Abstract
Objectives
We tested cytoplasmic HuR (cHuR) as a predictive marker for response to chemotherapy by examining tumor samples from the international European Study Group of Pancreatic Cancer-3 trial, in which patients with resected pancreatic ductal adenocarcinoma (PDA) received either gemcitabine (GEM) or 5-fluorouracil (5-FU) adjuvant monotherapy.
Background
Previous studies have implicated the mRNA-binding protein, HuR (ELAVL1), as a predictive marker for PDA treatment response in the adjuvant setting. These studies were, however, based on small cohorts of patients outside of a clinical trial, or a clinical trial in which patients received multimodality therapy with concomitant radiation.
Methods
Tissue samples from 379 patients with PDA enrolled in the European Study Group of Pancreatic Cancer-3 trial were immunolabeled with an anti-HuR antibody and scored for cHuR expression. Patients were dichotomized into groups of high versus low cHuR expression.
Results
There was no association between cHuR expression and prognosis in the overall cohort [disease-free survival (DFS), P = 0.44; overall survival, P = 0.41). Median DFS for patients with high cHuR was significantly greater for patients treated with 5-FU compared to GEM [20.1 months, confidence interval (CI): 8.3–36.4 vs 10.9 months, CI: 7.5–14.2; P = 0.04]. Median DFS was similar between the treatment arms in patients with low cHuR (5-FU, 12.8 months, CI: 10.6–14.6 vs GEM, 12.9 months, CI: 11.2–15.4).
Conclusions
Patients with high cHuR-expressing tumors may benefit from 5-FU-based adjuvant therapy as compared to GEM, whereas those patients with low cHuR appear to have no survival advantage with GEM compared with 5-FU. Further studies are needed to validate HuR as a biomarker in both future monotherapy and multiagent regimens.
Keywords: adjuvant therapy, BIOMARKER, cancer, ELAVL1, European Study Group of Pancreatic Cancer, HuR, pancreatic ductal adenocarcinoma, resected pancreatic
By the year 2030, pancreatic ductal adenocarcinoma (PDA) will become the second leading cause of cancer-related death in the United States.1 PDA is often asymptomatic at a curable stage, whereas the majority of patients present with local invasion or metastatic disease.2 Moreover, adjuvant chemotherapy after resection for PDA offers a proven, albeit limited, overall survival (OS) benefit.3–5 To date, 5-fluorouracil (5-FU), gemcitabine (GEM), and their derivatives (eg, capecitabine) are the best available drugs for PDA treatment in the adjuvant setting and represent the current standard of care.6,7 Previous studies have explored gene expression transcripts (ie, hENT1, CDA, dCK, RRM1, RRM2) as markers to guide adjuvant therapy.8–11 To date there is, however, no standardized predictive biomarker for adjuvant therapy approved for clinical use. Although better, targeted treatment options are in the pipeline, strategies to select resected patients for the optimal adjuvant therapy (ie, GEM vs 5-FU) may provide short-term strategies to favorably improve outcomes.
We have previously evaluated the mRNA binding protein, HuR (ELAVL1), as a predictive marker for PDA response to adjuvant 5-FU and GEM in 3 separate cohorts (Table 1).12–14 HuR is typically located in the nucleus of PDA cells. Under conditions of stress in the tumor microenvironment (eg, hypoxia, glucose deprivation, chemotherapy), HuR translocates from the nucleus to the cytoplasm and binds U- or AU-rich sequences in the 3’ untranslated region of select target mRNAs (eg, WEE1, PIM1, TRAIL) important for PDA cell survival and chemotherapy resistance.15–17 Generally, this binding stabilizes mRNAs and post-transcriptionally upregulates protein expression. One such identified target is deoxycytidine kinase (dCK), which is critical for DNA synthesis, but is also the rate-limiting GEM metabolizing enzyme.13 In the laboratory setting, enhanced HuR expression levels and/or cytoplasmic HuR (cHuR) leads to high protein levels of dCK, which can correlate with increased GEM efficacy.13 Our first 2 studies suggested that increased cHuR expression was a poor overall prognostic marker in a retrospective cohort of patients undergoing resection for PDA.12,13 However, in an ad-hoc review of data from a phase III clinical trial by McAllister et al. where patients received multimodality therapy, HuR was not predictive of survival after either adjuvant GEM or 5-FU (Table 1).14
TABLE 1.
Summary of Studies Evaluating HuR as a Predictive Marker for Pancreatic Ductal Adenocarcinoma in the Adjuvant Setting
| First Author | Year | Patient Population | No. Samples | Therapy | Findings |
|---|---|---|---|---|---|
| Costantino et al13 | 2009 | TJUH | 40 | GEM ±Xeloda ± XRT | High cHuR: negative prognostic marker (OS HR 7.34, CI 2.05–26.22, p = 0.0022) |
| Richards et al12 | 2010 | TJUH | 53 | GEM (n = 24) | High cHuR: negative prognostic marker (OS HR 2.84, CI 1.04–7.74, P = 0.04) |
| No adjuvant therapy or follow-up (n = 21) | High cHuR: higher AJCC T stage (P = 0.005) | ||||
| McAllister et al14 | 2014 | RTOG9704 | 165 | GEM + XRT (n = 76) | cHuR: no prognostic or predictive value |
| 5-FU + XRT (n = 89) | |||||
| This study | 2016 | ESPAC-3 | 379 | GEM (n = 193) 5-FU (n = 186) | High cHuR: positive predictive marker for 5-FU (DFS HR 0.51, CI 0.3–0.85, P = 0.01) |
| 5-FU (n = 186) |
Based on this previous work, we sought to study HuR in a larger cohort of patients from a 2-armed, multi-institutional, randomized controlled phase III trial. The European Study Group of Pancreatic Cancer (ESPAC)-3 trial was a National Cancer Institute-funded, phase III trial, which randomized patients with resected pancreatic cancer to 1 of 2 approved first-line adjuvant chemotherapies.18 The results of this trial demonstrated a very limited and nonsignificant difference in median progression-free survival (PFS) and OS between 5-FU and GEM (PFS, 14.1 vs 14.3 months, P = 0.53; OS, 23 vs 23.6 months, P = 0.39). Short of a prospective biomarker-driven randomized trial, the present study served as the best cohort of patients available to study HuR as a prognostic and predictive biomarker. The absence of adjuvant radiation in the present study was particularly compelling for this biomarker study, because external beam radiation treatment engages HuR biologically through mechanisms that are independent from those affected by chemotherapy.19
METHODS
After study approval from the institutional review board at Thomas Jefferson University, 1233 tissue microarray (TMA) samples were obtained from a total of 379 patients with resected PDA enrolled in the ESPAC-3 trial. In the EsPAC-3 trial, patients were randomly assigned to receive 6 months of either GEM (1000 mg/m2 intravenous infusion once weekly for 3 of every 4 weeks) (n = 537) or 5-FU + folinic acid (folinic acid, 20 mg/m2 intravenous injection + 5-FU, 425 mg/m2 intravenous bolus injection given for 5 days every 28 days) (n = 551).18 The observation arm of this trial was discontinued early due to statistical evidence demonstrating a significant survival benefit in patients receiving adjuvant chemotherapy (n = 61). The 379 patients included in the present study were selected based on the availability of TMA. All available TMA samples were used without additional filtering. All patients received either adjuvant GEM or 5-FU.
Tissue Microarray Preparation
An experienced pathologist evaluated tumor samples from patients enrolled in the ESPAC-3 trial after hematoxylin and eosin staining of sections. TMA samples were generated from a representative area of each tumor with 1 to 8 cores arrayed for each patient. In total, 1233 TMA samples were generated from the 379 patients, 589 from 186 patients who received 5-FU and 644 from 193 patients who received GEM.
Immunohistochemical Analyses
Antigen retrieval was performed using Discovery CCI (Ventana, Tuscon, AZ cat 950–500) for a total application time of 64 minutes. Primary immunolabeling was performed using HuR antibody (3A2) (Santa Cruz Biotechnology, Santa Cruz, CA sc-5261) at a 1:300 dilution with Ventana Antibody Dilution Buffer (cat ADB250) for a 44-minute incubation at room temperature. We have previously published that the HuR antibody is specific for the protein via immunoblotting techniques.13,17 Immune complexes were visualized using the ultraView Universal DAB (diaminobenzidine tetrahydrochloride) Detection Kit (Ventana cat 760–500), which uses a rabbit horseradish peroxidase multimer cocktail for secondary immunolabeling. Slides were then washed with a Tris-based reaction buffer (Ventana cat 950–300) and stained with Hematoxylin II (Ventana cat 790–2208) for 8 minutes.
Scoring of the Tissue Microarray
A pathologist (W.J.) with expertise in pancreatic pathology scored cHuR immunolabeling within tumor cells in a blinded manner using a previously published scoring system.13 The pathologist assessed labeling intensity (strong vs weak), percentage of cell labeling, and cellular localization of labeling (nuclear vs cytoplasmic). Each sample was then graded based on the following scale: 0, no labeling; 1, weak diffuse labeling; 2, strong labeling in less than 50% of tumor cells; 3, strong labeling in greater than 50% of tumor cells. All available cores were graded for each patient. To account for variability in the number of cores, the graded cytoplasmic scores for each patient were averaged to obtain a cHuR score. These were analyzed in dichotomized groups identified as low cHuR (score <1.5) and high cHuR (score ≥1.5), similar to previously published reports.13
Statistical Analyses
All analyses were carried out using SAS version 14.0 (SAS Institute Inc., Cary, NC). Univariate Cox regression was carried out for all control and treatment variables for the response variables of OS time and disease-free survival (DFS) time. Control variables included HuR labeling, age, sex, smoking status, lymph node status, margin status, and clinical stage, whereas treatment variables included 5-FU and GEM. Kaplan-Meier survival curves were used to explore the relation between HuR labeling, treatment, and survival arms. Multivariable Cox regression models were built to investigate associations that appeared in exploratory analysis. These models included HuR labeling levels, treatment, and relevant control variables as covariates. The predictive utility of HuR was evaluated by testing the significance of the interaction between treatment and dichotomized HuR labeling in a Cox proportional hazards regression model. Univariate tests for association of potential confounders with HuR labeling levels were performed using Student t test for normally distributed continuous variables, Wilcoxon Rank-Sum test for nonnormal continuous variables, and Pearson chi-squared test for categorical variables. Variables associated with HuR labeling at a threshold value of P ≤ 0.2 were considered for inclusion in multivariable models.
RESULTS
Evaluation of Cytoplasmic HuR Expression and Correlation with Clinicopathologic Features
We received TMAs that included specimens from 379 patients enrolled in the ESPAC-3 trial, of which 186 were treated with 5-FU and 193 with GEM. All samples had at least some trace reactivity to HuR antibody. For patients with more than 1 core sample, the mean variance in cHuR score was 0.228 (±0.377). Of the 379 patients, 75 (19.8%) had tumors with high cHuR expression and 304 (80.2%) had tumors with low cHuR expression (Fig. 1). Patient demographics, treatment group allocation, and clinicopathologic features were similar between patients with high and low cHuR expression (Table 2).
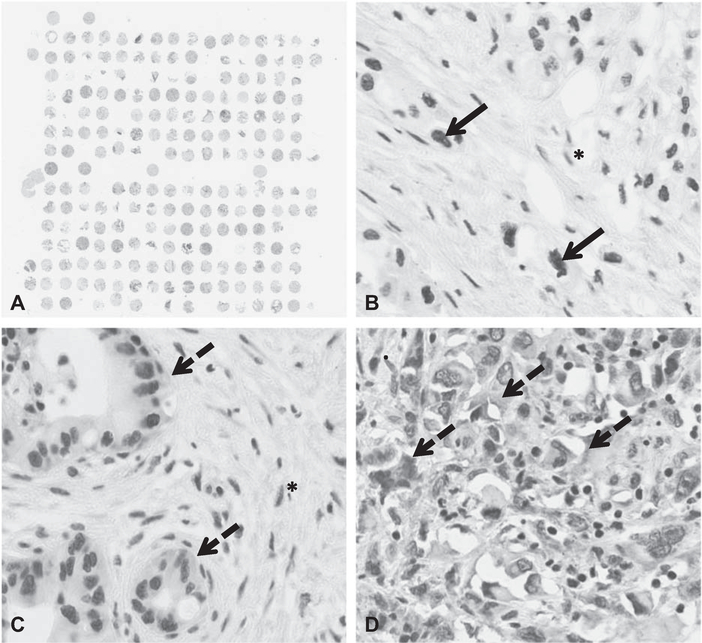
FIGURE 1.
Pancreatic ductal adenocarcinoma tissue microarray from the ESPAC-3 trial. Nuclear HuR labeling within tumor cells marked by solid arrow, cytoplasmic HuR labeling within tumor cells marked by dashed arrow. HuR labeling within tumor microenvironment (eg, stromal cells) identified by “*.” A, Sample TMA, low magnification. B, No cytoplasmic HuR (only nuclear HuR detected), high magnification. C, Low cytoplasmic HuR (weak labeling in <50% of cells), high magnification. D, High cytoplasmic HuR (strong labeling in almost 100% of cells), high magnification.
TABLE 2.
Summary of Patient Demographics and Clinicopa- thologic Features Stratified by cHuR Level
| Variable | Low cHuR (n = 304) | High cHuR (n = 75) | P |
|---|---|---|---|
| Age (yr) | |||
| Mean | 63.46 | 62.15 | 0.26 |
| Sex | |||
| Female | 127 (42%) | 35 (47%) | 0.44 |
| Male | 177 (58%) | 40 (53%) | |
| Smoker status | |||
| Never | 116 (38%) | 30 (40%) | 0.93 |
| Past | 108 (36%) | 25 (33%) | |
| Present | 48 (16%) | 12 (16%) | |
| Missing | 32 (10%) | 8 (11%) | |
| Treatment arm | |||
| 5-FU | 153 (50%) | 33 (44%) | 0.33 |
| GEM | 151 (50%) | 42 (56%) | |
| Lymph node | |||
| Negative | 61 (20%) | 18 (24%) | 0.45 |
| Positive | 243 (80%) | 57 (76%) | |
| Resection margins | |||
| Negative | 170 (56%) | 43 (57%) | 0.83 |
| Positive | 134 (44%) | 32 (43%) | |
| Stage | |||
| I | 20 (6%) | 6 (8%) | 0.45 |
| II | 75 (25%) | 19 (25%) | |
| III | 197 (65%) | 49 (66%) | |
| IV | 10 (3%) | 0 (0%) | |
| Missing | 2 (1%) | 1 (1%) | |
Correlation of Cytoplasmic HuR Expression and Response Outcomes
In evaluating the entire cohort, there was no association between cHuR expression and DFS [hazard ratio (HR) = 1.11, confidence interval (CI): 0.85–1.46, P = 0.44] or OS (HR = 0.89, CI: 0.68–1.18, P = 0.41), limiting HuR’s utility as a prognostic marker in a mixed population of patients treated with 5-FU and GEM. As expected based on results from the original ESPAC-3 trial, positive lymph nodes and positive resection margins were prognostic for worse DFS and OS by univariate analysis (Table 3).
TABLE 3.
Univariate Association Between Disease-free Survival, Overall Survival, and Various Prognostic Factors
| Variable | HR (95% CI) | P | |
|---|---|---|---|
| DFS | High vs low cHuR (n = 379) | 1.11 (0.85, 1.46) | 0.44 |
| N stage (n = 379) | 2.06 (1.54, 2.77) | <0.0001 | |
| Positive resection margins (n = 379) | 1.60 (1.28, 1.99) | <0.0001 | |
| OS | High vs low cHuR (n = 379) | 0.89 (0.68, 1.18) | 0.41 |
| N stage (n = 379) | 2.6 (1.58, 2.95) | <0.0001 | |
| Positive resection margins (n = 379) | 1.54 (1.23, 1.94) | 0.0002 |
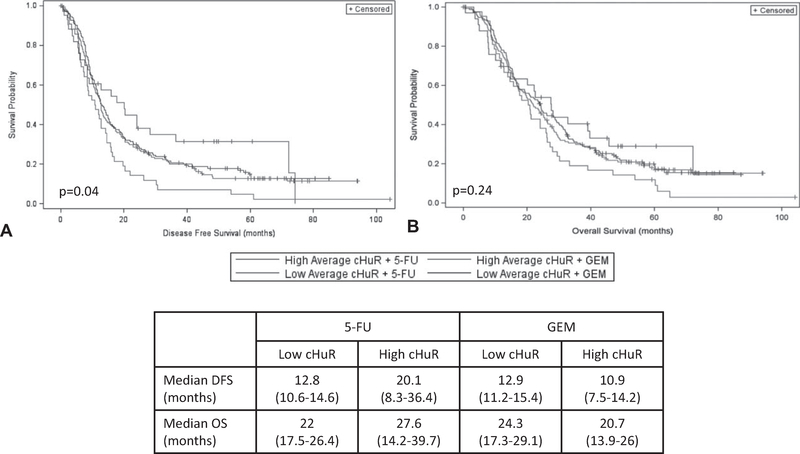
Kaplan-Meier survival curves demonstrated a differential response in DFS (Fig. 2A) when stratified by cHuR expression and treatment arm (P = 0.04). Patients with high cHuR who were treated with 5-FU had a near doubling of median DFS as compared to those treated with GEM (5-FU, 20.1 months, CI: 8.3–36.4 vs GEM 10.9 months, CI: 7.5–14.2, P = 0.012). Median DFS was, however, similar between the treatment arms in patients with low cHuR (5-FU, 12.8 months, CI: 10.6–14.6 vs GEM, 12.9 months, CI: 11.2–15.4, P = 0.44). Overall, the worst DFS was observed in patients with high cHuR who were treated with GEM. A significant differential response was not seen in OS (P = 0.24), although there was a persistent trend toward improved survival in patients with high cHuR who were treated with 5-FU as compared to GEM (5-FU, 27.6 months, CI: 14.2–39.7 vs GEM, 20.7 months, CI: 13.9–26, P = 0.08) (Fig. 2B).
FIGURE 2.
Kaplan-Meier survival curves for (A) DFS and (B) OS, stratified by average cHuR score group and treatment arm.
Unadjusted Cox regression was used to evaluate cHuR as a predictive marker for DFS and OS. This analysis found cHuR to be predictive of DFS when stratified by the treatment arm (P = 0.012) (Table 4). Specifically, high cHuR was associated with improved DFS in patients treated with 5-FU as compared to GEM (HR = 0.51, CI: 0.31–0.85, P = 0.01). In patients treated with GEM, high cHuR was associated with reduced DFS when compared to low cHuR (HR = 1.54, CI: 1.08–2.2, P = 0.02). Overall, cHuR was not a significant predictive marker for OS (P = 0.29). There was a trend toward improved OS in patients with high cHuR treated with 5-FU as opposed to GEM (HR = 0.63, CI: 0.38–1.06, P = 0.08). As seen with DFS, there was again a trend toward reduced DFS with GEM in patients with high cHuR (HR = 1.4, CI: 0.97–1.99, P = 0.07).
TABLE 4.
Univariate Model for cHuR and Treatment Arm Interaction
| Variable | HR (95% CI) | P | |
|---|---|---|---|
| DFS | cHuR, treatment arm interaction | 0.012 | |
| 5-FU vs GEM: high cHuR | 0.51 (0.3, 0.85) | 0.01 | |
| 5-FU vs GEM: low cHuR | 1.05 (0.82, 1.34) | 0.70 | |
| High vs low cHuR: GEM | 1.54 (1.09, 2.2) | 0.02 | |
| High vs low cHuR: 5-FU | 0.75 (0.49, 1.16) | 0.2 | |
| OS | cHuR, treatment arm interaction | 0.29 | |
| 5-FU vs GEM: high cHuR | 0.63 (0.38, 1.06) | 0.08 | |
| 5-FU vs GEM: low cHuR | 1.05 (0.82, 1.36) | 0.69 | |
| High vs low cHuR: GEM | 1.4 (0.98, 1.99) | 0.07 | |
| High vs low cHuR: 5-FU | 0.84 (0.43, 1.31) | 0.44 |
Multivariable Cox regression was performed, adjusting cHuR expression for lymph node status and resection margins (Table 5). After adjustment, cHuR expression was no longer a significant predictive marker for DFS (P = 0.11) or OS (P = 0.27). There was, however, a trend in the multivariate model toward improved DFS in patients with high cHuR treated with 5-FU (HR = 0.64, CI: 0.39–1.08, P = 0.09), and reduced survival with GEM (DFS, HR = 1.51, CI: 1.06–2.16, P = 0.02; OS, HR = 1.39, CI: 0.95–2.05, P = 0.09).
TABLE 5.
Multivariable Model for Disease-free Survival, Overall Survival, and Treatment Arm Interaction, Adjusted for Lymph Node Status and Resection Margins
| Variable | HR (95% CI) | P | |
|---|---|---|---|
| DFS | cHuR, treatment arm interaction | 0.11 | |
| 5-FU vs GEM: high cHuR | 0.64 (0.39,1.08) | 0.09 | |
| 5-FU vs GEM: low cHuR | 1.03 (0.80,1.32) | 0.82 | |
| High vs low cHuR: GEM | 1.51 (1.06, 2.16) | 0.02 | |
| High vs low cHuR: 5-FU | 0.95 (0.61, 1.47) | 0.81 | |
| Positive lymph nodes | 1.87 (1.37, 2.55) | <0.0001 | |
| Positive resection margins | 1.43 (1.14, 1.79) | 0.0023 | |
| OS | cHuR, treatment arm interaction | 0.27 | |
| 5-FU vs GEM: high cHuR | 0.73 (0.42, 1.27) | 0.27 | |
| 5-FU vs GEM: low cHuR | 1.03 (0.79, 1.35) | 0.82 | |
| High vs low cHuR: GEM | 1.39 (0.95, 2.05) | 0.09 | |
| High vs low cHuR: 5-FU | 0.99 (0.61, 1.59) | 0.95 | |
| Positive lymph nodes | 2.01 (1.43, 2.84) | <0.0001 | |
| Positive resection margins | 1.48 (1.16, 1.89) | 0.0016 |
DISCUSSION
The search for a robust reproducible predictive biomarker for pancreatic cancer has eluded investigators for decades. In a previous study of 40 resected PDA specimens from our group, low cHuR correlated with a 7-fold increased risk in mortality in patients receiving adjuvant GEM therapy (HR = 7.34, CI: 2.05–26.22, P = 0.0022).13 These findings were later supported in a smaller cohort from the same institution.12 In a group of 24 patients who received adjuvant GEM monotherapy, low cHuR was predictive of worse OS as compared to high cHuR (HR = 2.84, CI: 1.04–7.74, P = 0.04). These results were attributed to the role of HuR in upregulating and activating dCK, an enzyme responsible for metabolizing GEM to its active metabolites.13 These data were validated by in vitro experiments, and supported a model in which HuR sensitized tumor cells to the action of GEM by enhancing the prodrug’s conversion to its active form.13 Recently, McAllister et al14 assessed the predictive value of cHuR in 165 patients from the Radiation Therapy Oncology Group (RTOG) 9704 trial. Although the present study had the advantage of including a large sample of patients from a randomized trial, radiation therapy administration was interpreted as a confounder.20 Ionizing radiation complicates HuR biology by inducing phosphorylation of HuR by checkpoint kinase 2, which causes dissociation of HuR-mRNA complexes, altered downstream gene expression, and improved cell survival in vitro.19 In the RTOG 9704 cohort, there was no difference between low and high HuR expressing tumors with respect to DFS or OS in either arm. It was presumed that the radiation effect might have contributed to the lack of any interaction between cHuR and outcome.
Since the publication of these studies, we and others have, however, demonstrated through in vivo and in vitro models that HuR actually supports a therapeutic resistant phenotype in PDA through HuR-mediated overexpression of key prosurvival proteins.21–24 Most recently, Elebro et al24 reported an association between high cHuR expression and reduced OS in GEM-treated patients with pancreatobiliary-type periampullary tumors compared with untreated controls (HR 2.07, 95% CI: 1.03–4.17, P = 0.028). These later studies conflict with the earlier work, and indicate that HuR status is more likely to promote resistance to treatment than it is to enhance drug sensitivity. For this reason, we specifically sought to analyze a cohort of samples from a rigorously accrued phase III trial with robust patient follow-up and no radiation therapy arm.
Herein, we evaluated TMA samples generated from tumors of patients enrolled in the ESPAC-3 phase III trial in which patients received either GEM or 5-FU monotherapy, without any radiation. cHuR was evaluated objectively as a prognostic marker and as predictive marker for each specific treatment. cHuR expression alone was not an informative prognostic marker for DFS or OS in the total cohort. In the 20% of our cohort with high cHuR expression, a significant survival advantage was, however, seen in patients receiving 5-FU, whereas these patients’ tumors were relatively resistant to GEM. This translated to a near doubling in median DFS (20.1 vs 10.9 months, P = 0.04). Notably, these values were above and below the median DFS for the entire ESPAC-3 cohort, (14.3 months).7 Patients with low cHuR had no difference in median DFS with respect to treatment arm, and the values approximated the above-mentioned median value for the ESPAC-3 trial. This observation provides a measure of validation for the present study, because the subgroup with low cHuR comprised the majority of the current cohort (80.2%, n = 304).18 Once adjusted for lymph nodes status and positive resection margins, cHuR was no longer a significant predictive marker. There, however, remained a trend toward improved DFS with 5-FU treatment in patients with high cHuR. The loss of statistical significance is most likely due to the small sample size of patients with high cHuR (19.8%, n = 75) relative to the overall large cohort studied.
GEM and 5-FU (including capecitabine, a prodrug of 5-FU) therapy have remained the standard of care for PDA treatment in the adjuvant setting based on the results of ESPAC-3, and other phase III clinical trials.3–5,18,20,25 Results from the Charité Onkologie-001 trial demonstrated a significant improvement in both DFS and OS in patients with PDA who received adjuvant GEM versus observation (DFS, 13.4 vs 6.7 months, P < 0.001; OS, 22.8 vs 20.2 months, P = 0.01).4,5 Soon after, the RTOG9704 trial randomized patients to receive 5-FU-based chemoradiation sandwiched between GEM or 5-FU monotherapy and ultimately showed no survival benefit between the 2 treatment groups, but OS was on par with patients receiving adjuvant therapy in Charité Onkologie-001. ESPAC-3 evaluated 5-FU and GEM without radiation therapy in a large, phase III, randomized controlled trial and found there to be no significant difference in median PFS (14.1 vs 14.3 months, P = 0.53) or OS (23 vs 23.6 months, P = 0.39) between the 2 treatment groups.18 Recently reported, ESPAC-4 found a small benefit in median survival with combination GEM/capecitabine (a 5-FU derivative) compared with GEM alone (28 vs 25.5 months, P = 0.032).7 Additional GEM and 5-FU based multiagent regimens are currently being tested in phase II and III trials, providing alternative treatment strategies (, ). Thus, there remains an opportunity to better select patients for specific regimens, to optimize treatment efficacy and minimize treatment toxicity.
Compared to our previous studies, we strongly believe this current cohort represents the best population available to study in the adjuvant setting, because all patients were enrolled in a randomized trial with long-term follow-up and no radiation confounder. We acknowledge limitations of our analysis. One limitation is that our analysis remains underpowered to detect a true difference in survival with respect to HuR status. In this cohort, only 19.8% of patients had tumors with high cHuR expression. Underscoring this point, detecting high cHuR in less than 20% of patients from ESPAC-3 does not correlate with previous scoring patterns in which high cHuR was detected in closer to half the tumors evaluated in the study.12–14 Another limitation of using HuR as a biomarker is the reliance on immunohistochemistry and a subjective scoring system. Scoring is dependent on a trained pathologist and may be subject to variable interpretation by different pathologists. The present study only evaluated patients who received adjuvant 5-FU and GEM monotherapy and cannot be applied to patients receiving combination therapies (FOLFIRINOX, GEM/nab-paclitaxel), or to those with locally advanced or metastatic tumors. Still, the present study provides evidence that cHuR may be predictive for 5-FU efficacy in patients with resectable PDA and high cHuR. If validated, these findings challenge prior work highlighting HuR as promoting chemotherapyresistance.21,22 Perhaps HuR regulates specific transcripts that contribute to 5-FU sensitivity. Alternatively, high cHuR may be a marker of rapid cell division, and these cells may be particularly susceptible to 5-FU cytotoxic therapy.14
Regardless of the exact mechanism, there is an opportunity to optimize outcomes using the drugs that we currently have available, and offer an opportunity to personalize treatments for patients with PDA. As a next step, patients enrolled in the Adjuvant Pancreatic Adenocarcinoma Clinical Trial and ESPAC-4 trial would be the preferred cohorts to test the predictive value of cHuR in the context of relevant combination therapies, which are already 5-FU and GEM based, and provide a basis for a prospective, personalized trial in which treatments are based on cHuR expression.
Footnotes
The authors report no conflicts of interest.
REFERENCES
- 1.Society AC. Cancer Facts and Figures 2015. Available at: http://www.cancer.org/acs/groups/content/@editorial/documents/document/acspc-044552.pdf. Accessed January 14, 2016.
- 2.Hidalgo M Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos JP, Stocken DD, Friess H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–1210. [DOI] [PubMed] [Google Scholar]
- 4.Oettle H, Post S, Neuhaus P, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. [DOI] [PubMed] [Google Scholar]
- 5.Oettle H, Neuhaus P, Hochhaus A, et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310:1473–1481. [DOI] [PubMed] [Google Scholar]
- 6.Oncology NCPGi. Pancreatic Adenocarcinoma [NCCN Clinical Practice Guidelines in Oncology Web Site]. 2015. Available at: http://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf. Accessed January 14, 2016.
- 7.Neoptolemos JP, Palmer D, Ghaneh P, et al. ESPAC-4: a multicenter, international, open-label randomized controlled phase III trial of adjuvant combination chemotherapy of gemcitabine (GEM) and capecitabine (CAP) versus monotherapy gemcitabine in patients with resected pancreatic ductal adenocarcinoma. J Clin Oncol. 2016;34(suppl abstr):LBA4006. [Google Scholar]
- 8.Fujita H, Ohuchida K, Mizumoto K, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenhalf W, Ghaneh P, Neoptolemos JP, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst. 2014;106:djt347. [DOI] [PubMed] [Google Scholar]
- 10.Farrell JJ, Elsaleh H, Garcia M, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–195. [DOI] [PubMed] [Google Scholar]
- 11.Farrell JJ, Bae K, Wong J, et al. Cytidine deaminase single-nucleotide polymorphism is predictive of toxicity from gemcitabine in patients with pancreatic cancer: RTOG 9704. Pharmacogenomics J. 2012;12:395–403. [DOI] [PubMed] [Google Scholar]
- 12.Richards NG, Rittenhouse DW, Freydin B, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252: 499–505. [DOI] [PubMed] [Google Scholar]
- 13.Costantino CL, Witkiewicz AK, Kuwano Y, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAllister F, Pineda DM, Jimbo M, et al. dCK expression correlates with 5-fluorouracil efficacy and HuR cytoplasmic expression in pancreatic cancer: a dual-institutional follow-up with the RTOG 9704 trial. Cancer Biol Ther. 2014;15:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorospe M HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 16.Abdelmohsen K, Lal A, Kim HH, et al. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. [DOI] [PubMed] [Google Scholar]
- 17.Burkhart RA, Pineda DM, Chand SN, et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol. 2013;10:1312–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 19.Masuda K, Abdelmohsen K, Kim MM, et al. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. EMBO J. 2011;30:1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regine WF, Winter KA, Abrams R, et al. Fluorouracil-based chemoradiation with either gemcitabine or fluorouracil chemotherapy after resection of pancreatic adenocarcinoma: 5-year analysis of the U.S. Intergroup/RTOG 9704 phase III trial. Ann Surg Oncol. 2011;18:1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco FF, Jimbo M, Wulfkuhle J, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2016;35:2529–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal S, Burkhart RA, Beeharry N, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74:1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romeo C, Weber MC, Zarei M, et al. HuR contributes to TRAIL resistance by restricting death receptor 4 expression in pancreatic cancer cells. Mol Cancer Res. 2016;14:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elebro J, Ben Dror L, Heby M, et al. Prognostic effect of hENT1, dCK and HuR expression by morphological type in periampullary adenocarcinoma, including pancreatic cancer. Acta Oncol. 2016;55:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regine WF, Winter KA, Abrams RA, et al. Fluorouracil vs gemcitabine chemotherapy before and after fluorouracil-based chemoradiation following resection of pancreatic adenocarcinoma: a randomized controlled trial. JAMA. 2008;299:1019–1026. [DOI] [PubMed] [Google Scholar]