Abstract
Lysine crotonylation is a newly discovered post‐translational modification, which is structurally and functionally different from the widely studied lysine acetylation. Recent advances in the identification and quantification of lysine crotonylation by mass spectrometry have revealed that non‐histone proteins are frequently crotonylated, implicating it in many biological processes through the regulation of chromatin remodelling, metabolism, cell cycle and cellular organization. In this review, we summarize the writers, erasers and readers of lysine crotonylation, and their physiological functions, including gene transcription, acute kidney injury, spermatogenesis, depression, telomere maintenance, HIV latency and cancer process. These findings not only point to the new functions for lysine crotonylation, but also highlight the mechanisms by which crotonylation regulates various cellular processes.
Keywords: crotonylation, HCT, HDCR, PTM, reader
1. INTRODUCTION
Protein post‐translational modifications (PTMs), in which amino acid residues in a protein are covalently modified, have been increasingly recognized to play important roles in various biological pathways.1, 2 With the application of mass spectrometry‐based proteomics, novel histone PTMs have been documented, ranging from small chemical modifications (eg acetylation and phosphorylation) to the addition of complete proteins (eg ubiquitylation).3, 4 Post‐translational modification of proteins occurs in all living organisms. It governs many important cellular processes including the activation of enzymes, protein localization and protein degradation.5
Lysine is an amphipathic residue with a hydrophobic side chain. Acylation of lysine neutralizes the positive charge of the amino group and may change the conformation of proteins.6 According to the difference in hydrocarbon chain length, hydrophobicity and charge, the short‐chain lysine acylations include the well‐studied lysine acetylation and propionylation, butyrylation, 2‐hydroxyisobutyrylation, succinylation, malonylation, glutarylation, crotonylation and β‐hydroxybutyrylation.7, 8, 9, 10, 11, 12
Lysine crotonylation is a newly discovered histone PTM, which is specifically enriched at active gene promoters and potential enhancers in mammalian cell genomes.11 Crotonylation can be catalysed reversibly by protein crotonyltransferases and decrotonylases. The crotonylation of lysine was first identified on histones.11 Afterwards, more eukaryotic non‐histone proteins were identified as being crotonylated, and they were involved in cellular metabolism, cell cycle and cellular organization process.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 In this review, we concentrate mainly on recent studies about lysine crotonylation and discuss its implications.
2. THE DISCOVERY OF LYSINE CROTONYLATION
Tan et al11 first reported protein lysine crotonylation in 2011. They used an integrated, mass spectrometry‐based proteomics approach, which takes advantage of in vitro propionylation, efficient peptide separation using isoelectric focusing (OFFGEL) and the high sensitivity of the LTQ Orbitrap Velos mass spectrometer to carry out a comprehensive analysis of histone PTMs. With this approach, they identified lysine crotonylation as a novel histone mark type. A total of 28 human histone peptides were found to have lysine crotonylation in this study (Figure 1). In addition, they also generated a specific anti‐crotonyl‐lysine antibody and applied it in corroborating Western blot and immunostaining experiments. Interestingly, they confirmed the existence of 19 crotonylation marks in HeLa cells using isotopic labelling with D4‐crotonate. The crotonyl group contains a unique C‐C π‐bond, which results in a rigid and planar configuration. Following the initial discovery of lysine crotonylation, the landscape of these modifications is rapidly expanding.
Figure 1.
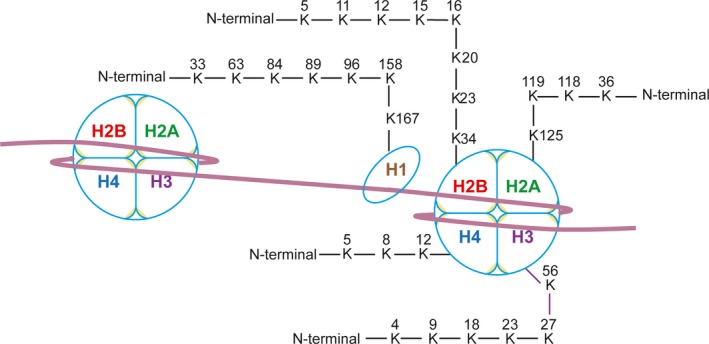
Illustrations of histone crotonylation sites in human. All reported lysine (K) crotonylation sites on histone H1, H2A, H2B, H3 and H4 are shown in different colours
3. PROTEIN CROTONYLOME
Although initially identified on histones, lysine crotonylation has expanded to large number of non‐histone proteins. To characterize the global crotonylation proteome, the proteomic method based on sensitive immune‐affinity purification and high‐resolution liquid chromatography‐tandem (LC‐MS/MS) was applied to identify new crotonylated proteins and modification sites. The utilization of antibodies with high specificity to the crotonylated peptides involved in immunoprecipitation significantly improved the ability to enrich and identify crotonylated lysine residues. In recent years, several landmark studies have revealed dramatically improved number of crotonylated lysine residues and crotonylated proteins (Table 1), and these proteins play roles in nearly all cellular processes including chromatin remodelling, metabolism, cell cycle and cellular organization.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Crotonylome mapping provides an important resource for discovering novel properties and regulatory functions of crotonylation.
Table 1.
Crotonylome studies discussed in this review
| Species | Crotonylated peptides | Crotonylated proteins | Year | Refs |
|---|---|---|---|---|
| Human H1299 cells | 2696 | 1024 | 2017 | 13 |
| Human HeLa cells | 558 | 453 | 2017 | 14 |
| Human A549 cells | 7765 | 2034 | 2017 | 15 |
| Human HCT116 cells | 816 | 392 | 2018 | 16 |
| Human peripheral blood | 1109 | 347 | 2018 | 17 |
| Nicotiana tabacum | 2044 | 637 | 2017 | 18 |
| Zebrafish | 557 | 218 | 2018 | 19 |
| Rice | 1265 | 690 | 2018 | 20 |
| Rhodotorula mucilaginosa | 1691 | 629 | 2018 | 21 |
| Papaya fruit | 5995 | 2120 | 2018 | 22 |
4. WRITERS OF LYSINE CROTONYLATION
Lysine crotonylation is enzymatically regulated by the dynamic balance between crotonyltransferases and decrotonylases.23 The crotonyltransferases were colloquially termed writers, which catalyse the covalent modification of lysine crotonylation. The histone acetyltransferases (HATs) were also shown to have histone crotonyltransferase (HCT) activities (Figure 2; Table 2). There are three major HATs families, which can be categorized into three major families by their sequence and structural features: p300/CBP (CREB‐binding protein), GNAT (Gcn5‐related N‐acetyltransferases) and MYST (MOZ, Ybf2/Sas3, Sas2 and Tip60) families.24 The p300/CBP and MYST families are identified only in eukaryotic cells, while the GNAT family is present and conserved in all domains of life.
Figure 2.
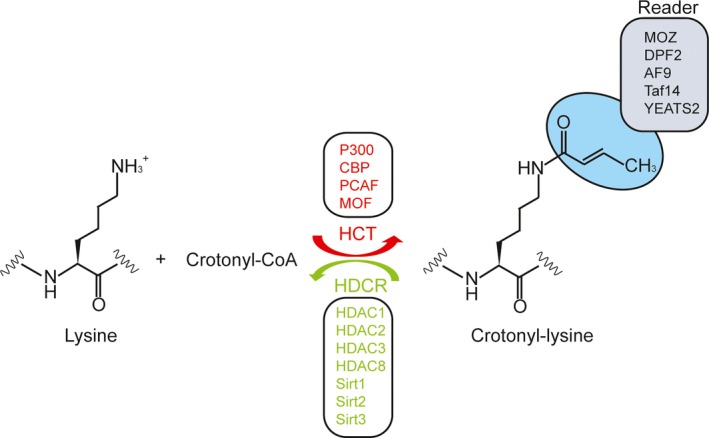
Protein crotonylation is balanced by HCT and HDCR, and recruits Readers. Protein crotonylation can be enzymatically catalysed by lysine crotonyltransferase (HCT) and removed by decrotonylase (HDCR). Crotonylations can also act as docking marks to recruit downstream readers
Table 2.
Writers, erasers and readers discussed in this review
Sabari et al25 first reported that p300 has both HAT and HCT activities. In the cell‐free assays, they demonstrated that p300‐catalysed histone crotonylation directly stimulated transcription to a greater degree than p300‐catalysed histone acetylation. Besides, the level of histone crotonylation was sensitive to changes in the cellular concentration of crotonyl‐CoA, due to p300's dual enzymatic activities. Afterwards, MOF was also identified to catalyse histone crotonylation, including histone H3 at lysine residues 4, 9, 18 and 23, and histone H4 at lysine 8 and 12.26 For the first time, Xu et al13 identified HAT can catalyse crotonylation on non‐histone proteins. In their study, they found non‐histone protein, NPM1, is strongly crotonylated by CBP and MOF, and moderately crotonylated by PCAF. However, the crotonylation of DEAD‐box RNA helicase, DDX5, can only be catalysed by CBP.
5. ERASERS OF LYSINE CROTONYLATION
Histone deacetylases (HDACs) were also reported to have histone decrotonylase (HDCR) activity (Figure 2; Table 2). The decrotonylases were colloquially termed erasers, which could remove the covalent modification of lysine crotonylation. Currently, two major families of KDACs have been divided into four classes: the NAD‐dependent sirtuin family (class III Sirt1‐7) and the zinc‐dependent Rpd3/Hda1 family (classes I [HDAC1, 2, 3 and 8], II [HDAC4, 5, 6, 7, 9 and 10] and IV [HDAC11]).27
HDAC3 was firstly reported to exhibit HDCR activity in vitro. By utilizing a collection of fluorogenic substrates, HDAC3‐NCoR1 was exhibited decrotonylase activity with a catalytic efficiency that is comparable to the deacetylase activity of other KDAC isoforms.28 After analysing the deacylase activity of Sirt1 through SIRT7 using catalytic amounts of enzyme and different expansive panel of acylated H3K9 peptides, Sirt1 and Sirt2 were also indicated act as efficient decrotonylases.29 With the application of an optimized CLASPI approach to comprehensively profile ‘eraser’ enzymes, Bao et al30 identified human Sirt1, Sirt2 and Sirt3 as decrotonylases in vitro, and using X‐ray crystallography, they examined the molecular basis of how the enzymes recognize crotonylated group. Afterwards, class I HDACs and Sirt1 were proved as active HDCR enzymes,31 by screening ectopically expressed HDAC and Sirt family proteins in HeLa cells, with immunofluorescence staining using an anti‐crotonyl‐lysine specific antibody. Kelly et al32 found that knock‐down HDAC1/2 in embryonic stem (ES) cells increased global histone crotonylation levels and caused a great reduction in total decrotonylase activity. Xu et al13 found that HDAC1 and HDAC3 decrotonylate NPM1, which can be reversed upon TSA treatment.
6. READERS OF LYSINE CROTONYLATION
Double PHD finger (DPF), bromodomain and YEATS are three major classes of acetylation and non‐acetyl acylation readers.6 Histone crotonylations can act as docking marks to recruit downstream readers (Figure 2; Table 2). Using ChIP‐qPCR and immunofluorescence assays, Xiong et al33 suggested the colocalization of MOZ and H3K14 crotonylation, and they also indicated that DPF domains of MOZ and DPF2 maintained high affinity for crotonylation at H3K14. A recent study reported that the binding affinity between AF9 YEATS domain and crotonyl‐lysine was higher than that to acetyl‐lysine.34 Furthermore, using heteronuclear NMR spectroscopy, they determined a 3D solution structure of the AF9 YEATS domain bound to an H3 crotonylated at Lys 18 (H3K18cr) peptide. Afterwards, Taf14 YEATS domain was identified as a reader of crotonyl‐lysine binding to histone H3 crotonylated at Lys 9 (H3K9cr) via a unique π‐π‐π stacking mechanism.35 Using a modified histone peptide array, Zhao et al36 found that YEATS2 bound to acylated histone peptides with the best preference for histone H3 crotonylated at Lys 27 (H3K27cr) peptide.
7. FUNCTIONAL ROLES OF LYSINE CROTONYLATION
7.1. Regulation of gene transcription
Tan et al11 firstly revealed the function of histone crotonylation in regulating gene transcription. They found histone crotonylation specifically labelled the enhancers and transcription starting site of active genes in human somatic and mouse male germ cell genomes. In addition, histone crotonylation was enriched on sex chromosomes and specifically marked X‐linked genes that escaped sex chromosome inactivation in male germinal cells immediately following meiosis. A recent study reported p300‐mediated histone crotonylation stimulates gene transcription to a greater degree than histone acetylation.37 With a cell‐based model of transcriptional activation, they also suggested different level of cellular crotonyl‐CoA concentration led the changes in histone crotonylation levels flanking the regulatory elements of activated genes and was correlated with gene expression. By using the novel CBP/p300 mutants (p300 I1395G and CBP I1432G) with deficient histone acetyltransferase but competent HCT activity, Liu et al26 demonstrated CBP/p300 can promote transcriptional activation in the absence of HAT and presence of HCT activity. Afterwards, their team colleagues found selective HDCR in mammalian cells correlates with a broad transcriptional repression, by using novel HDAC1 and HDAC3 mutants (HDAC1/3‐VRPP) with impaired HDAC but intact HDCR activity.31
7.2. Regulation of acute kidney injury
Acute kidney injury (AKI) is a potentially lethal condition with no available therapy beyond replacement of renal function.38 Ruiz‐Andres et al39 studied histone crotonylation level of acute kidney injury induced by folic acid or cisplatin, in murine proximal tubular cells and kidneys from mice. Furthermore, they indicated the histone crotonylation level of kidney tissue was increased during AKI. Beside, ChIP‐seq assays revealed histone crotonylation was enriched at the genes encoding the mitochondrial biogenesis regulator PGC‐1α and the sirtuin‐3 decrotonylase. When the crotonylation level was increased by adding crotonate in cultured tubular cells or in the kidneys in vivo, the expression of PGC‐1α and sirtuin‐3 was increased. In addition, crotonate administration protected from experimental AKI and prevented the decrease in renal function. For the first time, they suggested increased histone crotonylation level might have a beneficial effect on AKI.
7.3. Regulation of spermatogenesis
Genome‐wide removal of histones from chromatin and their replacement is a unique epigenetic event during spermatogenesis.40 Liu et al41 showed that chromodomain Y‐like transcription corepressor CDYL acted as a crotonyl‐CoA hydratase to convert crotonyl‐CoA to β‐hydroxybutyryl‐CoA, as a result destroying crotonyl‐CoA for histone crotonylation reaction. They also indicated histone crotonylation was associated with the reactivation of the sex chromosome‐linked genes of round spermatids. Using ChIP‐quantitative PCR, they suggested histone crotonylation levels in round spermatids cells were significantly higher than those in spermatocytes cells, indicating histone crotonylation levels during spermatogenesis are regulated by various complex mechanisms including both enzymatic and nonenzymatic reactions.
7.4. Regulation of depression
Histone crotonylation was found decreased in the medial prefrontal cortex of susceptible rodents exposed to chronic social defeat stress.42 In addition, when knocking down CDYL in prelimbic cortex, the histone crotonylation was increased. Using ChIP‐quantitative PCR, this finding also demonstrated that CDYL inhibits VGF transcription by its dual influence on histone crotonylation and H3K27me3 of VGF promoter. For the first time, the relationship between crotonylation and depression was identified, and CDYL‐mediated histone crotonylation may play an important role in regulating stress‐induced depressive behaviours.
7.5. Regulation of telomere maintenance
Telomere elongation with increasing passage depends on the mechanisms of both telomerase and recombination‐based alternative lengthening of telomeres.43 When crotonylation induced by crotonic acid, Zscan4 was found activated, and T‐SCE level increased, which maintained telomeres and reduced telomere damage during chemical induction.44 In addition, chemically induced pluripotent stem cells' (CiPSC) clone formation at stage II during middle induction was improved by crotonylation, indicating that extraembryonic endoderm state is primed for action by crotonic acid.
7.6. Regulation of HIV latency
Latent HIV reservoirs in the host are established early before viral infection.45, 46 ACSS2, the crotonyl‐CoA‐producing enzyme acyl‐CoA synthetase short‐chain family member 2, was identified to influence HIV replication and viral latency by regulating histone crotonylation at HIV long‐terminal repeat.47 After histone crotonylation was induced by ACSS2 in vitro and ex vivo, HIV latency was disrupted. Furthermore, when inhibiting histone crotonylation with suppressing ACSS2, latent HIV reactivation was dampened, suggesting a potential role of histone decrotonylation in HIV latency establishment. For the first time, this finding linked maintenance of viral latency to histone crotonylation by ACSS2 at the HIV long‐terminal repeat.
7.7. Regulation of cancer
Cancer is a life‐threatening malignancy that has become a global healthcare problem.48, 49, 50 Using immunohistochemical methods, we recently reported that levels of crotonylation in eight types of cancer.51 We found the expression level of crotonylation was down‐regulated in liver, stomach and kidney carcinomas, and up‐regulated in thyroid, oesophagus, colon, pancreas and lung carcinomas, suggesting that crotonylation may modulate different cancer progression. Besides, lysine crotonylation was identified involving in hepatoma cell motility and proliferation. For the first time, we indicated that status of lysine crotonylation may represent an important type of post‐translational modifications accounting for cancer progression.
8. CONCLUSIONS
Lysine crotonylation is recently identified as a novel evolutionarily conserved histone PTM.11 The recent discovery of a large panel of new histone PTMs, including lysine crotonylation, may change our vision of epigenome.6, 52 Lysine crotonylation is involved in many pathways that regulate diverse cellular functions ranging from gene expression to telomere maintenance (Table 3). It would be interesting in future to determine how their functions are mechanistically regulated by crotonylation.
Table 3.
Functional roles of lysine crotonylation discussed in this review
| Functional roles of lysine crotonylation | In vivo, ex vivo or cell culture studies | Descriptive or interventional studies | The model studied | Year | Refs |
|---|---|---|---|---|---|
| Regulation of gene transcription | In vivo | Descriptive study | Meiotic and postmeiotic male germ cells and tissues | 2011 | 11 |
| Cell culture | Interventional study | 293T cells | 2017 | 26 | |
| Cell culture | Interventional study | HeLa cells | 2017 | 31 | |
| Ex vivo and cell culture | Interventional study | Cell‐free system and LPS induced inflammatory response in the macrophage cell line RAW 264.7 | 2018 | 37 | |
| Regulation of acute kidney injury | In vivo and cell culture | Interventional study | Cultured murine proximal tubular cells and kidneys from mice with AKI | 2016 | 39 |
| Regulation of spermatogenesis | In vivo | Descriptive study | Mouse testes | 2017 | 41 |
| Regulation of depression | In vivo | Interventional study | Mice model | 2018 | 42 |
| Regulation of telomere maintenance | Cell culture | Interventional study | MEF cells | 2018 | 44 |
| Regulation of HIV latency | Cell culture | Interventional study | J‐Lat A1 and CD4+ T cells | 2018 | 47 |
| Regulation of cancer | In vivo and cell culture | Interventional study | HCC tissues and human hepatoma‐derived cell | 2019 | 51 |
The developments in high‐resolution LC‐MS/MS approaches have enabled the crotonylome‐wide mapping of lysine crotonylation.4 Thus, future crotonylome studies should be well designed. Identification of crotonylation on non‐histone proteins could expand our understanding of how non‐histone proteins are involved in diverse cellular functions and signalling pathways, and may facilitate the clarification of the precise modulation on protein functions.
In general, one protein may occur simultaneously multiple PTMs.53 Indeed, recent years gradually intensifying research on crotonylation and many publications were dedicated to only one of the many possible lysine crotonylations. Even for crotonylation, most crotonylated proteins have multiple crotonylated lysine residues. Therefore, the crosstalk between different PTMs and their contributions in regulating protein function should be investigated. Human disease in which multiple factors may be involved, is a complex phenotype and cannot be easily interpreted by analysing a single type PTM.54 Whether different PTMs could be functionally grouped or specific signalling to chromatin depends on each PTMs. The hypothesis would imply that diverse PTMs may cooperate with each other to provide a wealth of regulatory potential.
Over the past years, we have witnessed the tremendous advances in understanding of the mechanisms and cellular functions on protein crotonylation. However, it still exists many limitations in the published studies. Despite a growing understanding of crotonylation function in regulating diverse physiological function, it remains unclear how such biochemical changes occur and whether they play crucial roles in more disease progression. Besides, current crotonylation studies have mainly focused on a small number of histones, limiting the ability to clarify the relationship between crotonylation and human physiological processes. Moreover, the number of writers, erasers and readers of lysine crotonylation found so far is still very limited, more studies on the discovery and identification of writers, erasers and readers will be helpful for understanding the function of crotonylation.
In the further research, we should focus on how human physiological processes and related diseases mechanistically regulated by crotonylation. Beside, more studies should search for understanding the role of crotonylation in non‐histones. We can also discovery more sites about lysine crotonylation, including the mechanisms by which they occur. Therefore, future studies are needed to uncover the effects of crotonylation on regulating protein functions and to interpret the underlying mechanisms behind protein crotonylation's ability to modulate diverse physiological and pathological processes.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
AUTHOR CONTRIBUTIONS
JHW and HQZ provided direction and guidance throughout the preparation of this manuscript. JHW draw the graph. HYL and JC collected and prepared the related literature. All authors have read and approved the final manuscript.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Natural Science Foundation of China grants (Grant No. 81702780, 81730071 and 81621063). This work was also supported by grants from the Ministry of Science and Technology of China (2016YFC1302103 and 2015CB553906), the Beijing Natural Science Foundation 7171005 and the Youth Innovation Fund of the First Affiliated Hospital of Zhengzhou University. We also thank for the support from Academy of Medical Sciences of Zhengzhou University Translational Medicine platform.
Wan J, Liu H, Chu J, Zhang H. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019;23:7163–7169. 10.1111/jcmm.14650
Junhu Wan and Hongyang Liu contributed equally to this work.
DATA AVAILABILITY STATEMENT
Data will be available upon reasonable request.
REFERENCES
- 1. Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258‐264. [DOI] [PubMed] [Google Scholar]
- 2. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693‐705. [DOI] [PubMed] [Google Scholar]
- 3. Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458‐458.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu J, Wu M, Liu X. Proteomic approaches beyond expression profiling and PTM analysis. Anal Bioanal Chem. 2018;410:4051‐4060. [DOI] [PubMed] [Google Scholar]
- 5. Dutta A, Abmayr SM, Workman JL. Diverse activities of histone acylations connect metabolism to chromatin function. Mol Cell. 2016;63:547‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2017;18:90‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai L, Peng C, Montellier E, et al. Lysine 2‐hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol. 2014;10:365‐370. [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, Sprung R, Tang YI, et al. Lysine propionylation and butyrylation are novel post‐translational modifications in histones. Mol Cell Proteomics. 2007;6:812‐819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie Z, Dai J, Dai L, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan M, Peng C, Anderson K, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605‐617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie Z, Zhang D, Chung D, et al. Metabolic regulation of gene expression by histone lysine beta‐hydroxybutyrylation. Mol Cell. 2016;62:194‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu W, Wan J, Zhan J, et al. Global profiling of crotonylation on non‐histone proteins. Cell Res. 2017;27:946‐949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei W, Mao A, Tang B, et al. Large‐scale identification of protein crotonylation reveals its role in multiple cellular functions. J Proteome Res. 2017;16:1743‐1752. [DOI] [PubMed] [Google Scholar]
- 15. Wu Q, Li W, Wang C, et al. Ultradeep lysine crotonylome reveals the crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J Proteome Res. 2017;16:3664‐3671. [DOI] [PubMed] [Google Scholar]
- 16. Huang H, Wang DL, Zhao Y. Quantitative crotonylome analysis expands the roles of p300 in the regulation of lysine crotonylation pathway. Proteomics. 2018;18:e1700230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen W, Tang D, Xu Y, et al. Comprehensive analysis of lysine crotonylation in proteome of maintenance hemodialysis patients. Medicine. 2018;97:e12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun H, Liu X, Li F, et al. First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci Rep. 2017;7:3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwon OK, Kim SJ, Lee S. First profiling of lysine crotonylation of myofilament proteins and ribosomal proteins in zebrafish embryos. Sci Rep. 2018;8:3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu S, Xue C, Fang Y, et al. Global involvement of lysine crotonylation in protein modification and transcription regulation in rice. Mol Cell Proteomics. 2018;17:1922‐1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Q, Li Y, Apaliya MT, et al. The response of Rhodotorula mucilaginosa to patulin based on lysine crotonylation. Front Microbiol. 2018;9:2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu K, Yuan C, Li H, et al. A qualitative proteome‐wide lysine crotonylation profiling of papaya (Carica papaya L.). Sci Rep. 2018;8:8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao S, Zhang X, Li H. Beyond histone acetylation‐writing and erasing histone acylations. Curr Opin Struct Biol. 2018;53:169‐177. [DOI] [PubMed] [Google Scholar]
- 24. Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non‐histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20:156‐174. [DOI] [PubMed] [Google Scholar]
- 25. Sabari B, Tang Z, Huang HE, et al. Intracellular crotonyl‐CoA stimulates transcription through p300‐catalyzed histone crotonylation. Mol Cell. 2015;58:203‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X, Wei W, Liu Y, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase‐deficient and crotonyltransferase‐competent CBP/p300. Cell Discov. 2017;3:17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren J, Sang Y, Lu J, Yao YF. Protein acetylation and its role in bacterial virulence. Trends Microbiol. 2017;25:768‐779. [DOI] [PubMed] [Google Scholar]
- 28. Madsen AS, Olsen CA. Profiling of substrates for zinc‐dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew Chem Int Ed Engl. 2012;51:9083‐9087. [DOI] [PubMed] [Google Scholar]
- 29. Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long‐chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350‐31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bao X, Wang YI, Li X, et al. Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3:e02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei W, Liu X, Chen J, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898‐915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelly R, Chandru A, Watson PJ, et al. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Scientific Reports. 2018;8:14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiong X, Panchenko T, Yang S, et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12:1111‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Q, Zeng L, Zhao C, Ju Y, Konuma T, Zhou MM. Structural Insights into histone crotonyl‐lysine recognition by the AF9 YEATS domain. Structure. 2016;24:1606‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andrews FH, Shinsky SA, Shanle EK, et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12:396‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao D, Guan H, Zhao S, et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016;26:629‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabari BR, Tang Z, Huang HE, et al. Intracellular Crotonyl‐CoA stimulates transcription through p300‐catalyzed histone crotonylation. Mol Cell. 2018;69:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berger K, Moeller MJ. Mechanisms of epithelial repair and regeneration after acute kidney injury. Semin Nephrol. 2014;34:394‐403. [DOI] [PubMed] [Google Scholar]
- 39. Ruiz‐Andres O, Sanchez‐Niño MD, Cannata‐Ortiz P, et al. Histone lysine crotonylation during acute kidney injury in mice. Dis Model Mech. 2016;9:633‐645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483‐488. [DOI] [PubMed] [Google Scholar]
- 41. Liu S, Yu H, Liu Y, et al. Chromodomain protein CDYL acts as a crotonyl‐CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol Cell. 2017;67(853–66):e5. [DOI] [PubMed] [Google Scholar]
- 42. Liu Y, Li M, Fan M, et al. Chromodomain Y‐like protein‐mediated histone crotonylation regulates stress‐induced depressive behaviors. Biol Psychiat. 2019;85:635‐649. [DOI] [PubMed] [Google Scholar]
- 43. Liu L. Linking telomere regulation to stem cell pluripotency. Trends Genet. 2017;33:16‐33. [DOI] [PubMed] [Google Scholar]
- 44. Fu H, Tian C‐L, Ye X, et al. Dynamics of telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Reports. 2018;11:70‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dandekar S. Pathogenesis of HIV in the gastrointestinal tract. Current HIV/AIDS Reports. 2007;4:10‐15. [DOI] [PubMed] [Google Scholar]
- 46. Somsouk MA, Estes JD, Deleage C, et al. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS. 2015;29:43‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang G, Nguyen D, Archin NM, et al. HIV latency is reversed by ACSS2‐driven histone crotonylation. J Clin Invest. 2018;128:1190‐1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wan J, Liu H, Yang L, Ma L, Liu J, Ming L. JMJD6 promotes hepatocellular carcinoma carcinogenesis by targeting CDK4. Int J Cancer. 2019;144:2489-2500. [DOI] [PubMed] [Google Scholar]
- 49. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 50. Wan J, Liu H, Feng Q, Liu J, Ming L. HOXB9 promotes endometrial cancer progression by targeting E2F3. Cell Death Dis. 2018;9:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976‐982. [DOI] [PubMed] [Google Scholar]
- 52. Olsen CA. Expansion of the lysine acylation landscape. Angew Chem Int Ed Engl. 2012;51:3755‐3756. [DOI] [PubMed] [Google Scholar]
- 53. Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Millar AH, Heazlewood JL, Giglione C, Holdsworth MJ, Bachmair A, Schulze WX. The scope, functions, and dynamics of posttranslational protein modifications. Annu Rev Plant Biol. 2019;70(1):119‐151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon reasonable request.


