Abstract
Introduction
Only ~ 50% of hypertensive patients will respond to treatment.
Objective
This pilot study aims to identify clinical and metabolite markers that predict response to lisinopril.
Methods
Hypertensive patients (n = 45) received lisinopril (10 mg) at their baseline visit. Blood pressures were reevaluated one week later. Responders to lisinopril (n = 19) were defined by a 10% decline in systolic blood pressure. Plasma metabolites were evaluated with mass spectrometry.
Results
BMI (p = 0.009), GFR (p = 0.015) and 2-oxoglutarate were included in a logistic regression model to predict response to lisinopril.
Conclusions
Further validation cohorts are needed to confirm the predictive values of these clinical and metabolic markers.
Keywords: Hypertension, Precision medicine, Personalized medicine, Lisinopril, ACE/ARB, 2-Oxoglutarate
1. Introduction
The 2017 American College of Cardiology and American Heart Association hypertension (HTN) guidelines estimated that approximately 45% of United States adults have HTN (Bundy et al. 2018). Angiotensin converting enzyme inhibitors (ACEIs) are the first line therapy (Bundy et al. 2018; Gu et al. 2012) and one-third of all hypertensive patients take an ACEI (Gu et al. 2012), however response rates to therapy remain low (Gu et al. 2012). Precision medicine approaches to investigate individual response to ACEIs may elucidate personalized prescribing recommendations for hypertensive patients leading to greater response to therapeutic interventions.
The use of metabolomics to identify the expression patterns of small molecules within a biological system and elucidate associated pathways has the potential to aid in developing predictive precision medicine models (Azad and Shulaev 2018; Beger et al. 2016; Tzoulaki et al. 2018). For example, metabolomics have been used to metaboli-cally phenotype septic patients who responded to L-Carnitine treatment, leading to a more in-depth understanding of drug mechanism (Puskarich et al. 2015). Additionally, metabolomic analyses of pretreatment samples have been used to identify predictors of response to therapies treating major depressive disorder (Kaddurah-Daouk et al. 2011) and cervical cancer (Hou et al. 2014). To date, metabolomic approaches to identify biomarkers of response to ACEI are lacking.
The objective of this pilot study was to evaluate clinical variables and metabolomic markers that might predict response to lisinopril and to provide preliminary data for future prospective study designs on this topic.
2. Methods
2.1. Study design
This was a secondary analysis of a parent trial examining the pharmacogenomic effectiveness of metoprolol succinate in hypertensive patients (NCT02293096). See Supplemental Methods for details. Patients between 18 and 89 years of age with uncontrolled HTN (> 140/90 mmHg) were enrolled. Blood pressure was evaluated at baseline and follow-up [median (IQR): 8 days (7, 12)] via automated inflatable cuff after 10 min seated in a quiet room with a research assistant. At baseline, plasma samples were collected prior to patients receiving ACEI (lisinopril, 10 mg daily). Patients who responded to lisinopril therapy with at least a 10% decline in systolic blood pressure (SBP) at their follow-up visit were defined as “responders”. All others were defined as “non-responders”. Clinical parameters and a list of all current medications were collected at baseline. The day and time of last dose of lisinopril were recorded at follow-up. All subjects reported compliance to taking lisinopril at follow-up and 70% of the cohort had follow-up BP measurements performed within one half-life of their last lisinopril dose. Dietary records were not included.
2.2. Metabolite extraction and identification
We compared baseline provocative plasma samples. Samples were prepared for ultra high performance liquid chromatography MS (UHPLC-MS) metabolomics in the following manner (see Supplemental Methods for details). Polar metabolites were extracted from plasma in ice-cold lysis/ extraction buffer (methanol:acetonitrile:water 5:3:2) at a 1:25 dilution. Prior to extraction, isotopically labeled standards were added at expected biological concentrations to the lysis buffer for absolute quantitation. Samples were agitated (30 min, 4 °C) and centrifuged (18,213×g, 10 min, 4 °C). Non-polar metabolites were extracted from plasma in ice-cold methanol at a 1:10 dilution. Samples were vortexed, incubated at − 20 °C for 30 min, and centrifuged (18,213 g, 10 min, 4 °C). Protein pellets were discarded. Supernatants were diluted 1:1 using 10 mM ammonium and stored at − 80 °C prior to metabolomic analyses.
Plasma extracts were injected into a Thermo Vanquish UHPLC system (San Jose, CA, USA) coupled to a Thermo Q Exactive mass spectrometer with electrospray ionization (Bremen, Germany). Polar metabolites were separated on a Kinetex C18 column (150 × 2.1 mm, 1.7 μm—Phenomenex, Torrance, CA, USA) at 45 °C using a 5-min gradient method (Nemkov et al. 2019) at 450 pl/min and mobile phases (A: water/0.1% formic acid; B: acetonitrile/0.1% formic acid) for positive ion mode. Non-polar metabolites were separated on an Acquity HSS T3 column (150 × 2.1 mm, 1.8 μm—Waters, Milford, MA, USA) at 45 °C using a seventeen minute gradient method (Reisz et al. 2019) at 300 μl/min (unless where noted) and mobile phases containing IPA (isopropanol, A: 72:25 H2O:ACN 5 mM NH4OAc, B: 50:45:5 IPA:ACN:H2O 5 mM NH4OAc) for negative ion mode. See Supplemental Methods for details. Quality controls were generated from pooled aliquots of extracts and ran every 15 analytical runs to control for technical variability, as judged by coefficients of variation. Metabolites were identified and validated as previously described (Nemkov et al. 2015, 2017). The metabolomics data reported in this paper are available via https://www.ebi.ac.uk/metabolights/and study ID: MTBLS1021.
2.3. Metabolomic statistical analyses
Supervised targeted analyses of metabolomic data were performed using Metaboanalyst (Version 4.0) to identify significant differences in metabolite peak intensities between responders and non-responders. Independent t tests with Bonferroni multiple comparisons correction, fold changes, volcano plots, and partial least squares with discriminant analysis (PLS-DA) plots were created after generalized logarithm transformation. Fold changes were calculated from non-transformed peak intensities between responders and non-responders. Fold changes of at least a twofold difference where p < 0.05 were considered significant. PLS-DA plots identified and accounted for differences in the metabolomes between groups based on individual components. Permutation testing was used to cross-validate the PLS-DA model. A power analysis was completed to inform future studies.
2.4. Clinical variables statistical analyses
Mann-Whitney U tests were used to identify significant continuous clinical variables and Chi square tests were used to identify significant categorical clinical variables. Significant metabolites and clinical variables were investigated together using multinomial logistic regression tests to predict response to lisinopril. Bonferroni correction was used to account for multiple comparisons. All statistical analyses were completed in SPSS for Windows (Build 1.0.0.1174).
3. Results and discussion
3.1. Participant grouping and clinical characteristics
Of the total participant population (n = 45), 19 subjects (42.2%) were defined as responders to lisinopril therapy at their follow-up visit [median% SBP decline (IQR): − 16.5% (− 20.4%, − 14.0%) vs. − 4.8% (− 7.0%, 2.3%) responder vs. non-responder, respectfully, p < 0.001]. The absolute decrease in mean arterial pressures between baseline and follow-up visits was also significantly different between the responder and non-responder groups [median decrease (IQR): − 18.9 mmHg (− 22.3, − 13.6) vs. -1.45 mmHg (− 6.7, 3.8), responder vs. non-responder, respectively, p < 0.001].
Accounting for the power achieved in this study, significant differences in patient demographics, baseline blood pressures, smoking status or diabetes status between the responder and non-responder groups could not be confirmed. Responders and non-responders did not differ in the number of days on lisinopril between baseline and follow-up or the time since last lisinopril dose at follow-up. Some subjects were taking other antiHTN medications at their baseline visit (4 responders vs. 5 non-responders), however within our patient population, we found no significant correlation between lisinopril responsiveness and whether or not an individual was taking other antiHTN medications. Similarly, we did not observe any correlation between lisinopril responsiveness and whether or not subjects were taking NSAIDs or steroids.
BMI is the most common measure of whole body adiposity. As BMI approaches obese levels, cardiac output, sympathetic nervous system stimulation, and activity of the renin-angiotensin-aldosterone system (Vaneckova et al. 2014) also increase, driving the development of HTN. In this overweight/obese population, responders had lower BMIs than non-responders [median (IQR): 28.1 kg/m2 (26.6, 33.4) vs. 32.9 kg/m2 (29.2, 37.3), responder vs. non-responder, respectfully, p < 0.05]. Within our pilot cohort, there was a lack of difference in weight-based dosing (BMI/dose) between responders and non-responders. A larger validation cohort could more confidently conclude this lack of difference.
Responders also had lower GFRs [92 ml/min/1.73 m3 (76, 110) vs. 101 ml/min/1.73 m3 (92, 119), responder vs. non-responder, respectfully, p < 0.05] than non-responders, although GFR remained within the normal clinical range. Lisinopril is not metabolized after absorption and is excreted unchanged, therefore increased renal excretion (higher GFR) is expected to be associated with decreased lisinopril availability and decreased responsiveness. Checking lisinopril levels in future studies could verify the proposed mechanism of GFR on response to lisinopril.
3.2. Baseline plasma metabolites
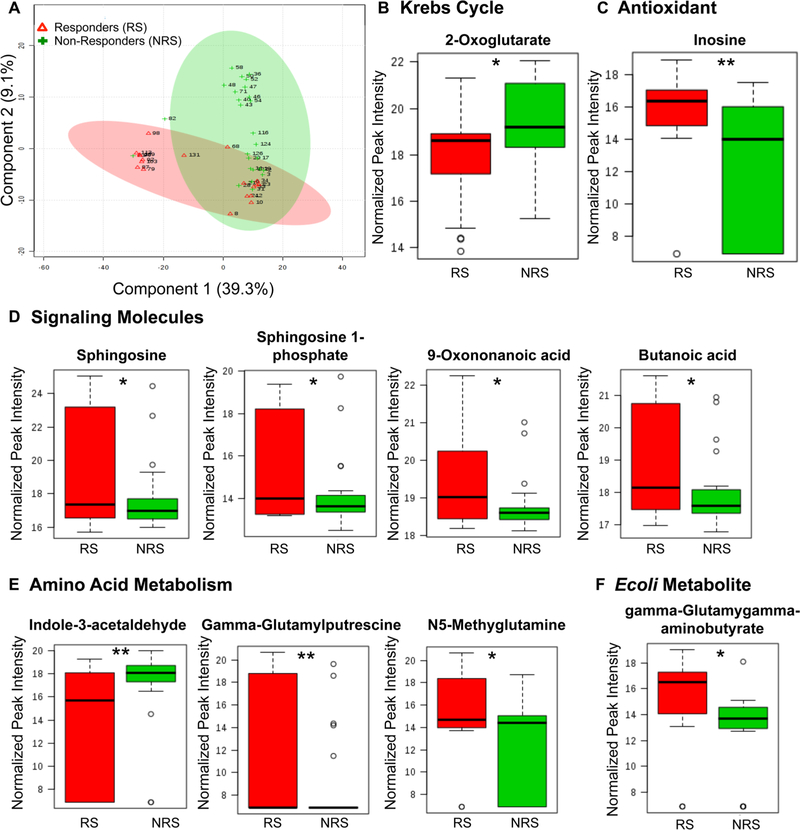
From this pilot cohort, metabolomic analyses identified 201 metabolites present across both groups. PLS-DAs of the represented metabolites accounted for almost 50% of the difference in the metabolome between the responder and non-responder study populations (Fig. 1a). Of the represented metabolites, 10 were differentially expressed (p < 0.05) between responders and non-responders (Fig. 1c–e).
Fig. 1.
Plasma metabolomic differences between responders and non-responders to lisinopril therapy at baseline. a PLS-DA 2D image—principal component analysis of metabolites in plasma samples from patients prior to the initiation of lisinopril therapy. Red = responders to lisinopril, Green = non-responders to lisinopril. b-f Relative quan-titation of peak intensities from differentially expressed metabolites between responders and non-responders. Responder (RS), n = 19, Non-Responder (NRS), n = 26. The (*) denotes a P value < 0.05, (**) denotes a P value < 0.01
Of note, a number of metabolites involved in the regulation of blood pressure and vascular health as a whole were identified. Butanoic acid was increased in responders (FC: 2.30, p = 0.041), which is a direct agonist of the G-protein coupled receptor (GPCR), hydroxycarboxylic acid receptor 2 (HCA2) (Offermanns et al. 2011) to induce vasodilation (Blad et al. 2011) and inhibit atherosclerotic activity (Blad et al. 2011). Additionally, levels of sphingosine 1-phosphate (S1P, FC: 2.61, p = 0.032) and the precursor to S1P, sphingosine (FC: 5.20, p = 0.033), were elevated in responders compared to non-responders. S1P signals through a GPCR to regulate angiogenesis (Swan et al. 2010), vascular stability (Swan et al. 2010), and vascular permeability (Swan et al. 2010). Finally, elevated levels of the lipid peroxidation product, 9-oxononanoic acid (FC: 2.17, p = 0.020), may indicate increased activation of secreted phospholipase A2 (PLA2) (Ren et al. 2013) and decreased liver de novo lipogenesis (Shinsuke Minamoto 1988) in responders. Taken together, the increased presence of metabolites involved in regulating blood pressure and vascular heath in hypertensive individuals who responded to lisinopril therapy might suggest that these subjects have the machinery in place to regulate their blood pressure whereas those who do not respond are less well positioned to do so.
Responders also had relatively lower levels of 2-oxoglutarate (2-OXO, FC: 0.46, p=0.017). 2-OXO is an intermediate in the Krebs cycle and plays a major role in carbon and nitrogen metabolism pathways, can be a marker for sensing cellular nutritional status, and has cellular signaling roles. Circulating levels of 2-OXO increase following periods of prolonged or intermittent exercise and cellular starvation (Lewis et al. 2010) and can act as an antioxidant (Liu et al. 2018). Considering that higher levels of 2-OXO have been associated with chronic heart failure (Chen et al. 2014) and that HTN is a risk factor for chronic heart failure, this could be an indication that non-responders have a more advanced level of cardiovascular disease.
3.3. Cross validation of metabolomics analyses
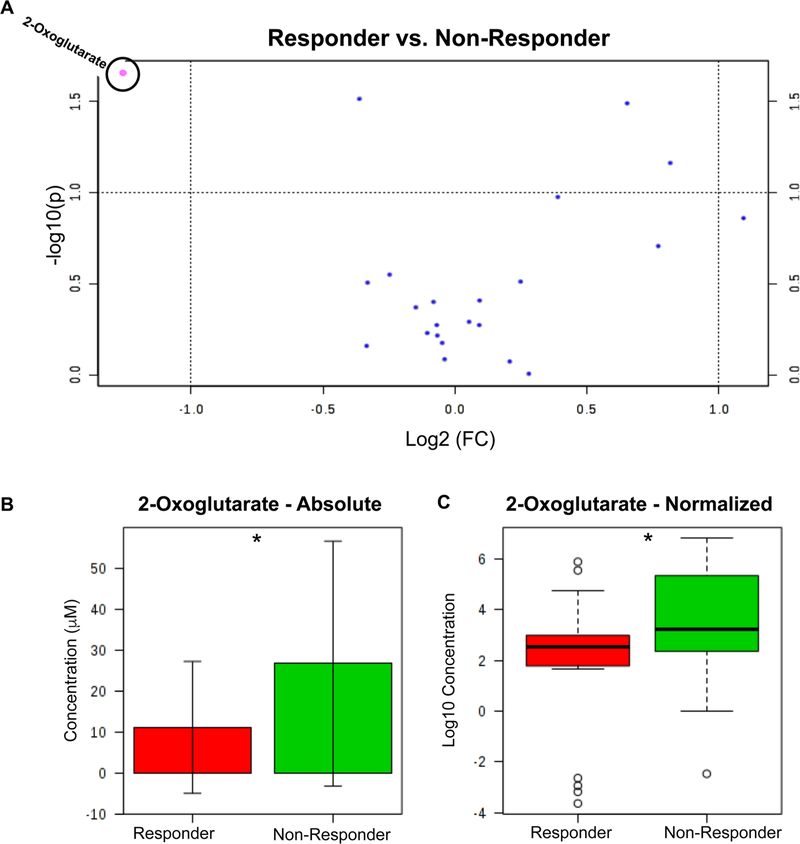
Absolute quantitation of a subset of metabolites (n = 23 metabolites, Supplemental Table 1) was used to cross validate the metabolomic findings. The volcano plot of the absolute quantitation data (Fig. 2a) suggests that only one of the tested metabolites, 2-OXO, was significant by both p value and fold change. Although both the absolute (Fig. 2b: median difference 3.59 μM) and normalized concentrations (Fig. 2c: median difference 0.68 μM) confirmed that responders had lower levels of 2-OXO at baseline compared to non-responders (p < 0.05). Permutation testing (see Supplemental Fig. 1) to validate the PLS-DA model and subsequent power analysis (see Supplemental Fig. 2) indicated that these data are not powered sufficiently to identify significant differences smaller than the ones observed (60 subjects required for power level of 0.8). However, the ROC curve (Supplemental Fig. 3, AUROC = 0.69, p = 0.03) suggests that 2-OXO may prove to be a significant biomarker predicting response to lisinopril with further validation and testing.
Fig. 2.
Absolute quantitation analysis of a subset of metabolites (n = 23 metabolites) a Volcano plot of absolute quantitation data from overall lisinopril cohort. b-c Absolute quantitation data for 2-oxoglutarate for original b and log-transformed concentrations c. Responder, n = 19, Non-Responder, n = 26. The (*) denotes a P value < 0.05
3.4. Modeling predictors of lisinopril effectiveness
A logistic regression analysis of clinical factors and metabolites was performed to identify a model that could be predictive of response to lisinopril in this pilot population. The subsequent model includes GFR (p = 0.023), log transformed BMI (p = 0.009), and log transformed 2-OXO (p = 0.015) absolute quantitation levels as predictive factors. This model demonstrates prediction of response to lisinopril with a Pseudo R-Square 0.278 at a significance of p < 0.001.
In summary, this pilot study provides preliminary evidence that suggests that increases in GFR, 2-OXO and BMI could predict poor response to lisinopril in treatment of HTN. Validating the predictive ability of these clinical and metabolic factors in a larger cohort may provide further evidence for the utility of metabolomics in precision prescribing of ACEIs.
Supplementary Material
Acknowledgments
Funding This study was funded by the National Institutes of Health (NIH R35GM124939, NIH K23 GM110516, and NIH CCTSI UL1 TR001082)
Footnotes
Compliance with ethical standards
Conflict of interest None of the authors have any conflicts of interest to disclose.
Ethical approval All procedures were performed in accordance with the ethical standards of the institutional research committee (COMIRB protocol# 13–3174) and with the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from all participants included in the study.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Azad RK, & Shulaev V. (2018). Metabolomics technology and bio-informatics for precision medicine. Briefings in Bioinformatics. 10.1093/bib/bbx170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M,… Pharmacometabolomics Task Group-Metabolomics Society, I. (2016). Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics, 12(10), 149 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blad CC, Ahmed K, AP IJ, & Offermanns S. (2011). Biological and pharmacological roles of HCA receptors. Advances in Pharmacology, 62, 219–250. 10.1016/B978-0-12-385952-5.00005-1. [DOI] [PubMed] [Google Scholar]
- Bundy JD, Mills KT, Chen J, Li C, Greenland P, & He J. (2018). Estimating the association of the 2017 and 2014 hypertension guidelines with cardiovascular events and deaths in us adults: An analysis of national data. JAMA Cardiol, 3(7), 572–581. 10.1001/jamacardio.2018.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PA, Xu ZH, Huang YL, Luo Y, Zhu DJ, Wang P,. Xu DL (2014). Increased serum 2-oxoglutarate associated with high myocardial energy expenditure and poor prognosis in chronic heart failure patients. Biochim Biophys Acta, 1842(11), 2120–2125. 10.1016/j.bbadis.2014.07.018. [DOI] [PubMed] [Google Scholar]
- Gu Q, Burt VL, Dillon CF, & Yoon S. (2012). Trends in anti-hypertensive medication use and blood pressure control among United States adults with hypertension: The National Health And Nutrition Examination Survey, 2001 to 2010. Circulation, 126(17), 2105–2114. 10.1161/CIRCULATIONAHA.112.096156. [DOI] [PubMed] [Google Scholar]
- Hou Y, Yin M, Sun F, Zhang T, Zhou X, Li H,… Li K. (2014). A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol Biosyst, 10(8), 2126–2133. 10.1039/c4mb00054d. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Boyle SH, Matson W, Sharma S, Matson S, Zhu H,… Delnomdedieu M. (2011). Pretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: A proof of concept. Transl Psychiatry, 1. 10.1038/tp.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC,… Gerszten RE. (2010). Metabolic signatures of exercise in human plasma. Sci Transl Med, 2(33), 33ra37. 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, He L, & Yao K. (2018). The antioxidative function of alpha-ketoglutarate and its applications. BioMed Research International, 2018, 3408467. 10.1155/2018/3408467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamoto S, Kanazawa K, Ashida H, & Natake M. (1988). Effect of orally administered 9-oxononanoid acid on lipogenesis in rat liver. Biochimica et Biophysica Acta, 959, 199–204. [DOI] [PubMed] [Google Scholar]
- Nemkov T, D’Alessandro A, & Hansen KC (2015). Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids, 47(11), 2345–2357. 10.1007/s00726-015-2019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemkov T, Hansen KC, & D’Alessandro A. (2017). A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Communications in Mass Spectrometry, 31(8), 663–673. 10.1002/rcm.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemkov T, Reisz JA, Gehrke S, Hansen KC, & D’Alessandro A. (2019). High-throughput metabolomics: Isocratic and gradient mass spectrometry-based methods In D’Alessandro A. (Ed.), High-throughput metabolomics (Vol. 1978). New York: Humana. [DOI] [PubMed] [Google Scholar]
- Offermanns S, Colletti SL, Lovenberg TW, Semple G, Wise A, & AP IJ (2011). International union of basic and clinical pharmacology. LXXXII: Nomenclature and classification of hydroxy-carboxylic acid receptors (GPR81, GPR109A, and GPR109B). Pharmacological Reviews, 63(2), 269–290. 10.1124/pr.110.003301. [DOI] [PubMed] [Google Scholar]
- Puskarich MA, Finkel MA, Karnovsky A, Jones AE, Trexel J, Harris BN, et al. (2015). Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Annals of the American Thoracic Society, 12(1), 46–56. 10.1513/AnnalsATS.201409-4150C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz JA, Zheng C, D’Alessandro A, & Nemkov T. (2019). Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics InDAlessandr A. (Ed.), High-throughput metabolomics (pp. 121–135). New York: Humana. [DOI] [PubMed] [Google Scholar]
- Ren R, Hashimoto T, Mizuno M, Takigawa H, Yoshida M, Azuma T, et al. (2013). A lipid peroxidation product 9-oxononanoic acid induces phospholipase A2 activity and thromboxane A2 production in human blood. Journal of Clinical Biochemistry and Nutrition, 52(3), 228–233. 10.3164/jcbn.12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan DJ, Kirby JA, & Ali S. (2010). Vascular biology: The role of sphingosine 1-phosphate in both the resting state and inflammation. Journal of Cellular and Molecular Medicine, 14(9), 2211–2222. 10.1111/j.1582-4934.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, Iliou A, Mikros E, & Elliott P. (2018). An overview of metabolic phenotyping in blood pressure research. Current Hypertension Reports, 20(9), 78 10.1007/s11906-018-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaneckova I, Maletinska L, Behuliak M, Nagelova V, Zicha J, & Kunes J. (2014). Obesity-related hypertension: Possible patho-physiological mechanisms. Journal of Endocrinology, 223(3), R63–78. 10.1530/JOE-14-0368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.