Abstract
While base-specific support is commonly used for single-column oligodeoxynucleotide synthesis, the universal linker is critical for high-throughput synthesis of potentially thousands of samples in a single run. Here, we report conditions for cleavage and complete dephosphorylation of two commercial universal linkers, UnySupport and UnyLinker, processed in the gas phase (NH3) using our custom device. First, we compared the average yield of T10mers over time (15, 30, 60, 120, and 240 minutes, 40 psi, 80°C and 90°C). For samples processed with water added prior to incubation, we discovered a substantial increase in yield compared to those left dry (up to 55%). This was also the case for samples subjected to increases in chamber pressure (10, 20, 30 and 40 psi, 120 minutes, 80°C and 90°C). Next, we compared the effects of increased temperature, pressure and incubation times on the rates of dephosphorylation. We found the optimum conditions to be either 10 psi, 120 minutes at 80°C or 60 minutes at 90°C; in both cases, water added to columns prior to incubation had a substantial effect on rate of reaction as well as overall yield compared with those left dry. Finally, performance between the two linkers was similar enough to conclude each fulfills the desired requirements for mainstream, high-throughput oligodeoxynucleotide cleavage/deprotection and dephosphorylation in the gas phase.
Keywords: DNA synthesis, DNA/RNA, DNA alkylation, solid phase synthesis
INTRODUCTION
In solid-phase oligodeoxynucleotide (ODN) synthesis, the substrate, in which ODNs are synthesized from 3′ to 5′ is available either as controlled-pore-glass (CPG) or polystyrene (PS). The standard support, for example, has the first base pre-attached at the 3′ end with a long-chain alkyl amine tethered to a succinate bridge for ODN separation postsynthesis. The other main type of support is the abasic universal linker, which may be an inverted ribonucleoside, or is dichloroacetyl,[1] vicinal diol,[2] or catechol based.[3] While standard support has its place in single-column ODN production on instruments such as the AB 394 and 3900, the universal linker is required for use in high-density titer plates during mainstream, high-throughput ODN production;[4,5] manually loading base-specific support is not only prone to human error when handling ≤ 96 samples, but is impractical when loading ≥384-well plates. Furthermore, the universal linker allows for a wide variety of specialized monomers to be coupled at the 3′ end that cannot be achieved with standard support. What also makes the universal linker unique is that once it is cleaved from the support it must go through a rate-limiting process of dephosphorylation or phosphate intramolecular cyclization[6] in order to dissociate completely from the ODN, generating a 3′ hydroxyl. Incomplete dephosphorylation, for example, results in a linker-bound ODN, which inhibits biological activity; applications such as PCR require that primers maintain this hydroxyl group for polymerase recognition and strand elongation. Therefore, a means in which to effectively remove the universal linker is critical. And while some linkers necessitate harsher conditions such as lithium chloride treatment for complete dephosphorylation, milder, more user-friendly conditions are desired for mainstream ODN production.
The current method for cleavage and deprotection of ODNs in both single column and 96-well plates involves use of ammonium hydroxide (NH4OH) in the liquid-phase.[6] Typically, the end-user manually pipettes NH4OH into each individual column/well; after which, the samples are capped off to prevent loss of ammonia through evaporation. With a minimum two-day turnaround, this entails cleavage from the support (1–2 hours at room temperature, nucleoside deprotection (17 hours at 55°C) and lyophilization of final product (4 hours) to remove residual ammonia. A faster approach employs a 1:1 mixture of NH4OH and methylamine (AMA), which reduces deprotection time to 2 hours at room temperature with use of highly base-labile nucleoside protecting groups.[7] However, one of the major bottlenecks in high-throughput ODN production for processing potentially thousands of small volume samples in the liquid-phase is evaporation. One solution has been to process ODNs in the gas phase where ammonia/methylamine vapors are contained inside a reaction vessel, under elevated pressure and temperature.
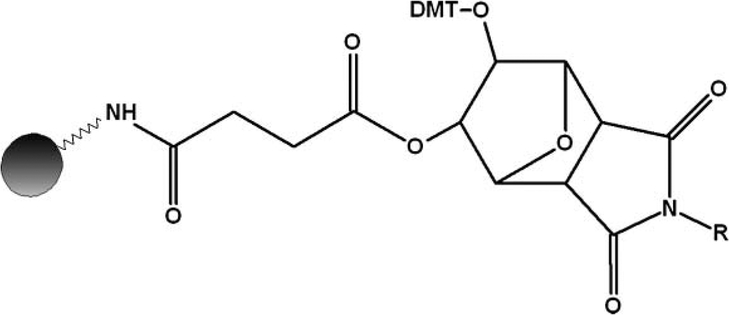
While a method and apparatus for rapid cleavage and deprotection of ODNs synthesized from standard support have been previously described in the gas phase,[8–10] we further define parameters for cleavage and dephosphorylation of ODNs synthesized from the universal linker. Here, with the use of our custom gas-phase device (Figure 1), we processed T10mers synthesized from two commercially available universal linker substrates, UnySupport (US), offered by Glen Research, and the UnyLinker (UL) developed by Isis Pharmaceuticals[2] (Figure 2). These two supports are structurally identical save for a unique side group (methyl (US) and phenyl (UL), respectively). In addition to ODNs synthesized from loose CPG, we also defined parameters for processing samples generated from fritted material with CPG embedded in a polymeric resin; this is simply a plug that is pressure-fitted into an empty synthesis column (e.g., 50–200 nanomole AB style). As a cleavage and dephosphorylation agent, we chose anhydrous ammonia; while both linkers and apparatus are compatible with methylamine, it does introduce greater hazard to the end-user due to the reagent’s extremely high flammability and toxicity, which accounts for its much stricter regulation in the laboratory. We then tested a series of T10mers under both “wet” and “dry” conditions, and compared their yields and rates of dephosphorylation over various temperature and pressure settings, and incubation periods; the results of which, may provide the end-user with a safer, more efficient, time-saving, and cost-effective method of processing ODNs in mainstream postsynthesis production using the universal linker as a starting substrate in the gas phase.
FIGURE 1.
Custom gas-phase device used for processing ODNs generated from the universal linker support, which includes the following setup: A) reaction chamber that can house two blocks of synthesis columns (50–200 nmole AB-style) or two 96, 384, and 1536-well titer plates back to back; B) top and bottom heating blocks span entire length of chamber; C) thermostat; D) top and bottom heating block temperature limits; E) vacuum manifold; F) ammonia gas lecture bottle; G) chamber pressure gauge; and H) ammonia gas traps.
FIGURE 2.
Chemical structure of universal linker moiety used for this research. The side-group R represents either methyl (UnySupport) or phenyl (UnyLinker).
METHODS AND MATERIALS
ODN Synthesis
ODN synthesis was performed in-house at the Stanford Genome Technology Center (Palo Alto, CA, USA) with an AB 3900 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA, USA). Solid substrates tested include: 500Å CPG UnySupport (US and US-Frit) (Glen Research, Sterling, VA, USA) and UnyLinker (UL; Chemgenes, Wilmington, MA, USA). Cycle conditions were similar to manufacturer’s recommended protocol, which included use of the following reagents: deblock 3% TCA/DCM (AiC, Framingham, MA, USA), acetonitrile, 0.02M oxidizing solution, cap A/B, 0.1M dT (Sigma-Aldrich), and 0.25 M 5-benzylthio-1H-tetrazole (Glen Research). Following synthesis of T10mers (TTTTTTTTTT (5′–3′)), samples for gas-phase optimization were taken directly from the instrument and set aside to dry. Controls in the liquid-phase were cleaved from the support with NH4OH (28–30%) (J.T. Baker, Phillipsburg, NJ, USA) at room temperature followed by dephosphorylation and cyanoethyl deprotection for 17 hours at 55°C (or comparable conditions used for the test samples). After lyophilization, ODNs were resuspended and the optical density for each was measured using the NanoDrop at 260 λ.
Sample Preparation
For this study, we weighed individual synthesis columns, then took the difference after CPG was added. Given an initial loading (μm/gm) for each support type, we were able to determine a theoretical yield in nanomoles (nmoles). Final product yield (actual) was then taken as a percent of its theoretical value, which became the adjusted target yield all test samples were compared to. Although we had initial loading of CPG embedded in the frit as well, we did not have an accurate weight of starting CPG (ratios of CPG to resin can vary from frit to frit); therefore, we only measured the raw yield of final product to get an estimate for comparing each condition.
Sample Handling in the Gas Phase
For wetted samples, 30 μl water (RNAse/DNAse free) were delivered to each column prior to incubation; nothing was added to the dry columns. Both “dry” and “wet” samples were placed inside a 96-column holder (stainless steel), which was positioned inside the reaction chamber. Three samples were synthesized per linker per condition, and placed in the middle and opposite corners of the column holder.
Gas-Phase Apparatus Design
As shown in Figure 1, the chamber is designed to house two high-density titer plates back to back, which are positioned along rails of the inner wall (A). Top and bottom heating blocks (B) are controlled by a digital temperature gauge (C); upper limits are set at a maximum temperature as a safety measure so that the desired setting is not exceeded (D). A vacuum manifold can also be used for evacuating the chamber prior to incubation (E). An auxiliary ammonia lecture bottle (F) feeds pressurized gas into the chamber that is monitored with a pressure gauge (G) housed directly on top. Following incubation, ammonia gas is percolated through a water trap (H) generating NH4OH for disposal. A number of shut-off valves are introduced throughout the plumbing to close off the system and prevent backflow. A port is also available on the side of the chamber to allow for additional equipment such as an inner fan for increased gas and heat circulation.
Apparatus Setup for ODN Cleavage and Dephosphorylation
After loading the samples into the rack, the door was sealed shut and the chamber evacuated. Top and bottom heat blocks were then turned on until target temperature was achieved (5 minutes); after which, the chamber was charged with ammonia gas (Sigma Aldrich, St. Louis, MO, USA) until it reached the desired pressure (psi) (<10 seconds); samples were then incubated based on the conditions tested. Following incubation, the heat blocks were switched off and the chamber evacuated of ammonia, which was percolated into a water trap (<2 minutes).
Postincubation
Following depressurization of the chamber, samples were removed, whereupon water was added to “wet” and “dry” columns (70 μl and 100 μl, respectively), then each was spun down to collect the processed ODNs. Samples were then taken immediately for analysis.
ODN Analysis
For yield comparisons, the optical density of each ODN was measured to determine actual quantity (nmoles). ODN purity and rates of dephosphorylation were further analyzed by reverse-phase high-performance liquid chromatography (HPLC) using a DNASep C-18 column at 60°C with ultraviolet (UV) detection at 260 λ; running buffers consisted of water, acetonitrile, triethyl ammonium acetate, and EDTA (HPLC unit and operating software used for sample analysis consisted of the Transgenomic Wave System).
RESULTS AND DISCUSSION
Because of the vast array of synthesis platforms, starting material can be prepared in a number of ways: 1) Loose CPG or PS can be made into a slurry (e.g., chloroform/adiponitrile and dibromomethane) and delivered manually by pipette to the individual wells of a titer plate; 2) dry CPG/PS can be packed into empty synthesis columns where it is sandwiched between top and bottom filters; or 3) where commercially available, CPG also comes in a fritted material with the support embedded within a polymeric resin. As such, for all the following experiments, we focused our testing on methods 2 and 3 since they are likely applicable to most ODN synthesis facilities.
Furthermore, to determine conditions for ODN release from the universal linker and for complete dephosphorylation in the gas phase using our custom device, we explored several parameters: 1) effects of temperature, 2) pressure, 3) incubation time, and 4) addition of water to specified samples prior to incubation compared with those left dry. While we initially compared four temperature settings (room temperature, 65°C, 80°C, and 90°C), ODNs processed at room temperature in our device gave very poor yields and inconsistent rates of dephosphorylation as did samples at 65°C; therefore, we focused primarily on data obtained at 80°C and 90°C.
ODN Cleavage from the Solid Substrate
Because the amount (mg) of CPG added by hand to each column varied, it was necessary to “normalize” final yields so that test and control samples could be more accurately compared. This was done by first calculating the target, theoretical yield (total nmoles produced from an ideal synthesis); actual yields were then measured and taken as a percentage of the theoretical values. For instance, if the theoretical yield of a control ODN was calculated at 100 nmoles, but we only obtained 80 nmoles from the actual synthesis, then 80 nmoles became the new target yield to which the test sample was compared.
To evaluate yield of ODN separation from the solid support tethered with the universal linker (US, US-Frit, and UL), we tested a series of T10mers in both “wet” and “dry” conditions in the gas phase. We then compared their yield to controls processed with NH4OH over time (15, 30, 60, 120, and 240 minutes at both 80°C and 90°C). While the average yield between “wet” test samples and the control was unremarkable, we observed little to no effect on “wet” product yield with increasing incubation time; however, “wet” samples were substantially higher in yield than those left dry (Table 1, line A). Furthermore, “dry” samples did show a gradual increase in yield within each condition. Test ODNs were also subjected to increased chamber pressure (10, 20, 30 and 40 psi, 120 minutes, 80°C and 90°C). Again, there was little to no difference between “wet” test and control samples, although there was a marked increase in yield over those left dry (Table 1, line B). An increase in chamber pressure also had an effect on “dry” product yield where we saw a gradual increase from 10–40 psi. Moreover, columns with water added showed little evaporation over time (<10% by weight after 120 minutes and <20% after 240 minutes, 90°C).
TABLE 1.
Percent of increases in yield for samples processed with water over that of samples left drya
| US | UL | US-Frit | ||
|---|---|---|---|---|
| 15–240 min | ||||
| 80°C | 42% | 34% | 55% | A |
| 90°C | 34% | 32% | 55% | |
| 10–40 psi | ||||
| 80°C | 40% | 55% | 41% | B |
| 90°C | 34% | 41% | 27% | |
Conditions included: A) 15, 30, 60, 120, and 240 minutes incubation periods at 40 psi, and B) varying chamber pressures at 10, 20, 30, and 40 psi (NH3) for 120 minutes, (both at 80°C and 90°C, respectively).
In addition to using CPG, we tested the cleavage properties of ODNs (T10mers) synthesized on PS (data not shown). Despite its increased hydrophobicity, we achieved similar results to those samples processed under the same conditions as stated above; product return from PS was substantially higher when wetted prior to incubation than when left dry.
As it is with the standard support, the mechanism of ODN release is a direct nucleophilic attack on the carbonyl group of the ester linkage by ammonia.[11] To further explain the increase in yield for wetted samples (as in the control, NH4OH), presence of water not only facilitates the reaction but acts as a conduit and ODN reservoir; while strands are released from the support, they are trapped by the water molecules until eluted from the column. Although we only used water during incubation and for eluting samples postincubation, there are additives that could potentially increase yield, which include alcohols, ethers, amides and nitriles.[12] Also, adding another round of water (or elution buffer) should further increase yield, although we have not defined an ideal number of elution steps for maximum product return.
ODN Universal Linker Dephosphorylation
The term “dephosphorylation” here refers to dissociation of the entire US and UL linker from the ODN 3′ terminus following succinate cleavage by means of phosphate intramolecular cyclization.[6] Figure 3 shows HPLC chromatogram data of incomplete dephosphorylation of the US and UL ODNs (panels A and B, respectively) where the linker arms are still attached to the 3′ end of each ODN. US-linker has a shorter retention time at 3.82 minutes compared with the UL-linker at 4.78 minutes, due to its less hydrophobic methyl side group.
FIGURE 3.
US and UL T10mers (A and B, respectively) showing both complete (T10) and partial dephosphorylation (T10-linker); Me = methyl, and Ph = phenyl. All samples have presence of T10 alkylation by acrylonitrile at 4.17 minutes (N3-cyanoethyl-dT).
To determine the optimum incubation time to achieve complete dephosphorylation (80°C or 90°C, 40 psi), we subjected a series of T10mers to the same time-points as above (15–240 minutes). Using HPLC analysis, the rate of dephosphorylation was measured by the disappearance of linker-bound ODN trailing peaks. Figure 4 shows that 100% dephosphorylation of all “wet” samples was achieved in our system at 120 minutes, 80°C (A) and at 60 minutes, 90°C (B). ODNs processed without addition of water showed only a gradual increase in dephosphorylation over time where a small percentage still retained the linker at 240 minutes: C) 87.01% (US), 99.07% (UL)98.46%, (Frit), and D) 93.91% (US), 93.40% (UL), 99.38% (Frit), 80°C and 90°C, respectively. For samples processed at 80°C, we believe there was no reason to test 15, 30 or 60 minutes time-points with anything <40 psi since at 60 minutes (40 psi, 80°C) we achieved at most 99.01% dephosphorylation under “wet” conditions (Figure 4A); the same is true at 30 minutes (40 psi, 90°C) where we reached only a maximum of 95.24% (Figure 4C).
FIGURE 4.
Rate of dephosphorylation over time. A and B) samples processed at 80°C, 40 psi at 15, 30, 60, 120, and 240 minutes (“wet” vs. “dry,” respectively); C and D) samples processed at 90°C under the same conditions (“Frit” material has US CPG embedded).
Figure 5 shows the effects of increasing NH3 chamber pressure on the rate of dephosphorylation. At 120 minutes, “wet” samples are fully dephosphorylated from 10 to 40 psi at both 80°C and 90°C; and again for “dry” samples, we see only a gradual increase in target product formation. Since data for both temperature sets including “wet” and “dry” conditions showed little variation between the two, we averaged the results into one graph for simplicity.
FIGURE 5.
Averaged effects of chamber pressure (10, 20, 30 and 40 psi) on rate of dephosphorylation at 80°C and 90°C (both “wet” and “dry” conditions, 120 minutes). “Frit” material has US CPG embedded, while “all wet” indicates water has been added to each US, UL, and US-Frit sample prior to incubation.
In addition, we compared the effects of temperature on rate of dephosphorylation at 65°C, 80°C and 90°C (“wet,” 120 minutes, 40 psi). While we achieved 100% removal of the linker adduct at 80°C and 90°C, 6–10% still retained the linker at 65°C (data not shown); maximum time required for complete dephosphorylation at this temperature has not been defined, but we believe it would take between 120 and 240 minutes to fully process. While increasing the temperature accelerates dephosphorylation, there may be instances where heat-sensitive monomers/substrates could be negatively affected; therefore, choosing a milder temperature (either at 65°C or 80°C) should prove beneficial despite longer reaction times. Furthermore, if one desired to use anhydrous conditions, it would require either an incubation time >240 minutes or chamber psi >40 (at either 80°C or 90°C). And though water is not necessary for linker removal by dephosphorylation, it does accelerate the process substantially.
As shown in Figure 3, N3-cyanoethyl-dT is present (4.17 minutes) in all test and control samples (NH4OH). During β-elimination, acrylonitrile is produced, which alkylates dT residues at their N3 positions.[13] We determined its presence by treatment with 10% diethylamine (DEA) in acetonitrile by flushing the column with 1 ml over 10 minutes. Furthermore, when we processed T10mers using AMA, we found no trace of this by-product. Therefore, it is reasonable to assume using an aggressive base such as methylamine in the gas phase[14,15] could eliminate N3-cyanoethyl-dT formation. As an alternative, ODNs could be treated with 10% DEA prior to incubation, followed by cleavage, deprotection and dephosphorylation using NH3 gas. It is worth mentioning too, that neither increases in temperature, pressure nor incubation times had any marked effect on relative yield of alkylated by-product in either the gas or liquid-phase processes.
Although we only tested US, US-Frit, and UL substrates, other commercially popular universal linkers are available such as the catechol-based linker offered by CTGen[3] and Universal Supports II and III[1] also from Glen Research. For linkers II and III, however, the manufacturer does not recommend using either in the gas phase due to incompatibilities with de-protection as well as incompatibilities with DEA treatment for acrylonitrile removal.
In conjunction with using a simple T10mer sequence in our efforts to examine only yields and rates of dephosphorylation of the universal linker in the gas phase, we did process samples with mixed-base composition (data not shown). These included sequences ranging from 20 to 30 bp in length with the highly stable nucleoside blocking groups, isobutyl and benzoyl. Our unpublished data shows that using the optimal conditions stated above for complete dephosphorylation, performance was similar to that of samples processed in the liquid-phase (results based on HPLC analysis and polymerase-based applications such as real-time-PCR). However, a more critical evaluation would be necessary to determine completeness of deprotection in our system by use of CE and LC-MS, and is beyond the scope of this article. Also a great deal of literature is available on the performance of ultra base-labile blocking groups such as acetyl, N,N -dimethylformadine and N -phenoxyacetyl for those interested in rapid cleavage/deprotection schemes using the gas-phase method.[14,15]
CONCLUSION
With the continued advances in high-throughput, low volume DNA synthesis automation, use of a gas-phase device for ODN cleavage/deprotection and dephosphorylation is one viable option to mainstream postsynthesis production. And while alternative means of processing bulk synthesis product may be available, no such method has been verified or tested by our lab. Therefore, with the optimum conditions for cleavage and dephosphorylation of ODNs synthesized on the universal linker provided here (either 10 psi, 120 minutes at 80°C or 60 minutes at 90°C), we hope our work will further compliment outside research detailing rapid deprotection schemes; the results of which, could ultimately define a complete package for high-throughput processing of potentially thousands of samples in just a few hours or less using either NH3 or methylamine. Furthermore, yields and rates of dephosphorylation between US/Frit and UL universal linker substrates tested here were very similar in performance; therefore, we believe both meet the desired requirements for high-throughput ODN cleavage/deprotection and dephosphorylation in the gas phase.
Acknowledgments
The authors wish to thank Certified Scientific and its affiliates for fabricating the gas-phase apparatus used in this research.
REFERENCES
- 1.Yagodkin A; Azhayev A Universal linker phosphoramidite. Arkivoc, 2009, 187–197. [Google Scholar]
- 2.Ravikumar VT; Kumar RK; Olsen P; Moore MN; Carty RL; Andrade M; Gorman D; Zhu X; Cedillo I; Wang Z; et al. UnyLinker: An efficient and scaleable synthesis of oligonucleotides utilizing a universal linker molecule: A novel approach to enhance the purity of drugs. Org Process Res Dev, 2008, 12, 399–410. [Google Scholar]
- 3.Anderson KM; Jaquinod L; Jensen MA; Ngo N; Davis RW A novel catechol-based universal support for oligonucleotide synthesis. J. Am. Chem. Soc 2008, 72, 9875–9880. [DOI] [PubMed] [Google Scholar]
- 4.Lashkari DA; Hunicke-Smith SP; Norgren RM; Davis RW; Brennan T An automated multiplex oligonucleotide synthesizer: development of high-throughput, low-cost DNA synthesis. Proc. Natl Acad. Sci. USA, 1995, 92, 7912–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng JY; Chen HH; Kao YS; Kao WC; Peck K High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer. Nucleic Acids Res, 2002, 30, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pon RT Solid-phase supports for oligonucleotide synthesis. Curr. Protoc. Nucleic Acid Chem, 2001, Chapter 3, Unit 3 1. [DOI] [PubMed] [Google Scholar]
- 7.Reddy MP; Farooqui F; Hanna NB Methylamine deprotection provides increased yield of oligonucleotides. Tetrahedron Letters, 1995, 36, 8929–8932. [Google Scholar]
- 8.Iyer RP; Yu D; Xie J; Zhou W; Agrawal S The use of gaseous ammonia for the deprotection and cleavage steps during the solid-phase synthesis of oligonucleotides, and analogs. Bioorganic & Medicinal Chemistry Letters, 1997, 7, 1443–1448. [Google Scholar]
- 9.Boal JH; Wilk A; Harindranath N; Max EE; Kempe T; Beaucage SL Cleavage of oligodeoxyribonucleotides from controlled-pore glass supports and their rapid deprotection by gaseous amines. Nucleic Acids Res, 1996, 24, 3115–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempe T Recovery of oligonucleotides by gas phase cleavage U.S. Patent, 1996, 5,514,789. [Google Scholar]
- 11.Palom Y; Grandas A; Pedroso E The mechanism of cleavage under basic conditions of succinylanchored oligonucleotides. Nucleosides and Nucleotides, 17, 1998, 1177–1182. [Google Scholar]
- 12.Chow F; Kempe T Process and reagents for processing synthetic oligonucleotides U.S. Patent 1997, 5,656,741. [Google Scholar]
- 13.Capaldi DC; Gans H; Krotz AH; Arnold J; Carty RL; Moore MN; Scozzari AN; Lowery K; Cole DL; Ravikumar VT Synthesis of high-quality antisense drugs. Addition of acrylonitrile to phosphorothioate oligonucleotides: adduct characterization and avoidance. Org Process Res Dev, 2003, 7, 832–838. [Google Scholar]
- 14.Reddy MP; Hanna NB; Farooqui F Fast cleavage and deprotection of oligonucleotides. Tetrahedron Letters, 1994, 35, 4311–4314. [Google Scholar]
- 15.Vu H; McCollum C; Jacobson K; Theisen P (Fast oligonucleotide deprotection phosphoramidite chemistry for DNA synthesis. Tetrahedron Letters, 1990, 31, 7269–7272. [Google Scholar]