Abstract
Cacnals encodes the α1S subunit (Cav1.1) of voltage-dependent calcium channels, and is required for normal skeletal and cardiac muscle function, where it couples with the ryanodine receptor to regulate muscle contraction. Recently CACNA1S was reported to be expressed on the tips of retinal depolarizing bipolar cells (DBCs) and colocalized with metabotropic glutamate receptor 6 (mGluR6), which is critical to DBC signal transduction. Further, in mGluR6 knockout mice, expression at this location is down regulated. We examined RNAseq data from mouse retina and found expression of a novel isoform of Cacnals. To determine if CACNA1S was a functional component of the DBC signal transduction cascade, we performed immunohistochemistry to visualize its expression in several mouse lines that lack DBC function. Immunohistochemical staining with antibodies to CACNA1S show punctate labeling at the tips of DBCs in wild type (WT) retinas that are absent in Gpr179nob5 mutant retinas and decreased in Grm6−/− mouse retinas. CACNA1S and transient receptor potential cation channel, subfamily M, member 1 (TRPM1) staining also colocalized in WT retinas. Western blot analyses for CACNA1S of either retinal lysates or proteins after immunoprecipitation with the CACNA1S antibody failed to show the presence of bands expected for CACNA1S. Mass spectrometric analysis of CACNA1S immunoprecipitated proteins also failed to detect any peptides matching CACNA1S. Immunohistochemistry and western blotting after expression of GPR179 in HEK293T cells indicate that the CACNA1S antibody used here and in the retinal studies published to date, cross-reacts with GPR179. These data suggest caution should be exercised in conferring a role for CACNA1S in DBC signal transduction based solely on immunohistochemical staining.
Keywords: CACNA1S, GPR179, Retina, Antibody cross-reactivity, Bipolar cells
Introduction
Visual processing in mammalian retinas starts with photon absorption by photoreceptor cells, rods, and cones, which are optimized for light detection over many orders of light intensity. The synapse between cone photoreceptors and bipolar cells (BCs) establishes two parallel pathways for visual processing, mediated by two classes of BCs. Hyperpolarizing or OFF BCs utilize ionotropic glutamate receptors to sense glutamate released from photoreceptors and hyperpolarize in response to increments in light intensity. Depolarizing or ON BCs (DBCs) utilize a metabotropic glutamate receptor 6 (mGluR6) that when activated closes the transient receptor potential cation channel, subfamily M, member 1 (TRPM1) channel, consequently light increments decrease mGluR6 activity, TRPM1 opens and the cells depolarize. Rod photoreceptors are responsible for low light vision and transfer the signal to a specialized DBC, the rod BC that connects to the cone BC network via an All amacrine cell. Because rods connect primarily to a single ON BC type, defects in the mGluR6 to TRPM1 signal transduction cascade result in a heterogeneous series of diseases characterized by impairment of low light vision and are called complete congenital stationary night blindness (cCSNB). Incomplete forms of CSNB (iCSNB) result from defects in glutamate release from rod photoreceptors (Bech-Hansen et al., 2000; Dryja et al., 2005; Gregg et al., 2007; van Genderen et al., 2009; Peachey et al., 2012). cCSNB and iCSNB can be classified using the standard flash electroretinogram and neither show retinal degeneration.
cCSNB causing mutations have been found in Nyx, which encodes a leucine rich repeat protein called nyctalopin (Bech-Hansen et al., 2000; Scholl et al., 2001; Gregg et al., 2003); Grm6, encoding the mGluR6 (Masu et al., 1995; Dryja et al., 2005; Zeitz et al., 2005); Trpm1, (nonselective melastatin-type transient receptor potential channel) (Morgans et al., 2009; Shen et al., 2009; van Genderen et al., 2009; Koike et al., 2010); Gpr179, encoding a seven transmembrane domain protein (Audo et al., 2012; Peachey et al., 2012); and Lrit3, that encodes another leucine rich repeat protein (Zeitz et al., 2013; Neuille et al., 2014). All the proteins encoded by these genes are expressed and colocalize on the dendritic tips of DBCs. Recent studies found the expression of CACNA1S, a L-type voltage-dependent calcium channel (VDCC), also is present on the dendrites of DBCs in mouse retina (Specht et al., 2009; Soto et al., 2012; Tummala et al., 2014), where it colocalizes with mGluR6. The stability and localization of CACNA1S depends on mGluR6 expression and its cascade components, suggesting that CACNA1S is part of the mGluR6/TRPM1 complex (Tummala et al., 2014). Specht et al. observed a decrease in immunolabeling of retinal CACNA1S in Cacna1f and Bassoon mutant mice (Specht et al., 2009). Cacna1s encodes the pore-forming α subunit of L-type voltage gated calcium channels that are expressed in skeletal muscle and heart where they are required for normal muscle contraction. In skeletal muscle, the CACNA1S containing VDCCs interact with ryanodine receptors. CACNA1S plays a role as a voltage sensor and generates signals that mediate the opening of the ryanodine receptors, releasing Ca2+ from the sarcoplasmic reticulum and initiating muscle contraction (Rios & Pizarro, 1991; Mosca et al., 2013).
The goal of this study was to determine the role, if any, of CACNA1S in retinal processing. We discovered a novel mRNA isoform, however we found that the antibody used to characterize its expression in retina both here and in previous studies (Specht et al., 2009; Soto et al., 2012; Tummala et al., 2014) cross-reacts with GPR179, which is known to be required for DBC function. Together, our results suggest that the CACNA1S protein is not expressed in mouse retina and that the CACNA1S antibody cross-reacts with GPR179.
Materials and methods
All procedures were performed in accordance with local Institutional Animal Care and Use Committees and the Society for Neuroscience policies for the use of animals in research. All mice were kept in a local Association for Assessment and Accreditation of Laboratory Animal Care approved facility, under a 12 h light/dark cycle. Descriptions of all mouse lines used in this study have been published previously (Pearring et al., 2011; Ray et al., 2014). Mice of either sex were used in the experiments.
Antibodies and reagents
Antibodies (and their dilutions) used in immunohistochemical and immunoblotting experiments were: mouse monoclonal anti-CACNA1S (1:800; MAB427; Millipore, Billerica, MA), sheep anti-GPR179 (1:1000; peptide KVQEETPGEDLDRPVLQKR; Ray et al., 2014), and rhodamine peanut agglutinin (PNA) conjugate 566 (1:1000; Vector Laboratories, Burlingame, CA). Secondary antibodies (1:1000; Invitrogen, Thermo Fisher Scientific, Waltham, MA) were: donkey anti-sheep Alexa 488, donkey anti-mouse Alexa 647, and donkey anti-rabbit Alexa 546.
Immunohistochemistry
Immunohistochemistry was performed as previously reported (Pearring et al., 2011; Ray et al., 2014). Mice were euthanized, their eyes were enucleated, and the lens was removed. Eyecups were fixed for 30 min in 4% paraformaldehyde in phosphate buffered saline (PBS), pH 7.4. After fixing, eyecups were washed three times in PBS then cryoprotected in increasing concentrations of sucrose in PBS (5, 10, 15% for 1 h each and 20% overnight). Eyecups were embedded in 2:1 OCT/20% sucrose PBS solution and frozen in a liquid nitrogen-cooled bath of isopentane. Eyecups were sectioned (18 μm) using a Leica 1850 cryostat (Leica Biosystems Inc. Buffalo Grove, IL), mounted on glass slides, and stored at −80°C. Before immunostaining, sections were warmed to 37°C and washed with PBS and PBS containing 0.5% Triton X-100 (PBX) for 5 min each. After blocking in PBX plus 5% normal donkey serum (blocking solution) for 1 h, sections were incubated with primary antibody diluted in blocking solution overnight at room temperature, and then washed three times for 10 min each with PBX. Sections were incubated with secondary antibody (1:1000) in PBX for 1 h at room temperature followed by washing for 10 min in PBX twice and in PBS once, then cover-slipped using Vectashield (Vector Laboratories, Burlingame, CA). Images were taken using an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) and universally adjusted for brightness using Photoshop (Adobe Systems, San Jose, CA).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
RNA was isolated from mouse retinal and skeletal tissues using TRIZOL reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA) and cDNA was synthesized using Superscript III First-Strand Synthesis System (Invitrogen, Thermo Fisher Scientific, Waltham, MA) as recommended by the manufacturer. TaqMan assays were used to quantify mRNA levels. Probes specific to exon boundaries 9/10 (Mm00489257_ml), 16a/16b (AJ1RUOX), 16/17 (Mm01285315_ml), and 17/18 (Mm00489265_ml) were used. TaqMan probes to 18S RNA were used to normalize the data for Cacna1s.
Mass spectrometry
Mass spectrometric analysis was performed as previously described (Ray et al., 2014). After euthanizing the mice, retinas were isolated from wild type (WT) mice in lysis buffer (1% Nonidet P-40, 2 mM ethylenediaminetetraacetic acid (EDTA), and 20 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO)) by rotating at 4°C for 45 min. Samples were centrifuged at 17,000 × g for 20 min to remove cell debris, and the supernatant was precleared with Dynabeads (Invitrogen, Thermo Fisher Scientific, Waltham, MA) for 1 h at 4°C. Samples (1 mg total protein in 500 μl) were incubated with 10 μg CACNA1S antibody overnight at 4°C. Lysates and antibody complexes were incubated with 100 μl Dynabeads for 1.5 h at 4°C, followed by five washes with Tris-buffered saline containing 0.3% Tween-20. After eluting with NuPAGE LDS sample buffer (Invitrogen, Thermo Fisher Scientific, Waltham, MA), protein complexes were electrophoresed on 4—12% NuPAGE gels (Invitrogen, Thermo Fisher Scientific, Waltham, MA) until the highest molecular weight standard (250 kDa) had moved ~5 mm into the gel. In-gel tryptic digestion was performed prior to mass spectrometry as described elsewhere (Rood et al., 2010).
The resulting peptide mixture was resolved by liquid chromatography using an EASY n-LC (Thermo Fisher Scientific, Waltham, MA) ultra high performance liquid chromatography system with buffer A (2% v/v acetonitrile/0.1% v/v formic acid) and buffer B (80% v/v acetonitrile/0.1% v/v formic acid) as mobile phases. The mass spectrometry data from liquid chromatography elutes were collected using an Orbitrap Elite ETD mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Proteome Discoverer v1.3.0.330 was used to analyze the data collected and Scaffold v3.6.5 was used to calculate the false discovery rate using the peptide and protein prophet algorithms.
Cell culture, transfection and immunoblotting
Human embryonic kidney (HEK293T) cells were cultured in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin. Cells were seeded on 60 mm culture dishes one day prior to transfection and transfected with a mouse Gpr179 expression plasmid tagged with FLAG and green fluorescent protein (GFP), using jetPrime reagent (Polyplustransfection, New York, NY) or Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. 24–48 h after transfection, cells were harvested in NP-40 lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM EDTA, and 1% Nonidet P40, pH 8.0, supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO)) and disrupted by rotating for 45 min at 4°C. Cell debris was removed by centrifugation at 17,000 × g for 15 min at 4°C and the supernatant was collected and protein quantified using the Bradford reagent (Bio-Rad, Hercules, CA). 30 μg total protein lysates were loaded per lane and analyzed on 4–12% NuPAGE gels (Invitrogen, Thermo Fisher Scientific, Waltham, MA), transferred to polyvinylidene difluoride (PVDF) membranes, and blocked with Odyssey Blocking Buffer. Membranes were incubated with primary antibodies (mouse anti-CACNA1S and sheep anti-GPR179; 1:1000), diluted in Odyssey Blocking Buffer, and washed four times with TBS (50 mM Tris-Cl, pH 7.6; 150 mM NaCl) containing 0.1% Tween-20 (TBST). After incubating with IRDye800 CW and IRDye680 CW-conjugated secondary antibodies diluted in Odyssey Blocking Buffer (LI-COR, Lincoln, NE), membranes were washed four times with TBST. Protein bands were visualized by scanning the membranes in an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) using both 700 and 800 nm channels.
Coimmunoprecipitation
Coimmunoprecipitation of proteins from mouse retina and heart, and HEK293T cell lysates was performed as described previously (Ray et al., 2014). WT and Gpr179nob5 mouse retinas and WT mouse heart were isolated, homogenized in lysis buffer (1% Nonidet P-40, 2 mM EDTA, and 20 mM HEPES, pH 7.4, supplemented with protease inhibitor cocktail) and lysed further by rotating at 4°C for 45 min. Homogenates were cleared by centrifugation at 17,000 × g for 20 min at 4°C. Retina and heart lysates (400 μg total protein in 200 μl) or transfected HEK cell lysates (500 μg total protein in 200 μl) were precleared by incubating with 12 μl Dynabeads protein G (Invitrogen, Thermo Fisher Scientific, Waltham, MA) at 4°C for 1 h. Precleared lysates were incubated with 2–5 μg of anti-CACNA1S or anti-GPR179 antibodies overnight at 4°C on an orbital rocker. 45 μl of Dynabeads protein G was added and incubated for 1–2 h at 4°C. Dynabeads were collected and washed four times with Tris-buffered saline containing 0.3% Tween 20. Protein complexes were eluted with 40 μl of 4X LDS loading buffer by incubation at 70°C for 10 min, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting.
Results and discussion
Skeletal muscle L-type voltage gated calcium channels (VGCCs) consist of CACNA1S, the pore-forming subunit, and three ancillary subunits, CACNA2D1, CACNB1, and CACNG1 (Pietri-Rouxel et al., 2010). CACNA1S also is expressed in heart and mutations result in hypokalemic periodic paralysis (Maffe et al., 2009). The CACNA1S containing channels in skeletal muscle and heart are part of large multifunctional complexes and as such impact functions beyond calcium entry (Pietri-Rouxel et al., 2010). The reports that CACNA1S was expressed on the dendritic tips of ON BCs raised the possibility that it has a unique role in ON BC function, which was further supported by the observation that it colocalized with several ON BC signal cascade components (Tummala et al., 2014). Further, its expression was decreased in Gnn6−/−, Gnao1−/−, Gnb3−/−, Gng13−/−, and Trpm1−/− mice (Tummala et al., 2014). The present study was initiated prior to creating the conditional Cacna1s knockout mouse needed for functional studies, because a regular knockout would be perinatal lethal (Chaudhari, 1992).
CACNA1S antibody stains tips of ON BCs and is disrupted in mouse models of CSNB
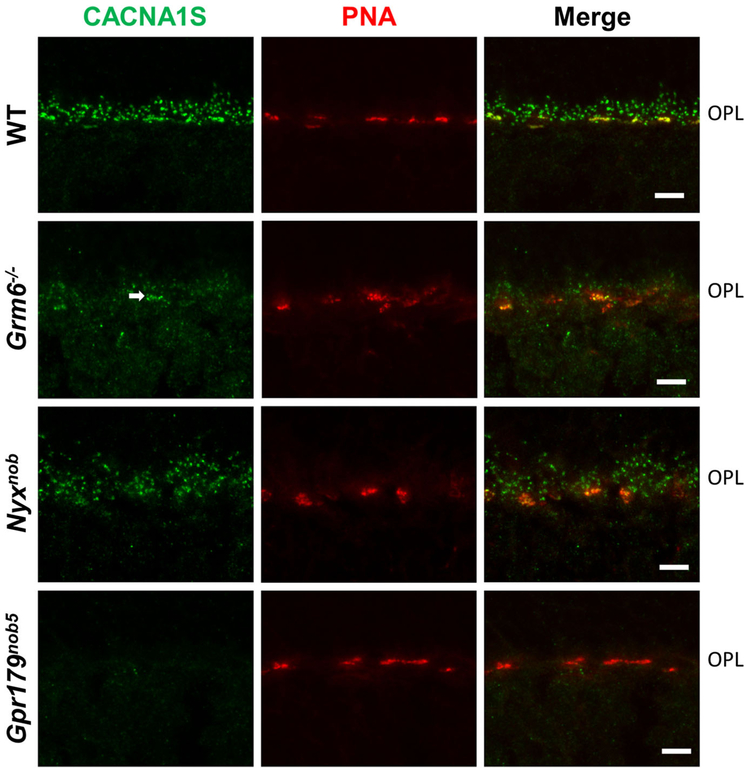
Previous studies showed that CACNA1S expression was localized to the dendritic tips of DBCs, where it co-localized with mGluR6 (Specht et al., 2009). To evaluate further the expression of CACNA1S at the photoreceptor to BC synapses, we examined its expression in, Nyxnob, Gpr179nob5, as well as Grm6−/− and Tipm1−/− mouse retinas. Consistent with earlier reports, we found punctate staining at the dendritic tips of DBCs in WT. In Grm6−/− retina sections staining for CACNA1S is present at the dendritic tips of all DBCs, although it appears to be reduced compared to WT, although the staining in the Gnn6−/− retinas appeals less effected on cone terminals (arrow. Fig. 1). This pattern is essentially identical to that of GPR179 staining in the Grm6−/− retina (Ray et al., 2014). Others reported that expression of CACNA1S was dependent on mGluR6 (Tummala et al., 2014). However, in that report there appeals to be faint staining by CACNA1S antibodies at the dendritic tips of all DBCs, which would be consistent with our data. The fact that the intensity of the staining in that report appears less than we observe may be due to slight differences in fixation conditions, which we have observed can greatly impact the sensitivity of many antibodies and lead to considerable variability.
Fig. 1.
CACNA1S staining is absent in Gpr179 mutant mice retina. Representative confocal images of WT, Grm6−/−, Nyxnob and Gpr179nob5 mice retinal slices stained with CACNA1S antibody (green) and PNA (red). PNA labels the cone pedicles and is used as a marker for the outer plexiform layer (OPL). The merged images show that CACNA1S and PNA colocalizes in WT, Grm6−/− and Nyxnob retinas. Scale bar represents 5 μm.
To extend these results, we examined the expression of CACNA1S in the Nyxnob and in Gpr179nob5 mouse retinas (Fig. 1). CACNA1S staining is similar to WT in the in Nyxnob retina, but is absent in the Gpr179nob5 retinas (Fig. 1), suggesting that CACNA1S expression and/or its localization is dependent on GPR179 expression.
These data and those published previously suggest a role for CACNA1S in DBC function. However, to date, there are no reports of a putative role for a VDCC in the DBC signal transduction. To examine this directly, a conditional knockout mouse for Cacna1s is required, because null alleles of the gene are peri-natal lethal because of a lack of skeletal muscle contraction (Chaudhari, 1992). Further the above data presupposes that the antibody to CACNA1S is specific, which has not been tested in the retina in a Cacna1s knockout mouse.
A novel mRNA isoform of Cacna1s gene is expressed in mouse retina
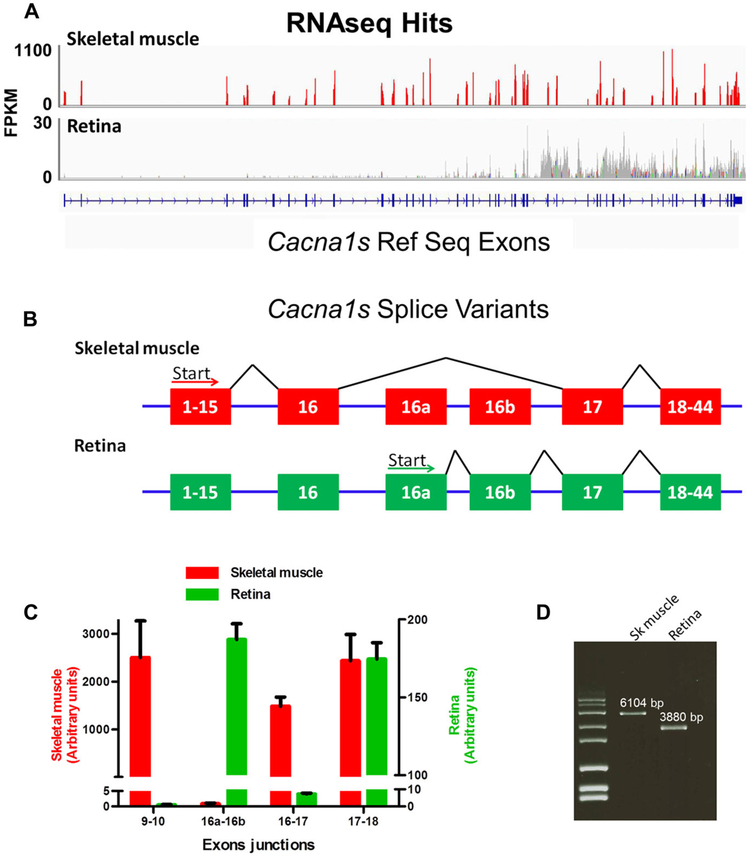
To obtain further evidence of CACNA1S expression in retina, we examined RNAseq data. CACNA1S is expressed abundantly in skeletal and cardiac muscle and polymerase chain reaction (PCR) products representing a small fragment of mRNA (exon 42-44) for Cacna1s were identified from retina by Vardi and colleagues (Tummala et al., 2014). To examine expression levels further and the possible existence of variant isoforms in retina, we utilized publically available RNAseq data obtained for skeletal muscle and retina (Brooks et al., 2011; Sandstrom, 2012). The RNAseq data representing skeletal muscle mRNA aligned with the known Cacna1s exons, and showed robust expression with fragments per kilobase of exon per million reads mapped (FPKM) values of ~1100 (Fig. 2A). RNAseq data obtained from retina showed alignments to exons 17—44 but at a much lower level, ~30 FPKM, compared to expression in skeletal muscle. There was a complete absence of any sequences aligning to exons 1–16 of Cacna1s in the retinal RNAseq data. Examination of the alignments of the retina sequences indicated that there were two previously undesciibed exons, referred to here as 16a and 16b, located between exon 16 and exon 17 expressed in the retina (Fig. 2B). To confirm the expression of different exons, we isolated total RNA from skeletal muscle and retinal tissue, reverse transcribed it into cDNA and used PCR to amplify and sequence specific regions representing the Cacna1s transcript. This novel transcript is predicted to encode a protein of 132.6 kDa. We quantihed the expression level of different regions of skeletal muscle and retina transcripts using real-time RT-PCR. cDNA fragments amplified by primer pairs flanking intron boundaries of exons 9–10, 16–17,16a–16b and 17–18 (Fig. 2B) were used to quantify expression of the various splice variants. Amplified products representing the exon 9–10, 16–17 and 17–18 junction fragments were present in skeletal muscle cDNA (Fig. 2C). However, the expression of junction fragments representing exons 9–10 and 16–17 were near background in retina, whereas the fragments representing the junction of exons 16a–16b were comparable to that for a fragment from exon 17–18 in retina (Fig. 2C). The relative levels of transcripts from skeletal muscle and retina as represented by the exon 17–18 junction normalized to 18S RNA levels showed that retina had ~14 fold less total expression than muscle. These data are consistent with the RNAseq alignments, confirming that there is essentially no expression of exons 1–16 of Cacna1s in retina and the absolute level of expression in retina is very low compared to skeletal muscle. To further characterize the transcripts in muscle and retina, we amplified full-length cDNA fragments from each tissue (Fig. 2D). Cloning and sequencing the amplified products revealed the expected cDNA sequence from skeletal muscle. All attempts to amplify any fragment that included exons 1–16 from retina failed. However, we were able to amplify a 3,880 bp cDNA fragment from retina that was consistent with expression of exons 16a–44 (Fig. 2D). This novel transcript is predicted to encode a protein of 132.6 kDa containing 71 novel amino acids from exons 16a and 16b, which included a signal sequence.
Fig. 2.
Cacna1s expression in the retina. (A) Schematic of genome coverage of RNAseq data (Brooks et al. 2011 and Sandstrom, 2012) at the Cacna1s locus. The y-axis represents expression level as FPKM. (B) Schematic diagram of skeletal muscle and retina splicing pattern for Cacna1s, as indicated by analyses of the RNAseq alignment data. The retina uses two previously undescribed exons, referred to here as 16a and 16b. (C) qRT-PCR analyses of Cacna1s expression in skeletal muscle and retina. Assays were located at the indicated exon junctions. There is no expression of Cacna1s exons 1-16 in retina. Data were normalized to 18S expression. Note the difference in scales used for skeletal muscle and retina. (D) Amplified full-length transcript from skeletal muscle and retina revealed that the retinal isoform of Cacna1s is 2,224 bp smaller than the skeletal muscle isoform.
Absence of CACNA1S protein in mouse retina
To determine if the CACNA1S protein was expressed in mouse retina, we did western blots from retinal lysates. The CACNA1S antibody detects robust expression of the expected size (190 kDa) protein in heart lysates (Fig. 3). In contrast, we were unable to detect a protein representing the full-length form of CACNA1S in retina, although there was a faint band (box in Fig. 3) at ~130 kDa, the predicted size of the retinal isoform (132.6 kDa). To evaluate whether this band was CACNA1S, we excised the region from the gel (box in Fig. 3), extracted the proteins, and performed mass spectrometry. The mass spectrometric analyses failed to reveal the presence of any peptides that matched CACNA1S (data not shown) suggesting the band was due to nonspecific staining.
Fig. 3.
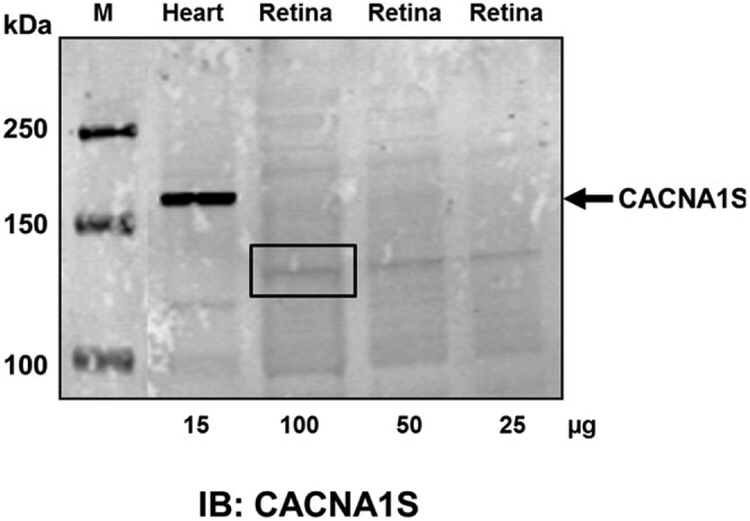
CACNA1S protein expression is not detected in mouse retina. Western blot analysis from retinal lysates (25–100μg) failed to detect CACNA1S expression. Heart lysates were used as a positive control for CACNA1S expression. The boxed area corresponds to ~125–140 kDa and this region was excised from the gel and the proteins therein analyzed by mass spectrometry. There were no peptides to CACNA1S identified (data not shown), suggesting the faint band is nonspecific staining.
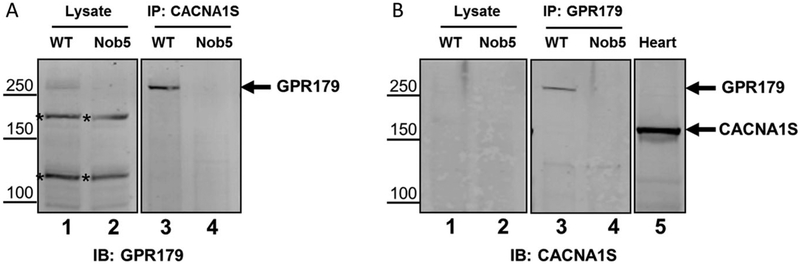
The previous experiments indicate CACNA1S, if present, is expressed at low levels. This is not unreasonable because we used whole retina lysates, and the immunohistochemical staining shows expression only on the tips of DBCs. Therefore, to enrich for CACNA1S, we used immunoprecipitation from mouse retinal lysates and attempted to identify the presence of CACNA1S using mass spectrometry. The mass spectrometric analysis revealed peptides to ~275 proteins, but failed to detect any to CACNA1S. Table 1 shows the top 10 hits. These include immunoglobulin G (IgG) molecules and several proteins that are part of the cytoskeleton. The only known DBC transduction molecule in this list is GPR179, for which 96 unique peptides with >95% identification probability mapped to GPR179 with a protein coverage of 58% (Table 1, Fig. 4). These data indicate either strong interaction between CACNA1S and GPR179 or that the CACNA1S antibody recognizes mouse GPR179. To examine these possibilities further we immunoprecipitated proteins from retinal lysates with antibodies to CACNA1S and GPR179 and then immunoblotted for GPR179 and CACNA1S, respectively (Fig. 5). Given our suspicion that the CACNA1S antibody was cross-reacting with GPR179 we used retinal lysates from Gprl79nob5 (Nob5) retinas as a negative control. As a positive control for the CACNA1S we included lysates from heart. The immunoblots of retinal lysates with antibody to GPR179 show the presence of a protein in the WT retinas at ~250 kDa, consistent with the molecular weight of GPR179 as expected and its absence from the Nob5 retinas. There also are two nonspecific bands present in both WT and Nob5 samples (Fig. 5A, * in lanes 1 and 2). After immunoprecipitation with the CACNA1S antibody and immunoblotting with GPR179 antibodies, the most prominent band is at ~250 kDa, the predicted size for GPR179, and it is absent from the Nob5 sample (Fig. 5A, in lanes 3 and 4). Note also that the nonspecific bands are absent after immunoprecipitation. Immunoblotting of WT or Nob5 retina lysates for CACNA1S (Fig. 5B, lanes 1 and 2) failed to detect any bands, although a robust band is present in heart lysates from WT mice (Fig. 5B, lane 5). After immunoprecipitation with the GPR179 antibody, immunoblotting for CACNA1S showed a band at ~250 kDa, the predicted size of GPR179 (Fig. 5B, lane 3) from WT retinas, but absent from Nob5 retinas. Combined these data further suggest that the antibody to CACNA1S cross-reacts with GPR179.
Table 1.
Proteins identified by mass spectrometry in CACNA1S antibody immunoprecipitates from retina lysates
| Identified proteina | Number of unique peptides |
% Coverage |
|---|---|---|
| Ig gamma-1 chain C region secreted form | 353 | 63 |
| Ig kappa chain C region | 211 | 100 |
| Protein Spna2 | 171 | 56 |
| Microtubule-associated protein 1 A | 161 | 46 |
| Spectrin beta chain, non-erythrocytic 1 | 157 | 56 |
| Ig kappa chain V-III region PC 6684 | 153 | 96 |
| Fibronectin | 152 | 52 |
| Protein Gprl79 | 127 | 58 |
| Microtubule-associated protein 1B | 126 | 49 |
| Ig gamma-3 chain C region | 124 | 64 |
| Ig gamma-2A chain C region, A allele | 103 | 47 |
| Ig gamma-2B chain C region | 101 | 51 |
| CACNA1S | 0 | 0 |
Only the top 10 hits are shown. CACNA1S is included for completeness.
Fig. 4.
Amino acids coverage of GPR179 from unique peptides detected in mass spectrometric analysis after immunoprecipitation with CACNA1S antibody.
Fig. 5.
CACNA1S antibody immunoprecipitates and detects GPR179 in mouse retina. (A) Western blot for GPR179 from retinal lysates (lanes 1 and 2) and after immunoprecipitation with CACNA1S antibody (lanes 3 and 4). GPR179 is present in WT retina lysates, as well as two other nonspecific bands (*). After immunoprecipitation with CACNA1S antibody, immunoblotting for GPR179 detects a protein at the predicted size of GPR179 (note the nonspecific bands are now absent). Lysates from Gpr179Nob5 mice (Nob5) are used as a negative control for GPR179. (B) Immunoblotting for CACNA1S in retina lysates and after immunoprecipitation with GPR179 antibodies. The CACAN1S antibody fails to detect any robust expression in WT or Nob5 (lanes 1 and 2) retina lysates. After immunoprecipitation with antibodies to GPR179, the CACNA1S antibody detects a band of ~250 kDa (the predicted size of GPR179) in WT retina but not Nob5 retinas (lanes 3 and 4). Lysates from heart were used as a positive control for the CACNA1S antibody (lane 5).
CACNA1S antibody recognizes GPR179
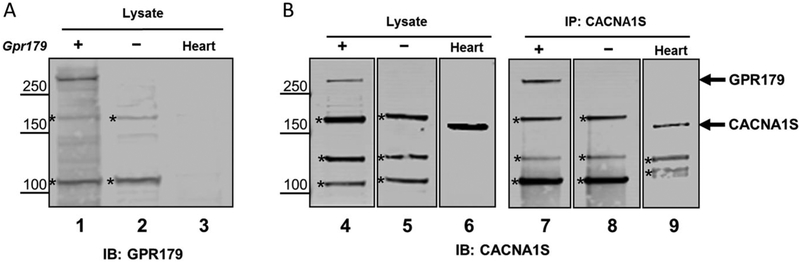
To examine the potential cross-reactivity of CACNA1S antibody with GPR179, we expressed mouse GPR179 in HEK293T cells and analyzed the lysates from transfected cells. Western blots from Gpr179-transfected cell lysates using GPR179 antibody show the expression of protein at the expected size of ~250 kDa (Fig. 6A, lane 1), which is absent in untransfected cells and heart lysates (Fig. 6A, lanes 2 and 3). Western blots using the CACNA1S antibody on these same lysates show that this antibody also recognizes GPR179 (Fig. 6B, lane 4 and 5) as well as two nonspecific bands (asterisks) in both transfected and untransfected cell lysates. Mouse heart lysate was used as a positive control for CACNA1S expression. Finally, we immunoprecipitated proteins using the CACNA1S antibody from Gpr179-transfected cells and heart lysates and immunoblotted for CACNA1S (Fig. 6B, lanes 7 and 9). The CACNA1S antibody immunoprecipitates GPR179 from transfected cell lysates and is detected by CACNA1S immunoblotting at the expected size (Fig. 6B, lane 7). This in vitro finding further confirms the cross-reactivity of the CACNA1S antibody with GPR179.
Fig. 6.
CACNA1S antibody detects and immunoprecipitates GPR179 after expression in HEK293T cells. (A) Western blot for GPR179 in lysates from HEK293T cells after transfection with mouse GPR179 expression plasmid (+, lane 1), negative control (−, lane 2) and heart lysate (lane 3). GPR179 is only present in the transfected cells. (B) Western blot for CACNA1S in lysates from HEK293T cells after transfection with GPR179 expression plasmid (+, lane 4), negative control (−, lane 5) and heart lysate as positive control (lane 6). Note that the CACNA1S anybody detects 3 nonspecific proteins in HEK293T cells (*, lanes 4 and 5). After immunoprecipitation of transfected cells with CACNA1S antibody, a band representing GPR179 is present (lane 7), absent in the negative control (lane 8) and present in the positive control (lane 9). Nonspecific bands also are present in the immunoprecitates (*, lanes 7, 8, 9). These data indicate that the CACNA1S anybody detects both GPR179 and CACNA1S proteins.
In conclusion, we were unable to find any evidence for the expression of CACNA1S protein in retinal lysates, even though the antibody recognizes a protein localized at the tips of ON BCs in immunohistochemical experiments. Collectively, our data suggest that the CACNA1S protein is not expressed at the dendritic tips of mouse DBCs; rather the antibody is cross-reacting with GPR179. Final confirmation of this hypothesis would require the generation of a conditional knockout mouse line of Cacna1s, which, given the weight of evidence we present, would seem to be an unwise investment.
References
- Audo I, Bujakowska K, Orhan E, Poloschek CM, Defoort-Dhellemmes S, Drumare I, Kohl S, Luu TD, Lecompte O, Zrenner E, Lancelot ME, Antonio A, Germain A, Michiels C, Audier C, Letexier M, Saraiva JP, Leroy BP, Munier FL, Mohand-Said S, Lorenz B, Friedburg C, Preising M, Kellner U, Renner AB, Moskova-Doumanova V, Berger W, Wissinger B, Hamel CP, Schorderet DF, De Baere E, Sharon D, Banin E, Jacobson SG, Bonneau D, Zanlonghi X, Le Meur G, Casteels I, Koenekoop R, Long VW, Meire E, Prescott K, de Ravel T, Simmons I, Nguyen H, Dollfus H, Poch O, Leveillard T, Nguyen-Ba-Charvet K, Sahel JA, Bhattacharya SS & Zeitz C (2012). Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. American Journal of Human Genetics 90, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Hansen NT, Naylor MJ, Maybaum TA, Sparkes RL, Koop B, Birch DG, Bergen AA, Prinsen CF, Polomeno RC, Gal A, Drack AV, Musarella MA, Jacobson SG, Young RS & Weleber RG (2000). Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nature Genetics 26, 319–323. [DOI] [PubMed] [Google Scholar]
- Brooks MJ, Rajasimha HK, Roger JE & Swaroop A (2011). Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(−/−) retinal transcriptomes. Molecular Vision 17, 3034–3054. [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N (1992). A single nucleotide deletion in the skeletal muscle-specific calcium channel transcript of muscular dysgenesis (mdg) mice. Journal of Biological Chemistry 267, 25636–25639. [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Berson EL, Fishman GA, Sandberg MA, Alexander KR, Derlacki DJ & Rajagopalan AS (2005). Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proceedings of the National Academy of Sciences of the United States of America 102, 4884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg R, Lukasiewicz P, Peachey N, Sagdullaev B & McCall M (2003). Nyctalopin is required for signaling through depolarizing bipolar cells in the murine retina. Investigative Ophtalmology and Visual Science 44, 4180. [Google Scholar]
- Gregg RG, Kamermans M, Klooster J, Lukasiewicz PD, Peachey NS, Vessey KA & McCall MA (2007). Nyctalopin expression in retinal bipolar cells restores visual function in a mouse model of complete X-linked congenital stationary night blindness. Journal of Neurophysiology 98, 3023–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, Ueda H, Kondo M, Mori Y, Tachibana M & Furukawa T (2010). TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proceedings of the National Academy of Sciences of the United States of America 107, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffe S, Signorotti E, Perucca A, Bielli M, Hladnik U, Ragazzoni E, Maduli E, Paffoni P, Dellavesa P, Paino AM, Zenone E, Parravicini U, Pardo NF, Cucchi L & Zanetta M (2009). Atypical arrhythmic complications in familial hypokalemic periodic paralysis. Journal of Cardiovascular Medicine 10, 68–71. [DOI] [PubMed] [Google Scholar]
- Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y & Shigemoto R (1995). Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell 80, 757–765. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Zhang J, Jeffrey BG, Nelson SM, Burke NS, Duvoisin RM & Brown RL (2009). TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proceedings of the National Academy of Sciences of the United States of America 106, 19174–19178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca B, Delbono O, Laura Messi M, Bergamelli L, Wang ZM, Vukcevic M, Lopez R, Treves S, Nishi M, Takeshima H, Paolini C, Martini M, Rispoli G, Protasi F & Zorzato F (2013). Enhanced dihydropyridine receptor calcium channel activity restores muscle strength in JP45/CASQ1 double knockout mice. Nature Communications 4, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuille M, El Shamieh S, Orhan E, Michiels C, Antonio A, Lancelot ME, Condroyer C, Bujakowska K, Poch O, Sahel JA, Audo I & Zeitz C (2014). Lrit3 deficient mouse (nob6): A novel model of complete congenital stationary night blindness (cCSNB). PLoS One 9, e90342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peachey NS, Ray TA, Florijn R, Rowe LB, Sjoerdsma T, Contreras-Alcantara S, Baba K, Tosini G, Pozdeyev N, Iuvone PM, Bojang P Jr., Pearring JN, Simonsz HJ, van Genderen M, Birch DG, Traboulsi EI, Dorfman A, Lopez I, Ren H, Goldberg AF, Nishina PM, Lachapelle P, McCall MA, Koenekoop RK, Bergen AA, Kamermans M & Gregg RG (2012). GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. American Journal of Human Genetics 90, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Bojang P Jr., Shen Y, Koike C, Furukawa T, Nawy S & Gregg RG (2011). A role for nyctalopin, a small leucine-rich repeat protein, in localizing the TRP melastatin 1 channel to retinal depolarizing bipolar cell dendrites. Journal of Neuroscience 31, 10060–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri-Rouxel F, Gentil C, Vassilopoulos S, Baas D, Mouisel E, Ferry A, Vignaud A, Hourde C, Marty I, Schaeffer L, Voit T & Garcia L (2010). DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. The EMBO Journal 29, 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Heath KM, Hasan N, Noel JM, Samuels IS, Martemyanov KA, Peachey NS, McCall MA & Gregg RG (2014). GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. Journal of Neuroscience 34, 6334–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios E &; Pizarro G (1991). Voltage sensor of excitation-contraction coupling in skeletal muscle. Physiological Reviews 71, 849–908. [DOI] [PubMed] [Google Scholar]
- Rood IM, Deegens JK, Merchant ML, Tamboer WP, Wilkey DW, Wetzels JE & Klein JB (2010). Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney International 78, 810–816. [DOI] [PubMed] [Google Scholar]
- Sandstrom R (2012). UW_RnaSeq_SkMuscle_adult-8wks_C57BL/6. NCBI GEO dataset, NCBI GEO dataset. [Google Scholar]
- Scholl HP, Langrova H, Pusch CM, Wissinger B, Zrenner E & Apfelstedt-Sylla E (2001). Slow and fast rod ERG pathways in patients with X-linked complete stationary night blindness carrying mutations in the NYX gene. Investigative Ophthalmology & Visual Science 42, 2728–2736. [PubMed] [Google Scholar]
- Shen Y, Heimel JA, Kamermans M, Peachey NS, Gregg RG & Nawy S (2009). A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. Journal of Neuroscience 29, 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto F, MA X, Cecil JL, Vo BQ, Culican SM & Kerschensteiner D (2012). Spontaneous activity promotes synapse formation in a cell-type-dependent manner in the developing retina. Journal of Neuroscience 32, 5426–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht D, Wu SB, Turner R, Dearden R, Koentgen E, Wolfrum U, Maw M, Brandstatter JH & tom Dieck S (2009). Effects of presynaptic mutations on a postsynaptic Cacna1s calcium channel colocalized with mGluR6 at mouse photoreceptor ribbon synapses. Investigative Ophthalmology &; Visual Science 50, 505–515. [DOI] [PubMed] [Google Scholar]
- Tummala SR, Neinstein A, Fina ME, Dhingra A & Vardi N (2014). Localization of Cacna1s to ON bipolar dendritic tips requires mGluR6-related cascade elements. Investigative Ophthalmology & Visual Science 55, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen MM, Bijveld MM, Claassen YB, Florijn RJ, Pearring JN, Meire FM, McCall MA, Riemslag FC, Gregg RG, Bergen AA & Kamermans M (2009). Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. American Journal of Human Genetics 85, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, Jacobson SG, Hamel CP, Bujakowska K, Neuille M, Orhan E, Zanlonghi X, Lancelot ME, Michiels C, Schwartz SB, Bocquet B, Antonio A, Audier C, Letexier M, Saraiva JP, Luu TD, Sennlaub F, Nguyen H, Poch O, Dollfus H, Lecompte O, Kohl S, Sahel JA, Bhattacharya SS & Audo I (2013). Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. American Journal of Human Genetics 92, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, van Genderen M, Neidhardt J, Luhmann UF, Hoeben F, Forster U, Wycisk K, Matyas G, Hoyng CB, Riemslag E, Meire F, Cremers FP & Berger W (2005). Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15 Hz flicker electroretinogram. Investigative Ophthalmology & Visual Science 46, 4328–4335. [DOI] [PubMed] [Google Scholar]