Abstract
BACKGROUND
Circular RNAs (circRNAs) are a newly discovered class of endogenous non-coding RNAs that may have roles in cancer genesis and development. In the recent literature, dysregulated circRNAs have been extensively investigated in hepatocellular carcinoma (HCC). Whether or not circRNAs are of clinical value for the management of HCC has not been characterized.
AIM
To meta-analyze the diagnostic and prognostic value of abnormally expressed circRNAs in HCC.
METHODS
Eligible studies were sourced from PubMed, EMBASE, and CNKI online databases. Data on patients’ clinical characteristics, including diagnostic efficacy and overall survival, were extracted. The diagnostic and prognostic parameters were respectively synthesized using the bivariate meta-analysis model and multivariate Cox hazard regression analysis based on Stata 12.0. The trim and fill method was adopted to assess the possible effects from publication bias.
RESULTS
A total of 21 eligible studies were included. The pooled sensitivity, specificity, and area under the curve of abnormally expressed circRNAs in distinguishing HCC from non-cancer controls were 0.78 (95%CI: 0.69–0.85), 0.80 (95%CI: 0.74–0.86), and 0.86, respectively. Survival analyses showed that the down-regulated circRNA expression signature correlated perfectly with HCC survival [hazard ratio (HR) = 0.42, 95%CI: 0.19–0.91, P = 0.028; I2 = 92.7%, P = 0.000], whereas the HCC cases with high circRNA levels had significantly poorer prognoses than those of patients with low circRNA levels (HR = 2.22, 95%CI: 1.50–3.30, P = 0.000; I2 = 91%, P = 0.000). Moreover, abnormally expressed circRNAs were intimately associated with tumor size, differentiation grade, microvascular invasion, metastasis, TNM stage, and serum alpha fetal protein level in patients with HCC. Stratified analysis based on sample type, control source, and expression status also yielded robust results.
CONCLUSION
Abnormally expressed circRNA signatures show immense potential as novel non-invasive biomarker(s) for HCC diagnosis and prognosis.
Keywords: CircRNA, Hepatocellular carcinoma, Diagnosis, Prognosis, Meta-analysis
Core tip: Current studies investigating the clinical significance of circular RNAs (circRNAs) in hepatocellular carcinoma (HCC) were conducted using single-center and small-scale design, and the findings were controversial. We collected and analyzed the up-to-date clinical data on the significance of circRNAs in the diagnosis and prognosis of HCC. The results indicated that circRNAs may be novel indicators for the prognosis and diagnosis of HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC), one of the most common cancers of the digestive system, remains one of the leading causes of cancer deaths worldwide[1]. In China, the incidence of HCC was shown to have increased remarkably over the past decades, which has resulted in great health and economic burdens[2]. Although the technological advances for HCC treatment in recent years have vastly improved the clinical outcomes of patients with HCC, the 5-year survival rate is very low[3]. Particularly for patients with advanced HCC, the median survival time was shown to be only 3–9 mo[4]. The sensitivity and specificity of the currently used blood biomarkers such as alpha fetoprotein (AFP) and protein induced by vitamin K absence or antagonist-II (PIVKA-II) are not satisfactory for HCC detection[5]. For prognosis monitoring, no biomarkers were well developed. Therefore, it is necessary to investigate novel effective biomarkers for HCC.
Non-coding RNAs play important roles in cancer biology, providing potential targets for cancer intervention. As a new class of endogenous noncoding RNAs, circular RNAs (circRNAs) are a series of functional non-coding transcripts generated from the backsplicing of exons, introns, or both[6]. Unlike linear RNAs, circRNAs form covalently closed continuous loop structures, characterized by stability, abundance, and specific expression in different tissues and cells during development[6,7]. CircRNAs act as key regulators in a wide range of biological processes, including the initiation and progression of several types of cancer[8,9]. CircRNAs are aberrantly expressed in cancer tissues, especially in HCC, suggesting that these molecules could be novel biomarkers for HCC diagnosis and prognosis[10-30]. Whether or not circRNAs are of clinical value for the diagnosis of HCC must be clarified. Herein, we conducted this meta-analysis, aiming to assess the diagnostic and prognostic utility of circRNA expression signature in HCC.
MATERIALS AND METHODS
Study selection
The international online databases including PubMed, EMBASE, EBSCO, Biomed central, and CNKI were searched for eligible studies indexed until May 1, 2018. The searching items were: (“liver cancer” or “liver neoplasms [MeSH Terms]” or “hepatocellular carcinomas [MeSH Terms]”) and (“circular RNA [MeSH Terms]” or “circRNA” or “hsa circ”) and (“prognosis” or “prognoses [MeSH Terms]” or “prognostic factors [MeSH Terms]” “HR” or “hazard ratio” or “overall survival” or “OS” or “survival [MeSH Terms]” or “disease-free survival” or “DFS” or “EFS” or “event-free survival” or “progression-free survival” or “PFS”) or (“diagnosis [MeSH Terms]” or “diagnoses [MeSH Terms]” or “sensitivity and specificity [MeSH Terms]” or “ROC” or “ROC curve [MeSH Terms]” or “AUC”). The attached reference list of literature was also manually searched to increase the search sensitivity.
Selection criteria
Studies were selected in compliance with the following criteria: (A) Studies were limited to those which evaluated the diagnostic or prognostic or clinicopathological features of circRNA(s) in HCC patients; (B) the true positive, false positive, false negative, and true negative values for diagnosis, or estimated hazard ratio (HR) values with 95%CIs for survival, were either available among studies or could be extracted indirectly; (C) cases were definitely diagnosed with pathological evidence; and (D) the specimens were obtained prior to any radiotherapy or chemotherapy treatments. Irrelevant papers were excluded according to the following criteria: (A) Studies with insufficient data to form the 2 × 2 table for diagnosis, or the HRs with 95%CIs for survival were unavailable; (B) Studies were rated as low quality; and (C) Basic studies, reviews, meta-analyses, comments, letters or case reports, etc. were also excluded.
Data extraction
The baseline contents were collected independently by two trained authors. The information covered included data such as the name of the first author, year of publication, study design, ethnicity, sample size, pathologic data of the study population, circRNA signature, test methods, sensitivity, specificity, cut-off value setting, HR values with 95%CIs for survival, follow-up time, etc. Any disagreements which appeared during data summarization were resolved by group consensus, or the articles’ authors were reached out to.
Study quality grading
Study quality for diagnostic articles was evaluated by the Quality Assessment for Studies of Diagnostic Accuracy II checklist[31]. The tool comprises two domains including “risk of bias” and “applicability concerns”, containing seven questions regarding patient selection, index tests, reference standards, flow, and timing. The answer of risk for bias could be rated as “no” (0 points), “yes” (1 point), or “unclear” (0 points). The study quality for the case-control study was judged in line with the Newcastle-Ottawa Quality Assessment Scale (NOS) checklist[32], in which the assessment focuses on a total of eight items categorized in terms of study selection, comparability, and outcome, with a maximum judgment score of “9”. An answer of “yes” receives a score of “1”; otherwise, no sores were awarded.
Statistical analysis
Statistical analyses were conducted based on the Stata 12.0 program (Stata Corporation, College Station, TX, United States). Heterogeneity among studies was assessed using chi-square and I2 tests. Either P < 0.05 in the chi-square test or I2 > 50% was regarded as significant heterogeneity. The diagnostic parameters were synthesized using the bivariate meta-analysis model, and HRs with 95%CIs were combined using multivariate Cox hazard regression analysis. A random-effects model was chosen when significant heterogeneity appeared in the pooled effect size. Sensitivity analysis was performed to trace the underlying outlier studies included in the pooled effects. The bias due to publication was detected by Deek’s funnel plot and Begg’s and Egger’s tests, and P < 0.05 was set to indicate statistically significant differences. If publication bias appeared, the trim and fill method was adopted to assess the possible effects of bias on the overall pooled effects[33].
RESULTS
Study enrollment
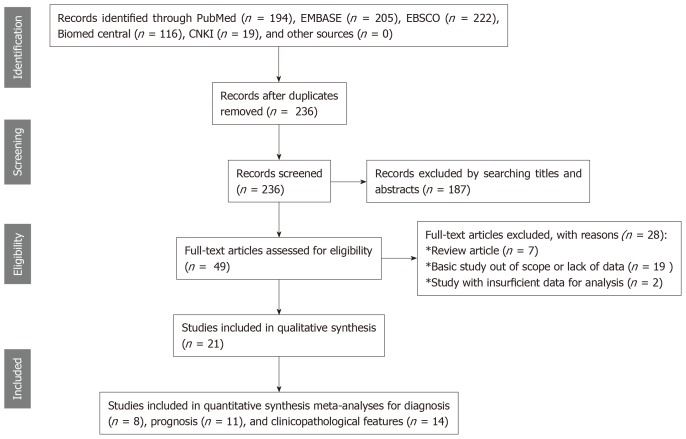
Figure 1 presents the flowchart of the literature search procedure. Searching PubMed, EMBASE, EBSCO, Biomed central, and CNKI databases, as well as other sources, resulted in an initial inclusion of 236 records after duplicates were removed. Two authors independently screened the titles and abstracts of the 236 publications, and 187 records were excluded because their study contents were unrelated to cirRNAs in HCC. The remaining articles were intensively evaluated for the full-text contents, and 28 of them were evaluated as review articles, or basic studies with irrelevant data, or relevant articles with insufficient information, which therefore were all eliminated. In the final stage, only 21 studies, including 8 publications for diagnosis[11-13,19-21,25,29], 11 for prognosis[10,11,14-18,22,26,28,30], and 14 for clinicopathological feature[11-13,15,17,18,22-24,26-30], were included in the quality assessment and quantitative synthesis.
Figure 1.
Study flow diagram for the diagnostic and prognostic meta-analyses.
Characteristics of included studies
Study characteristics are shown in Tables 1 and 2. All of the included studies were identified as case-control studies, in which eight studies with 712 HCC cases and 788 controls assessed the diagnostic performance of circRNAs in HCC, and eleven studies including 2719 cohorts focused on the evaluation of the prognostic value of cirNRAs. All HCC cases were reliably diagnosed based on histopathological methods. The control sources included chronic hepatitis, liver cirrhosis, para-tumorous tissues[11,20,21,25], and non-cancer/healthy individuals[19,29]. Amplification of circRNAs was enabled by using the qPCR test, and GAPDH or β-actin was used for normalization. The circRNA signatures for diagnosis included hsa_circ_0003570, circZKSCAN1, hsa_circ_0005075, Hsa_circ_0001649, hsa_circ_0091582, hsa_circ_0128298, hsa_circ_0004018, hsa_circ_0001445, and circRNAs panel sets. CircRNA profiles for prognosis contained hsa_circ_0001649, circ-ITCH, circMTO1, cSMARCA5, circC3P1, hsa_circRNA_100338, hsa_circ_0064428, circRNA101368, hsa_circ_0103809, and circ-ZEB1.33.
Table 1.
Characteristics of the included studies for diagnosis and clinicopathologic features
| Study | Ethnicity | Patient size | Control size | Control source | Sample type | CircRNA Name | Expression status | Method | Cut-Off value | Control gene | AUC | QUADAS score |
| Fu et al[12], 2017 | Chinese | 107 | 107 | CH & LC | Tissue | hsa_circ_0003570 | Decreased | qRT-PCR | 12.24 | GAPDH | 0.70 | 4 |
| Yao et al[25], 2017 | Chinese | 102 | 102 | Adjacent non-cancerous liver tissue | Tissue | circZKSCAN1 | Decreased | qRT-PCR | Unclear | GAPDH | 0.834 | 4 |
| Shang et al[21], 2016 | Chinese | 30 | 30 | Adjacent nontumorous tissue | Tissue | hsa_circ_0005075 | Increased | qRT-PCR | 0.000586 | GAPDH | 0.94 | 6 |
| Qin et al[20], 2016 | Chinese | 89 | 89 | Paired adjacent liver tissues | Tissue | Hsa_circ_0001649 | Decreased | qRT-PCR | 0.00079 | β-actin | 0.63 | 6 |
| Chen et al[11], 2018 | Chinese | 30 | 30 | Para-tumorous tissues | Tissue | hsa_circ_0091582 | Increased | qRT-PCR | Unclear | GAPDH | 0.679 | 5 |
| Chen et al[11], 2018 | Chinese | 30 | 30 | Para-tumorous tissues | Tissue | hsa_circ_0128298 | Increased | qRT-PCR | Unclear | GAPDH | 0.664 | 5 |
| Chen et al[11], 2018 | Chinese | 48 | 48 | Para-tumorous tissues | Tissue | hsa_circ_0128298 | Increased | qRT-PCR | Unclear | GAPDH | 0.668 | 5 |
| Huang et al[17], 2017 | Chinese | 102 | 129 | Para-tumorous and CH tissues | Tissue | hsa_circ_0004018 | Decreased | qRT-PCR | 0.531 | GAPDH | 0.848 | 5 |
| Zhang et al[28],2018 | Chinese | 104 | 52 | Healthy control | Plasma | hsa_circ_0001445 | Decreased | qRT-PCR | Unclear | GAPDH | 0.862 | 5 |
| Zhang et al[29], 2018 | Chinese | 104 | 57 | LC | Plasma | hsa_circ_0001445 | Decreased | qRT-PCR | Unclear | GAPDH | 0.672 | 5 |
| Zhang et al[29], 2018 | Chinese | 104 | 44 | CH | Plasma | hsa_circ_0001445 | Decreased | qRT-PCR | Unclear | GAPDH | 0.764 | 5 |
| Han et al[16], 2017 | Chinese | 80 | 80 | Non-cancer tissue | Tissue | CircRNA pattern | / | qRT-PCR | Unclear | Unclear | 0.988 | 3 |
| Han et al[16], 2017 | Chinese | 20 | 20 | Non-cancer tissue | Tissue | CircRNA pattern | / | qRT-PCR | Unclear | Unclear | 0.976 | 3 |
AUC: Area under the curve; circRNA: Circular RNA; CH: Chronic hepatitis; LC: Liver cirrhosis; qRT-PCR: Quantitative real-time PCR; GAPDH: Reduced glyceraldehyde-phosphate dehydrogenase; QUADAS: Quality Assessment for Studies of Diagnostic Accuracy.
Table 2.
Characteristics of the included studies for prognosis and clinicopathologic features
| Study | Locale | Patient size | TNM stage (I/II/III/IV) | Sample type | CircRNA signature | Expression status | Survival indicator | Follow-up time | HR and 95%CI extraction | P-value (survival) | NOS scores |
| Cai et al[10], 2018 | China | 78 | Unclear | Tissue | hsa_circ_0103809 | Increased | OS | Unclear | Indirectly | 0.001 | 6 |
| Zhong et al[30], 2018 | China | 47 | 7, 15, 16, 9 | Tissue | circC3P1 | Decreased | OS | Unclear | Indirectly | 0.030 | 6 |
| Li et al[18], 2018 | China | 51 | I–II: 24, III–IV: 27 | Tissue | circRNA101368 | Increased | OS | Unclear | Directly | 0.001, 0.033 | 7 |
| Weng et al[22], 2018 | China | 120 | I–III: 60, 14, 46 | Tissue | hsa_circ_0064428 | Increased | OS | Unclear | Indirectly | 0.033 | 7 |
| Chen et al[11], 2018 | China | 78 | Unclear | Tissue | hsa_circ_0128298 | Increased | OS | Median: 37 months | Directly | 0.009, 0.014 | 8 |
| Gong et al[14], 2018 | China | 64 | 12, 22, 17, 13 | Tissue | circ-ZEB1.33 | Increased | OS | Unclear | Indirectly | 0.015, 0.019 | 7 |
| Yu et al[26], 2018 | China | 208 | I: 62, II–III: 101 | Tissue | cSMARCA5 | Decreased | OS | Unclear | Directly | 0.001, 0.021 | 7 |
| Huang et al[17], 2017 | China | 80 | I–II: 43, III–IV: 37 | Tissue | hsa_circRNA_100338 | Increased | OS | 5 years | Indirectly | <0.01 | 8 |
| Han et al[16], 2017 | China | 116 | Unclear | Tissue | circMTO1 | Decreased | OS | Unclear | Indirectly | 0.0023 | 7 |
| Guo et al[15], 2017 | China | 1800 | Unclear | Tissue | circ-ITCH | Decreased | OS | Unclear | Directly | <0.001 | 6 |
| Zhang et al[27], 2018 | China | 77 | I–II: 34, III–IV: 43 | Tissue | hsa_circ_0001649 | Decreased | OS | Unclear | Directly | 0.015, 0.011 | 6 |
| Xu et al[23], 2018 | China | 76 | I–II: 23, III–IV: 53 | Tissue | hsa_circ_0001649 | Decreased | / | / | / | / | / |
| Zhang et al[28], 2018 | China | 86 | Early: 38, Late: 48 | Tissue | circsMaD2 | Decreased | / | / | / | / | / |
circRNA: Circular RNA; OS: Overall survival; HR: Hazard ratio; NOS: Newcastle-Ottawa Scale; TNM: Tumor-node-metastasis.
Methodological quality assessment
The quality and bias of all diagnostic studies were independently appraised by two authors in accordance with the QUADAS-II criteria, whereby studies were assessed for patient selection, index test, reference standard, flow, and timing[31]. As reported in Figure 2, all included eight publications for diagnosis were judged as low risk for applicability concerns, and three studies were assessed with bias in patient selection, or index test, or reference standard, and received rated QUADAS scores equal to three points. Evaluation of the quality of all case-control studies was enabled by applying the NOS checklist[32]. As shown in Table 3, all the included prognostic studies received rated NOS scores higher or equal to six, and thus they were all included in the final synthesis.
Figure 2.
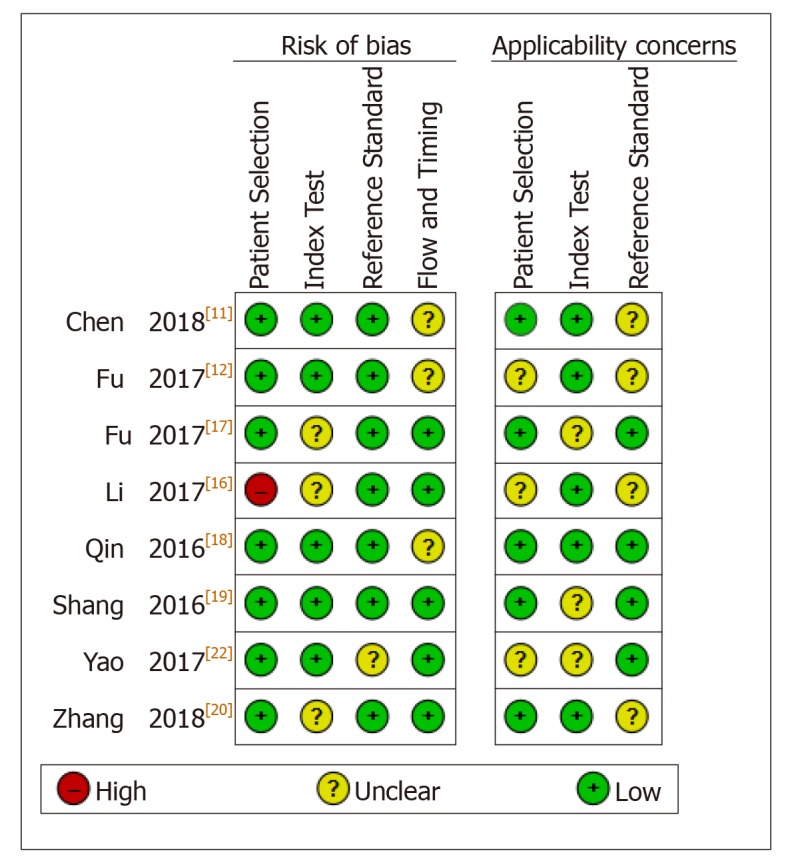
Study quality regarding the risk of bias and applicability concerns as assessed by the QUADAS II tool.
Table 3.
Study quality and bias in the retrospective cohort studies assessed via the Newcastle-Ottawa Scale checklist
| Study |
Cohort selection |
Comparability |
Outcome ascertainment |
|||||
| Representativeness of the exposed cohort | Selection of the non-exposed cohort | Ascertain-ment of exposure | Demonstration that outcome of interest was not present at start of study | Comparability of cases and controls on the basis of the design or analysis | Assessment of outcome | Was follow-up long enough for outcomes to occur | Adequacy of follow up of cohorts | |
| Cai et al[10], 2018 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Zhong et al[30], 2018 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Li et al[18], 2018 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 |
| Weng et al[22], 2018 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 |
| Chen et al[11], 2018 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Gong et al[14], 2018 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 |
| Yu et al[26], 2018 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 |
| Huang et al[17], 2017 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Han et al[16], 2017 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 0 |
| Guo et al[15], 2017 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Zhang et al[27], 2018 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
Investigations of heterogeneity
In the overall diagnostic meta-analysis, the chi-square and I2 tests revealed significant substantial heterogeneity among pooled effects (Q = 49.403, df = 2.00, P = 0.000; I2 = 95.95, 95%CI: 92.85–99.05). In line with the diagnostic effects, clear heterogeneity was also observed in the pooled prognostic effects for both the elevated (P = 0.000; I2 = 91%) and down-regulated circRNA profiles (P = 0.000; I2 = 92.7%). Thus, all weights were synthesized using a random-effects model.
Overall diagnostic performance
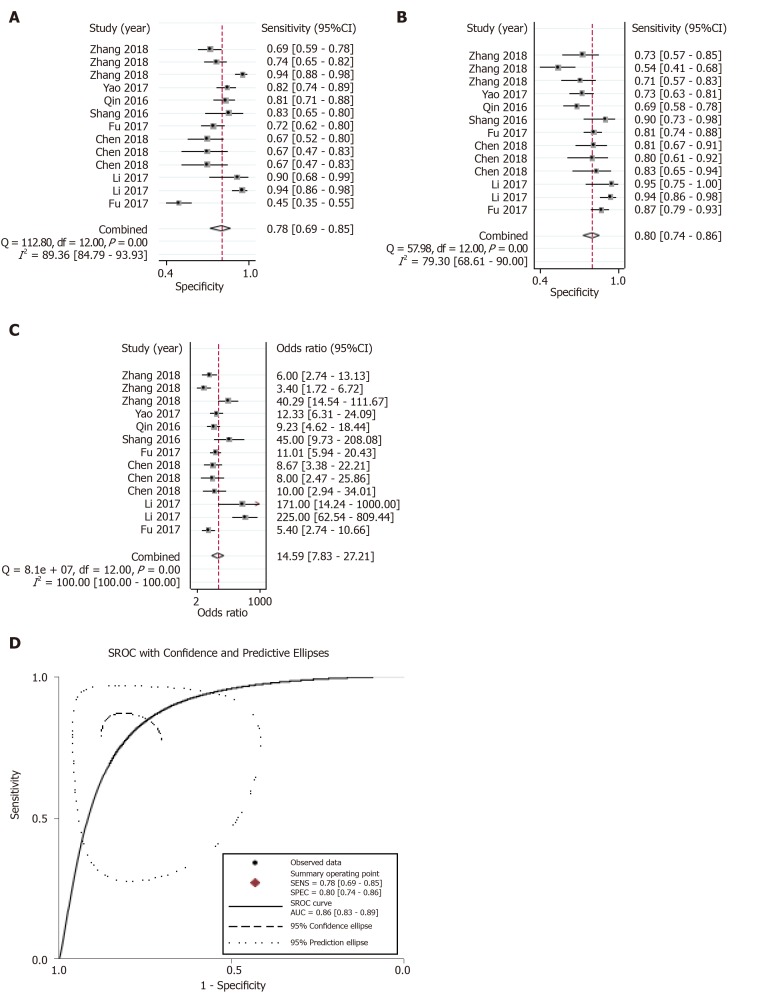
The summary receiver operating characteristic curve was employed to assess the diagnostic efficacy of circRNA profiles in distinguishing HCC from non-tumorous controls. The pooled sensitivity (Figure 3A), specificity (Figure 3B), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) (Figure 3C), and area under the curve (AUC) (Figure 3D) were estimated to be 0.78 (95%CI: 0.69–0.85), 0.80 (95%CI: 0.74–0.86), 3.97 (95%CI: 2.85–5.54), 0.27 (95%CI: 0.19–0.39), 14.59 (95%CI: 7.83–27.21), and 0.86, respectively.
Figure 3.
Overall diagnostic performance. A-D: Forest plots of the combined (A) sensitivity, (B) specificity, (C) diagnostic odds ratio, and (D) area under the curves among the eight diagnostic studies.
Prognostic value
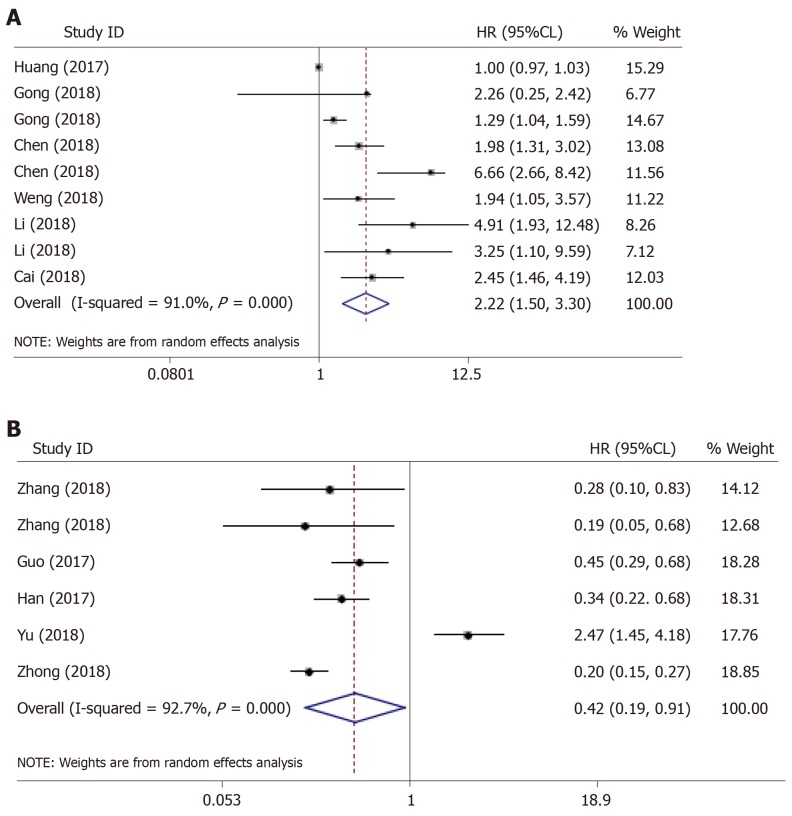
We found distinct prognostic value of the abnormally expressed circRNA signature in HCC, wherein the signature covered up-regulated circRNAs and was negatively correlated with the overall survival (OS) of patients with HCC (HR = 2.22, 95%CI: 1.50–3.30, P = 0.000) (Figure 4A), hinting that these circRNAs could be considered as independent prognostic biomarkers in HCC. Meanwhile, the significantly higher survival time (OS) was found in HCC patients with down-regulated circRNA profile (HR = 0.42, 95%CI: 0.19–0.91, P = 0.028) (Figure 4B), suggesting that circRNAs with decreased expression status were more prone to act as tumor suppressor genes in HCC.
Figure 4.
The outlier in the pooled prognostic effects of down-regulated circular RNAs in hepatocellular carcinoma. A and B: Forest plots of the combined hazard ratios with 95% confidence intervals for the (A) up-regulated and (B) down-regulated circular RNA profiles in predicting the overall survival of hepatocellular carcinoma patients.
Clinicopathological association
Analysis of the association between circRNA expression and clinicopathological factors in HCC also yielded robust results. As shown in Table 4, significant associations were observed between the circRNA expression and alcoholism (pooled P = 0.0323), tumor size (pooled P = 0.00012), differentiation grade (pooled P = 0.000), microvascular invasion (pooled P = 0.003744), TNM stage (pooled P = 0.000), metastasis (pooled P = 0.000), and serum AFP level (pooled P = 0.0115).
Table 4.
Associations between circular RNA expression and clinicopathological factors in patients with hepatocellular carcinoma
| Class | Included studies | χ2 | Pooled P-value |
| Gender | 18 | 36.426 | 0.4487 |
| Age | 18 | 32.517 | 0.635 |
| Smoking (yes vs no) | 5 | 8.597 | 0.5707 |
| Alcoholism | 5 | 19.684 | 0.0323 |
| Tumor size | 13 | 57.979 | 0.00012 |
| Tumor number (single vs multiple) | 7 | 14.3614 | 0.4231 |
| Encapsulation (incomplete/complete) | 3 | 3.8078 | 0.7026 |
| Differentiation grade (well/moderate/poor) | 11 | 66.9698 | 1.97 × 10−6 |
| Microvascular invasion | 3 | 19.261 | 0.003744 |
| TNM stage | 13 | 76.1066 | 2.51 × 10−7 |
| HBsAg | 7 | 14.4284 | 0.418306 |
| Serum AFP | 12 | 42.4249 | 0.0115 |
| Metastasis | 12 | 79.8852 | 6.35 × 10−8 |
| ALT | 3 | 5.4896 | 0.4827 |
| AST | 4 | 12.3545 | 0.1361 |
| GGT | 3 | 14.3614 | 0.4231 |
| Cirrhosis (yes/no) | 5 | 5.8236 | 0.8298 |
ALT: Alanine transaminase; AST: Aspartate transaminase; GGT: Gamma-glutamyl transpeptidase; AFP: Alpha fetoprotein; TNM: Tumor-node-metastasis; HbsAg: Hepatitis B surface antigen.
Stratified analysis
The stratified analysis depending on sample type revealed that the tissue-based circRNA testing had higher diagnostic efficacy in confirming HCC than plasma-based analysis (AUC: 0.88 vs 0.72; DOR: 15.17 vs 8.93). Different effects were also observed in differentially expressed circRNAs, wherein up-regulated circRNA profile yielded a better diagnostic performance than down-regulated circRNAs (AUC: 0.97 vs 0.81; DOR: 11.48 vs 8.75). Moreover, the analysis grouped by control type showed that circRNA profiles could differentiate chronic hepatitis or cirrhosis from HCC, with an AUC of 0.84, sensitivity of 0.77, and specificity of 0.76; additionally, the circRNA expression signature was able to distinguish adjacent non-cancerous liver tissues from HCC samples, with an AUC of 0.73 and specificity of 0.75 (Table 5).
Table 5.
Subgroup analysis conducted based on sample type, control type, and expression status among the diagnostic studies
| Analysis | Included individual studies | Sensitivity 95%CI | Specificity 95%CI | PLR 95%CI | NLR 95%CI | DOR 95%CI | AUC | Heterogeneity |
| Sample type | ||||||||
| Tissue | 10 | 0.73 (0.70–0.77) | 0.82 (0.78–0.84) | 4.03 (2.98–5.46) | 0.29 (0.19–0.43) | 15.17 (8.42–27.34) | 0.88 | I2 = 73.9%, P = 0.0001 |
| Plasma | 3 | 0.79 (0.74–0.84) | 0.65 (0.57–0.73) | 2.33 (1.47–3.7) | 0.28 (0.12–0.63) | 8.93 (2.37–33.64) | 0.72 | I2 = 87.3%, P = 0.0004 |
| Expression status | ||||||||
| Up-regulated circRNAs | 4 | 0.70 (0.62–0.78) | 0.83 (0.76–0.89) | 4.00 (2.71–5.91) | 0.37 (0.28–0.50) | 11.48 (5.90–22.33) | 0.97 | I2 = 21.2%, P = 0.2832 |
| Down-regulated circRNAs | 7 | 0.74 (0.70–0.77) | 0.75 (0.71–0.78) | 2.75 (2.16–3.5) | 0.33 (0.22–0.49) | 8.75 (5.31–14.43) | 0.81 | I2 = 70.4%, P = 0.0025 |
| Control type | ||||||||
| Chronic hepatitis/cirrhosis vs HCC | 3 | 0.77 (0.72–0.81) | 0.76 (0.71–0.81) | 3.08 (2.49–3.80) | 0.32 (0.25–0.41) | 10.89 (7.51–15.78) | 0.84 | I2 = 0.0%, P = 0.5262 |
| Adjacent non-cancerous liver tissue vs HCC | 6 | 0.63 (0.57–0.68) | 0.75 (0.69–0.81) | 2.33 (1.44–3.75) | 0.52 (0.40–0.69) | 4.7 (3.12–7.08) | 0.73 | I2 = 0.0%, P = 0.4953 |
AUC: Area under the curve, PLR: Positive likelihood ratio; NLR: Negative likelihood ratio; DOR: Diagnostic odds ratio; HCC: Hepatocellular carcinoma.
Sensitivity analysis
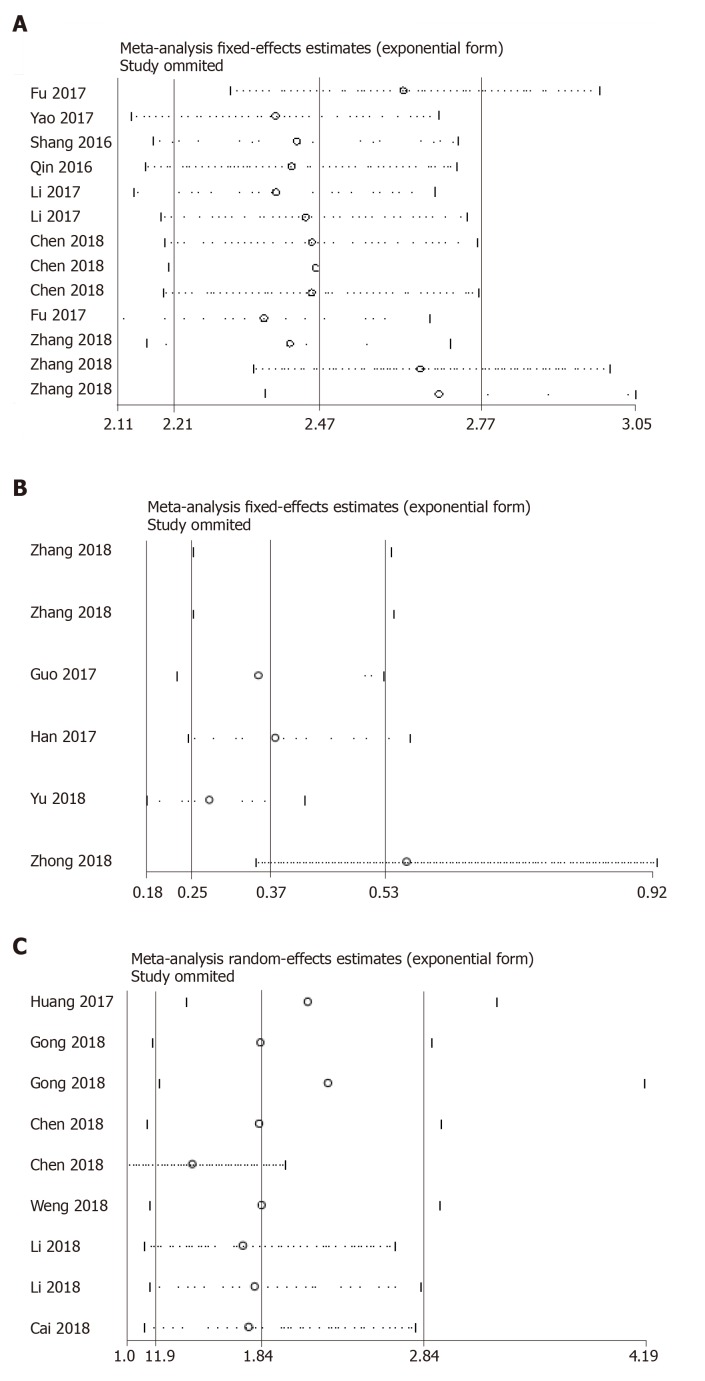
Sensitivity analysis was performed in both the diagnostic and prognostic effect sizes. As exemplified by Figure 4, one study[30] was identified as the outlier in the pooled prognostic effects of down-regulated circRNAs in HCC. After elimination of the outlier data and re-analysis of the effect, the I2 dropped from 92.3% to 90%, indicating that included heterogeneous studies were a substantial cause of study heterogeneity. No outliers were detected in other pooled effects (Figure 5).
Figure 5.
Sensitivity analysis of the outlier data. A: Diagnostic studies; B: Down-regulated; and C: Up-regulated circular RNA profiles in predicting the overall survival in hepatocellular carcinoma.
Publication bias
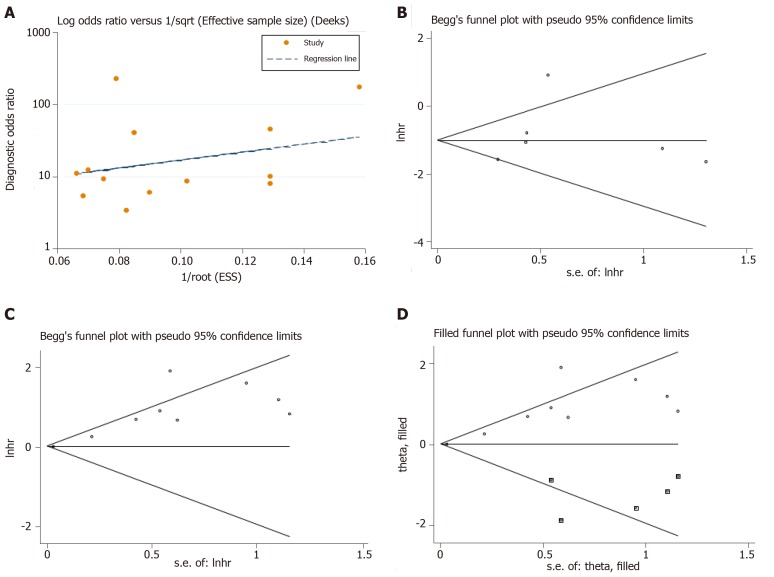
Publication bias was judged using different methods for different pooled effects. As shown in Figure 6A, no clear publication bias was detected in the combined diagnostic effects (Deek’s funnel plot, P = 0.446), nor in the analysis of down-regulated circRNA profile (Egger’s test, P = 0.606, Figure 6B). Nevertheless, the funnel plot showed evidence of publication bias in the effects of up-regulated circRNA profile (Egger’s test, P = 0.001, Figure 6C), and the trim and fill method was applied to trace the possible impacts from bias[33]. As indicated in Figure 6D, the filled funnel plots identified five imputed studies, but the effect was slightly altered before and after adjustment (variance = 0.187, P = 0.005 vs variance = 0.287, P = 0.000).
Figure 6.
Publication bias judged by the Deek’s funnel plot for the diagnostic meta-analysis. A-C: Begg’s test for the down-regulated and up-regulated circular RNA (circRNA) signatures in predicting the overall survival in hepatocellular carcinoma; D: The trim and fill method performed to assess the possible effects of bias on the overall pooled effects of the up-regulated circRNA signature. The hollow circles in squares indicate the imputed studies.
DISCUSSION
HCC is among the most frequent causes of cancer death in digestive system tumors[1-3]. There are many investigated biomarkers for HCC, such as AFP, PIVKA-II, and the ratio of lens culinaris agglutinin-reactive alpha-fetoprotein to total AFP (AFP-L3/AFP)[34]. However, these biomarkers retain several limitations on their overall diagnostic efficacies[5,34]. In this respect, ideal noninvasive biomarkers are urgently needed to reinforce HCC detection. Circular RNAs (circRNAs), which are a group of covalently closed circular non-coding RNAs, have been recently identified as key regulators in cell development and function in HCC[35]. Accumulating investigations have shown that a large number of circRNAs are dysregulated in HCC[10-30], giving rise to the differential expression status and association in tumor diagnosis and prognosis. In the present study, we conducted diagnostic and prognostic meta-analyses and assessed the clinical significance of circRNA expression profiles in HCC.
A recently published meta-analysis showed that circRNAs are promising diagnostic biomarkers for tumors[36]. In our diagnostic meta-analysis, a total of eight studies were included, covering 712 HCC cases. The combined ROC curve showed that the circRNA expression profile had favorable sensitivity (0.78), specificity (0.80), and AUC (0.86) values in confirming HCC. Moreover, the respective PLR and NLR values were 3.97 and 0.27, which means that the circRNA signature achieved a ratio of nearly 4 between the true positive and false positive rates, and the probability of HCC patients that tested negative for circRNAs versus the probability of cases that tested positive had a ratio of 0.27. Importantly, the pooled DOR, a key parameter used in meta-analyses of diagnostic test accuracy studies, was estimated to be 14.59 and suggested the powerful capability of circRNA signatures in discriminating HCC from non-cancer cases. These encouraging findings suggest that circRNA expression signatures could be considered as important potential biomarkers for the diagnosis of patients with HCC.
An increasing number of single studies have documented the prognostic value of circRNAs in HCC[10,11,14,16-18,22,26,28,30]. In our pooled analysis, we found that circRNAs with different expression statuses in HCC displayed distinct prognostic features. The down-regulated circRNA profile (hsa_circ_0001649, circ-ITCH, circMTO1, cSMARCA5, and circC3P1) was closely associated with favorable OS in patients with HCC, whereas the up-regulated circRNA signature (hsa_circRNA_100338, hsa_circ_0064428, circRNA101368, hsa_circ_0103809, and circ-ZEB1.33) was related to worse OS time in HCC. A newly published study has reviewed the oncogenic (tumor suppression) roles of single circRNAs in HCC[37], further evidencing our findings. These encouraging results showed that circRNA expression signatures may be developed as potential indicator(s) for predicting the OS of HCC patients. The clinicopathological value of the circRNA expression profile also manifested robust results; circRNAs were found to be markedly associated with alcoholism, tumor size, differentiation grade, microvascular invasion, TNM stage, metastasis, and serum AFP level, suggesting that abnormally expressed circRNAs are likely to be implicated in tumor progression in HCC as well.
Our study still retains many limitations. The overriding problem is the substantial heterogeneity which appeared among studies. The sensitivity analysis identified one study[30] as the outlier in the pooled prognostic effects of down-regulated circRNAs in HCC. Our analysis further confirmed the impact of heterogeneous studies on the generation of heterogeneity among combined effects. Additionally, biases from publications appeared in one of our pooled prognostic analyses. Nevertheless, our further assessment using a nonparametric trim and fill procedure confirmed that the combined accuracy is not an artifact of unpublished negative studies. Consequently, the accuracy of all the pooled effects was shown to be relatively reliable.
In summary, our study shows evidence that abnormally expressed circRNAs may play a critical role in HCC progression and could serve as diagnostic and prognostic biomarkers for cases of HCC. Future in-depth research is required to further evaluate the utilities of single or panel circRNA(s) for HCC diagnosis and prognosis.
ARTICLE HIGHLIGHTS
Research background
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide. At present, reliable biomarkers for HCC are unavailable, so it is necessary to investigate novel effective ones. The application of circular RNAs (circRNAs) in numerous tumors has been drawing considerable attention. However, the clinical value of circRNAs in HCC has not been determined.
Research motivation
We sought to provide evidence on the potential clinical value of abnormal circRNAs in HCC from the perspective of evidence-based medicine.
Research objectives
This meta-analysis was designed to reveal the clinicopathological, prognostic, and diagnostic features of circRNAs in HCC.
Research methods
We searched for articles in PubMed, EMBASE, and CNKI databases before May 2019. Studies reporting on the clinicopathologic, diagnostic, or prognostic significance of circRNAs in HCC were eligible for inclusion. The meta-analysis was performed with Stata and Meta-DiSc software, and the study quality was assessed in accordance with the Quality Assessment of Diagnostic Accuracy Studies-2 Checklist and the Newcastle-Ottawa Scale. According to the heterogeneity of the studies, a fixed- or random-effects model was used for pooling analysis.
Research results
A total of 21 studies were eligible for the meta-analysis. The results showed that the abnormality in the expression of circRNAs was of good significance in the diagnostic determination of HCC. The down-regulation of circRNAs was negatively correlated with HCC prognosis, while the up-regulated circRNAs were positively related to the overall survival. The circRNAs were significantly associated with poor clinicopathological features in patients with HCC.
Research conclusions
This meta-analysis suggested that circRNAs may be a promising biomarker for the determination of diagnosis and prognosis of HCC in clinical practice.
Research perspectives
The results of this meta-analysis may contribute to a better understanding of the potential clinical application of abnormal circRNAs in HCC.
Footnotes
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
PRISMA 2009 Checklist statement: The authors have read the PRISMA 2009 Checklist, and the manuscript was prepared and revised according to the PRISMA 2009 Checklist.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Peer-review started: June 19, 2019
First decision: July 31, 2019
Article in press: September 4, 2019
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): A
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hann HW S-Editor: Zhang L L-Editor: Wang TQ E-Editor: Qi LL
Contributor Information
Ya-Lin Jiang, Department of Clinical Laboratory, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang 471003, Henan Province, China.
Meng-Meng Shang, Department of Clinical Laboratory, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang 471003, Henan Province, China; Department of Clinical Laboratory, The Family Planning Guidance Station of Luanchuan County, Luoyang 471500, Henan Province, China.
Shi-Zhen Dong, Department of Clinical Laboratory, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang 471003, Henan Province, China.
Yong-Chao Chang, Department of Clinical Laboratory, The First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology, Luoyang 471003, Henan Province, China. zero6666888@163.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Medavaram S, Zhang Y. Emerging therapies in advanced hepatocellular carcinoma. Exp Hematol Oncol. 2018;7:17. doi: 10.1186/s40164-018-0109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SJ, Jang JY, Jeong SW, Cho YK, Lee SH, Kim SG, Cha SW, Kim YS, Cho YD, Kim HS, Kim BS, Park S, Bang HI. Usefulness of AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5811. doi: 10.1097/MD.0000000000005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng JQ, Liu L, Wang MR, Li PY. Circular RNAs in digestive system cancer: potential biomarkers and therapeutic targets. Am J Cancer Res. 2018;8:1142–1156. [PMC free article] [PubMed] [Google Scholar]
- 9.Cui X, Wang J, Guo Z, Li M, Li M, Liu S, Liu H, Li W, Yin X, Tao J, Xu W. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17:123. doi: 10.1186/s12943-018-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Hu B, Ji L, Ruan X, Zheng Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10:1690–1702. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Zhang C, Lin J, Song X, Wang H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1275–1283. doi: 10.2147/CMAR.S166740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S, Chen Z, Xiao B, Hu Y. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018:32. doi: 10.1002/jcla.22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, Song R. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18:116. doi: 10.1186/s12935-018-0602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, Wang Z, Wen P, Yang H, Shi X, Pan J, Ye H. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169–48177. doi: 10.18632/oncotarget.18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 17.Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, Zhou J, Tang ZY, Huang XY. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P, Chen R, Huang Z, Hu X, Huang Y, Tang D. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17:2349–2359. doi: 10.1080/15384101.2018.1526599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Dong Y, Huang Z, Kuang Q, Wu Y, Li Y, Li M. Computational identifying and characterizing circular RNAs and their associated genes in hepatocellular carcinoma. PLoS One. 2017;12:e0174436. doi: 10.1371/journal.pone.0174436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 21.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng Qiaoyou, Chen Minjiang, Li Maoquan, Zheng Yong-Fa, Shao Guoliang, Fan Weijun, Xu Xi-Ming, Ji Jiansong. Global microarray profling identifed hsa_ circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2018;0:1–7. doi: 10.1136/jmedgenet-2018-105440. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L, Cui Y, Jiang X. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461. doi: 10.1016/j.bbrc.2018.01.077. [DOI] [PubMed] [Google Scholar]
- 25.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, Liu B, Yang Y. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, Sun SH, Yang F, Zhou WP. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 2018;11:2853–2863. doi: 10.2147/OTT.S158008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Qiu S, Luo P, Zhou H, Jing W, Liang C, Tu J. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22:135–142. doi: 10.3233/CBM-171109. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu X, Zhu M, Liang C, Yu M, Tu J. The Circular RNA hsa_circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and May Serve as a Diagnostic Biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong L, Wang Y, Cheng Y, Wang W, Lu B, Zhu L, Ma Y. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044–1049. doi: 10.1016/j.bbrc.2018.03.221. [DOI] [PubMed] [Google Scholar]
- 31.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 32.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 33.Lin N, Lin Y, Fu X, Wu C, Xu J, Cui Z, Lin D. MicroRNAs as a Novel Class of Diagnostic Biomarkers in Detection of Oral Carcinoma: A Meta-Analysis Study. Clin Lab. 2016;62:451–461. doi: 10.7754/clin.lab.2015.150802. [DOI] [PubMed] [Google Scholar]
- 34.Huang TS, Shyu YC, Turner R, Chen HY, Chen PJ. Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: a systematic review and meta-analysis protocol. Syst Rev. 2013;2:37. doi: 10.1186/2046-4053-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao R, Zou H, Liao W. Prospect of Circular RNA in Hepatocellular Carcinoma: A Novel Potential Biomarker and Therapeutic Target. Front Oncol. 2018;8:332. doi: 10.3389/fonc.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18:303. doi: 10.1186/s12885-018-4213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP. The Emerging Role of Circular RNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:1548–1559. doi: 10.7150/jca.24566. [DOI] [PMC free article] [PubMed] [Google Scholar]