Abstract
While both cis and trans (adhesive)-interactions cooperate in the assembly of intercellular adhesions, computational simulations have predicted that two-dimensional confinement may promote cis-oligomerization, in the absence of trans-interactions. Here, single-molecule tracking of cadherin extracellular domains on supported lipid bilayers revealed the density-dependent formation of oligomers and cis-clusters in the absence of trans-interactions. Lateral oligomers were virtually eliminated by mutating a putative cis (lateral) binding interface. At low cadherin surface coverage, wild-type and mutant cadherin diffused rapidly, consistent with the motion of a lipid molecule within a cadherin-free supported bilayer and with cadherins diffusing as monomers. Although the diffusion of mutant cadherin did not change appreciably with increasing surface coverage, the average short-time diffusion coefficient of wild-type cadherin slowed significantly above a fractional surface coverage of ~0.01 (~1100 molecules/μm2). A detailed analysis of molecular trajectories suggested the presence of a broad size distribution of cis-cadherin oligomers. These findings verify predictions that two-dimensional confinement promotes cis-oligomerization, in the absence of trans-interactions.
Graphical Abstract
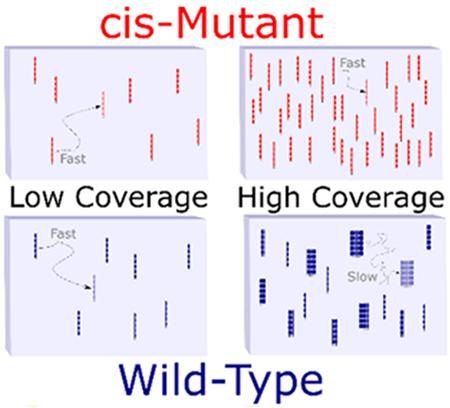
Cadherins are transmembrane adhesion proteins responsible for cell-to-cell cohesion in all tissues. Cadherin-mediated intercellular adhesion regulates biological processes such as cell proliferation, tissue rearrangement, tissue boundary formation, and macromolecular transport across tissue boundaries.1–4 This cohesion is facilitated by binding between cadherin molecules on opposing cells—referred to as trans bonds. The structure of cadherin consists of an intracellular domain, transmembrane domain, and five tandem, structurally similar extracellular domains, EC1–EC5, numbered from the N-terminus.3,5 The assembly of adherens junctions has been demonstrated to involve the formation of both trans bonds between molecules on adjacent cells and cis bonds between molecules on the same membrane.6–21 Cis-interactions were not detected in traditional solution-phase binding assays, even at high concentrations.22,23 However, molecular dynamics and Monte Carlo simulations predicted that 2D confinement within intermembrane adhesive zones reduces configurational entropy (by decreasing the availability of rotational, translational, and conformational states) and protein–protein repulsion, compared to cadherin molecules in the solution phase.24,25 This was postulated to increase the thermodynamic driving force for cis-interactions and to facilitate subsequent cadherin clustering at cell–cell adhesions by reducing the entropic penalty of association.24–28 However, experimental verification of the predicted effects of confinement on cadherin interactions has remained elusive. While both cis and trans bonds appear to influence the assembly of cadherin-mediated intercellular adhesions,10,21,24,29 single-molecule Förster Resonance Energy Transfer (FRET) measurements of cadherin extracellular domains bound to supported bilayers failed to detect cis-interactions.30 Similarly, photon counting histogram analysis measurements suggested that cadherins on supported bilayers outside of the cell adhesion zone diffuse as monomers.29 Recent experiments using ensemble-averaged FRET reported the formation of cis-cadherin dimers but not the formation of larger aggregates.31
Here, we used single-molecule tracking to quantitatively characterize the dynamics of wild-type and mutant cadherin bound to a supported lipid bilayer at various cadherin surface coverages. The underlying hypothesis of this approach was that cadherin clusters that formed through cis-interactions would diffuse more slowly than monomeric cadherin. A previously described cadherin mutant that is incapable of cis-interactions was used in negative control experiments.10 Through the analysis of a large number of molecular trajectories, the systematic effect of surface coverage on the dynamics of cadherin bound to the supported lipid bilayer was quantified, providing new insights into cadherin interaction-induced cluster formation when associated with a 2D fluid lipid bilayer.
To determine how increasing surface coverage affected the dynamics of E-cad on supported lipid bilayers, ~7500 trajectories composed of ~200 000 molecular displacements were analyzed at each of the 14 protein concentrations examined. Similar statistics were obtained for control experiments that tracked fluorescently labeled lipids within supported lipid bilayers, both in the presence and absence of bound E-cad. Complementary cumulative squared displacement distributions (CCSDDs) were obtained from these data and fitted to eq S3, and the weighted average short-time diffusion coefficient (DShort) was calculated according to eq S4. The fits and resulting parameters are shown in Figure S2 and Table S2 in the Supporting Information for samples with E-cad, and Figure S3 and Table S3 show the results for control samples with fluorescent lipid tracers. It was expected that a slowing of molecular diffusion would provide a signature of cis-interaction-induced cluster formation because a large cluster of proteins diffuses more slowly than an individual protein due to multiple protein-lipid interactions in the cluster; these interactions are the dominant source of drag at high Ca2+ concentration.32 Thus, we hypothesized that diffusion of wild-type E-cad might slow down more dramatically compared to the mutant with increased protein coverage. As shown in Figure 1A, the resulting general trend supported these expectations as DShort observed for wild-type E-cad at high surface coverage decreased significantly compared to that of the mutant and of fluorescent lipids in a cadherin-free bilayer. At low surface coverage, DShort (see Tables S2 and S3) for both wild-type and mutant E-cad was similar to that of a single fluorescent lipid in a cadherin-free bilayer (i.e., ~0.7 μm2/s), with no apparent systematic trend. Moreover, the diffusion of mutant E-cad did not deviate significantly from that of a lipid in a cadherin-free bilayer across all surface coverages investigated. Because each E-cad domain was conjugated to a single lipid molecule via hexahistidine-NTA binding, the behavior under these conditions was consistent with that expected for E-cad monomers. As expected, we observed that lipid diffusion slowed modestly from 0.666 ± 0.006 to 0.470 ± 0.005 μm2/s at a wild-type E-cad fractional surface coverage of ~0.06 (~6700 molecules/μm2), which represented a smaller decrease than that observed for the wild-type E-cad itself. This decrease in average lipid diffusion was presumably due to the fact that a fraction of lipid molecules was associated or confined within E-cad clusters or was obstructed by clusters.
Figure 1.

(A) Symbols indicate DShort for E-Cad as a function of fractional surface coverage. For comparison, DShort for fluorescently labeled lipids in a cadherin-free bilayer was ~0.67 μm2/s. Vertical error bars correspond to the standard deviation of fitting 100 subsamples using a bootstrap method with replacement. (B) DTA as a function of fractional surface coverage. Vertical error bars represent the parameter standard deviation from fitting. (C) Anomalous diffusion exponent (〈r2(τ)〉 α values) as a function of surface coverage. Vertical error bars represent the parameter standard deviation from fitting. (D) Mean residence time, 〈RT〉 as a function of fractional surface coverage. Vertical error bars correspond to the standard deviation of fitting 100 subsamples using a bootstrap method with replacement. All horizontal error bars represent the standard deviation between movies. The dashed vertical line is a guide to the eye, representing an approximate threshold between noninteracting and interacting fractional surface coverage regimes.
At sufficiently high surface coverage, above a fractional surface coverage threshold of approximately 0.01 (~1100 molecules/μm2), the short-time diffusion of wild-type E-cad decreased systematically while the diffusion of mutant E-cad was not substantially affected. This suggested that the slowing of wild-type E-cad diffusion was specifically due to the formation of clusters mediated by cis-interactions, as opposed to simple crowding. For example, at a fractional surface coverage of 0.0115 ± 0.0008 (1280 ± 90 molecules/μm2), DShort for wild-type E-cad fell to 0.407 ± 0.004 μm2/s, whereas at a similar fractional surface coverage of 0.019 ± 0.006 (2100 ± 700 molecules/μm2), DShort for mutant E-cad remained high at 0.72 ± 0.01 μm2/s. This suggested the onset of significant cis-interactions between wild-type E-cad molecules. With increasing fractional surface coverage, DShort for wild-type E-cad decreased further to 0.235 ± 0.003 μm2/s at a fractional surface coverage of 0.16 ± 0.02 (18 000 ± 2000 molecules/μm2), consistent with the ubiquitous presence of large E-cad clusters. For context, the average fractional surface coverage of E-cad expressed on MDCK cells has been estimated as ~1.5 × 10−4 (~17 molecules/μm2) via quantitative flow cytometry.33 Although this value is much lower than the fractional surface coverage threshold observed for cis-oligomerization of ~0.01 (~1100 molecules/μm2), the local fractional surface coverage by area within cell–cell adhesions is estimated to reach as high as ~0.44 (~49 000 molecules/μm2), indicating the physiological relevance of this observed threshold.15,21 Furthermore, simulations predicted cis dimer and oligomer formation at a fractional surface coverage by area of ~0.16 (~18 000 molecules/μm2).24
The calculation and fitting of the ensemble time-averaged and ensemble-averaged mean-squared displacement ( and 〈r2(τ)〉, as conventionally defined by eqs S6 and S5, respectively) permitted the determination of the time-averaged diffusion coefficient (DTA) and the anomalous diffusion exponent (α) to complement the average short-time diffusion coefficient extracted from fitting of the CCSDDs (see eqs S7 and S8). Interestingly, was consistent with Fickian diffusion for all conditions, as α ≅ 1. This result allowed determination of DTA. Figure S4 and Table S4 show fits to eq S8 and the resulting parameter (DTA) associated with each fit. Values of DTA as a function of fractional surface coverage for both wild-type and mutant E-cad are shown in Figure 1B. The trend exhibited was very similar to that observed for DShort, with absolute values that were systematically larger.
The fact that the values of DTA were greater than DShort is due to the fact that they were determined using independent methods and represented different aspects of the complex diffusive motion. For example, the calculation of gives equal weighting to each trajectory, while CCSDDs put equal emphasis on each diffusive step; therefore, longer trajectories have a greater impact on the distribution. Differences between effective diffusion coefficients determined using independent methods are expected for nonergodic, anomalous diffusion.34–36 Importantly, like DShort, DTA values for wild-type and mutant E-cad were similar at low surface coverage, and DTA was insensitive to surface coverage for mutant E-cad. Again, consistent with DShort, once above a surface coverage threshold, DTA decreased significantly for wild-type E-cad, reaching a minimum of 0.435 ± 0.007 μm2/s at a fractional surface coverage of 0.16 ± 0.02 (18 000 ± 2000 molecules/μm2). This trend, which was identical to that observed for DShort, provided additional support for the presence of cluster formation at high surface coverage.
〈r2(τ)〉 was consistent with subdiffusion at all fractional surface coverages considered for wild-type and mutant E-cad, with α < 1. This deviation from Fickian behavior may be due to membrane heterogeneity, crowding, binding to obstacles, lipid rafts, among other confining environments.37–39 Graphs and resulting parameters from 〈r2(τ)〉 fits to eq S7 are shown in Figure S5 and Table S5. Figure 1C shows the value of the anomalous diffusion exponent, α, as a function of fractional surface coverage for wild-type and mutant E-cad. Although it can be challenging to identify the precise nature of the physical effects that cause subdiffusive behavior, the reduction of the anomalous diffusion exponent α from the value of unity expected for Brownian motion is a quantitative measure of the degree of subdiffusion that can be used to characterize the complexity of the environment. Again, the trend observed in α was similar to that observed for DShort and DTA, where there was no significant difference between wild-type and mutant E-cad until a surface coverage threshold was crossed. In this case, once this threshold was crossed, α for wild-type and mutant E-cad deviated, namely, α decreased significantly for the wild-type and there was no significant change for the mutant. This decrease in α indicated increased subdiffusive behavior for the wild-type E-cad. This behavior was consistent with the formation of protein clusters because a protein within a cluster that diffused slowly on the lipid bilayer was essentially confined within that cluster for a period of time. An increased presence of clusters at high E-cad surface coverage would increase the likelihood that a given molecule would become associated with a cluster during the course of a measured trajectory. Thus, the apparent value of α decreased for wild-type E-cad at high surface coverage, compared to the noninteracting mutant. At low fractional coverage (less than ~6 × 10−3), wild-type E-cad and mutant E-cad at all surface coverages were still modestly subdiffusive. It is hypothesized that this was due to a combination of transient confinement at defect sites and apparent subdiffusion inherent to single-molecule tracking.40 This trend in subdiffusive behavior for wild-type and mutant E-cad with increasing surface coverage was supported by identical systematic trends in the degree of velocity anticorrelation, as indicated by the velocity autocorrelation functions as a function of lag time and velocity-averaging time (Figure S8).41
Compared to the average parameter values described above, step-size distributions, such as van Hove correlation functions, provide more insight into the details of diffusion. Figure 2 shows representative van Hove correlation functions (defined by eq S9) at a time lag of 0.21 s for wild-type and mutant E-cad at representative low, intermediate, and high surface coverages corresponding to fractional surface coverages of ~5 × 10−6 (~0.6 molecules/μm2), ~9 × 10−3 (~1000 molecules/μm2), and ~0.3 (~39 000 molecules/μm2), respectively. For simple Brownian diffusion, the van Hove correlation function would be Gaussian. Here, the measured functions consisted of a central peak with heavy tails that represent large displacements. The central peak was determined to represent displacements less than the localization error of the system. These small displacements correspond to confined or slow diffusing periods because the width of the central peak did not increase significantly with lag time (Figure S6).42 The tails correspond to longer displacements during mobile time intervals. Interestingly, the fraction of displacements within the central peak systematically increased from low to high surface coverage for wild-type E-cad but not for the mutant. Consistent with the discussion above, this systematic change was hypothesized to be due to increased cluster formation, which increased the probability of small or confined displacements and decreased the probability of a large displacement under high surface coverage. The mutant distributions showed no significant difference in central peak or tails with changing surface coverage, and the mutant distributions were indistinguishable from those of the wild-type cadherin at low coverage. The non-Gaussian nature of the step-size distribution provides support for the fact that diffusion within the bilayer, even at low coverage, was complex and anomalous, as previously suggested by the values of the anomalous diffusion exponent for 〈r2(τ)〉.
Figure 2.

Representative van Hove correlation functions for wild-type (WT) and mutant (M) E-cad at low, intermediate, and high fractional surface coverages at a time lag of 0.21 s.
In investigating the relationship between surface coverage and average molecular residence time, distributions composed of >280 000 molecular residence times were analyzed at 7 different surface coverages for mutant and wild-type E-cad. Fits of these distributions to eq S10 and the resulting parameters are shown in Figure S7 and Table S6, respectively. Figure 1D shows the average residence time, 〈RT〉, as a function of surface coverage for wild-type and mutant E-cad. At low surface coverage, the average residence time was ~0.07 s for both the wild-type and mutant E-cad, and there was no significant dependence on surface coverage in the range of 4 × 10−6−1 × 10−2. However, above an apparent threshold coverage of ~0.01, the average wild-type residence time increased significantly for higher values of surface coverage. No significant trend was observed for the mutant E-cad, even as the fractional surface coverage increased to 0.53 ± 0.09. The increase in average residence time for the wild-type E-cad was hypothesized to be due to an increase in lateral intermolecular interactions. In the absence of lateral interactions, the residence time is primarily dictated by the binding strength of the His6–NTA coupling. However, a given E-cad ectodomain within a cluster that is stabilized by lateral cis-interactions will appear to be bound more strongly and will remain bound for a longer time on average. Thus, as the fractional surface coverage crossed the threshold where lateral interactions became significant, the wild-type E-cad molecules were more likely to encounter clusters, resulting in an increased average residence time. This did not occur for mutant E-cad, which does not form cis bonds. The trend seen for 〈RT〉 is consistent with that seen for DShort, DTA and α.
Inspection of the trajectory friction factor distributions allows for direct observation of protein cluster formation and the estimation of apparent cluster sizes. Figure 3 shows representative friction factor (f) distributions for wild-type and mutant E-cad at low, intermediate, and high fractional surface coverages. The friction factor is inversely proportional to the effective diffusion coefficient (eq S13) and is related to the amount of drag experienced by a moving object. Under the conditions of the experiments, differences in drag were primarily expected to be due to differences in the magnitude of the interactions with the lipid bilayer, which was related to the size of a cluster. As expected, these distributions decayed rapidly for wild-type E-cad at low fractional surface coverage and for mutant E-cad at all fractional surface coverages, indicating a low probability of observing a trajectory with a large friction factor. This was consistent with the expectation that the vast majority of objects were E-cad monomers in these experiments. Specifically, in previous work, we found that at the Ca2+ concentrations employed here, the majority of E-cad monomers diffused rapidly, consistent with a “standing-up” configuration where the E-cad domain interacted with only a single lipid. Only a small fraction of trajectory segments exhibited slow diffusion, consistent with a lying-down configuration.32 However, as the fractional surface coverage for wild-type E-cad increased, the tail of the probability distribution systematically broadened, indicating an increased probability of observing objects that experienced higher friction factors. For the mutant E-cad, the tails of the probability distributions did not change significantly with surface coverage. This observation was consistent with the presence of protein clusters for wild-type E-cad at medium and high surface coverage and provides indirect information about the size distribution of these clusters. For example, as the monomeric species in a “standing-up” configuration was represented by f = 0.5 s/μm2, at high surface coverage, a significant number of trajectories were observed with friction factors at least 150× as large. These data strongly suggest that the protein clusters formed consisted not only of dimers but also of an increasing number of much higher order oligomers. This direct observation of large cluster formation provides the framework to support the hypotheses discussed above, regarding the relationship between protein dynamics and surface coverage.
Figure 3.

Representative probability distribution functions of trajectory friction factors for wild-type and mutant E-cad at low, intermediate, and high fractional surface coverages.
The results shown here demonstrate that at high fractional surface coverage wild-type E-cad extracellular domains bound to lipids in supported lipid bilayers exhibited behavior consistent with cis-interaction-induced cluster formation. There appeared to be a fractional surface coverage threshold separating noninteracting and interacting regions. Once this threshold was crossed, wild-type E-cad behavior deviated from that of the cis-binding mutant, in terms of residence time, diffusion, and degree of subdiffusion. Upon the basis of these observed differences, we hypothesize that above the threshold intermolecular cis-interactions facilitated protein cluster formation that in turn caused this deviation in behavior. We note that this deviation of wild-type E-cad behavior from that of the cis-mutant, while mainly due to cis-interactions, may be slightly enhanced by stronger nonspecific interactions between wild-type E-cad proteins relative to the cis-mutant.
By exploring a wide range of surface coverages using an approach that is extremely sensitive to the presence of oligomers, these studies successfully made connections with computational simulations that were not possible in previous measurements. In particular, cis-interactions have not previously been observed experimentally in solution-based experiments or on supported planar membranes at low surface coverage; the current results therefore provide important experimental support for the theoretical prediction that 2D confinement promotes the assembly of cis-oligomers.22,23,25,43 A recent report of cis-interactions on cell membranes at lower coverages reported dimer formation, but findings presented here indicate the formation of higher-order oligomers at higher E-cad surface coverages.31 Previous photon counting histogram analysis of E-cad on supported lipid bilayers also failed to detect cis-interactions.29 However, the fractional surface coverage used in those experiments was in the range of ~9 × 10−4−6 × 10−3 (~100–700 molecules/μm2). The latter is in the low to intermediate range of surface coverages investigated here, where both the mutant and wild-type diffused similarly as monomers.29 E-cad on free cell membranes dimerizes at concentrations below the threshold for clustering identified in this study. However, high cadherin concentrations observed at cell–cell adhesions exceeds the threshold,15,21 such that cis-interactions between cadherins under two-dimensional confinement are expected to facilitate the assembly of intercellular adhesions. This work sets the stage for the quantitative investigations of E-cad cis-interactions on a lipid bilayer via single-molecule FRET that may provide details about the nature and mechanism of these lateral interactions between extracellular domains.
EXPERIMENTAL METHODS
Total internal reflection fluorescence (TIRF) microscopy was used to observe the density-dependent translational dynamics of wild-type and mutant E-cad extracellular domains on 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)-supported lipid bilayers containing 5% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS-NTA(Ni)). A mixture of Alexa 555-labeled E-cad and either wild-type or mutant unlabeled E-cad at varying concentration was incubated with the bilayer to allow binding of the hexahistidine-tagged E-cad to the DGS-NTA lipids. Individual E-cad molecules were detected, localized, and tracked using methods described previously.44 Further sample preparation details are described in the Supporting Information.
The effective short-time average diffusion coefficient (DShort) was determined using the parameters acquired by fitting the complementary cumulative squared displacement distribution for each experimental condition to a four-mode Gaussian mixture model (eq S3). Using the best parameters of the Gaussian mixture model fit, the effective short-time average diffusion coefficient was calculated as the weighted average of the four diffusive modes (eq S4). The time-averaged diffusion coefficient (DTA) and the anomalous diffusion exponent (α) were determined using the ensemble time-averaged and ensemble-averaged mean-squared displacements, respectively. DTA was calculated by fitting the ensemble time-averaged mean-squared displacement to the Brownian diffusion model (eq S8). As the ensemble-averaged mean-squared displacement was subdiffusive, fitting was performed using an anomalous diffusion model (eq S7) to extract α. Distributions of molecular displacements, decomposed into their one-dimensional components, were determined using the self-part of the van Hove correlation function (eq S9).45 The influence of cis-interactions was further characterized using the average molecular residence time (〈RT〉), which was determined for all conditions by fitting complementary cumulative residence time distributions to a two-mode exponential mixture model (eq S10). Lastly, an effective diffusion coefficient for each trajectory (DT) was calculated using eq S12, and probability distribution functions of trajectory friction factors (f) were constructed, where the friction factor was related to the diffusion coefficient by the Einstein relation (eq S13).46 The friction factor for a given cadherin cluster was related to the drag that it experienced and therefore provided insight into the size of the cluster. Additional equations, definitions, and other details of trajectory analysis are provided in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 1R01GM117104.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.9b01500.
Detailed sample preparation, imaging techniques, data analysis, trajectory analysis, and additional tables and figures showing bulk protein concentrations, representative molecular trajectories, complementary cumulative squared displacement distributions and their Gaussian mixture model fits and parameters, time-averaged mean-square displacement curves, time-averaged diffusion coefficient values, ensemble-averaged mean-squared displacement curves and power law fitting parameters, Van Hove correlation functions, complementary cumulative residence time distributions and their mixture model fits and parameters, and velocity autocorrelation functions for lag times and velocity averaging times (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Gumbiner BM Cell Adhesion: The Molecular Basis of Tissue Architecture and Morphogenesis. Cell 1996, 84, 345–357. [DOI] [PubMed] [Google Scholar]
- (2).Gumbiner BM Regulation of Cadherin-Mediated Adhesion in Morphogenesis. Nat. Rev. Mol. Cell Biol 2005, 6, 622–634. [DOI] [PubMed] [Google Scholar]
- (3).Niessen CM; Leckband D; Yap AS Tissue Organization by Cadherin Adhesion Molecules: Dynamic Molecular and Cellular Mechanisms of Morphogenetic Regulation. Physiol. Rev 2011, 91, 691–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Takeichi M Cadherin Cell Adhesion Receptors as a Morphogenetic Regulator. Science 1991, 251, 1451–1455. [DOI] [PubMed] [Google Scholar]
- (5).Yap AS; Brieher WM; Gumbiner BM Molecular and Functional Analysis of Cadherin-Based Adherens Junctions. Annu. Rev. Cell Dev. Biol 1997, 13, 119–146. [DOI] [PubMed] [Google Scholar]
- (6).Shapiro L; Fannon AM; Kwong PD; Thompson A; Lehmann MS; Grubel G; Legrand JF; Als-Nielsen J; Colman DR; Hendrickson WA Structural Basis of Cell-Cell Adhesion by Cadherins. Nature 1995, 374, 327–337. [DOI] [PubMed] [Google Scholar]
- (7).Leckband D; Sivasankar S Cadherin Recognition and Adhesion. Curr. Opin. Cell Biol 2012, 24, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Klingelhofer J; Laur OY; Troyanovsky RB; Troyanovsky SM Dynamic Interplay between Adhesive and Lateral E-Cadherin Dimers. Mol. Cell. Biol 2002, 22, 7449–7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Troyanovsky RB; Sokolov E; Troyanovsky SM Adhesive and Lateral E-Cadherin Dimers Are Mediated by the Same Interface. Mol. Cell. Biol 2003, 23, 7965–7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Harrison OJ; Jin X; Hong S; Bahna F; Ahlsen G; Brasch J; Wu Y; Vendome J; Felsovalyi K; Hampton CM; Troyanovsky RB; Ben-Shaul A; Frank J; Troyanovsky SM; Shapiro L; Honig B The Extracellular Architecture of Adherens Junctions Revealed by Crystal Structures of Type I Cadherins. Structure 2011, 19, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Brieher WM; Yap AS; Gumbiner BM Lateral Dimerization Is Required for the Homophilic Binding Activity of C-Cadherin. J. Cell Biol 1996, 135, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yap AS; Brieher WM; Pruschy M; Gumbiner BM Lateral Clustering of the Adhesive Ectodomain: A Fundamental Determinant of Cadherin Function. Curr. Biol 1997, 7, 308–315. [DOI] [PubMed] [Google Scholar]
- (13).Yap AS; Niessen CM; Gumbiner BM The Juxtamembrane Region of the Cadherin Cytoplasmic Tail Supports Lateral Clustering, Adhesive Strengthening, and Interaction with P120ctn. J. Cell Biol 1998, 141, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Leckband DE; de Rooij J Cadherin Adhesion and Mechanotransduction. Annu. Rev. Cell Dev. Biol 2014, 30, 291–315. [DOI] [PubMed] [Google Scholar]
- (15).Indra I; Choi J; Chen CS; Troyanovsky RB.; Shapiro L.; Honig B.; Troyanovsky SM. Spatial and Temporal Organization of Cadherin in Punctate Adherens Junctions. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E4406–E4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhu B; Chappuis-Flament S; Wong E; Jensen IE; Gumbiner BM; Leckband D Functional Analysis of the Structural Basis of Homophilic Cadherin Adhesion. Biophys. J 2003, 84, 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).du Roure O; Buguin A; Feracci H; Silberzan P Homophilic Interactions between Cadherin Fragments at the Single Molecule Level: An AFM Study. Langmuir 2006, 22, 4680–4684. [DOI] [PubMed] [Google Scholar]
- (18).Hong S; Troyanovsky RB; Troyanovsky SM Binding to F-Actin Guides Cadherin Cluster Assembly, Stability, and Movement. J. Cell Biol 2013, 201, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Troyanovsky RB; Indra I; Chen CS; Hong S; Troyanovsky SM Cadherin Controls Nectin Recruitment into Adherens Junctions by Remodeling the Actin Cytoskeleton. J. Cell Sci 2015, 128, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Troyanovsky RB; Laur O; Troyanovsky SM Stable and Unstable Cadherin Dimers: Mechanisms of Formation and Roles in Cell Adhesion. Mol. Biol. Cell 2007, 18, 4343–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wu Y; Kanchanawong P; Zaidel-Bar R Actin-Delimited Adhesion-Independent Clustering of E-Cadherin Forms the Nano scale Building Blocks of Adherens Junctions. Dev. Cell 2015, 32, 139–154. [DOI] [PubMed] [Google Scholar]
- (22).Haussinger D; Ahrens T; Aberle T; Engel J; Stetefeld J; Grzesiek S Proteolytic E-Cadherin Activation Followed by Solution Nmr and X-Ray Crystallography. EMBO J. 2004, 23, 1699–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Koch AW; Bozic D; Pertz O; Engel J Homophilic Adhesion by Cadherins. Curr. Opin. Struct. Biol 1999, 9, 275–281. [DOI] [PubMed] [Google Scholar]
- (24).Wu Y; Jin X; Harrison O; Shapiro L; Honig BH; Ben-Shaul A Cooperativity between Trans and Cis Interactions in Cadherin-Mediated Junction Formation. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 17592–17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wu Y; Vendome J; Shapiro L; Ben-Shaul A; Honig B Transforming Binding Affinities from Three Dimensions to Two with Application to Cadherin Clustering. Nature 2011, 475, 510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang F; Marcus WD; Goyal NH; Selvaraj P; Springer TA; Zhu C Two-Dimensional Kinetics Regulation of Alphalbeta2-Icam-1 Interaction by Conformational Changes of the Alphal-Inserted Domain. J. Biol. Chem 2005, 280, 42207–42218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wu J; Fang Y; Zarnitsyna VI; Tolentino TP; Dustin ML; Zhu C A Coupled Diffusion-Kinetics Model for Analysis of Contact-Area Frap Experiment. Biophys. J 2008, 95, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Xie ZR; Chen J; Wu Y Linking 3d and 2d Binding Kinetics of Membrane Proteins by Multiscale Simulations. Protein Sci. 2014, 23, 1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Biswas KH; Hartman KL; Yu CH; Harrison OJ; Song H; Smith AW; Huang WY; Lin WC; Guo Z; Padmanabhan A; Troyanovsky SM; Dustin ML; Shapiro L; Honig B; Zaidel-Bar R; Groves JT E-Cadherin Junction Formation Involves an Active Kinetic Nucleation Process. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 10932–10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhang Y; Sivasankar S; Nelson WJ; Chu S Resolving Cadherin Interactions and Binding Cooperativity at the Single Molecule Level. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Singh DR; Ahmed F; Sarabipour S; Hristova K Intracellular Domain Contacts Contribute to Ecadherin Constitutive Dimerization in the Plasma Membrane. J. Mol. Biol 2017, 429, 2231–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cai Y; Shashikanth N; Leckband DE; Schwartz DK Cadherin Diffusion in Supported Lipid Bilayers Exhibits Calcium-Dependent Dynamic Heterogeneity. Biophys. J 2016, 111, 2658–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tabdili H; Langer M; Shi Q; Poh YC; Wang N; Leckband D Cadherin-Dependent Mechanotransduction Depends on Ligand Identity but Not Affinity. J. Cell Sci 2012, 125, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schütz GJ; Schindler H; Schmidt T Single-Molecule Microscopy on Model Membranes Reveals Anomalous Diffusion. Biophys. J 1997, 73, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Metzler R; Jeon JH; Cherstvy AG; Barkai E Anomalous Diffusion Models and Their Properties: Non-Stationarity, Non-Ergodicity, and Ageing at the Centenary of Single Particle Tracking. Phys. Chem. Chem. Phys 2014, 16, 24128–24164. [DOI] [PubMed] [Google Scholar]
- (36).Ritchie K; Shan XY; Kondo J; Iwasawa K; Fujiwara T; Kusumi A Detection of Non-Brownian Diffusion in the Cell Membrane in Single Molecule Tracking. Biophys. J 2005, 88, 2266–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Saxton MJ Anomalous Diffusion Due to Obstacles: A Monte Carlo Study. Biophys. J 1994, 66, 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Saxton MJ Anomalous Diffusion Due to Binding: A Monte Carlo Study. Biophys. J 1996, 70, 1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Schutz GJ; Kada G; Pastushenko VP; Schindler H Properties of Lipid Microdomains in a Muscle Cell Membrane Visualized by Single Molecule Microscopy. EMBO J 2000, 19, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Martin DS; Forstner MB; Käs JA Apparent Subdiffusion Inherent to Single Particle Tracking. Biophys. J 2002, 83, 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Weber SC; Thompson MA; Moerner WE; Spakowitz AJ; Theriot JA Analytical Tools to Distinguish the Effects of Localization Error, Confinement, and Medium Elasticity on the Velocity Autocorrelation Function. Biophys. J 2012, 102, 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hofling F; Franosch T Anomalous Transport in the Crowded World of Biological Cells. Rep. Prog. Phys 2013, 76, No. 046602. [DOI] [PubMed] [Google Scholar]
- (43).Katsamba P; Carroll K; Ahlsen G; Bahna F; Vendome J; Posy S; Rajebhosale M; Price S; Jessell TM; Ben-Shaul A; Shapiro L; Honig BH Linking Molecular Affinity and Cellular Specificity in Cadherin-Mediated Adhesion. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 11594–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Faulón Marruecos D; Kienle DF; Kaar JL; Schwartz DK Grafting Density Impacts Local Nanoscale Hydrophobicity in Poly(Ethylene Glycol) Brushes. ACS Macro Lett. 2018, 7, 498–503. [DOI] [PubMed] [Google Scholar]
- (45).Hansen J-P Theory of Simple Liquids: With Applications to Soft Matter; 2006. [Google Scholar]
- (46).Edward JT Molecular Volumes and the Stokes-Einstein Equation. J. Chem. Educ 1970, 47, 261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


