaction relationship among these genes (Figure 1(e)).
The GO analysis confirmed that the 62 DEGs are involved in host defense to bacterial infection and cell growth pathways, such as cellular process, response to stimulus, regulation of biological process, immune system process, response to bacterium, and cell proliferation (Supplementary ), and the top five enriched biological process GO terms for the 62 DEGs are shown in Supplementary .
3.2. F. nucleatum Inhibits the Proliferation of GFs
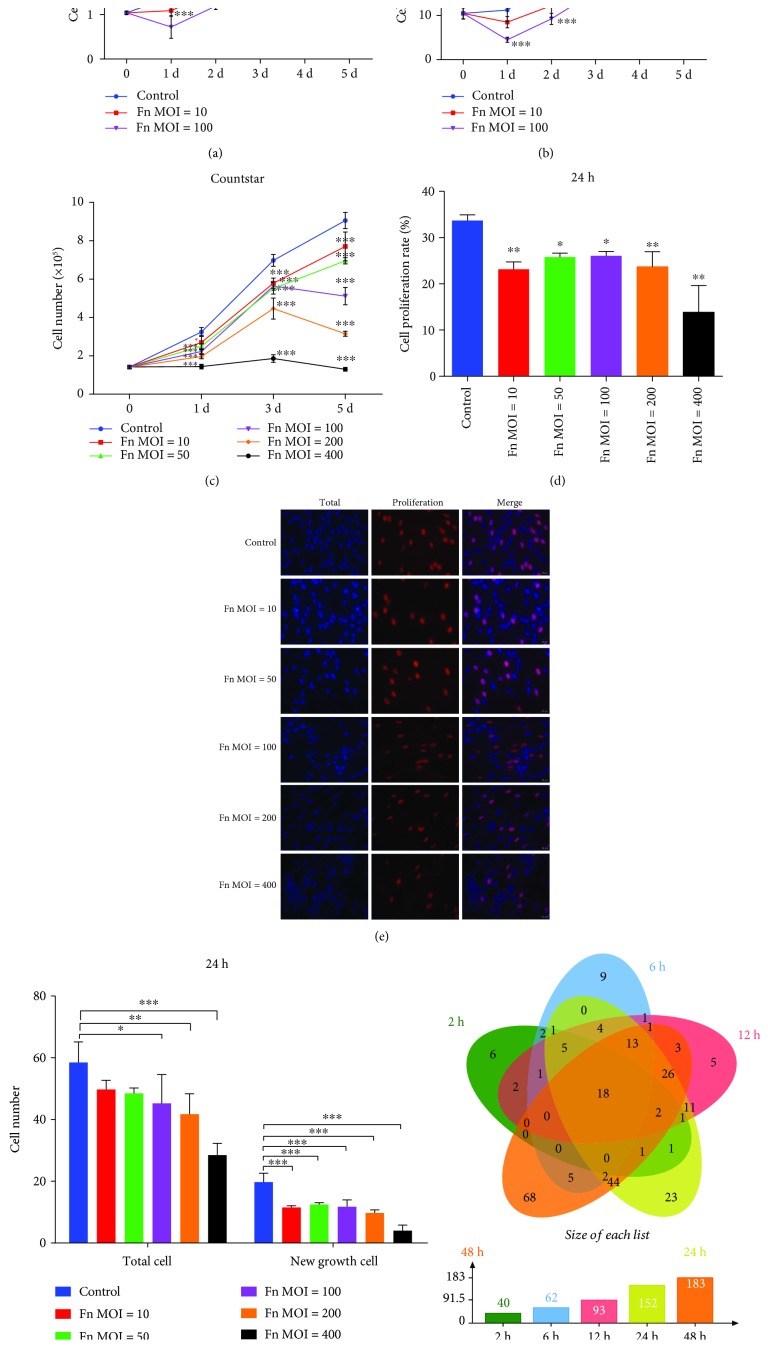
To study the effect of F. nucleatum on the proliferation of GFs, we applied a cell counting assay and EdU-labeling assay to quantify the cell proliferation rate over 5 days of F. nucleatum stimulation at MOIs of 10 and 100. The cell counting assay showed that F. nucleatum significantly inhibited the proliferation of GFs in a time- and dose-dependent manner (Figures 2(a) and 2(b)). To further test the inhibitory effect of different F. nucleatum concentrations, GFs stimulated with F. nucleatum at MOIs of 10, 50, 100, 200, and 400 were quantified at 3 time points over 5 days. The results showed that the inhibitory effect aggrandized with the increase of F. nucleatum concentration, and the high concentration of F. nucleatum (MOI of 400) directly blocked the cell growth (Figure 2(c)). The EdU-labeling assay demonstrated that F. nucleatum significantly decreased the number of GFs and reduced the cell proliferation rate after 24 h of stimulation (Figure 2(d)–2(f)). The DEGs of the 54 samples enriched in the cell proliferation pathway (GO: 0008283) were compared using an overlap analysis, and the results indicated that the number of cell proliferation-related DEGs continuously increased as the F. nucleatum stimulation time increased; 40, 62, 93, 152, and 183 genes were detected at 2 h, 6 h, 12 h, 24 h, and 48 h, respectively, and 18 DEGs, including CCL2, SOD2, and NFKBIA, were consistently upregulated across all the time points (Figure 2(g) and Supplementary ). The above results suggest that F. nucleatum dose- and time-dependently regulates cell proliferation-related genes and inhibits the proliferation of GFs.
Figure 2.
Effects of F. nucleatum on cell proliferation. Cell numbers of GFs detected by an electronic cell counter (a) and hemocytometer (b) with or without F. nucleatum stimulation at MOIs of 10 and 100 (n = 6). (c) Number of GFs detected by electronic cell counter after F. nucleatum stimulation (MOIs of 0, 10, 50, 100, 200, and 400, n = 6). (d) Cell proliferation rate of GFs detected by EdU assay (n = 6). (e) EdU assay of GFs after F. nucleatum stimulation (MOIs of 0, 10, 50, 100, 200, and 400) at 24 h. Scale bar: 20 μm. NC: negative control; PC: positive control. (f) Statistical results of total and new cell growth after EdU labeling (n = 6). (g) Venn diagram of cell proliferation-related DEGs from the GO biological process analysis among the five time points. All data are shown as the mean ± SD. Statistical analyses were performed by one-way (d) and two-way (a, b, c, f) ANOVA with Tukey's multiple-comparison test. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 compared with the Control.
3.3. F. nucleatum Promotes the Apoptosis of GFs
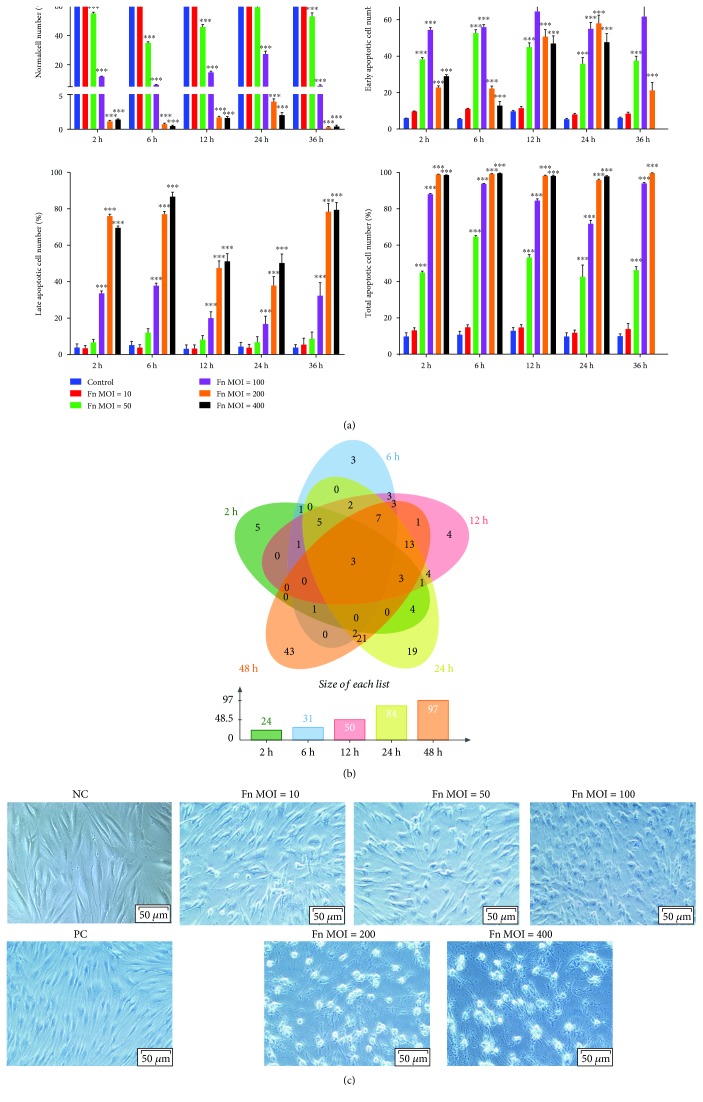
To further study the reason for the proliferation inhibition of GFs, the apoptosis rates of F. nucleatum-stimulated GFs were detected by flow cytometry. The results indicated that the low concentration of F. nucleatum (MOIs of 10) showed no influence on GF apoptosis, while high concentration of F. nucleatum (MOIs of 50, 100, 200, and 400) significantly decreased the number of normal cells and increased the number of apoptosis cells at 2 h stimulation and maintained the similar apoptosis trend to 36 h. At MOIs of 50 and 100, the number of early apoptotic cells was greater than the number of late apoptotic cells at the 2, 6, 12, 24, and 36 h F. nucleatum stimulation, while at the MOIs of 200 and 400, the number of late apoptotic GFs was larger than the early apoptotic cells (Figure 3(a) and Supplementary ). The DEGs of the 54 samples enriched in the apoptotic process (GO: 0006915) were compared using an overlap analysis, and the results suggested that 24, 31, 50, 84, and 97 apoptosis-related genes were differentially expressed at 2 h, 6 h, 12 h, 24 h, and 48 h, respectively (Figure 3(b)). Three apoptosis-related genes, CSF2, PTGS2, and SOD2, were continuously upregulated across the 5 time points (Figure 3(b) and Supplementary ). A trypan blue staining assay indicated that F. nucleatum dose-dependently increased the number of nonviable cells, which were stained blue, and the cell morphology changed from long spindle to round, and fewer live cells were detected after F. nucleatum stimulation at MOIs of 200 and 400 (Figure 3(c)). These results suggest that F. nucleatum might inhibit the proliferation of GFs by dose-dependently promoting GF apoptosis and time-dependently upregulating a group of apoptosis process-related genes.
Figure 3.
Effect of F. nucleatum on GF apoptosis. (a) Statistical analysis of GF apoptosis with F. nucleatum stimulation (MOIs of 0, 10, 50, 100, 200, and 400) at 2 h, 6 h, 12 h, 24 h, and 36 h. The histogram represents the mean ± SD (n = 3). (b) Venn diagram of apoptosis-related DEGs from the GO biological process analysis of the five paired comparisons. (c) Trypan blue staining results of GFs without or with F. nucleatum stimulation at MOIs of 10, 50, 100, 200, and 400 at 24 h. Scale bar: 50 μm. NC: negative control; PC: positive control. Statistical analyses were performed by two-way ANOVA with Turkey's multiple-comparison test. ∗∗∗P < 0.001 compared with the control at each time point.
3.4. F. nucleatum Promotes Intracellular ROS Generation in GFs
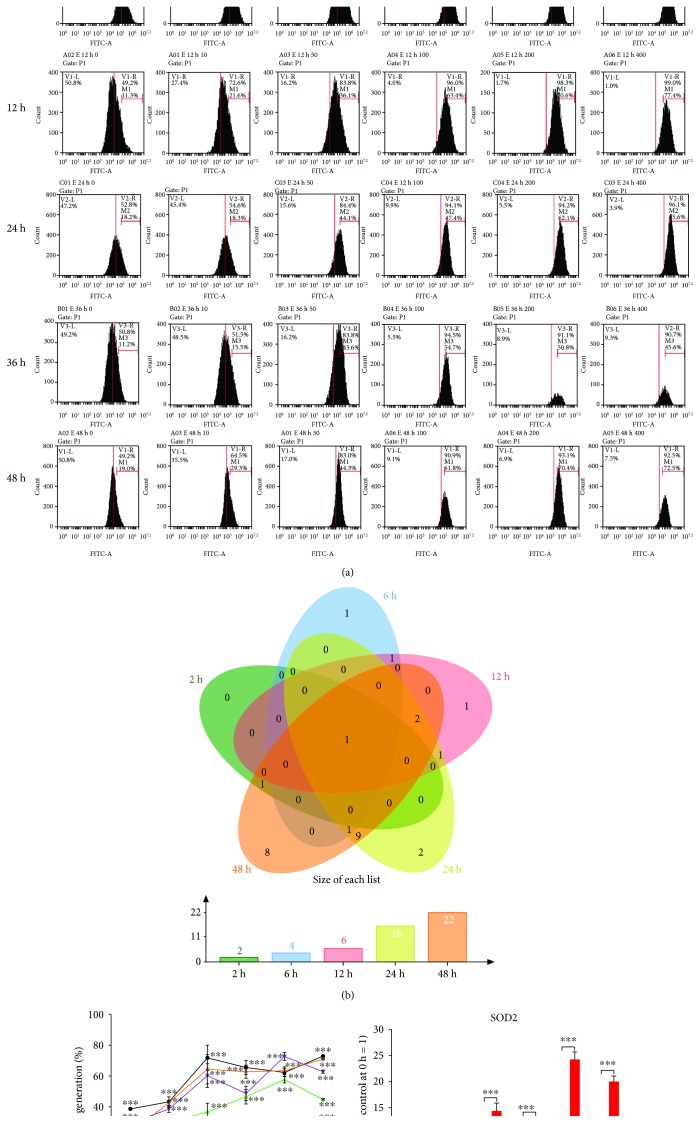
To investigate whether the alteration in GF biological properties is associated with intracellular ROS production, GFs were treated with F. nucleatum (MOIs of 10, 50, 100, 200, and 400) for 2, 6, 12, 24, 36, and 48 h, and the level of ROS was detected at the different stimulation times and doses. The flow cytometry analysis results showed that the level of intracellular ROS was significantly elevated in a dose- and time-dependent manner (Figures 4(a) and 4(c)). The DEGs of the 54 samples enriched in the response to reactive oxygen species (GO: 0000302) were compared using an overlap analysis, and the results indicated that the number of differentially expressed ROS production-related genes also increased in a time-dependent manner, and 2, 4, 6, 16, and 22 DEGs were detected after F. nucleatum stimulation (MOI of 100) at 2, 6, 12, 24, 36, and 48 h (Figure 4(b)). SOD2 is a unique DEG that was continuously upregulated across the 5 time points. The relative gene expression level of SOD2 is shown in Figure 4(d). In addition, SOD2 also acts as the main contributor to the regulation of the proliferation and apoptosis of GFs (Supplementary and Supplementary ).
Figure 4.
Effects of F. nucleatum on ROS generation in GFs. (a) Flow cytometry analysis of ROS production with F. nucleatum stimulation (MOIs of 0, 10, 50, 100, 200, and 400) at 2 h, 6 h, 12 h, 24 h, and 36 h. (b) Venn diagram of ROS production-related DEGs from the GO biological process analysis after the five paired comparisons. (c) Statistical results of ROS generation by flow cytometry (n = 3). (d) The relative gene expression level of SOD2 by qRT-PCR. The histogram represents the mean ± SD. Statistical analyses were performed by two-way ANOVA with Turkey's multiple-comparison test (c) and multiple t-test (d). ∗P < 0.05 and ∗∗∗P < 0.001 compared with the control at each time point.
3.5. F. nucleatum Promotes GF Inflammatory Cytokine Production
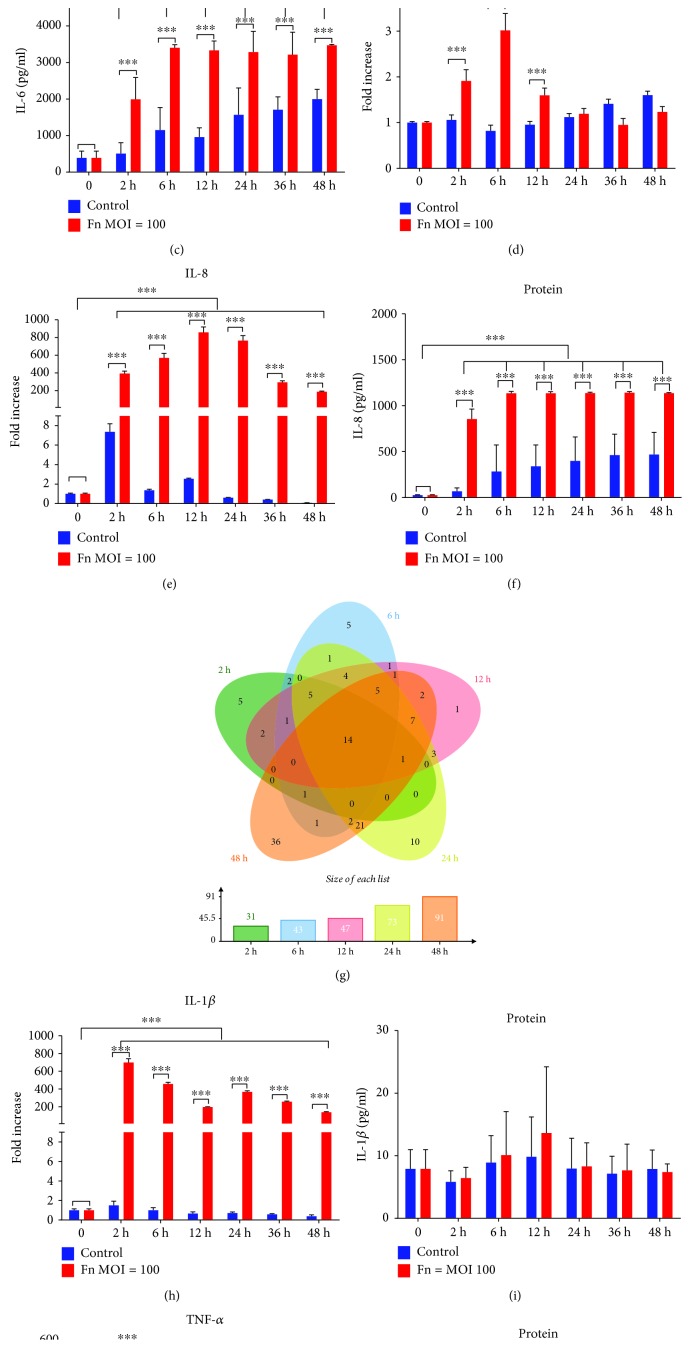
The expression of Toll-like receptor- (TLR-) 2 and TLR4, which are closely related to inflammatory cytokine production, was analyzed by qRT-PCR. The expression of the TLR4 gene was elevated after F. nucleatum stimulation at 2 h, 12 h, 24 h, and 36 h, while showing no significant differences compared with the TLR4 expression in normal cells at 48 h (Figure 5(a)). The TLR2 gene expression level was increased in F. nucleatum-stimulated GFs at 2 h, 6 h, and 12 h, while showing no significant differences at 24 h and 48 h compared with that in nonstimulated GFs (Figure 5(d)). Compared with nonstimulated GFs at each time point, F. nucleatum-stimulated GFs showed significantly elevated gene expression levels of IL-6, IL-8, IL-1β, and TNF-α across the whole time course (Figures 5(b), 5(e), 5(h), and 5(j)); however, at the protein level, IL-6 and IL-8 were significantly increased after F. nucleatum stimulation, while IL-1β and TNF-α were not (Figures 5(c), 5(f), 5(i), and 5(k)). The DEGs of the 54 samples enriched in the defense response (GO: 0006952) were compared using an overlap analysis, which indicated that 31, 43, 47, 73, and 91 DEGs related to defense response were time-dependently increased after F. nucleatum stimulation; 14 overlapping DEGs were significantly upregulated among the 5 time points in the F. nucleatum stimulation groups (Figure 5(g) and Supplementary ).
Figure 5.
Effects of F. nucleatum on inflammatory cytokine production in GFs. TLR4 (a), TLR2 (d), IL-6 (b), IL-8 (e), IL-1β (h), and TNF-α (g) gene expression with F. nucleatum stimulation (MOIs of 0 and 100) from 0 to 48 h (n = 3). IL-6 (c), IL-8 (f), IL-1β (i), and TNF-α (k) protein expression with F. nucleatum stimulation (MOIs of 0 and 100) from 0 to 48 h (n = 3). (g) Venn diagram of the defense response-related DEGs among the five time points. The histogram represents the mean ± SD. Statistical analyses were performed by multiple t-test. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6. F. nucleatum Activates GF Nuclear Factor-κB (NF-κB), Mitogen-Activated Protein Kinase (MAPK), and Protein Kinase B (AKT) Signaling Pathway
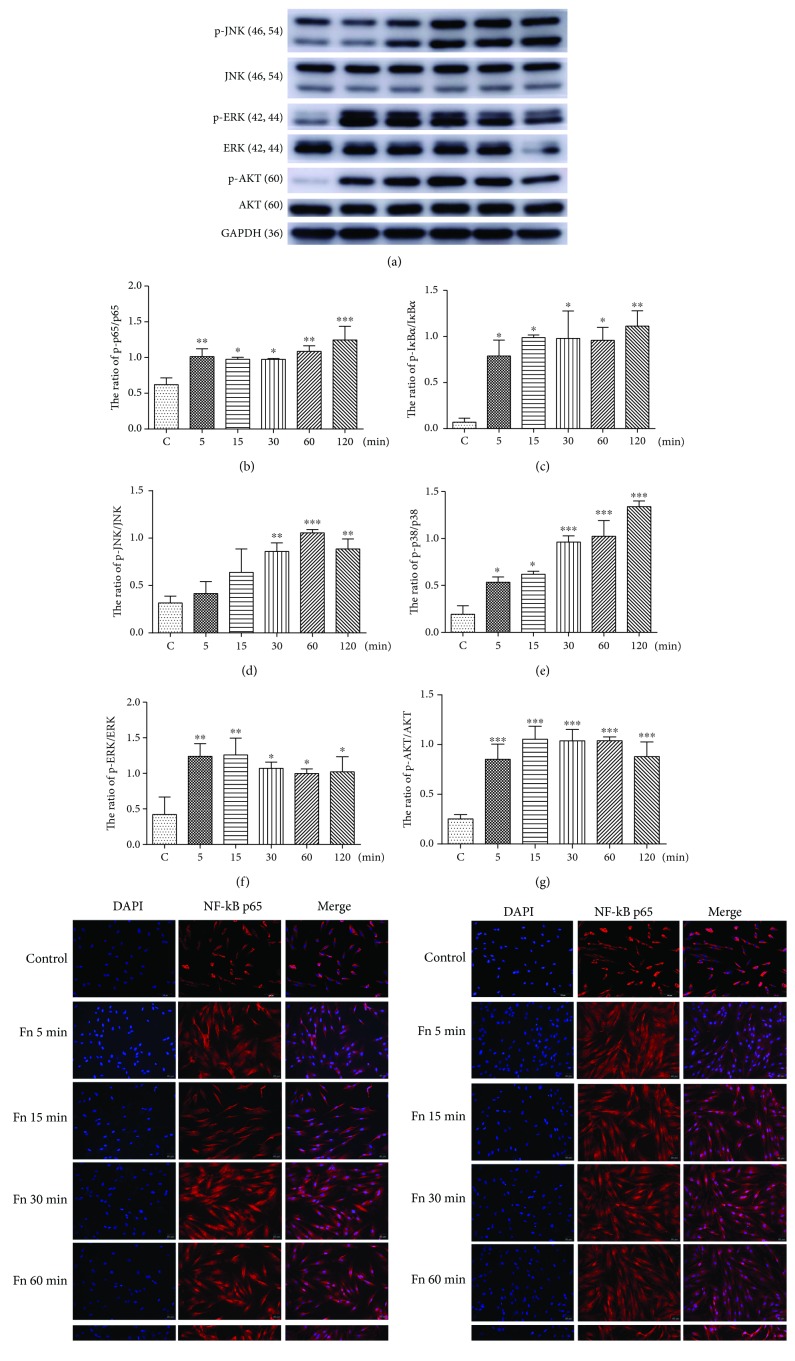
To further explore the mechanisms of F. nucleatum-induced biological process alteration in GFs, the cell proliferation, apoptosis, and immune response-related NF-κB, MAPK, and AKT signaling pathways were analyzed by western blotting and immunofluorescence. The results showed that F. nucleatum increased the proportion of p-p65 to p65 and p-IκBα to IκBα after 5 min of stimulation and that this increase was maintained for 120 min (Figure 6(a)–6(c)). The immunofluorescence analysis of cells showed that p65 and p-p65 translocation from the cytoplasm to the nucleus also increased continuously after F. nucleatum stimulation from 5 min to 120 min (Figures 6(h) and 6(i)). These results indicate that F. nucleatum activates the NF-κB signaling pathway by increasing the phosphorylation levels of p65 and IκBα and increasing the nuclear-to-cytoplasmic ratio of NF-κB p65 and NF-κB p-p65 levels. Furthermore, we confirmed that F. nucleatum significantly activated AKT/MAPK signaling pathways by increasing the phosphorylation levels of AKT, ERK, JNK, and p38 (Figures 6(a) and 6(d)–6(g)). The protein expression ratio of p-AKT/AKT and p-ERK/ERK was elevated after F. nucleatum stimulation at 5 min and maintained at a high level until 120 min (Figures 6(f) and 6(g)). In addition, the relative value of p-JNK/JNK was increased after 30 min of F. nucleatum stimulation (Figure 6(d)), and the phosphorylation level of p38 was significantly increased after 5 min of stimulation with F. nucleatum (Figure 6(e)). Furthermore, the protein level of p53 and phosphorylated p53 was detected, and the results indicated that F. nucleatum could not increase the protein expression ratio of p-p53/p53 after stimulation for 5 min to 120 min (Supplementary ).
Figure 6.
Effects of F. nucleatum on the activation of the NF-κB, MAPK, and PI3K-AKT pathways in GFs. (a) The protein levels of p38, phosphorylated p38, JNK, phosphorylated JNK, ERK, phosphorylated ERK, NF-κB p65, phosphorylated NF-κB p65, IκBα, phosphorylated IκBα, AKT, and phosphorylated AKT were detected by western blotting. The relative level of phosphorylated NF-κB p65/NF-κBp65 (b), phosphorylated IκBα/IκBα (c), phosphorylated JNK/JNK (d), phosphorylated p38/p38 (e), phosphorylated ERK/ERK (f), and phosphorylated AKT/AKT (g) was detected (n = 3). Immunofluorescence images of NF-κB p65 (h) and phosphorylated NF-κB p65 (i) in cells. The histograms represent means ± SD. Statistical analyses were performed using one-way ANOVA with Tukey's multiple-comparison test. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 compared with the Control.
3.7. Whole-Transcriptome Analysis of NF-κB, MAPK, and PI3K-AKT Signaling Pathways in F. nucleatum-Stimulated GFs
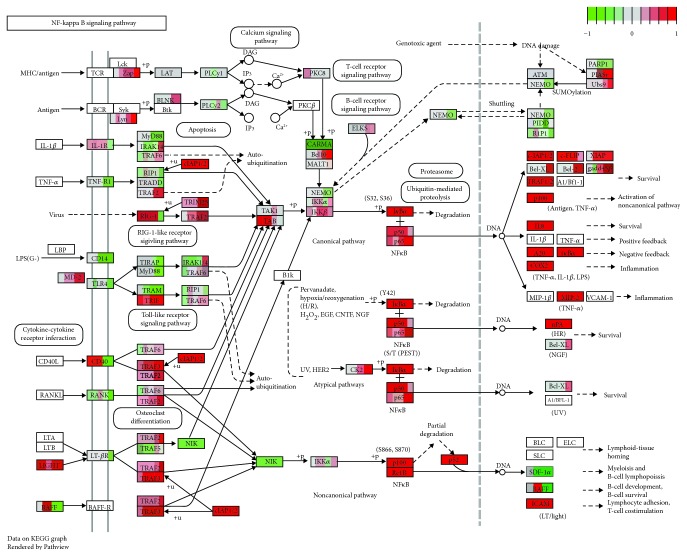
To further clarify the correlation among cell proliferation, apoptosis, oxidative stress, cytokine production and AKT/MAPK, and NF-κB signaling pathways, the DEGs induced by F. nucleatum stimulation from the 54 samples at each time point were integrated and visualized by Pathview. The results indicated that the DEGs were significantly enriched in the NF-κB, PI3K-AKT, and MAPK signaling pathways, which suggest that these pathways are significantly activated by F. nucleatum stimulation at the gene expression level and are consistent with the results of our biological experiments (Figure 7, Supplementary and Supplementary ). For the NF-κB signaling pathway, we confirmed that most DEGs downstream of the NF-κB signaling pathway were significantly upregulated by F. nucleatum stimulation, which is critical in regulating cell proliferation, apoptosis, ROS generation, and cytokine production (Figure 7). For example, cIAP1/2 and TRAF1/2 were elevated after F. nucleatum stimulation at 2 h, and Bcl-2 was upregulated at 12 h after F. nucleatum stimulation; these genes play key roles in cell proliferation and apoptosis. The gene expression levels of IL-8, cyclooxygenase 2 (COX2), A20, IκBα, and MIP2 were significantly upregulated throughout the whole F. nucleatum stimulation (from 2 to 48 h); these genes are closely associated with GF intracellular ROS generation and inflammatory cytokine production (Figure 7 and Supplementary ). In addition, we found that a large number of F. nucleatum-stimulated DEGs were enriched in the PI3K-AKT signaling pathway (Supplementary ) and MAPK signaling pathways (Supplementary ), which are mainly involved in cell proliferation and apoptosis. Furthermore, we found that the PI3K-AKT and MAPK signaling pathways were upstream of the NF-κB signaling pathway, which plays a synergetic role in regulating cell proliferation, apoptosis, ROS generation, and the inflammatory response.
Figure 7.
The Pathview analysis of the NF-κB signaling pathway. Every box is divided into five parts on average, and each part represents the relative gene expression level of F. nucleatum-stimulated GFs at 2 h, 6 h, 12 h, 24 h, and 48 h from left to right. The red color indicates that the gene expression level is upregulated after F. nucleatum stimulation. The green color indicates that the gene level is downregulated after F. nucleatum stimulation. The white color indicates that the gene expression level is not influenced by F. nucleatum stimulation. The gene expression level is calculated by the log fold change in the F. nucleatum stimulation group relative to the control group at each time point.
4. Discussion
Characterized by gingival inflammation and periodontal supporting tissue destruction, periodontitis is prevalent among adults over 30 years old [44, 45]. Understanding the pathogenic mechanisms of oral pathogens in periodontitis and identifying risk factors are important for maintaining human periodontal health. Previous studies have provided in-depth insights into the pathogenic effects of P. gingivalis [46]. F. nucleatum often appears simultaneously with P. gingivalis at periodontal infection sites [47], while its pathogenic role in periodontal disease has not been comprehensively reported. This study is the first to illuminate the comprehensive effects of F. nucleatum on cell proliferation, apoptosis, ROS generation, and inflammatory cytokine production in GFs.
Our study confirms that F. nucleatum inhibits the proliferation and promotes the apoptosis of GFs, which is in contrast to the role F. nucleatum plays in colorectal cancer cell (CRC) proliferation. F. nucleatum has been reported to promote E-cadherin-expressing CRC proliferation by modulating the E-cadherin/β-catenin signaling pathway or activating TLR4 signaling to NF-κB and upregulating the expression of microRNA-21 [24, 48]. Our transcriptomic analysis demonstrates that F. nucleatum alters the expression of proapoptotic genes in GFs by upregulating TRAILR2, NOXA, and PUMA expression and downregulating the substrates α-tubulin, actin, fodrin, and PARP (Supplementary ), which are crucial for the regulation of cell proliferation and apoptosis [49–55]. We corroborated the gene expression levels of TRAILR2, NOXA, PUMA, α-tubulin, actin, fodrin, and PARP by qRT-PCR, and the results indicated that F. nucleatum significantly upregulated TRAILR2 and NOXA at 2 to 48 h, while increased PUMA gene expression at 2, 24, and 48 h; F. nucleatum downregulated α-tubulin, actin, fodrin, and PARP gene expression at the long time stimulation at 12 h, 24 h, and 48 h, which were consistent with those of the RNA-seq analysis. However, at the 2 h and 6 h F. nucleatum stimulation, some disagreements between qRT-PCR and RNA-seq analysis were discovered in evaluating the gene level of actin and fodrin (Supplementary ). Moreover, F. nucleatum elevated the expression of Mcl-1, which ultimately induced apoptosis through bim-mediated mitochondrial apoptotic events (Supplementary ). F. nucleatum differentially regulates cell proliferation in GFs and CRCs, which indicates that the alteration of biological properties by F. nucleatum in normal human cells is quite different from that in tumor cell lines.
It has been reported that F. nucleatum stimulates ROS production and induces the tissue inflammatory response in TLR4-competent macrophages [56]. F. nucleatum induces IL-8, IL-1β, TNF-α, and ROS generation in Caco-2 colorectal adenocarcinoma cells by impairing autophagic flux [35]. In addition, recent studies have reported that in GFs, two NADPH oxidase isoforms, NOX1 and NOX2, are activated in response to F. nucleatum infection [57, 58]. However, the current study lacks the mechanism of ROS and inflammatory cytokine production in GFs after F. nucleatum stimulation. In this study, we confirm that F. nucleatum increases the production of ROS, activates TLR2 and TLR4 gene expression at early stimulation time points, and promotes the release of proinflammatory cytokines, such as IL-6 and IL-8 in GFs, which are concordant with the previous study. In addition, we found that F. nucleatum elevated the gene expression levels of IL-1β and TNF-α, but not the protein level. In gingival epithelial cells (GECs), F. nucleatum has been shown to activate the NLRP inflammasome and promote IL-1β secretion, which is different from our results in GFs [59, 60]. F. nucleatum differentially regulates IL-1β production in GFs and GECs, which indicates that the pathogenic mechanism of F. nucleatum in oral cells is quite complex, and it is necessary to clarify the mechanism of F. nucleatum in GFs.
Our genome-wide transcriptome analysis showed that F. nucleatum activates the NF-κB, MAPK, and PI3K-AKT signaling pathways, which is consistent with our biological validation results through western blotting assays and immunofluorescence. In addition, we demonstrated that a large number of DEGs are enriched in the MAPK signaling pathway and that the JNK and p38 MAPK signaling pathways are upstream of the NF-κB signaling pathway, which plays a key role in regulating cell proliferation, apoptosis, and inflammatory response [61–63]. The ERK MAPK and PI3K-AKT signaling pathways are significantly activated in GFs, and the expression levels of proliferation-related genes, such as Myc and Bcl-2, are dramatically varied after F. nucleatum stimulation, which are crucial in deciding the cell survival [64–67].
According to the Pathview analysis, we found that a larger number of DEGs that were induced by F. nucleatum stimulation downstream of NF-κB and AKT/MAPK signaling pathways (Figure 7 and Supplementary Figures , ) are associated with cell proliferation, apoptosis, ROS generation, and production of inflammatory cytokines, such as bcl2, IL-8, COX2, JNK, and Mcl-1. We validated DEGs including COX2, IL-8, and bcl2 by qRT-PCR, and the results were consistent with those of the RNA-seq analysis. IL-8 is an important chemokine and plays key roles in the acute inflammatory response and various inflammatory diseases [68, 69]. COX2 is critical for the formation of prostaglandins and maintains ROS homeostasis under inflammatory conditions, which are pivotal in the pathogenesis of periodontitis [70, 71]. In the future, we will construct some gene knockout strains of F. nucleatum to further explore the mechanism of F. nucleatum in GFs.
5. Conclusions
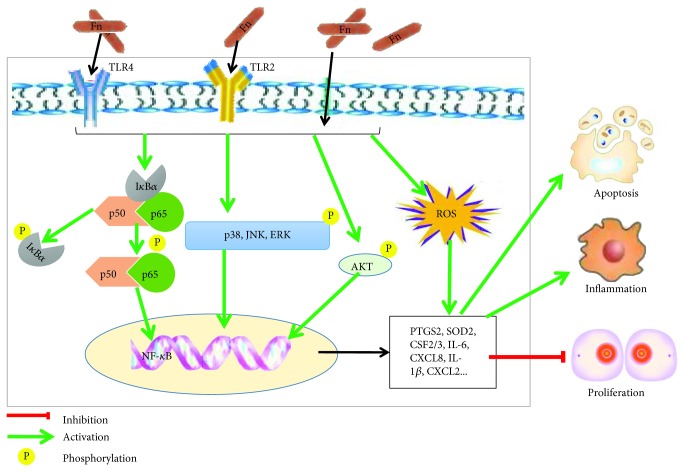
Our study indicates that F. nucleatum alters the gene expression profiles of GFs in a time-dependent manner and plays multidimensional roles in regulating GF biological properties. F. nucleatum inhibits cell proliferation and facilitates cell apoptosis, ROS generation, and inflammatory cytokine production partly through the activation of AKT/MAPK and the NF-κB signaling pathways in GFs. Our study opens a new window for understanding the pathogenic effects of periodontal pathogens on the host oral system. A schematic diagram of the effects of F. nucleatum on GFs and the potential pathogenic mechanism is shown in Figure 8.
Figure 8.
Putative mechanism for the effects of F. nucleatum on the biological processes in GFs. Schematic diagram depicting how F. nucleatum activates NF-κB, MAPK, and AKT signaling pathways and induces cell proliferation, apoptosis, and inflammatory response. TLR4: Toll-like receptor 4; TLR2: Toll-like receptor 2; IκBα: inhibitors of NF-κB; AKT/PKB: protein kinase B; NF-κB: nuclear factor kappa B; MAPK: mitogen-activated protein kinase; JNK: c-Jun N-terminal kinase; ERK: extracellular signal-regulated kinase.
Acknowledgments
We appreciate the volunteers who provided the samples. We thank Professor Shili Liu and other colleagues in our laboratory for all of the kind advice and support. We thank the Natural Science Foundation of Shandong Province (Nos. ZR201702190185 and 2018GSF118231), the National Natural Science Foundation of China (No. 81630072), the Construction Engineering Special Foundation of “Taishan Scholars” of Shandong Province (No. tsqn20161068), and the Program of Qilu Young Scholars of Shandong University for the financial support.
Abbreviations
- F. nucleatum:
Fusobacterium nucleatum
- GFs:
Gingival fibroblasts
- ROS:
Reactive oxygen species
- IL:
Interleukin
- TNF:
Tumor necrosis factor
- DMEM:
Dulbecco's modified Eagle's medium
- FBS:
Fetal bovine serum
- RNA-seq:
RNA sequencing
- QC:
Quality control
- DEGs:
Differentially expressed genes
- GO:
Gene ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- PCA:
Principal component analysis
- PBS:
Phosphate-buffered saline
- MOI:
Multiplicity of infection
- EdU:
5-Ethynyl-2′-deoxyuridine
- DCFH-DA:
2′,7′-dichlorofluorescein diacetate
- qRT-PCR:
Quantitative real-time polymerase chain reaction
- ELISA:
Enzyme-linked immunosorbent assay
- BCA:
Bicinchoninic acid
- SDS-PAGE:
Sulfate-polyacrylamide gel electrophoresis
- PVDF:
Polyvinylidene fluoride
- DAPI:
2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride
- SD:
Standard deviation
- ANOVA:
Analysis of variance
- TLRs:
Toll-like receptors
- NF-κB:
Nuclear factor-κB
- MAPK:
Mitogen-activated protein kinase
- AKT:
Protein kinase B
- CRCs:
Colorectal cancer cells
- GECs:
Gingival epithelial cells
- HSD:
Honestly significant difference.
Data Availability
All raw RNA-seq data are accessible through GEO series accession number, GSE118691. Other data from Materials and Methods used to support the findings of this study are included in the article. If any other data are needed, please contact the corresponding author.
Conflicts of Interest
The authors indicate no potential conflicts of interest.
Authors' Contributions
FQ and HKL conceived, designed, and supervised this study, and KWY and ZZW collected the samples. KWY, TD, and GH performed the experiments. KWY and JZL analyzed and interpreted the data. KWY and FQ wrote the paper. All authors commented on the manuscript.
Supplementary Materials
Table 1: primer sequences for quantitative real-time PCR (qRT-PCR).
Table 2: the clean reads of 54 samples after RNA-seq.
Table 3: genome mapping ratio summary of 54 samples.
Table 4: gene mapping ratio summary of 54 samples.
Table 5: top five enriched GO terms in the biological process for the 62 DEGs.
Figure 1: PCA of 54 samples of RNA-seq analyses.
Figure 2: GO analysis of the 62 overlapped DEGs after F. nucleatum stimulation at 2 h, 6 h, 12 h, 24 h, and 48 h.
Figure 3: heat map summarizing the 18 overlapped DEGs from the GO biological process analysis after the five paired comparisons of cell proliferation.
Figure 4: (a) Flow cytometry analysis of cell apoptosis. (b) Heat map of the 3 overlapped DEGs from the GO biological process analysis after the five paired comparisons of cell apoptosis.
Figure 5: heat map of the 14 overlapping DEGs from the GO biological process analysis after the five paired comparisons of cell defense response.
Figure 6: (a) The protein levels of p53, phosphorylated p53 (p-p53). (b) The ratio of p-p53 and p53.
Figure 7: the Pathview analysis of the PI3K-AKT signaling pathway.
Figure 8: the Pathview analysis of MAPK signaling pathway.
Figure 9: the relative gene expression level of Bcl-2 (a), COX2(b), A20 (c), IκBα (d), and MIP2 (e) by qRT-PCR.
Figure 10: the Pathview analysis of the apoptosis signaling pathway.
Figure 11: the relative gene expression level of TRAILR2 (a), NOXA (b), PUMA (c), tubulin (d), actin (e), fodrin (f), and PARP (g) by RT-PCR.
References
- 1.Albandar J. M., Tinoco E. M. B. Global epidemiology of periodontal diseases in children and young persons. Periodontology 2000. 2002;29(1):153–176. doi: 10.1034/j.1600-0757.2002.290108.x. [DOI] [PubMed] [Google Scholar]
- 2.Thornton-Evans G., Eke P., Wei L., et al. Periodontitis among adults aged≥30 years - United States, 2009-2010. MMWR Supplement. 2013;62:129–135. [PubMed] [Google Scholar]
- 3.Di Benedetto A., Gigante I., Colucci S., Grano M. Periodontal disease: linking the primary inflammation to bone loss. Clinical and Developmental Immunology. 2013;2013:17. doi: 10.1155/2013/503754.503754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darveau R. P. Periodontitis: a polymicrobial disruption of host homeostasis. Nature Reviews Microbiology. 2010;8(7):481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 5.Saini R., Marawar P. P., Shete S., Saini S. Periodontitis, a true infection. Journal of Global Infectious Diseases. 2009;1(2):149–150. doi: 10.4103/0974-777X.56251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emani S., Gunjiganur G. V., Mehta D. S. Determination of the antibacterial activity of simvastatin against periodontal pathogens, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans: an in vitro study. Contemporary Clinical Dentistry. 2014;5(3):377–382. doi: 10.4103/0976-237X.137959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Zhao J., Li C., Yu N., Zhang D., Pan Y. Clinical and microbiologic effect of nonsurgical periodontal therapy on patients with chronic or aggressive periodontitis. Quintessence International. 2013;44(8):575–583. doi: 10.3290/j.qi.a29752. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien-Simpson N. M., Veith P. D., Dashper S. G., Reynolds E. C. Antigens of bacteria associated with periodontitis. Periodontology 2000. 2004;35(1):101–134. doi: 10.1111/j.0906-6713.2004.003559.x. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi Y., Umeda M., Sakamoto M., Benno Y., Huang Y., Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. Journal of Periodontology. 2001;72(10):1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 10.Signat B., Roques C., Poulet P., Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Current Issues in Molecular Biology. 2011;13(2):25–36. [PubMed] [Google Scholar]
- 11.Diaz P. I., Zilm P. S., Rogers A. H. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(2):467–472. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- 12.Moore W. E. C., Moore L. V. H. The bacteria of periodontal diseases. Periodontology 2000. 1994;5(1):66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 13.Kai A., Cooke F., Antoun N., Siddharthan C., Sule O. A rare presentation of ventriculitis and brain abscess caused by Fusobacterium nucleatum. Journal of Medical Microbiology. 2008;57(5):668–671. doi: 10.1099/jmm.0.47710-0. [DOI] [PubMed] [Google Scholar]
- 14.Wijarnpreecha K., Yuklyaeva N., Sornprom S., Hyman C. Fusobacterium nucleatum: atypical organism of pyogenic liver abscess might be related to sigmoid diverticulitis. North American Journal of Medical Sciences. 2016;8(4):197–199. doi: 10.4103/1947-2714.179961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bytyci F., Khromenko E. Hepatic abscess caused by Fusobacterium necrophorum after a trip to the dentist. BMJ Case Reports. 2016;2016 doi: 10.1136/bcr-2015-210235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagaoka K., Yanagihara K., Morinaga Y., Kohno S. Detection of Fusobacterium nucleatum in two cases of empyema and lung abscess using paromomycin-vancomycin supplemented Brucella HK agar. Anaerobe. 2017;43:99–101. doi: 10.1016/j.anaerobe.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Kroon E., Arents N. A., Halbertsma F. J. Septic arthritis and osteomyelitis in a 10-year-old boy, caused by Fusobacterium nucleatum, diagnosed with PCR/16S ribosomal bacterial DNA amplification. BMJ Case Reports. 2012;2012(1, article bcr1220115335) doi: 10.1136/bcr.12.2011.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinnon A., Black A. Y., Lortie K., Fleming N. A. A case of adolescent pelvic inflammatory disease caused by a rare bacterium: Fusobacterium nucleatum. Journal of Pediatric and Adolescent Gynecology. 2013;26(6):e113–e115. doi: 10.1016/j.jpag.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Kruse A. B., Kuerschner A. C., Kunze M., et al. Association between high risk for preterm birth and changes in gingiva parameters during pregnancy-a prospective cohort study. Clinical Oral Investigations. 2018;22(3):1263–1271. doi: 10.1007/s00784-017-2209-9. [DOI] [PubMed] [Google Scholar]
- 20.Han Y. W., Fardini Y., Chen C., et al. Term stillbirth caused by oral Fusobacterium nucleatum. Obstetrics & Gynecology. 2010;115:442–445. doi: 10.1097/AOG.0b013e3181cb9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y. W., Shen T., Chung P., Buhimschi I. A., Buhimschi C. S. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. Journal of Clinical Microbiology. 2009;47(1):38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeng J. H., Chan C. P., Ho Y. S., Lan W. H., Hsieh C. C., Chang M. C. Effects of butyrate and propionate on the adhesion, growth, cell cycle kinetics, and protein synthesis of cultured human gingival fibroblasts. Journal of Periodontology. 1999;70(12):1435–1442. doi: 10.1902/jop.1999.70.12.1435. [DOI] [PubMed] [Google Scholar]
- 23.Bartold P. M., Gully N. J., Zilm P. S., Rogers A. H. Identification of components in Fusobacterium nucleatum chemostat-culture supernatants that are potent inhibitors of human gingival fibroblast proliferation. Journal of Periodontal Research. 1991;26(4):314–322. doi: 10.1111/j.1600-0765.1991.tb02069.x. [DOI] [PubMed] [Google Scholar]
- 24.Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji S., Shin J. E., Kim Y. S., Oh J. E., Min B. M., Choi Y. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infection and Immunity. 2009;77(3):1044–1052. doi: 10.1128/IAI.00449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin L., Chung W. O. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunology. 2011;4(4):409–419. doi: 10.1038/mi.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y. W., Shi W., Huang G. T. J., et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infection and Immunity. 2000;68(6):3140–3146. doi: 10.1128/IAI.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fawzy El-Sayed E. K., Dörfer C. E. Gingival mesenchymal stem/progenitor cells: a unique tissue engineering gem. Stem Cells International. 2016;2016:16. doi: 10.1155/2016/7154327.7154327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillipson M., Kubes P. The neutrophil in vascular inflammation. Nature Medicine. 2011;17(11):1381–1390. doi: 10.1038/nm.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P. L., Ohura K., Fujii T., et al. DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochemical and Biophysical Research Communications. 2003;305(4):970–973. doi: 10.1016/S0006-291X(03)00821-0. [DOI] [PubMed] [Google Scholar]
- 31.Morandini A. C. F., Sipert C. R., Gasparoto T. H., et al. Differential production of macrophage inflammatory protein-1α, stromal-derived factor-1, and IL-6 by human cultured periodontal ligament and gingival fibroblasts challenged with lipopolysaccharide from P. gingivalis. Journal of Periodontology. 2010;81(2):310–317. doi: 10.1902/jop.2009.090375. [DOI] [PubMed] [Google Scholar]
- 32.Ahn S.-H., Chun S. M., Park C., Lee J. H., Lee S. W., Lee T. H. Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection. PLoS One. 2017;12(11, article e188755) doi: 10.1371/journal.pone.0188755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ara T., Kurata K., Hirai K., et al. Human gingival fibroblasts are critical in sustaining inflammation in periodontal disease. Journal of Periodontal Research. 2009;44(1):21–27. doi: 10.1111/j.1600-0765.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 34.Rasul A., Bao R., Malhi M., et al. Induction of apoptosis by costunolide in bladder cancer cells is mediated through ROS generation and mitochondrial dysfunction. Molecules. 2013;18(2):1418–1433. doi: 10.3390/molecules18021418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang B., Wang K., Jia Y. P., et al. Fusobacterium nucleatum-induced impairment of autophagic flux enhances the expression of proinflammatory cytokines via ROS in Caco-2 cells. PLoS One. 2016;11(11, article e165701) doi: 10.1371/journal.pone.0165701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leavy O. Inflammation: regulating ROS. Nature Reviews Immunology. 2014;14(6):p. 357. doi: 10.1038/nri3685. [DOI] [PubMed] [Google Scholar]
- 37.Kim D., Langmead B., Salzberg S. L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B., Dewey C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):p. 323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye J., Fang L., Zheng H., et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research. 2006;34(Web Server):W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Mering C., Jensen L. J., Snel B., et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Research. 2005;33(Database issue):D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinane D. F. Causation and pathogenesis of periodontal disease. Periodontology 2000. 2001;25(1):8–20. doi: 10.1034/j.1600-0757.2001.22250102.x. [DOI] [PubMed] [Google Scholar]
- 45.Eke P. I., Dye B. A., Wei L., et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. Journal of Periodontology. 2015;86(5):611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mysak J., Podzimek S., Sommerova P., et al. Porphyromonas gingivalis: major periodontopathic pathogen overview. Journal of Immunology. 2014;2014:8. doi: 10.1155/2014/476068.476068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metzger Z., Lin Y. Y., Dimeo F., Ambrose W. W., Trope M., Arnold R. R. Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. Journal of Endodontics. 2009;35(1):86–94. doi: 10.1016/j.joen.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y., Weng W., Peng J., et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siegmund D., Lang I., Wajant H. Cell death-independent activities of the death receptors CD95, TRAILR1 and TRAILR2. The FEBS Journal. 2017;284(8):1131–1159. doi: 10.1111/febs.13968. [DOI] [PubMed] [Google Scholar]
- 50.Wang J., Cui D., Gu S., et al. Autophagy regulates apoptosis by targeting NOXA for degradation. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2018;1865(8):1105–1113. doi: 10.1016/j.bbamcr.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Panda P. K., Naik P. P., Meher B. R., et al. PUMA dependent mitophagy by Abrus agglutinin contributes to apoptosis through ceramide generation. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2018;1865(3):480–495. doi: 10.1016/j.bbamcr.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Chang W. T., Liu W., Chiu Y. H., et al. A 4-phenoxyphenol derivative exerts inhibitory effects on human hepatocellular carcinoma cells through regulating autophagy and apoptosis accompanied by downregulating α-tubulin expression. Molecules. 2017;22(5):p. 854. doi: 10.3390/molecules22050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aida N., Ushikubo T., Kobayashi F., et al. Actin stabilization induces apoptosis in cultured porcine epithelial cell rests of Malassez. International Endodontic Journal. 2016;49(7):663–669. doi: 10.1111/iej.12494. [DOI] [PubMed] [Google Scholar]
- 54.Lee J., Mun S., Park A., Kim D., Heun Cha B., Kang H. G. Bicalutamide enhances fodrin-mediated apoptosis through calpain in LNCaP. Experimental Biology and Medicine. 2018;243(10):843–851. doi: 10.1177/1535370218779780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo H., Liang H., Chen J., et al. Hydroquinone induces TK6 cell growth arrest and apoptosis through PARP-1/p53 regulatory pathway. Environmental Toxicology. 2017;32(9):2163–2171. doi: 10.1002/tox.22429. [DOI] [PubMed] [Google Scholar]
- 56.Tonetti M. S., Imboden M. A., Gerber L., Lang N. P., Laissue J., Mueller C. Localized expression of mRNA for phagocyte-specific chemotactic cytokines in human periodontal infections. Infection and Immunity. 1994;62(9):4005–4014. doi: 10.1128/iai.62.9.4005-4014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn S. H., Cho S. H., Song J. E., et al. Caveolin-1 serves as a negative effector in senescent human gingival fibroblasts during Fusobacterium nucleatum infection. Molecular Oral Microbiology. 2017;32(3):236–249. doi: 10.1111/omi.12167. [DOI] [PubMed] [Google Scholar]
- 58.Ahn S. H., Song J. E., Kim S., et al. NOX1/2 activation in human gingival fibroblasts by Fusobacterium nucleatum facilitates attachment of Porphyromonas gingivalis. Archives of Microbiology. 2016;198(6):573–583. doi: 10.1007/s00203-016-1223-7. [DOI] [PubMed] [Google Scholar]
- 59.Bui F. Q., Johnson L., Roberts J., et al. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cellular Microbiology. 2016;18(7):970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung S.-C., Huang P. R., Almeida-da-Silva C. L. C., Atanasova K. R., Yilmaz O., Ojcius D. M. NLRX1 modulates differentially NLRP3 inflammasome activation and NF-κB signaling during Fusobacterium nucleatum infection. Microbes and Infection. 2018;20(9-10):615–625. doi: 10.1016/j.micinf.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wada M., Canals D., Adada M., et al. P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene. 2017;36(47):6649–6657. doi: 10.1038/onc.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z. F., Zheng D., Fan G. C., Peng T., Su L. Heat stress prevents lipopolysaccharide-induced apoptosis in pulmonary microvascular endothelial cells by blocking calpain/p38 MAPK signalling. Apoptosis. 2016;21(8):896–904. doi: 10.1007/s10495-016-1263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L. B., Man Z. T., Li W., Zhang W., Wang X. Q., Sun S. Calcitonin protects chondrocytes from lipopolysaccharide-induced apoptosis and inflammatory response through MAPK/Wnt/NF-κB pathways. Molecular Immunology. 2017;87:249–257. doi: 10.1016/j.molimm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Drukker L., Margulis A., Chaouat M., Levitzki R., Maiorenko E., Ben Bassat H. Changes of PI3K/AKT/BCL2 signaling proteins in congenital giant nevi: melanocytes contribute to their increased survival and integrity. Journal of Receptor and Signal Transduction Research. 2013;33(6):359–366. doi: 10.3109/10799893.2013.838785. [DOI] [PubMed] [Google Scholar]
- 65.Zeldich E., Koren R., Nemcovsky C., Weinreb M. Enamel matrix derivative stimulates human gingival fibroblast proliferation via ERK. Journal of Dental Research. 2007;86(1):41–46. doi: 10.1177/154405910708600106. [DOI] [PubMed] [Google Scholar]
- 66.Chae H. J., Ha M. S., Yun D. H., et al. Mechanism of cyclosporine-induced overgrowth in gingiva. Journal of Dental Research. 2006;85(6):515–519. doi: 10.1177/154405910608500607. [DOI] [PubMed] [Google Scholar]
- 67.Heng B. C., Ye X., Liu Y., Dissanayaka W. L., Cheung G. S. P., Zhang C. Effects of recombinant overexpression of Bcl2 on the proliferation, apoptosis, and osteogenic/odontogenic differentiation potential of dental pulp stem cells. Journal of Endodontia. 2016;42(4):575–583. doi: 10.1016/j.joen.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 68.Eftang L. L., Esbensen Y., Tannæs T. M., Bukholm I. R. K., Bukholm G. Interleukin-8 is the single most up-regulated gene in whole genome profiling of H. pylori exposed gastric epithelial cells. BMC Microbiology. 2012;12(1):p. 9. doi: 10.1186/1471-2180-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baron V. T., Pio R., Jia Z., Mercola D. Early growth response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. British Journal of Cancer. 2015;112(4):755–764. doi: 10.1038/bjc.2014.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morton R. S., Dongari-Bagtzoglou A. I. Cyclooxygenase-2 is upregulated in inflamed gingival tissues. Journal of Periodontology. 2001;72(4):461–469. doi: 10.1902/jop.2001.72.4.461. [DOI] [PubMed] [Google Scholar]
- 71.Noguchi K., Shitashige M., Ishikawa I. Involvement of cyclooxygenase-2 in interleukin-1α-induced prostaglandin production by human periodontal ligament cells. Journal of Periodontology. 1999;70(8):902–908. doi: 10.1902/jop.1999.70.8.902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: primer sequences for quantitative real-time PCR (qRT-PCR).
Table 2: the clean reads of 54 samples after RNA-seq.
Table 3: genome mapping ratio summary of 54 samples.
Table 4: gene mapping ratio summary of 54 samples.
Table 5: top five enriched GO terms in the biological process for the 62 DEGs.
Figure 1: PCA of 54 samples of RNA-seq analyses.
Figure 2: GO analysis of the 62 overlapped DEGs after F. nucleatum stimulation at 2 h, 6 h, 12 h, 24 h, and 48 h.
Figure 3: heat map summarizing the 18 overlapped DEGs from the GO biological process analysis after the five paired comparisons of cell proliferation.
Figure 4: (a) Flow cytometry analysis of cell apoptosis. (b) Heat map of the 3 overlapped DEGs from the GO biological process analysis after the five paired comparisons of cell apoptosis.
Figure 5: heat map of the 14 overlapping DEGs from the GO biological process analysis after the five paired comparisons of cell defense response.
Figure 6: (a) The protein levels of p53, phosphorylated p53 (p-p53). (b) The ratio of p-p53 and p53.
Figure 7: the Pathview analysis of the PI3K-AKT signaling pathway.
Figure 8: the Pathview analysis of MAPK signaling pathway.
Figure 9: the relative gene expression level of Bcl-2 (a), COX2(b), A20 (c), IκBα (d), and MIP2 (e) by qRT-PCR.
Figure 10: the Pathview analysis of the apoptosis signaling pathway.
Figure 11: the relative gene expression level of TRAILR2 (a), NOXA (b), PUMA (c), tubulin (d), actin (e), fodrin (f), and PARP (g) by RT-PCR.
Data Availability Statement
All raw RNA-seq data are accessible through GEO series accession number, GSE118691. Other data from Materials and Methods used to support the findings of this study are included in the article. If any other data are needed, please contact the corresponding author.