Abstract
Melanoma is the most aggressive type of skin cancer, which readily metastasizes through lymph nodes to the lungs, liver, and brain. Since the repeated administration of most chemotherapeutic drugs develops chemoresistance and severe systemic toxicities, herein we synthesized 2-(4-hydroxy-1H-indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) methanone (abbreviated as QW-296), a novel tubulin destabilizing agent with little susceptible to transporter-mediated drug resistance. QW-296 disturbed the microtubule dynamics at the nanomolar concentration in A375 and B16F10 melanoma cells. QW-296 binding to colchicine-binding site on tubulin protein was confirmed by molecular modeling and tubulin polymerization assay. QW-296 significantly inhibited A375 and B16F10 cell proliferation, induced G2/M cell cycle arrest and led to apoptosis and cell death. To improve its aqueous solubility, QW-296 was encapsulated into methoxy poly(ethyleneglycol)-b-poly(carbonate-co-lactide) [mPEG-b-P(CB-co-LA)] polymeric nanoparticles by solvent evaporation, with the mean particle size of 122.0±2.28 nm and drug loading of 3.70% (w/w). Systemic administration of QW-296 loaded nanoparticles into C57/BL6 albino mice bearing lung metastatic melanoma at the dose of 20 mg/kg 4 times a week for 1.5 weeks resulted in significant tumor regression and prolonged mouse median survival without significant change in mouse body weight. In conclusion, QW-296 loaded nanoparticles have the potential to treat metastatic melanoma.
Keywords: Microtubule/tubulin inhibitor, melanoma, nanoparticles, tubulin polymerization
INTRODUCTION
Microtubules are an attractive target for chemotherapy due to their critical role in mitosis, cellular shape maintenance, and intracellular transportation.1,2 Microtubule has the following three distinct binding domains: vinca binding domain, taxane binding domain, and colchicine binding domain.2 Due to the dynamic nature of tubulin, chemotherapeutic agents that promote microtubule polymerization are classified as microtubule stabilizers, whereas agents that inhibit microtubule polymerization are designated as tubulin destabilizing agents. Chemotherapeutic agents that bind to taxane binding site are microtubule stabilizers, whereas agents that bind to vinca or colchicine binding site are tubulin destabilizers.3 Clinical potential of tubulin binding agents has been proven by many Food and Drug Administration (FDA) approved anticancer drugs including paclitaxel (PTX), docetaxel (DTX), and epothilones that bind to taxane binding domain, whereas vinblastine, vincristine, and vinorelbine bind to vinca binding domain. Combretastatin (CA) and colchicine bind to the colchicine binding domain.4–7 Clinical efficiency of drugs that bind to vinca or taxane binding domain is limited due to the development of chemoresistance by overexpressing transmembrane efflux pumps {P-glycoprotein (P-gp), multidrug resistance-associated protein (MRP) and/or breast cancer resistance protein (BCRP)} in cancer cells.8–10 Interestingly, the compounds interacting with the colchicine binding site has been reported to be less sensitive to clinically observed mechanisms of resistance.11 Despite the clinical use of colchicine for the treatment of gout, its use as an anticancer drug is limited due to toxic side effects.12
Among various malignant cancers, melanoma has a very high metastatic tendency in the late stages where cancer cells metastasize to the lymph nodes, lungs, liver, brain, and heart. This metastatic property of melanoma leads to the low survival time of patients typically less than a year.13 Dacarbazine (DTIC) is a first-line FDA-approved DNA alkylating agent used for treating advanced metastatic melanoma. However, it has not significantly improved the patient overall survival time14, and only 1–2% of patients respond well.15 The effectiveness of PTX and carboplatin combination, a second-line FDA-approved therapy, has only less than 40% of all the patients responded well while receiving this combination.16,17 Treatment of metastatic melanoma with the FDA approved BRAF inhibitors such as vemurafenib or dabrafenib with advanced BRAFV600 mutated melanoma has shown high response rate initially, however the effect last for a limited period. Similar results were observed when treated with MEK inhibitors such as trametinib.18 Further, the treatment of metastatic melanoma with immunomodulating drugs such as anti-PD-1 drugs (nivolumab, pembrolizumab) or anti-CTLA-4 antibody ipilimumab resulted in improved progression-free survival. However, treatment with these drugs results in the adverse effect of grade 3–4, including severe elevation in liver injury markers, life-threatening skin rashes, nephritis, and pneumonitis.19
Clinical efficiency of chemotherapeutic drugs is limited by off-target toxicity and development of chemoresistance by multiple mechanisms.20,21 Aggressively metastasizing cancers like melanoma may be dealt by addressing the following issues: (1) discovery of potent molecules acting on vital cell proliferation mechanism and block the cell migration/invasion (2) overcome the cellular resistance mechanism and (3) reduced off-target toxicity at the therapeutic dose. In this context, limited available therapeutic options for the treatment of metastatic melanoma, the high therapeutic efficiency of tubulin inhibitors, and relatively low sensitivity to clinical resistance, colchicine binding site offers a potential target to inhibit the tubulin protein in rapidly dividing and metastasizing cells.
Our group is continuously involved in the discovery and development of new small molecules to target the colchicine binding site of tubulin. Previously, we reported the discovery of diaryl-ketone chemotypes, with various linkers such as a phenyl ring,22 4-substituted methoxybenozyl arylthiazoles,23 phenylaminothiazoles,2 arylbenzoylimidazoles,24 reverse arylbenzoylimidazoles,25 and 1H-imidazo[4,5-c]pyridine,3 that selectively binds to the tubulin at colchicine binding site and stabilizes the tubulin at nanomolar concentration.
Nanoparticulate delivery systems have the potential to deliver drugs to the target site and prevent off-target toxicity26 by a passive delivery mechanism utilizing the Enhanced Permeability and Retention (EPR) effect.27 Beside this, nanoparticles also improve pharmacokinetic profiles by improving the aqueous solubility of the hydrophobic drug, extending circulation time and improving bioavailability. We successfully applied micellar delivery for 4-substituted methoxybenzoyl-aryl-thiazole-100 (SMART-100)28 and bicalutamide29 to improve their aqueous solubility using methoxy poly(ethylene glycol)-b-poly(D, L-lactide) (mPEG-PLA) polymer. Despite improved solubility, low drug loading in micelles prepared from mPEG-PLA based polymer shifted our interest to develop new, safe and biocompatible polymers. We utilized methoxy poly(ethyleneglycol)-bpoly(carbonate-co-lactide) [mPEG-b-P(CB-co-LA)] based nanoparticles to deliver (2-(1 H-indol-5-yl) thiazol-4-yl) 3, 4, 5-trimethoxyphenyl methanone (LY293) to treat melanoma. LY293-loaded nanoparticles significantly inhibited the proliferation of metastasized melanoma in a lung metastatic melanoma mouse model without toxicity to vital organs.30,31
In our recent studies, we synthesized a new SMART analog, SMART-OH (Figure 1A) and conjugated with methoxy poly(ethylene glycol)-b-poly(2-methyl-2-carboxyl-propylene carbonate-g-dodecanol) (mPEG-b-PCC-g-DC) to improve the drug payload and prevent premature drug release. The polymeric micelles with 14.3% drug payload, showed significant inhibition of tumor growth in the lung metastatic melanoma mouse model.32 In another study, we conjugated amino derivative of indocyanine green (ICG-NH2) with mPEG-b-PCC-g-DC for photodynamic and photothermal effects on melanoma cells. ICG conjugated micelles exhibited excellent in vivo stability and significant improvement in the therapeutic efficacy of ICG mediated phototherapy.33
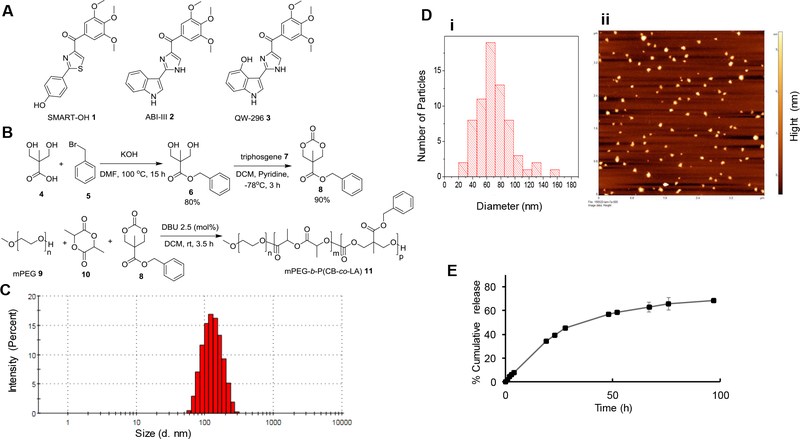
Figure 1.
Structure of colchicine inhibitors, mPEG-based polymer synthesis, and nanoparticle characterization. A) Chemical structure of SMART-OH, ABI-III and QW-296, B) Synthesis of mPEG-poly(carbonate-co-lactide). C) The particle size distribution of polymeric nanoparticles by w/o emulsification as determined by dynamic light scattering (DLS). D) Surface morphology and particle size distribution of nanoparticles by AFM. E) In vitro release of QW-296 from nanoparticles at pH 7.4. Drug concentrations were measured by HPLC. Results are expressed as the mean ± SD (n = 3).
Herein, we synthesized a new potent tubulin inhibitor 2-(4-hydroxy-1H-indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl)methanone (abbreviated QW-296 3) bearing a 4-hydroxyl group on the indole ring of ABI-III 2 (Figure 1A). Because of the low resistance index of QW-296 in taxane-resistant prostate cancer cells, compared to ABIIII, we pursued QW-296 for further in vivo efficacy. QW-296 loaded nanoparticles were prepared using our previously optimized {mPEG-b-P(CB-co-LA)} polymer and demonstrated its promising therapeutic efficiency in lung metastatic melanoma bearing C57/BL6 albino mice.
Materials and Methods
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM) was purchased from Invitrogen (Carlsbad, CA). All primary antibodies were purchased from Cell Signaling (Beverly, MA) and Santa Cruz Biotechnology (Dallas, TX). B16F10 mouse melanoma cell line and A375 human melanoma cell line were acquired from the American Type Culture Collection (ATCC). Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), and 1.0% antibiotic/antimycotic mixture (Invitrogen) PC3 and PC3-TXR cells were kindly provided by Dr. Evan T. Keller from the University of Michigan. These cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator containing 5% CO2 at 37 °C and their resistance to taxane was maintained by adding 200 nM paclitaxel to growth media biweekly. 2,2-Bis(hydroxymethyl)propionic acid, methoxy poly(ethylene glycol) (mPEG, 5,000 Da), 8-diazabicycloundec-7-ene (DBU), benzyl bromide, L-lactide, and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO) and used as received.
Synthesis of QW-296
Synthesis of QW-296 (Figure 1A) was achieved as described previously.34 Briefly, QW-296 was synthesized from compound 1 that reacts with POCl3 and DMF to give compound 2 followed by reaction with benzene sulfonyl chloride to afford compound 3 as a major product. Compound 3 will react with ammonium hydroxide and glyoxal to form compound 4. Subsequent compound 4 reacts with NBS in THF provided the dibromo imidazole derivative 5. Protection of the imidazole in the presence of SEMCl and NaH afforded intermediate 6 which could be smoothly reacted with 3,4,5-Trimethoxybenzoyl chloride and isopropylmagnesium chloride-lithium chloride complex to give 7, and then removal of benzenesulfonyl and bromo group provided the intermediate 8. Subsequent deprotection of the SEM group with 1 M HCl provided the compound 9, which after removal of benzyl group provided the target compound QW-296 as a yellow solid. The chemical structures of the synthesized compounds were characterized and confirmed by NMR and high-resolution mass spectrometry. The characterization data of QW-296 by 1H NMR, 13CNMR, HRMS, and HPLC are shown in supplementary Figures 3S and 4S.
Docking studies
Molecular Modeling.
The molecular modeling studies were performed with the published crystal structure of DJ101 in complex with tubulin at a resolution of 2.8Å (PDB code: 5H7O). Schrödinger Molecular Modeling Suite 2018 (Schrödinger LLC, New York, NY) was used for the modeling studies with similar procedures described before.3,35 Briefly, the structures of the protein-ligand complexes were prepared using the Protein Preparation Wizard workflow. The tubulin receptor grid was generated based on the native ligand using the Receptor Grid Generation. Both ABI-III and QW-296 tubulin inhibitors were built and prepared for docking using the Ligprep module before they were docked into 5H7O. The Glide docking score was used to show the estimated free energy of binding (kcal/mol); a lower (more negative) number indicates more favorable interaction. Hydrogen bonds and data analysis were carried out using the Maestro interface of Schrödinger software.
Tubulin Polymerization Assay
In vitro tubulin polymerization assay was performed using the fluorescence-based tubulin polymerization kit purchased from Cytoskeleton, Inc. (Denver, CO). The polymerization assay is a one-step assay, which uses a half area 96-well plate (Corning Costar, Corning, NY) for measuring tubulin polymerization. A 5 μL aliquot of a 10× concentration of the test compound in DI was pipetted into each well of the pre-warmed 96-well plates at 37 °C. Following the addition of compound, 50 μL of tubulin MIX was added (as per manufacturer’s protocol). Then, the plate was immediately subjected to kinetic measurements on a SpectraMAx M5e fluorescence microplate reader (Molecular Device, San Jose, CA). The kinetics of tubulin polymerization reaction was monitored for every 30 s over a period of 90 min at 37 °C with the medium shaking of the plate for 5 s before taking the first reading. The change in the intensity of fluorescence due to polymerization or depolymerization of tubulin was determined by setting the excitation wavelength at 360 nm and emission wavelength at 450 nm using medium gain. Reaction without any compound (DMSO, 2%) and tubulin protein served as negative controls. All experiments were performed in triplicates.
P-gp ATPase Activity Assay
Pgp-Glo assay system (Promega, Madison, WI) was used to evaluate the effect of QW-296 on Pgp ATPase activity according to the manufacturer’s protocol. In a 96-well untreated white plate, QW-296 was incubated at the dose of 10, 100 and 1000 nM with 25 μg of recombinant human Pgp membrane. Pgp-GIo assay buffer was used as the untreated control, 200 μM verapamil was used as the positive control of drug-induced Pgp ATPase activity, and 100 μM sodium orthovanadate was used as the selective inhibitor of Pgp ATPase activity. ATPase activity was initiated by the addition of 5 mM Mg ATP and incubated at 37 °C for 40 min. Further, luminescence was initiated by adding 50 μL of ATP detection reagent; after 20 min of incubation at room temperature, the plate was read on SpectraMAx M5e microplate reader (Molecular Device, San Jose, CA).
Synthesis of mPEG-b-P(CB-co-LA)
Benzyl-2,2-bis(methylol)propionate was synthesized as reported previously28 by reacting 2,2-bis(hydroxymethyl)propionic acid 4 with benzyl bromide 5 at 100 °C for 15 h. The monomer 5-methyl-5-benzyloxycarbonyl-1,3-dioxane-2-one (MBC) 8 was synthesized by reacting benzyl-2,2-bis(methylol)propionate 6 with triphosgene 7 at −70 °C in the presence of pyridine in dry dichloromethane. Random MBC and lactide block copolymer mPEG-b-P(CB-co-LA) 11 with target average molecular weight of 15,000 Da was synthesized by the ring-opening polymerization of L-lactide 10 and MBC 8 using mPEG 9 as a macroinitiator and DBU (2.5 mol% relative to mPEG) as a catalyst in dry DCM at room temperature for 3.5 h. Following synthesis, the copolymer was characterized by 1H NMR and gel permeation chromatography (GPC). The molecular weight and polydispersity index (PDI) of mPEG-b-P(CB-co-LA) polymers were determined by GPC using a Shimadzu gel permeation chromatography (GPC) system equipped with a Styragel HR 4E GPC column and a differential refractive index detector. Dimethylformamide (DMF) with 0.1% LiBr was used as eluent at a flow rate of 0.6 mL/min at 40 °C. Narrow PEG standards (3,770–71,000 g/mol) from American Polymer Standards Corp. were used for generating a standard curve, and the data was processed using Lab Solutions software ver. 5.84.
Preparation and characterization of nanoparticles
Blank and QW-296 loaded nanoparticles were prepared by oil/water (o/w) emulsification using 1.0% polyvinyl alcohol as an emulsifier. mPEG-b-P(CB-co-LA) and QW-296 (an amount equivalent to 10% theoretical drug loading) were dissolved in a solvent mixture of dichloromethane and acetone (1:1) and were emulsified using a sonica probe sonicator (Newtown, CT) at an amplitude of 20 W for 10 min. The organic phase was removed using a rotary evaporator, followed by hydration and centrifugation at 5000 rpm for 5.0 min. The supernatant was concentrated by ultracentrifugation at 20,000 rpm for 30 min followed by washing with PBS. Then, the sediment was suspended in the desired volume of PBS. The particle size distribution and ζ potential of drug-loaded nanoparticles were determined by dynamic light scattering (DLS) using a Malvern Zetasizer (Worcestershire, UK). Surface morphology and particle size of nanoparticles were determined by Atomic Force Microscopy (AFM). Briefly, nanoparticle samples were diluted in PBS, deposited on freshly cleaved mica modified with 1-(3-aminopropyl)-silatrane (APS), rinsed with de-ionized (DI) water, and dried with a gentle flow of argon. Images were collected with MultiMode Nanoscope IV system (Bruker Instruments, Santa Barbara, CA) in Tapping Mode at ambient conditions. Silicon probes RTESPA-300 (Bruker Nano Inc., CA) with a resonance frequency of ~300 kHz and a spring constant of ~40 N/m were used for imaging at scanning rate for about 2.0 Hz. Images were processed using the FemtoScan software package (Advanced Technologies Center, Moscow, Russia).
Drug Loading and Encapsulation
The encapsulation efficiency was estimated by reverse-phase high-performance liquid chromatography (RP-HPLC) at λmax of 222 nm using a C18 column (250 mm × 4.6 mm, 5μ, Alltech, Deerfield, IL), a solvent mixture of acetonitrile and water (55:45 v/v) was used as a mobile phase. QW-296 concentration was calculated using the peak area from a standard curve with R2 = 0.9992 and a quantification limit of 1.95 μg/mL. Drug content was determined by taking 500 μl of nanoparticles in 1 ml of dichloromethane (DCM) and sonicated in a bath sonicator for 15 min at room temperature to extract the drug content in DCM. Then, 500 μl DCM was taken in a glass vial, and DCM was removed under reduced pressure on a rotary evaporator and reconstituted in 1.0 ml of acetonitrile. The acetonitrile solution of QW-296 was filtered through a 0.2 μm syringe filter (Acrodisc, Waters, USA) and 20 μl of the filtered solution was injected into the HPLC system.
In vitro release of QW-296 from nanoparticles
QW-296 release from mPEG-b-P(CB-co-LA) nanoparticles was determined by placing 3.0 ml of formulated nanoparticles in a dialysis bag (1,000 Da cutoff) and dialyzed against 50 mL PBS (pH 7.4) containing 1.0% Tween 80 as a nonionic surfactant and emulsifier at 37°C in a temperature controlled shaker at the speed of 100 rpm. Sample (1.0 mL) was taken between 0 – 97 h and replaced with 1.0 mL PBS containing 1.0% Tween 80. The sample was dissolved in the mobile phase after removing the solvent using an evaporator, followed by determination of drug concentration using HPLC (mobile phase: Acetonitrile:water (55:45), C18 column (250 × 4.6 mm, 5μ, Alltech, Deerfield, IL), flow rate 1 mL/min). All experiments were performed in triplicate at different days, and the data reported as the mean ± SEM of three individual experiments.
Cell viability assay
A375 and B16F10 cells were seeded overnight in 96-well plates at a density of 2,500 cells per well and thereafter treated with DMSO, QW-296 dissolved in DMSO, and SMART-OH dissolved in DMSO for 48 h. At the end of 48 h, the cell culture medium was replaced with 100 μL of 0.5 mg/mL 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution in PBS and incubated for 4 h at 37 °C. Cell culture media was removed, and formazan crystals were dissolved in 200 μL of DMSO. The absorbance was measured at 560 nm. Similarly, PC3 and PC3-TXR cells were treated with PTX, QW-296, and ABI-III to calculate the IC50 values.
Cell cycle analysis
A375 and B16F10 cells were cultured in a six-well plate at a density of 0.2×106 cells per well. After reaching the confluency to 80%, the cells were treated with DMSO, QW-296 cosolvent and QW-296 loaded nanoparticles for 48 h. Cells were harvested, fixed in 70% ice-cold ethanol for 1 h and washed with PBS. A cell pellet containing 1×106 cells was suspended in 0.5 ml of FxCycle™ PI/RNase staining solution for 15 min at room temperature. Cell cycle was measured by a flow cytometer (BD FACS Calibur, NJ). Each experiment was carried in triplicate at different days and the data reported as the mean ± SEM.
Apoptosis Signaling by Annexin-V
A375 and B16F10 cells were seeded in 6 well plates overnight at the density of 0.2×106 cells per well and treated with DMSO, QW-296 cosolvent and QW-296 loaded nanoparticle for 48 h. Then, cells were washed twice with cold PBS, and the pellets were suspended in 500 μL binding buffer containing 5.0 μL of Annexin V-FITC and 5 μL of propidium iodide (Annexin V-FITC Apoptosis Kit, BioVision Inc., Milpitas, CA). The mixture was incubated for 5 min in the dark at room temperature and analyzed by flow cytometry (Ex = 488nm; Em = 530 nm). Each experiment was performed in triplicate at different days, and the data reported as the mean ± SEM.
Western blot analysis
A375 and B16F10 cells were cultured inoculated in a 6-well plate at the density of 0.2×106 cells per well and after 80% confluency, cells were treated with DMSO, QW-296 cosolvent, colchicine and QW-296 loaded nanoparticle for 48 h. Then, cells were washed twice with cold PBS, lysed with RIPA buffer, and protein concentrations were measured with bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL). The lysate was boiled for 5 min, subjected to a 15 % SDS-PAGE, and transferred to a PVDF membrane using the iBlot™system (Invitrogen, Carlsbad, CA). Membranes were blocked with 3.0 % BSA in 1× Tris-buffered saline (TBS) at room temperature for 1.0 h and then incubated with primary antibodies at 4 °C overnight, followed by incubation with anti-mouse, anti-goat or anti-rabbit IRDye 800CW secondary antibodies for 1.0 h at RT in dark conditions. All the blots were probed with total β-actin or GAPDH antibody as a control. The signal of target proteins was detected Li-COR Odyssey® infrared imaging system (Li-COR, Lincoln, NE).
Confocal Microscopy/Immunofluorescence
A375 and B16F10 cells were seeded on eight culturewell-chambered cover glass coverslips at a density of 2000 cells/well and were incubated overnight. After treatment of cells with DMSO, QW-296, and colchicine, for 24h, cells were washed twice with ice-cold PBS. Cells were fixed with 4% formaldehyde solution overnight at 4 °C. Then, cells were washed three times with PBS to remove the formaldehyde completely. Cells were then incubated in the mixture (1:1) of 2.0% BSA (in PBS) and 0.1% Triton X-100 (in PBS) for 1.0 h at room temperature. Then, cells were washed three times with PBS and incubated the cells with anti-α,β-tubulin antibody (Thermo Scientific, Rockford, IL) at 4 °C overnight. Cells were then washed three times with PBS to remove the primary antibody completely. After this, cells were incubated with secondary antibody (Goat anti-Rabbit IgG-FITC, Santa Cruz Biotechnology, Dallas, TX) for 1.0 h at room temperature. Then, cells were washed three times with PBS to remove the secondary antibody completely. Cells were then stained with a solution of DAPI (1.0 μl of DAPI in 200 μl of 0.1% Triton X-100) and incubated for 10 min at room temperature. Finally, cells were washed three times with PBS and images were acquired with a Zeiss 710 confocal microscope and Zen imaging software (Zeiss, Thornwood, NY).
Scratch migration assay
A375 and B16F10 cells were seeded in 24-well plates (2×105 cells/well) in replicates of three and incubated overnight. Using a 200 μL pipette tip, a straight line was scratched through the cell monolayer to remove an area of cells, and cells were washed several times to remove any debris and uprooted cells. Media was replaced with media containing vehicle (DMSO), and 60 nM of colchicine or QW-296. Images were obtained after 0, and 24 hours with an inverted microscope at 10× magnification.
Terminal Deoxynucleotidyl Transferase Nick End Labeling (TUNEL) Staining
Terminal deoxynucleotidyl transferase (TdT) mediated-16-deoxyuridine triphosphate (dUTP) Nick-End Labelling (TUNEL) system (Promega, USA) was used to detect apoptosis in lung tumor tissue sections. Briefly, paraffin-embedded tumor tissue sections were deparaffinized by immersing in xylene and rehydrated by sequentially immersing the slides through graded ethanol. After washing with PBS, sections were fixed with 4% formaldehyde, washed with PBS, and permeabilized by incubating with Proteinase K solution (20 μg/mL) for 15 min. Afterward, slides were covered with 100 μL of equilibration buffer for 15 min and washed with PBS. Slides were coated with rTdT incubation buffer (equilibration buffer + nucleotide mix + rTdT enzyme) and incubated at 37 °C for 60 min. Endogenous peroxidases were blocked using 0.3% hydrogen peroxide. For the positive control, the sample was treated with DNase I (5–10 units/ml) in DNase I buffer for 15 min. Tissue sections were incubated with streptavidin-conjugated horseradish peroxide (HRP) at room temperature for 30 min. Then, the visualization of fragmented DNA was performed by treating the samples with a solution of hydrogen peroxide and diaminobenzidine (DAB).
In vivo study
All animal experiments were performed following the NIH animal use guidelines and protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Nebraska Medical Center (UNMC), Omaha, NE. A mouse model of metastatic melanoma to the lung was established in 8–10 weeks old female C57BL/6 albino mice by injecting 2×105 B16F10-Luc cells suspended in 100 μl PBS in the tail vein. Mice were randomly divided into three groups of six animals per group when the radiance of the tumor had reached 106 p/sec/cm2sr. QW-296 in cosolvent and QW-296 loaded nanoparticles were administered intravenously to mice once every three days for a total of five times. Group 1 was kept as the control and received blank nanoparticles, group 2 received 20 mg/kg QW-296 in 35% of cosolvent (50% propylene glycol, 30% Cremophor® EL, and 20% ethanol) and 65% of dextrose solution, and group 3 received nanoparticle formulation equivalent to free 20 mg/kg of QW-296. Bioluminescent radiance of tumor was measured every other day using the IVIS® Spectrum imaging system (PerkinElmer Inc., MA). Mice were sacrificed, tumors and other major organs such as liver, spleen, kidney and heart were isolated. Three representative tumor tissues were collected per group and fixed with 10% buffered formalin for 24 h. The fixed samples were embedded in paraffin and thin sections of 4.0 μm were obtained and immunostained for hematoxylin and eosin (H&E), Ki67 and cleaved Caspase 3.
Statistical analysis
Data represented as the mean ± S.D. The statistical comparisons of the experiments were performed by two-tailed Student’s t-test. P<0.01 was considered statistically significant.
Results
Synthesis of QW-296
QW-296 (Figure 1A) was synthesized using the starting 4-(benzyloxy)-1H-indole as a starting material, which was formylated at 3rd position. Subsequently, after protecting indole nitrogen, imidazole group was introduced at the 3rd position. Imidazole was then brominated, and the imidazole nitrogen was protected followed by displacement of the 3-bromo group with the trimethoxybenzoly group. Subsequently, a remaining bromine atom and both protecting groups were removed to get QW-296 as a yellow solid. LCMS, NMR, and HRMS confirmed the purity and chemical structures of QW-296. 1H NMR (400 MHz, DMSO-d6) δ 14.02 − 13.06 (m, 2H), 11.59 (dd, J = 65.1, 2.9 Hz, 1H), 8.86 − 8.07 (m, 1H), 8.07 − 7.81 (m, 1H), 7.60 − 7.12 (m, 2H), 7.01 (td, J = 7.9, 5.2 Hz, 1H), 6.87 (dd, J = 8.1, 4.5 Hz, 1H), 6.41 (dd, J = 13.9, 7.6 Hz, 1H), 3.91 (d, J = 4.9 Hz, 6H), 3.79 (d, J = 4.4 Hz, 3H). 13C NMR (101 MHz, DMSO-d6) δ 185.08, 182.08, 152.79, 152.54, 151.76, 151.54, 148.71, 145.69, 141.07, 140.83, 138.57, 138.54, 137.56, 136.29, 133.22, 132.86, 130.25, 126.11, 125.15, 124.29, 124.25, 123.29, 113.93, 113.73, 106.57, 105.96, 105.47, 105.28, 105.04, 104.73, 102.53, 102.43, 60.11, 55.97, 55.73, 54.89. HRMS [C21H20N3O5+]: calcd 394.1403, found 394.1414. HPLC purity 99.1% (tR = 3.03 min). (Supplementary Figure 3S–4S).
Copolymer Synthesis and preparation of QW-296 loaded nanoparticles
We utilized our previously developed mPEG-b-P(CB-co-LA) copolymer30,31 to prepare QW-294 loaded nanoparticles (Figure 1B) owing to ease in synthesis, biocompatibility, and ability to form stable nanoparticles. The degree of polymerization and molecular weight of this copolymer was confirmed by 1H NMR spectra (Figure 1S). From 1H NMR, the calculated molecular weight of this copolymer was 13,000 Da with a low polydispersity index (PDI) of 1.118. The mean particle size of QW-294 loaded nanoparticles was 122.0 ± 2.28 nm with PDI of 0.153 as determined by DLS (Figure 1C). Surface morphology and particle size of these nanoparticles were determined by AFM, and ζ potential by Malvern Zetasizer. These nanoparticles were of spherical shape and had narrow particle size distribution, with the mean particle size of 70.02 nm (Figure D) and ζ potential of zero mV. These results were included in Figure 1 and Supplementary Figure 2S. The Drug loading determined by HPLC was 3.70% w/w at the theoretical loading of 10.0%. There was no batch to batch variation in particle size and drug loading.
In vitro release profiles of QW-296 loaded nanoparticles
The release profile of QW-296 from nanoparticles was determined by dialysis and results are shown in Figure 1E. There was no burst effect and only 7.9% of QW-296 was released at pH 7.4 after 4.0 h of dialysis, but almost 68.3% release was achieved after 97.0 h suggesting sustained drug release over a long period.
Molecular modeling and effect on tubule polymerization
To confirm the binding interaction of QW-296 with colchicine binding site, we used molecular modeling studies using high-resolution crystal structure of DJ101 in complex with tubulin. Molecular modeling confirmed that QW-296 has similar binding interactions as the native compound ABI-III, and binds to the same binding site in tubulin protein (Figure 2A).
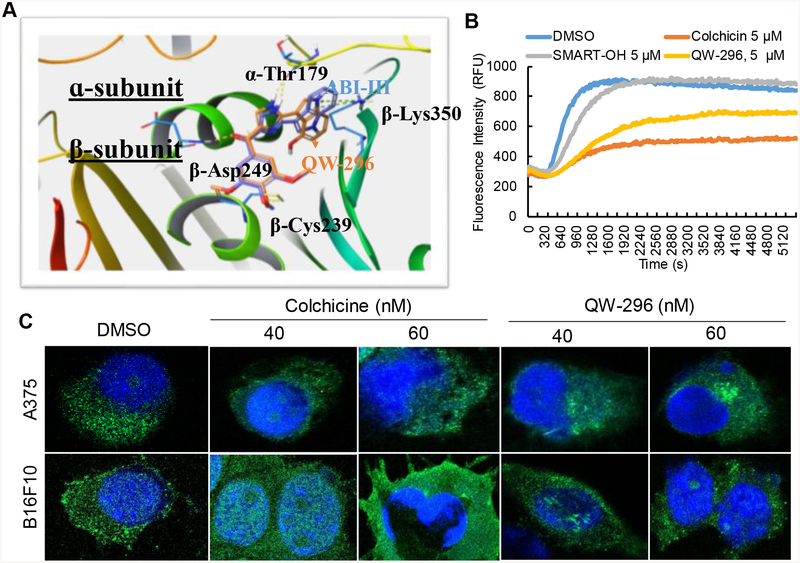
Figure 2.
Confirmation of the mechanism of action of QW-296. A) Proposed binding poses of ABI-III and QW-296 in the tubulin crystal structure (PDB code: 5H7O). Superposition of ABI-III (purple tube model; glide docking score −11.2) with QW-296 (orange tube model; glide docking score −11.9). B) Effect of QW-296 on tubulin polymerization in in vitro. C) Confocal images of A375 and B16F10 melanoma cells exposed to colchicine and QW-296 or the vehicle (DMSO) for 24 h.
To confirm the binding site of QW-296 on tubulin, we conducted in vitro tubulin polymerization assay. Tubulin (3 mg/ml) was exposed to 5 μM QW-296, colchicine, SMART-100 or vehicle control (2% DMSO), respectively, and incubated in tubulin buffer. Both QW-296 and colchicine effectively inhibited polymerization, while robust polymerization was observed in SMART-OH-treated group (Figure 2B). These results indicated that QW-296 is a strong tubulin depolymerizing agent. We also determined the effect of QW-296 on microtubule networks by confocal microscopy of A375 and B16F10 cells treated for 18 h with QW-296, using colchicine as a strong inhibitor of tubulin polymerization as a positive control. There was a significant alteration in microtubule arrangement in the treated cells (Figure 2C). Control cells exhibited well-dispersed microtubule networks throughout the cytoplasm, whereas the cells treated with QW-296 had scattered microtubule networks with weaker fluorescent signal due to an increase in dispersed cytoplasmic fluorescence, like colchicine-treated cells. These observations further confirm that QW-296 exhibits strong tubulin binding affinity at colchicine binding site.
Anticancer activity
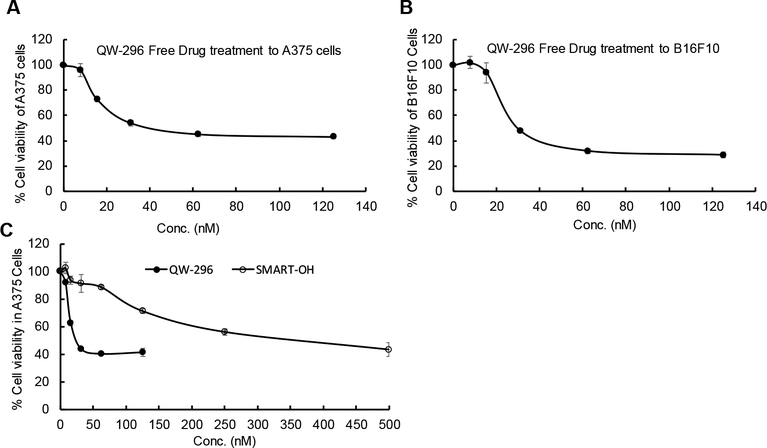
Cytotoxicity of QW-296 was determined in A375 and B16F10 cells by incubating cells at different drug concentrations. As shown in Figure 3A and 3B, there was dose-dependent cell killing, with the IC50 values of 30 and 35 nM, respectively. Since QW-296 was more effective in inhibiting tubulin than SMART-OH, we decided to compare QW-296 and SMART-OH for their dose-dependent cytotoxic effect on A375 cells. As shown in Figure 3C, QW-296 was significantly more effective in cell killing resulting in IC50 value of 30 nM compared to that of 350 nM for SMART-OH. Therefore, we decided to focus on QW-296 for all subsequent studies.
Figure 3.
Cell viability assay using QW-296 in melanoma cells. A) Cytotoxicity of QW-296 in A375 cells, B) in B16F10 cells and C) comparison of cytotoxicity of QW-296 and SMART-OH in A375 cells after 48 h of treatment.
Cell cycle arrest and apoptosis
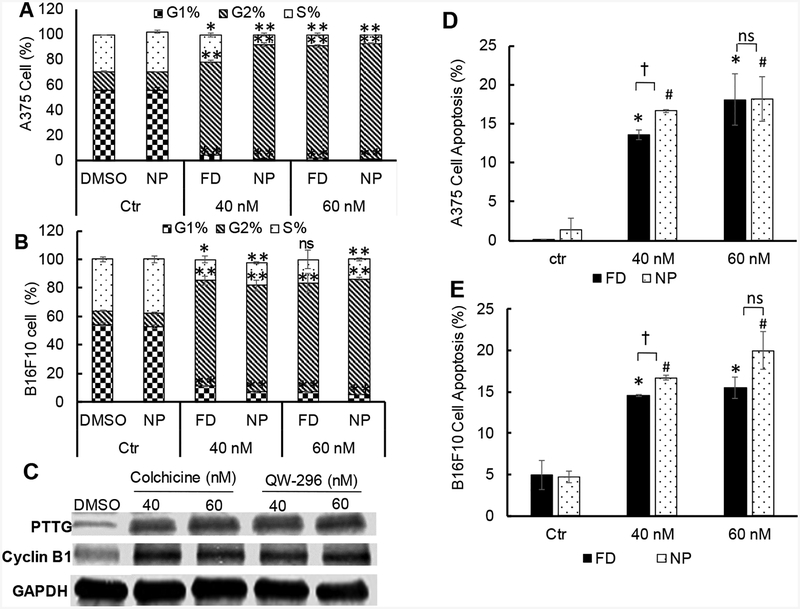
Cell cycle arrest was determined by propidium iodide (PI) staining after 24 h of treatment with QW-296 in cosolvent and nanoparticle formulations in A-375 and B16F10 cells. For A375 cells, results showed that treatment with free QW-296 at 40 and 60 nM caused 74.4 and 88.7% cell arrest in G2 phase after 24 h of treatment, whereas small number of cells were arrested in S-phase (21.5% and 9.0%, respectively) and G1 phase (4.1 and 2.3%, respectively). Treatment of these cells with QW-296 loaded nanoparticles (40 and 60 nM equivalent concentration) resulted in enhanced cell arrest in G2 phase 90.9% vs 74.4 % at 40 nM and 92.1% vs 88.7% at 60 nM. Further, the number of cells in S-phase (7.8% and 6.9%, respectively) and G1 phase (1.3% and 1.0%, respectively) were reduced. (Figure 4A)
Figure 4.
Effect of QW-296 on cell cycle analysis and induction of apoptosis in melanoma cells. A) Cell cycle analysis after treatment with QW-296 (free drug and loaded nanoparticles) to A375 cells, and B) B16F10 cells. C) Effect of the QW-296 treatment on the expression of G2/M related proteins PTTG and Cyclin B1. D) Induction of apoptosis by QW-296 as free drug and loaded in nanoparticles in A375 cells and E) B16F10 cells after 24 h of incubation. *, p<0.05 (comparison between control vs free drug), #, p<0.05 (comparison between control vs NP), †, p<0.05 (comparison between free QW-296 vs QW-296 loaded NP), ns, p>0.05.
Treatment of B16F10 cells with free QW-296 at 40 and 60 nM for 24h caused 74 and 76.3% cell arrest in G2 phase, whereas only 14.9.5% and 16.8% of these cells were arrested in S-phase, and 11.1 and 6.9%, respectively in G1 phase (Figure 4B). Treatment of these cells with QW-296 loaded nanoparticles at 40 and 60 nM equivalent doses resulted in an equal number of cell arrest in G1 phase (74.5% vs. 74 % at 40 nM concentration and whereas at 60 nM concentration increased the number of cells were arrested in the G2 phase (81.4% vs. 76.3%). Further, similar numbers of cells were observed in S-phase (15.8% and 13.7% respectively) and G1 phase (7.1% and 4.8% respectively) (Figure 4B).
To determine whether QW-296 binds to tubulin and arrests the cell cycle in G2/M phase,35 A375 and B16F10 cells were treated with QW-296 and probed for cyclin B and PTTG.36 Treatment with QW-296 and colchicine upregulated PTTG and cyclin B1 expression compared to non-treated cells (Figure 4C), suggesting QW-296 could block the cell cycle at the G2/M phase.37
Total apoptotic cell populations significantly increased from 0.1% in the non-treated control to 13.55% and 20.4% when A375 cells were incubated with QW-296 formulated in cosolvent at 40 and 60 nM, respectively. A similar number of cells were found apoptotic when treated with an equivalent concentration of QW-296 loaded nanoparticles (Figure 4D). For B16F10 cell, apoptotic cell population was significantly increased when the cells were treated with QW-296 formulated in cosolvent at 40 and 60 nM compared to the control group (4.95% vs. 14.55 and 15.5%, respectively), Further, the apoptotic cell population increased to 16.7% and 20%, respectively when the cells were treated with QW-296 loaded nanoparticles (Figure 4E). However, there was statistically no difference between cosolvent and nanoparticle formulation treated groups.
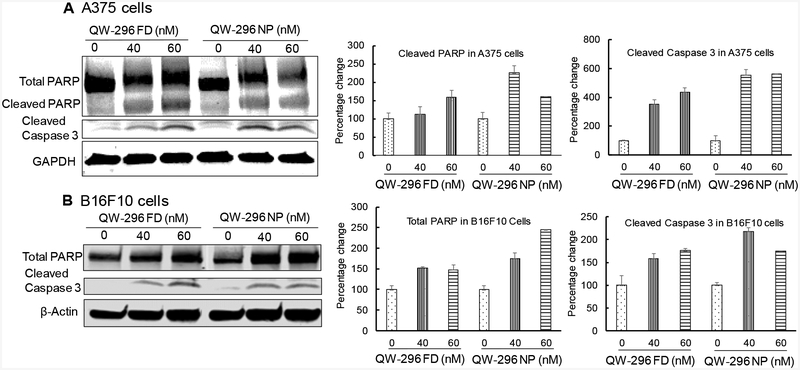
Poly(ADP-ribose) polymerase (PARP) helps cells to maintain their viability, but its cleavage facilitates cellular disassembly and serves as a marker of cells undergoing apoptosis.38 Therefore, we measured PARP and cleaved caspase 3 by Western blot analysis after treating A375 and B16F10 cells with QW-296 formulated in the cosolvent system or loaded into nanoparticles. Treatment of A375 cells with QW-296 resulted in increased expression of cleaved caspase 3 and subsequent PARP cleavage (Figure 5A). Similarly, increased expression of cleaved caspase 3 was observed when B16F10 cells were treated with QW-296 formulated in cosolvent or into nanoparticles (Figure 5B). These observations provide a reasonable mechanistic explanation of the observed cell death induction by treatment with QW-296.
Figure 5.
Estimation of apoptotic protein expression (PARP and Cleaved Caspase 3) by WB in A) A375 cells and B) B16F10 cells after treatment with QW-296 in cosolvent and QW-296 loaded nanoparticles after 48 h of treatment.
Effect of QW-296 on cell migration
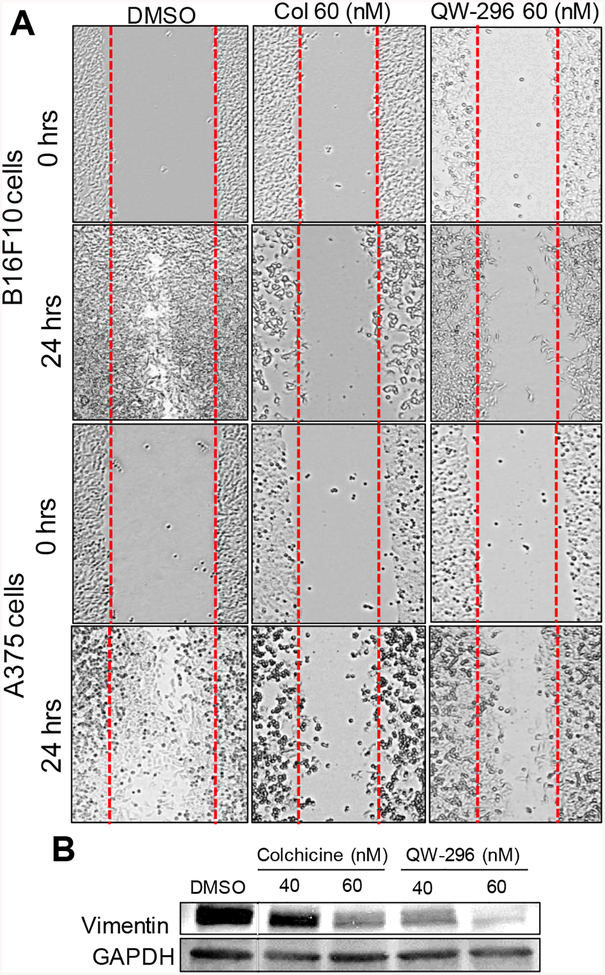
To determine the effect of QW-296 on cell migration through microtubule destabilization, we performed a scratch migration assay. After 24 hours, the vehicle (DMSO) treated A375 and B16F10 cells had nearly achieved complete closure of the wound by migrating to the scratch area. In contrast, QW-296 inhibited of cell migration and equivalent to colchicine (Figure 6A). These results demonstrate that QW-296 strongly reduces aberrant cancer cell proliferation and hinders cell migration efficiently as colchicine in metastatic melanoma. We also estimated the levels of vimentin in A375 cells after treatment with different concentrations of colchicine and QW-296. There was a significant decrease in vimentin protein expression level in dose dependent manner after treatment with QW-296 compared to control group. (Figure 6B).
Figure 6.
Effect of QW-296 and colchicine on cell migration and EMT marker. A) Scratch assay in B16F10 and A375 cells. B) Dose-dependent decrease in vimentin in A375 cells.
Effects of QW-296 on taxane-resistant cells
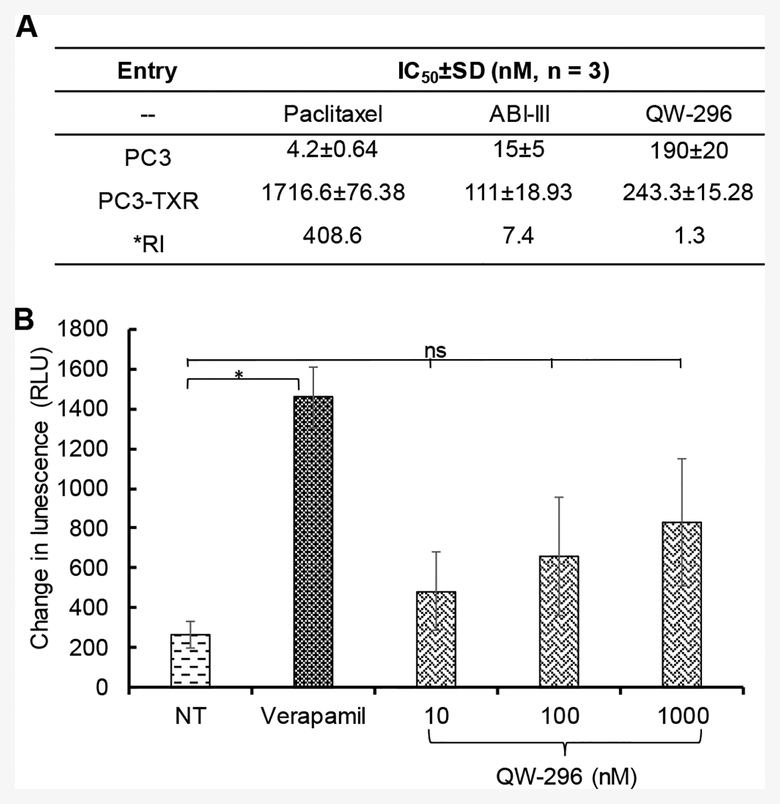
Paclitaxel (PTX) and vinblastine are known to develop chemoresistance to cancer cells.39,40 ABI-III showed high potency for TXR cell lines.24 Since ABI-III and QW-296 belong to the same family of compounds, we hypothesized that QW-296 would retain its ability to circumvent TXR mechanisms. To support this, we screened QW-296 against TXR cell line, such as PC3-TXR. QW-296 was found to be equipotent in both PC3 and PC3-TXR cells with resistance index only 1.3, whereas PTX showed significantly low potency in PC3-TXR cells with a resistance index of 408.6. Interestingly, the parent compound ABI-III also showed high potency in resistant lines with resistance Index of 7.4 (Figure 7A).
Figure 7.
Determination of resistance index and possible mechanism to overcome chemoresistance by QW-296. A) Anti-proliferative activity of paclitaxel (PTX), ABI-III and QW-296 on paclitaxel-resistant PC3-TXR cells and non-resistant PC3 cells. B) Effect of QW-296 on Pgp ATPase activity. Change in luminescence compared to 100 μM Na3VO4 treated samples was plotted (mean ± SD, n = 3). QW-296 at lower concentrations (10 and 100 nM) showed a non-significant effect compared to NT control on Pgp ATPase activity. *, p < 0.05.
Effect of QW-296 on Pgp activity
To identify the potential mechanism for low resistance index of QW-296 in TXR cells, we determined the effect of QW-296 on Pgp-ATPase activity using Pgp-Glo™ assay system. In this assay, a specific amount of ATP was incubated with recombinant human P-gp and then unmetabolized ATP was detected as a luciferase-generated luminescent signal. The decrease in luminescence signal after drug treatment reflect ATP consumption by Pgp. Changes in the luminescence of QW-296 treated samples from Na3VO4 treated samples were plotted to illustrate the stimulation or inhibition of P-gp ATPase activity due to QW-296 treatment (Figure 7B). Verapamil, a well-known P-gp substrate, stimulated P-gp ATPase activity, resulting in significantly decreased luminescence. Although there was no statistically significant difference in luminescence changes between the vehicle control treated group and QW-296 treated groups at 10 and 100 nM concentrations, suggesting that QW-296 is a mild stimulator of P-gp ATPase. However, at high concentration (1000 nM) QW-296 exhibited effects like that of an activator of Pgp. These results strongly indicate that QW-296 like verapamil, is a substrate of P-gp and leads to the inhibition of P-gp efflux. This may partially explain the mechanism of QW-296 to overcoming TXR.
In vivo efficacy of QW-296 loaded nanoparticles
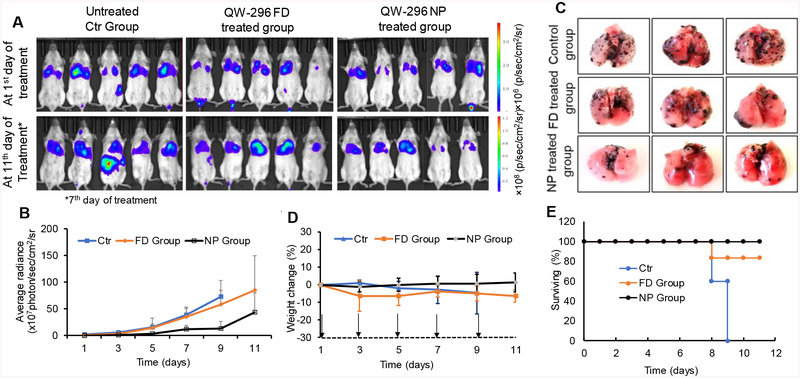
Preclinical efficacy of QW-296 loaded nanoparticles was evaluated in a lung metastatic melanoma model generated by intravenous injection of B16-F10-Luc cells into C57BL/6 albino mice. The mice with the bioluminescent radiance of 106 p/sec/cm2/sr were randomized into three groups: 1) control, 2) QW-296 free drug treatment group, and 3) QW-296 loaded nanoparticles treatment group. Mice treated with QW-296 formulated in cosolvent or loaded into nanoparticles showed inhibition of tumor growth compared to the control group. Higher tumor growth inhibition was observed in the group treated with QW-296 loaded nanoparticles treated group compared to QW-296 in cosolvent and control groups (Figures 8A and 8B). In addition, treatment with QW-296 loaded nanoparticles significantly reduced the number of lung tumor nodules compared to the control and cosolvent formulation groups (Figure 8C). We observed that body weights (Figure 8D) and physical activities of mice were normal in both the treatment groups whereas control group animals did not even survive during all treatment days. All control animals died at the end of the 9th day of the study. This further confirms that mice well tolerated QW-296 treatment (Figure 8E).
Figure 8.
In vivo therapeutic effect of QW-296 as FD and QW-296 loaded nanoparticles (NPs) in lung tumor metastasis mice model. A) In vivo representative bioluminescent images at day 1 and day 11 of treatment. Bioluminescent images of mice from control (blank NP), QW-296 FD group and QW-296 loaded in NP groups were taken every alternate day during the treatment (n = 6), *indicates bioluminescent images of control group on day 7th of the study. B) Radiance intensity plot of all three treatment groups measured from day 1 of treatment to the end of the study. C) Representative lungs showing tumor nodules of each group excised after sacrificing the mice at the end of the study. D) Mouse % body weight change during treatment for all the three groups. E) Survival plot of all the animals used using QW-296 in cosolvent and QW-296 loaded in NPs. Data represented as the mean ± SD (n=6). Arrow indicates the day of injection.
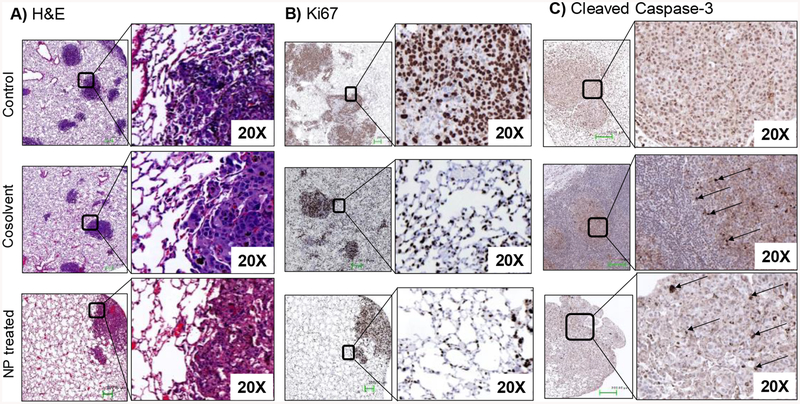
Hematoxylin and eosin (H&E) staining of lung tissues confirmed the extensive metastasis throughout the lung lobe in the control group and the inhibition of proliferation of tumor cells in the treated groups. Compared to the control and cosolvent group, the lung samples from QW-296 loaded nanoparticles treated group exhibited alveolar lumen with a limited mass of metastatic cells (Figure 9A). Ki67 protein is an excellent marker for determining the cell proliferation as Ki-67 protein is present during all active phases of the cell cycle but is absent in resting cells. Ki67 staining of lung tissues confirmed the extensive cell proliferation in the non-treated control group compared to the cosolvent and nanoparticles treated groups. Lung tissues treated with QW-296 loaded nanoparticles showed a significantly low number of cells Ki67 positive cells (Figure 9B). Furthermore, cleaved caspase-3 staining of lung tissues indicated the induction of significant apoptosis by treatment of QW-296 in the cosolvent or nanoparticles compared to the control group (Figure 9C).
Figure 9.
Effect of QW-296 treatment on tumor growth and tumor cell apoptosis. Lung tumor nodules from control, QW-296 in cosolvent and QW-296 loaded nanoparticles (NPs) treated groups were stained by A) hematoxylin and eosin (H&E), B) proliferation marker Ki67, and C) apoptosis marker cleaved Caspase 3.
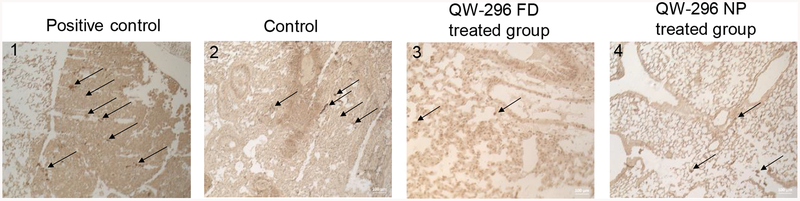
The ability of QW-296 to induce apoptosis was evaluated by treatment of lung tumors of different treatment groups by TUNEL colorimetric assay, which detects DNA fragments resulted from DNA damaging agents. Treatment with QW-296 caused very few cells positive for DNA damage compared to the positive control group (Figure 10). These results further confirm that QW-296 in specifically binds to the tubulin without any nonspecific DNA damage to the tissue.
Figure 10.
DeadEnd™ colorimetric TUNEL assay of mouse lung tissues to indicate DNA fragmentation in 1) positive control, 2) control group, 3) mice treated with 20mg/kg QW-296 in cosolvent system, and 4) mice treated with 20 mg/kg equivalent dose of QW-296 loaded NPs. Slides were observed under the light microscope and arrows show dark stained nuclei which indicate DNA fragmentation and nuclear condensation.
Additionally, we determined the systemic toxicity of QW-296 cosolvent and nanoparticle formulations by histological analysis of the major organs after systemic administration at the dose of 20mg/kg. There were no obvious histological changes observed in the livers, spleens, kidneys, and hearts from the treatment groups, suggesting that the mice tolerated QW-296 treatment well (Figure 5S).
Discussion
Poor prognosis to treat melanoma presents a major challenge and FDA approved drugs such as dacarbazine or its combination with PTX did not improve the overall patient’s survival. The use of new therapeutic options such as BRAF inhibitors, MEK inhibitors or immunomodulating drugs is limited due to the development of chemoresistance and severe life-threatening side effects. Microtubule binding drugs present a validated approach to treat many forms of cancer.41 The therapeutic efficiency of drugs that binds to taxane or vinca binding site on tubulin is restricted due to overexpression of drug efflux pump or expression of mutated tubulin βIII subtype.42 To deal with these challenges, the development of new drugs that bind to the colchicine-binding site on tubulin is a promising approach. Previously, we reported the several thiazole and indole derivatives that inhibit tubulin by interacting at colchicine binding site and exhibited high potency against many cancer cells.3,22–24 QW-296 is an advanced version of one of our previously reported compounds, ABI-III, by introducing a hydroxyl group on the indole nucleolus.
Since most anticancer agents including QW-296 are hydrophobic, we and others use lipid and polymer-based drug delivery systems to help them reach tumors after systemic administration. To further enhance its therapeutic efficiency, we encapsulated QW-296 into nanoparticles using mPEG-b-P(CB-co-LA) by o/w emulsification, with drug loading of 3.70% due to the presence of a polar phenolic group on the indole moiety of QW-296. Hydrophilic mPEG shell could prevent the aggregation of particles due to steric hindrance and possibly reduce uptake by the reticuloendothelial system (RES) and improve their circulation time in the blood stream. We observed that nanoparticles were stable at room temperature for at least two weeks without a significant change in their size and no sigh of precipitation of the drug was observed, suggesting good stability of the particles at room temperature. The nanoparticles efficiently encapsulated the drug, and we did not observe any burst drug release. There was sustained drug release from the nanoparticles, with approximately 70% of drug release in 100 hours (Figure 1E). This concludes that polymeric nanoparticles efficiently encapsulate the drug and release slowly for an extended period.
We previously reported the discovery of ABI-III, which exerted its anticancer effects by inhibiting tubulin polymerization by binding to tubulin at colchicine binding site.24 To better understand how the 4-OH-substituted ABI-III analog interacts with tubulin, ABI-III and QW-296 were investigated in the tubulin crystal structure (PDB code: 5H7O). The overview of the binding site of ABI-III and QW-296 in 5H7O was located at the interface between α- and β-subunits of tubulin dimer and extended slightly into β-subunit (Figure 2A). ABI-III (purple tube model) and QW-296 (orange tube model) showed very similar binding poses and overlapped very well. As anticipated, the TMP moiety in ABI-III and QW-296 was close to SH of Cys239 in β-subunit and allowed the formation of one hydrogen bond. A similar hydrogen bond was also observed between the carbonyl and β -Asp249. The imidazole NH moiety in ABI-III and QW-296 formed another hydrogen bond to Thr-179 in the α-subunit. The indole NH of ABI-III and QW-296 with β-Lys350 contributed to another hydrogen bond. The glide docking score of QW-296 was comparable to that of the prototype compound ABI-III (−11.9 vs. −11.2, respectively), suggesting that they may have similar effects on tubulin binding.
Microtubule stabilizing drugs promote microtubule polymerization and keep them in the polymerized state, whereas microtubule destabilizing agents such as colchicine and combretastatin, inhibit the tubulin polymerization43 and subsequently lead to cell cycle arrest and apoptosis. To confirm the mode of QW-296 action, we allowed polymerization of tubulin in the presence of QW-296. As shown in Figure 2B, QW-296 strongly inhibited the tubulin polymerization and very low fluorescent signals were recorded in the case of colchicine treated sample. However, SMART-OH showed poor inhibition of tubule polymerization at the same concentration, suggesting QW-296 is a more potent microtubule destabilizer than SMART-OH. To further confirm the QW-296 effect on microtubules of melanoma cells, we evaluated the microtubule disruptive effect on A375 and B16F10 melanoma cells. Cells treated with different concentration of QW-296 and colchicine showed dispersed tubulin protein with widespread cytoplasmic fluorescence suggesting the presence of depolymerized tubulin (Figure 2C).
Treatment of cells with tubulin binding drugs resulted in change in the cell morphology and accumulated cells in G2/M phase.37,44 However, such mitotic arrest persists for varying length of time depending on the drug potency. Subsequently, cells exit from cell cycle arrest and undergo apoptosis most probably through Bcl-2, activation of PARP and caspase activity. However, DNA damage and alkylating agents did not show such apoptosis induction pattern.37 Further, DNA damaging agents activate cell cycle checkpoint that prevents the progress of cells to M phase during DNA repair. Thus, to confirm this fact, we analyzed A375 cells after treatment with QW-296 for cell cycle-related proteins by Western blot. The cell cycle is regulated by a set of regulatory enzymes such as cyclins and cyclin-dependent kinases. The cell cycle arrest could be detected by the stabilization of corresponding kinase substrates whose degradation is a prerequisite to complete the cell cycle. Accumulation of G2/M phase proteins such as cyclin B1 and PTTG confirm that QW-296 inhibited mitosis (Figure 4C).
Previously, we used mPEG-b-P(CB-co-LA) polymeric nanoparticles to deliver microtubule destabilizer where treatment with drug-loaded nanoparticles showed improved cytotoxicity to melanoma cells.30,33 Treatment of A375 and B16F10 cells with QW-296 cosolvent or QW-296 loaded nanoparticles lead to cell cycle arrest in the G2/M phase (Figures 4A & 4B). These results also confirm that QW-296 nanoformulations potentiate the cell cycle arrest in the G2/M phase and subsequently the chromosomes failed to segregate after replication. Subsequently, cell cycle arrest in the G2/M phase, cells undergo apoptosis by activation of apoptotic markers which is a typical characteristic of drugs inhibiting the microtubules. We observed a significant population of A375 and B16F10 cells where apoptosis was induced by QW-296 treatment. QW-296 loaded nanoparticles induced apoptosis in these cells to a slightly greater extent (Figures 4D & 4E). Western blot analysis of protein samples isolated from A375 and B16F10 cell showed higher levels of cleaved Caspase 3, which is one of the key executioners of apoptosis. Further, the activation of caspase 3 resulted in the cleavage of total PARP and the subsequent appearance of the cleaved fragment of PARP (Figures 5A & 5B). No band of cleaved PARP in B16F10 cells may be due to non-specificity of the primary antibody against murine cells used in this experiment. These results confirmed that QW-296 inhibits the tubulin at colchicine binding site, resulting in cell cycle arrest in the G2/M phase and subsequently induced cell apoptosis.
Involvement of microtubules in cell migration and motility is well known.45 Microtubule binding drugs disturbed the tubulin dynamics and restrained the cells resulting in immobility and inability to respond to required cellular shape in an altered environment.46 The anti-migratory effects of QW-296 on A375 and B16F10 cells is demonstrated by its ability to inhibit the cells to migrate to heal the scratch wound which essentially required for cell mobility and remodeling in cellular structure. QW-296 inhibited cell migration, which is attributed to its ability to disrupt the microtubule dynamics (Figure 6A)
Epithelial to mesenchymal transition (EMT) plays an important role in the cell migration and invasion of cancer cells. We evaluated the effect of QW-296 treatment on an important biomarker associated with the mesenchymal form, vimentin. The high expression level of vimentin is associated with cancer invasion of various cancers, including melanoma.47 The reduction in vimentin expression impairs cell attachment, migration, and invasion in various cancer cell lines.48 Also, vimentin contributes to the construction of cellular cytoskeleton architecture by interacting with microfilaments and microtubules to provide cellular mechanical strength.49 Our results show that treatment of A375 cells with QW-296 significantly reduced vimentin expression compared to the control group and equivalent dose of colchicine (Figure 6B). These results strongly suggest that QW-296 has the potential to inhibit the migration and invasion of melanoma cells.
TXR is the major cause of clinical failure of PTX and vinblastine chemotherapy.24 Since QW-296 is an analog of ABI-III, it was effective in both PC3 and PC3-TXR cells, with a resistance index of only 1.3, whereas PTX showed high resistance index of 408.6. Importantly, the parent compound ABI-III showed resistance index of 7.4 suggesting that even though the potency of QW-296 is slightly lower than ABI-III, it is more effective in TXR cells with very low resistance index (Figure 7A). Low resistance index of QW-296 guided us to perform pgp-glo ATPase assay. It is known that PC3-TXR cell line has more than 200 genes upregulated in addition to P-gp overexpression.50 The results from Pgp-glo experiment suggested that QW-296 is a stimulator of P-gp even at the low dose, whereas at the higher dose (1000 nM), like verapamil, QW-296 showed a significant difference in the luminescence signal relative to the control sample (Figure 7B). These results suggest that QW-296 is a substrate for P-gp and inhibits the P-gp efflux and partially explain the mechanism to overcome TXR.
After the demonstration of excellent efficacy of QW-296 loaded nanoparticles over cosolvent treatment on melanoma cells, we tested the efficacy of QW-296 loaded nanoparticles in B16F10_luc lung metastatic mouse model and compared to the equivalent dose of a cosolvent of QW-296. The lung tumor burden was significantly low in the nanoparticle-treated group compared to the control group and the cosolvent solution treated. The tumor burden is indicated by the melanoma nodule on isolated lungs of the treatment groups (Figure 8C). Further, treatment with QW-296 loaded nanoparticles induced apoptosis as suggested by caspase 3 immunostaining of lung tumor tissue (Figure 9C). These results are in line with apoptosis induction experiment using melanoma cells.
Conclusion
We have synthesized a novel microtubule destabilizer QW-296 and encapsulated into polymeric nanoparticles. Our results demonstrate that QW-296 efficiently binds to microtubules and suppresses tubulin polymerization. QW-296 inhibits melanoma cell proliferation and invasion. Systemic administration of QW-296 loaded nanoparticles significantly inhibited metastatic lung melanoma compared to the control and the cosolvent formulation of QW-296. Future work will focus on designing additional microtubule inhibitors to further enhance their potencies and optimization of our polymeric delivery system by including targeting moieties.
Supplementary Material
Highlights.
Discovery of potent colchicine binding agent, QW-296, with IC50 in nanomolar range in melanoma cells.
QW-296 retains high potency in paclitaxel-resistant cells and inhibits cell migration.
Nanoparticulate delivery of QW-296 effectively regresses lung metastatic melanoma in mice.
Mice well tolerate QW-296 treatment without any significant toxicity to vital organs.
Acknowledgment
The faculty start-up fund supported this work from UNMC to Ram I. Mahato and NCI grant R01CA148706 to WL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kaur R, Kaur G, Gill RK, Soni R, Bariwal J. Recent developments in tubulin polymerization inhibitors: An overview. Eur J Med Chem. 2014;87:89–124. [DOI] [PubMed] [Google Scholar]
- 2.Li CM, Chen J, Lu Y, et al. Pharmacokinetic optimization of 4-substituted methoxybenzoyl-aryl-thiazole and 2-aryl-4-benzoyl-imidazole for improving oral bioavailability. Drug Metab Dispos. 2011;39(10):1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnst KE, Wang Y, Hwang DJ, et al. A potent, metabolically stable tubulin inhibitor targets the colchicine binding site and overcomes taxane resistance. Cancer Res. 2018;78(1):265–277. [DOI] [PubMed] [Google Scholar]
- 4.Gigant B, Wang C, Ravelli RB, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435(7041):519–522. [DOI] [PubMed] [Google Scholar]
- 5.Dorleans A, Gigant B, Ravelli RB, Mailliet P, Mikol V, Knossow M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc Natl Acad Sci U S A. 2009;106(33):13775–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravelli RB, Gigant B, Curmi PA, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. [DOI] [PubMed] [Google Scholar]
- 7.Perez EA. Microtubule inhibitors: Differentiating tubulin-inhibiting agents based on mechanisms of action, clinical activity, and resistance. Mol Cancer Ther. 2009;8(8):2086–2095. [DOI] [PubMed] [Google Scholar]
- 8.Chen ZS, Hopper-Borge E, Belinsky MG, Shchaveleva I, Kotova E, Kruh GD. Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10). Mol Pharmacol. 2003;63(2):351–358. [DOI] [PubMed] [Google Scholar]
- 9.Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8(5):411–424. [DOI] [PubMed] [Google Scholar]
- 10.Risinger AL, Jackson EM, Polin LA, et al. The taccalonolides: Microtubule stabilizers that circumvent clinically relevant taxane resistance mechanisms. Cancer Res. 2008;68(21):8881–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stengel C, Newman SP, Leese MP, Potter BV, Reed MJ, Purohit A. Class III beta-tubulin expression and in vitro resistance to microtubule targeting agents. Br J Cancer. 2010;102(2):316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Echteld I, Wechalekar MD, Schlesinger N, Buchbinder R, Aletaha D. Colchicine for acute gout. Cochrane Database Syst Rev. 2014;(8):CD006190 doi(8):CD006190. [DOI] [PubMed] [Google Scholar]
- 13.Garbe C, Eigentler TK, Keilholz U, Hauschild A, Kirkwood JM. Systematic review of medical treatment in melanoma: Current status and future prospects. Oncologist. 2011;16(1):5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coit DG, Andtbacka R, Bichakjian CK, et al. Melanoma. J Natl Compr Canc Netw. 2009;7(3):250–275. [DOI] [PubMed] [Google Scholar]
- 15.Eigentler TK, Caroli UM, Radny P, Garbe C. Palliative therapy of disseminated malignant melanoma: A systematic review of 41 randomised clinical trials. Lancet Oncol. 2003;4(12):748–759. [DOI] [PubMed] [Google Scholar]
- 16.Rao RD, Holtan SG, Ingle JN, et al. Combination of paclitaxel and carboplatin as second-line therapy for patients with metastatic melanoma. Cancer. 2006;106(2):375–382. [DOI] [PubMed] [Google Scholar]
- 17.Hauschild A, Agarwala SS, Trefzer U, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J Clin Oncol. 2009;27(17):2823–2830. [DOI] [PubMed] [Google Scholar]
- 18.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: Latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lugowska I, Teterycz P, Rutkowski P. Immunotherapy of melanoma. Contemp Oncol (Pozn). 2018;22(1A):61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad SS, Reinius MA, Hatcher HM, Ajithkumar TV. Anticancer chemotherapy in teenagers and young adults: Managing long term side effects. BMJ. 2016;354:i4567. [DOI] [PubMed] [Google Scholar]
- 21.Alfarouk KO, Stock CM, Taylor S, et al. Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell Int. 2015;15:71-015-0221-1 eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn S, Duke CB 3rd, Barrett CM, et al. I-387, a novel antimitotic indole, displays a potent in vitro and in vivo antitumor activity with less neurotoxicity. Mol Cancer Ther. 2010;9(11):2859–2868. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Li CM, Wang Z, et al. Design, synthesis, and SAR studies of 4-substituted methoxylbenzoyl-aryl-thiazoles analogues as potent and orally bioavailable anticancer agents. J Med Chem. 2011;54(13):4678–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Ahn S, Wang J, et al. Discovery of novel 2-aryl-4-benzoyl-imidazole (ABI-III) analogues targeting tubulin polymerization as antiproliferative agents. J Med Chem. 2012;55(16):7285–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao M, Ahn S, Wang J, et al. Discovery of 4-aryl-2-benzoyl-imidazoles as tubulin polymerization inhibitor with potent antiproliferative properties. J Med Chem. 2013;56(8):3318–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta PK. Drug targeting in cancer chemotherapy: A clinical perspective. J Pharm Sci. 1990;79(11):949–962. [DOI] [PubMed] [Google Scholar]
- 27.Hossen S, Hossain MK, Basher MK, Mia MNH, Rahman MT, Uddin MJ. Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: A review. Journal of Advanced Research. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Danquah M, Mahato RI. Synthesis and characterization of amphiphilic lipopolymers for micellar drug delivery. Biomacromolecules. 2010;11(10):2610–2620. [DOI] [PubMed] [Google Scholar]
- 29.Danquah M, Li F, Duke CB 3rd, Miller DD, Mahato RI. Micellar delivery of bicalutamide and embelin for treating prostate cancer. Pharm Res. 2009;26(9):2081–2092. [DOI] [PubMed] [Google Scholar]
- 30.Mundra V, Lu Y, Danquah M, Li W, Miller DD, Mahato RI. Formulation and characterization of polyester/polycarbonate nanoparticles for delivery of a novel microtubule destabilizing agent. Pharm Res. 2012;29(11):3064–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundra V, Peng Y, Kumar V, Li W, Miller DD, Mahato RI. Systemic delivery of nanoparticle formulation of novel tubulin inhibitor for treating metastatic melanoma. Drug Deliv Transl Res. 2015;5(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang R, Mondal G, Ness RA, et al. Polymer conjugate of a microtubule destabilizer inhibits lung metastatic melanoma. J Control Release. 2017;249:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mundra V, Peng Y, Rana S, Natarajan A, Mahato RI. Micellar formulation of indocyanine green for phototherapy of melanoma. J Control Release. 2015;220(Pt A):130–140. [DOI] [PubMed] [Google Scholar]
- 34.Yang R, Chen H, Guo D, et al. Polymeric micellar delivery of novel microtubule destabilizer and hedgehog signaling inhibitor for treating chemoresistant prostate cancer. J Pharmacol Exp Ther. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang DJ, Wang J, Li W, Miller DD. Structural optimization of indole derivatives acting at colchicine binding site as potential anticancer agents. ACS Med Chem Lett. 2015;6(9):993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pray TR, Parlati F, Huang J, et al. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5(6):249–258. [DOI] [PubMed] [Google Scholar]
- 37.Gururaja TL, Goff D, Kinoshita T, et al. R-253 disrupts microtubule networks in multiple tumor cell lines. Clin Cancer Res. 2006;12(12):3831–3842. [DOI] [PubMed] [Google Scholar]
- 38.Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. lesson from an uncleavable mutant. J Biol Chem. 1998;273(50):33533–33539. [DOI] [PubMed] [Google Scholar]
- 39.Geney R, Ungureanu l, Li D, Ojima I. Overcoming multidrug resistance in taxane chemotherapy. Clin Chem Lab Med. 2002;40(9):918–925. [DOI] [PubMed] [Google Scholar]
- 40.Verrills NM, Kavallaris M. Improving the targeting of tubulin-binding agents: Lessons from drug resistance studies. Curr Pharm Des. 2005;11(13):1719–1733. [DOI] [PubMed] [Google Scholar]
- 41.van Vuuren RJ, Visagie MH, Theron AE, Joubert AM. Antimitotic drugs in the treatment of cancer. Cancer Chemother Pharmacol. 2015;76(6):1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Chen J, Xiao M, Li W, Miller DD. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res. 2012;29(11):2943–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanton RA, Gernert KM, Nettles JH, Aneja R. Drugs that target dynamic microtubules: A new molecular perspective. Med Res Rev. 2011;31(3):443–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Shuai W, Sun H, et al. Design, synthesis and biological evaluation of quinoline-indole derivatives as anti-tubulin agents targeting the colchicine binding site. Eur J Med Chem. 2019;163:428–442. [DOI] [PubMed] [Google Scholar]
- 45.Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: Cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171(24):5507–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H, Ganguly A, Cabral F. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J Biol Chem. 2010;285(42):32242–32250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McInroy L, Maatta A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun. 2007;360(1):109–114. [DOI] [PubMed] [Google Scholar]
- 49.Liu CY, Lin HH, Tang MJ, Wang YK. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6(18):15966–15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda M, Mizokami A, Mamiya K, et al. The establishment of two paclitaxel-resistant prostate cancer cell lines and the mechanisms of paclitaxel resistance with two cell lines. Prostate. 2007;67(9):955–967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.