Abstract
X-linked inhibitor of apoptosis protein (XIAP) is an important regulator of cancer cell survival whose BIR3 domain (XIAP-BIR3) recognizes the Smac N-terminal tetrapeptide sequence (AVPI), making it an attractive protein-protein interaction (PPI) target for cancer therapies. We used the fragment molecular orbital (FMO) method to study the binding modes and affinities between XIAP-BIR3 and a series of its inhibitors (1–8) that mimic the AVPI binding motif; the inhibitors had common interactions with key residues in a hot spot region of XIAP-BIR3 (P1–P4 subpockets) with increased binding affinity mainly attributed to specific interactions with the P1 and P4 subpockets. Based on the structural information from FMO results, we proposed a novel XIAP natural product inhibitor, neoeriocitrin 10, which was derived from our preciously reported XIAP-BIR3 inhibitor 9, can be used as a highly potent candidate for XIAP-BIR3 inhibition. We also performed pair interaction energy decomposition analysis to investigate the binding energies between specific binding residues and individual ligands, showing that the novel natural product neoeriocitrin 10 had a higher binding affinity than epicatechin gallate 9. Molecular docking and dynamics simulations were performed to explore the mode of binding between 10 and XIAP-BIR3, demonstrating that 10 binds more strongly to the P1 and P4 pockets than 9. Overall, we present a novel natural product, neoeriocitrin 10, and demonstrate that the FMO method can be used to identify hot spots in PPIs and design new compounds for XIAP inhibition.
Keywords: Protein-protein interaction, Natural product, X-linked inhibitor of apoptosis protein, XIAP-BIR3 inhibitor, Molecular docking, MD simulation, Fragment molecular orbital method, Drug discovery
Graphical Abstract
1. Introduction
X-linked inhibitor of apoptosis protein (XIAP) is a member of the inhibitors of apoptosis protein (IAP) family which inhibits caspases via its three baculoviral IAP repeat (BIR) domains [1]. The deregulation of XIAP has been implicated in a variety of disorders, including neurodegeneration, cancer, and autoimmune disorders [2]. Interactions between XIAP and caspases are regulated by second mitochondria-derived activator of caspases (Smac), which is pro-death signaling protein released from the mitochondria during apoptosis signal transduction [3].
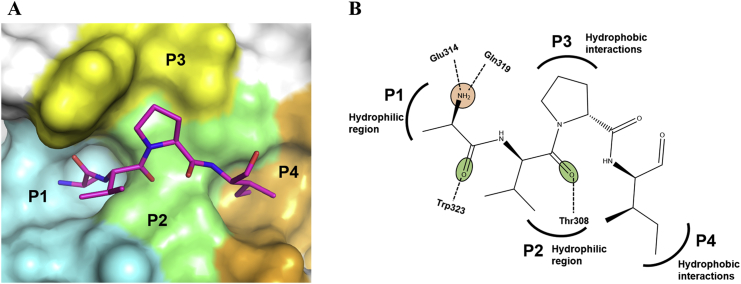
The crystal structure of XIAP-BIR3-Smac revealed that four N-terminal Smac residues, Ala1-Vla2-Pro3-Ile4 (AVPI), bind to a surface groove in XIAP-BIR3 [4]. Structure-based studies of XIAP-BIR3-AVPI [5] have been essential for developing small molecules in XIAP-targeted drug discovery and have allowed the binding pocket of XIAP-BIR3 to be divided into four subpockets (P1–P4 in Fig. 1A) [6]. Furthermore, the XIAP-BIR3 residues that interact with AVPI have also been revealed: Ala1 fits tightly in the P1 pocket formed by Asp309, Trp310, Glu314, Gln319, and the side chain of Leu307; Val2 forms hydrogen bonds with Thr308 and interacts with Leu307 in the P2 pocket; Pro3 makes van der Waals contacts with Trp323 and Tyr324 in the P3 pocket; and Ile4 inserts into the P4 pocket formed by Leu292, Val298, and the hydrophobic portions of the Lys297 and Lys299 side chains (Fig. 1B) [7].
Fig. 1.
Conformation of AVPI in complex with XIAP-BIR3. A. Crystal structure of AVPI in complex with XIAP-BIR3 (PDB: 1G73). B. Key interactions between AVPI and XIAP-BIR3. The surface of the residues in each XIAP-BIR3 subpocket that interact with AVPI are colored: P1-cyan; P2-light green; P3-yellow; and P4-orange.
The N-terminal peptide sequence of Smac binds to the BIR3 domain of XIAP (XIAP-BIR3) in a protein-protein interaction (PPI), hence Smac regulates apoptosis by releasing caspases [8,9]. PPI targets have been deemed intractable by small molecules due to the large, flat interface [10]. However, not all residues at the PPI interface are critical; there is a “hot spot” that confers most of the binding energies [11] and hot-spot residues are clustered in the small binding pockets of the PPI interface [12]. Understanding the XIAP-BIR3 hot spot residues may identify novel potent compounds to promote cancer cell apoptosis.
Understanding the structure of protein-ligand complexes is critical for rational drug discovery [13]. The fragment molecular orbital (FMO) method and pair interaction energy decomposition analysis (PIEDA) have been used to analyze interactions in the protein-ligand complex and PPIs [13,14]. The FMO method, proposed by Kitaura et al. [15], offers faster computational speeds than the traditional quantum-mechanical (QM) method [16] and has been shown to provide accurate information for investigating the nature of chemicals and binding information for protein-ligand interactions [17]. This information is essential for chemists to optimize and modify lead compounds to increase the binding affinities of protein-ligand interactions [18]. Pair interaction energies (PIEs) obtained using the FMO method provide comprehensive details of the interactions between a ligand and a protein [19].
Chessari etal. [20] and Tamanini etal. [21] used FBDD to identify lead compounds targeting XIAP-BIR3; weakly binding fragment 1 was optimized to form early lead 6 and further optimized to produce lead 8, an orally bioavailable drug that exhibited potent target engagement in cells and robust XIAP pharmacodynamic effects in vivo [21].
In this study, we used the FMO method to quantitatively evaluate specific interactions within the XIAP-BIR3-AVPI and XIAP-BIR3-ligand complexes, comparing the molecular interactions between AVPI and inhibitors 1–8, analyzing the energy components of the molecular interactions, and assessing the involvement of water molecules in ligand binding. In particular, we elucidated which XIAP-BIR3 pockets can be further exploited to improve binding affinity.
Energy decomposition analysis indicated that the P1 and P4 pockets were important. We screened a natural product, neoeriocitrin 10, which has a higher binding affinity than the previously reported XIAP-BIR3 inhibitor, epicatechin gallate 9 [22], as a potent candidate XIAP-BIR3 inhibitor. Molecular docking and molecular dynamic (MD) simulation [23] and the FMO method were used to investigate the binding mode and detailed interactions between 10 and XIAP-BIR3, while surface plasmon resonance (SPR) analysis [24] was used to evaluate the binding of 10 to XIAP-BIR3.
2. Experimental Section
2.1. Structure Preparation
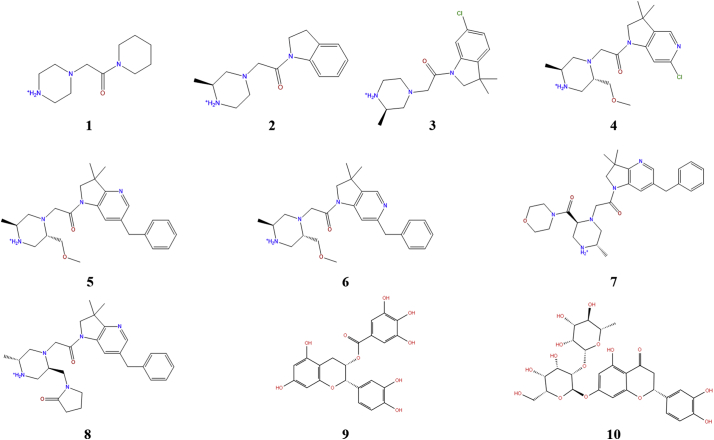
The crystal structures used in this work were retrieved from the Protein Data Bank (PDB); the PDB codes, chemical structures, and IC50 values of the ligands are shown in Table 1 and Fig. 3. Water molecules were identified from the crystal structures (if resolved) and treated explicitly to explore their role in protein-ligand binding. Hydrogen atoms were added to the protein-ligand complex structures at a physiological pH (7.4) and optimized using Prime in Maestro (Prime, Schrödinger, LLC, New York, NY, 2017) [25].
Table 1.
IC50 values of XIAP-BIR3-AVPI and XIAP-BIR3-ligand complexes.
| Ligand | XIAP-BIR3 IC50 (μM) | PDB | Resolution (Å) | Referencea |
|---|---|---|---|---|
| AVPI | 0.32 | 1G73 | 2.0 | 6, 9 |
| 1 | >5000 | 5C3H | 2.65 | 20 |
| 2 | >495 | 5C7A | 2.36 | 20 |
| 3 | 5.5 | 5C7C | 2.32 | 20 |
| 4 | 0.64 | 5C84 | 2.36 | 20 |
| 5 | 0.22 | 5M6F | 2.39 | 21 |
| 6 | 0.16 | 5C83 | 2.33 | 20 |
| 7 | 0.15 | 5M6H | 2.5 | 21 |
| 8 | 0.044 | 5M6M | 2.37 | 21 |
Crystal structures and experimental IC50 values were obtained from the listed references.
Fig. 3.
Chemical structures of eight XIAP-BIR3 inhibitors.
2.2. FMO Calculations
The FMO method was as follows:
-
(a)
During fragmentation, each residue of XIAP-BIR3 and its inhibitor was considered as a fragment. All protein and peptide residues were divided into one-residue fragments at the Cα site based on the hybrid orbital projection method, which was used to reduce the computational cost and correct errors from the projection operator [26].
-
(b)
All molecular orbitals on each fragment (monomer) were optimized in the electrostatic field of the whole system by self-consistent field (SCF) theory. All monomer energies and electron densities were self-consistently optimized at the same time through self-consistent charge (SCC) stage.
-
(c)
SCF cycles for fragment pairs (dimers) were calculated in the electrostatic field of all the remaining fragments. The difference between steps (b)and (c)was that the electrostatic field was formed of N–1 fragments in step (b) and N–2 fragments in step (c), where N refers to the total number of fragments. The energy of each monomer and dimer was obtained and was influenced by the electrostatic field of all fragments, namely the system.
-
(d)
The sum of the energies of the monomers and dimers was used to evaluate the total energy of the system; the binding affinity of the protein-ligand interaction was approximated as the sum of the PIEs. These interaction energies were decomposed by pair interaction energy decomposition analysis (PIEDA), which provided more physical details [27]. The PIE (∆Eijint) between fragments i and j consisted of four or five energy terms depending on whether or not implicit solvation models were considered: electrostatic (∆Ees), exchange repulsion (∆Eex), charge transfer with higher order mixed term (∆Ect+mix), dispersion (∆Edi), and solvation energy (∆Gsol). The PIEs in the FMO calculations were defined using Eq. (1) [27,28].
| (1) |
PIEDA is the sum of four energy terms: electrostatic, exchange repulsion, charge transfer with higher order mixed term, and dispersion [29]. The electrostatic term arises from the Coulomb interaction between the polarized charge distributions of the fragments, while charge transfer arises from the interaction between the occupied orbitals of a donor and unoccupied orbitals of an acceptor. Dispersion arises from the interaction between the induced dipole moments of two fragments, while exchange repulsion is derived from the interaction between fragments located in close proximity and is always repulsive [30]. The electrostatic and charge transfer terms are important in salt-bridges, hydrogen bonds, and polar interactions, whereas the dispersion term can be referred to as hydrophobic in nature and the exchange repulsion term describes steric repulsion [31]. The PIEs and PIEDA contributions of all XIAP-BIR3 residues with AVPI and the inhibitors are presented in the Supporting Information.
All ab initio FMO calculations were performed using the General Atomic and Molecular Electronic Structure System (GAMESS) [32] and energy decomposition analysis was performed at the second order Møller-Plesset perturbation theory (MP2) [33] and polarizable continuum model (PCM) [34] level with the 6-31G** basis set. In FMO calculation, we used all the residues in the crystal structures in both energy calculation and structure optimization. In PCM calculation, we used one body PCM (PCM[1]) and conductor-like PCM (C-PCM) with iterative solver. In SCF calculation, we used cutoff option using ‘RESDIM = 2.0’ for approximating the SCF energy by electrostatic interactions.
2.3. Virtual Screening
Molecular-docking-based virtual screening was conducted to find a novel natural product from our in-house natural product database by using glide in Prime in Schrödinger suite (Schrödinger LLC, New York, NY, 2011). The top 45 virtual hits (XP Gscore >3) based on the XIAP-BIR3 hot spot features are presented in the Supporting Information.
2.4. Molecular Docking
Molecular docking was conducted to investigate the interactions between compounds (9 and 10) and XIAP-BIR3 (PDB: 5M6M) by using glide-XP in Prime in Schrödinger suite (Schrödinger LLC, New York, NY, 2011). The docking pose of compound 10 was selected by e-model scores and visual inspection, while the docking pose of compound 9 was derived from our previous work [22].
2.5. Structural Optimization
The structures of the top-hit XIAP-BIR3-ligand (9 and 10) docking poses were optimized using the FMO/RHF/3-21G and frozen domain dimer (FDD) methods [35]. The protein was fixed during optimization and only the ligands were fully flexible. The polarizable buffer included protein residues within 5.2 Å of the ligands, while the active domain included the ligands. In FMO calculation, we used several options using ‘RESDIM = 2.0’ and ‘RESPPC = 2.0’, and ‘MODGRD = 42’. The FMO/FDD optimization of 9 and 10 took 100 and 120 steps, respectively.
2.6. MM-GBSA/MD Simulation
The docking pose of compound 10, which was selected in the Molecular Docking procedure, were used to investigate the interaction between 10 and XIAP-BIR3. Water molecules were removed, all hydrogen atoms were added, and the binding site was defined by the original binding ligand, 8, using Discovery Studio 4.1. The structure and initial conformation of 10 docked to XIAP-BIR3 were generated using CDOCKER in Discovery Studio 4.1. The top-hit docking pose for 10, which was selected from the molecular mechanics generalized Born surface area (MM-GBSA) calculation [36], was structurally refined using a MD simulation in Prime (Schrödinger LLC, New York, NY, 2011). The MM-GBSA method is one of the most popular approaches for estimating the relative binding affinities of protein-ligand complexes [37]. The relative binding free energy ∆Gbind [38] was estimated using Eq. (2),
| (2) |
where ∆Gbind is the calculated relative free energy including both ligand and protein energy, Gcomplex is the MM-GBSA energy of the minimized complex, Gligand is the MM-GBSA energy of the unbound ligand after removing it from the complex, and Gprotein is the MM-GBSA energy of the unbound protein after separating it from the ligand.
The MD simulation of the XIAP-BIR3-10 complex was performed using Desmond 4.1 in Maestro (Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2017. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2017). The protein-ligand complex was inserted into an orthorhombic box filled with 4579 water molecules (TIP3P model) and with a buffer distance of 10 Å. The MD simulation was studied suing an OPLS3 force field was used. Ions (Na+ and Cl−) were added to simulate a physiological concentration of monovalent ions (0.15 M). NPT (a constant number of particles, pressure, and temperature) used a constant temperature (300K) and pressure (1.01325 bar) as an ensemble class. The particle-mesh Ewald method [39] was used to calculate long-range electrostatic interactions, with a cutoff for van der Waals and short-range electrostatic interactions of 9 Å. Nose-Hoover thermostats [40] were used to maintain a constant simulation temperature, while the Martina-Tobias-Klein method [41] was used to control the pressure. An RESPA integrator [42] was used to integrate the equations of motion with an inner time step of 2.0 fs for bonded interactions and non-bonded interactions within the short-range cutoff. The default protocol in Desmond was used to reach system equilibration, then a 50 ns simulation was performed and the confirmations and energies were saved at each 4 ps interval. All analyses were also carried out with an MD simulation of 50 ns, in which the NPT ensemble was applied.
2.7. SPR Assay
The purified XIAP (BIR3 domain) protein with His-tag was prepared from BPS Bioscience, catalog number 75001. The interaction between 10 and XIAP-BIR3 was investigated using the ProteOnTM XPR36 Protein Interaction Array System (Bio-Rad Laboratories, California, USA). Purified XIAP-BIR3 was immobilized on the ProteOn GLH sensor chip. The ligand was diluted with 1× Phosphate-Buffered Saline with Tween 20 and 1% DMSO at six different concentrations (0, 6.25, 12.5, 25, 50, and 100 μM), and then passed over the chip at a flow rate of 100 μL/min. The data were analyzed by ProteOn Manager Software 2.0 using standard Langmuir models to fit the kinetic data. The association constant (kain M/s) represented the rate of complex formation and the dissociation constant (kdin s−1) represented the rate of complex decay. High-affinity interactions were characterized by low KD values, while the rapid recognition and binding of the interactants (rapid “on rate,” or high ka) and the stability of complex formation (slow “off rate,” or low kd) was represented by the equation KD = kd/ka.
3. Results and Discussion
3.1. FMO Study of the XIAP-BIR3-AVPI Complex
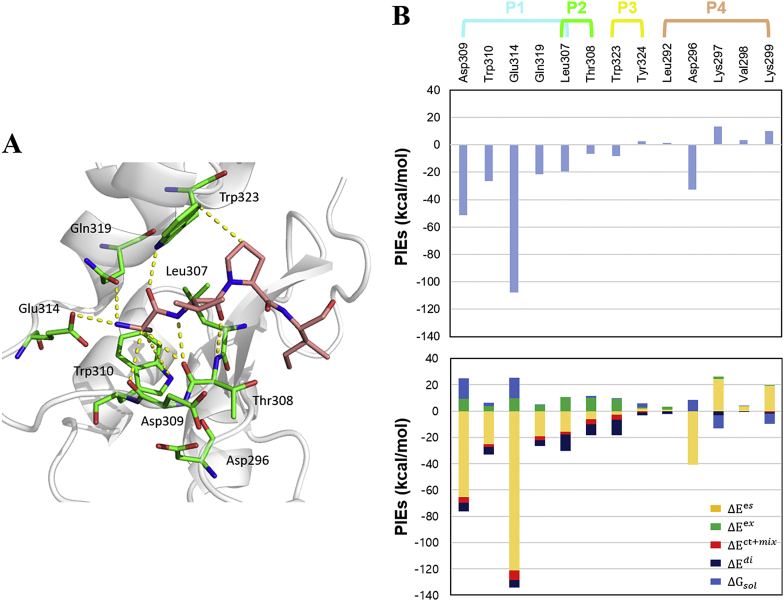
The P1 pocket of XIAP-BIR3 is the main binding region for substrate recognition and is more hydrophilic than the other three subpockets, while the P4 pocket is quite hydrophobic and further improves the ligand activity of XIAP-BIR3 [20]. We performed FMO calculations for the XIAP-BIR3-AVPI complex and detected eight strong interactions between AVPI and eight residues of XIAP-BIR3: Asp296, Leu307, Thr308, Asp309, Trp310, Glu314, Gln319, and Trp323 (Fig. 2). The strong electrostatic interactions between the oxygen atoms or NH2 group of AVPI and the charged residues of XIAP-BIR3 created significant attractive or repulsive interactions between AVPI and these residues. These results agree with published mutagenesis studies which demonstrated the central role of Asp296, Leu307, Trp310, Glu314, and Trp323 in the binding affinities of AVPI to XIAP-BIR3 and cIAP-BIR3 [6,[43], [44], [45], [46], [47]]. However, Asp296 was not directly involved in binding AVPI but was important for stabilizing the structure of the P1 pocket by interacting with the backbone NH of Asp309 [6]. Several hydrogen bond interactions formed by AVPI and Thr308, Asp309, and Gln319 have been previously reported by analyzing the crystal structure [48] with the FMO results revealing similar contributions. It has been demonstrated that Leu307 and Thr308 form hydrogen bonds with AVPI in the P2 pocket; however, the FMO results revealed that these interactions were not only dominated by electrostatic interactions but also by dispersion (nonpolar) energies (Fig. 2) [6,9,20,48].
Fig. 2.
FMO results for the AVPI in complex with XIAP-BIR3. A. The structure of AVPI binding to the XIAP-BIR3 subpockets. AVPI is shown in light pink, the protein residues are shown in green, and the nitrogen and oxygen atoms are shown in blue and red, respectively. Key interactions according to the FMO results are shown as yellow dashed lines. B. The upper bar plot describes the PIEs of the significant residues in the four XIAP-BIR3 subpockets, while the lower bar plot describes the PIEDA of these key interactions. The electrostatic, exchange repulsion, charge transfer with higher order mixed term, dispersion, and solvation energy terms are shown in yellow, green, red, dark blue, and light blue, respectively.
The FMO results also detected three repulsive interactions between AVPI and Lys297, Val298, and Lys299 in the P4 pocket (Supporting Information, Table S1); however, the dispersion energies of Lys297 (−2.98 kcal/mol) and Lys299 (−1.19 kcal/mol) contributed to binding between Ile4 and XIAP-BIR3. The total PIE between AVPI and the P1 pocket residues was −208.59 kcal/mol, while it was only −37.48 kcal/mol for the P2, P3, and P4 pockets combined, indicating that the P1 pocket of XIAP-BIR3 makes the largest contribution towards AVPI binding. These FMO results agree with an FBDD study in which the P1 pocket of XIAP-BIR3 played a central role in the design of alanine-based Smac mimetic inhibitors [20]. Therefore, the FMO method can be used to explore the key XIAP-BIR3 residues to gain a more detailed understanding of the nature of the PPIs in specific subpockets and to help design new XIAP-BIR3 inhibitors. Furthermore, the FMO method can provide an accurate and comprehensive list of the strong, weak, and repulsive interactions between XIAP-BIR3 inhibitors and the residues surrounding the four subpockets.
3.2. FMO Study of XIAP-BIR3-Ligand Complexes
We used the public data to investigate changes in the PIEs and specific interactions between XIAP-BIR3 and the inhibitors from fragment 1 (IC50 > 5 mM for XIAP-BIR3) to lead 8 (IC50 = 44 nM for XIAP-BIR3) (Table 1 and Fig. 3).
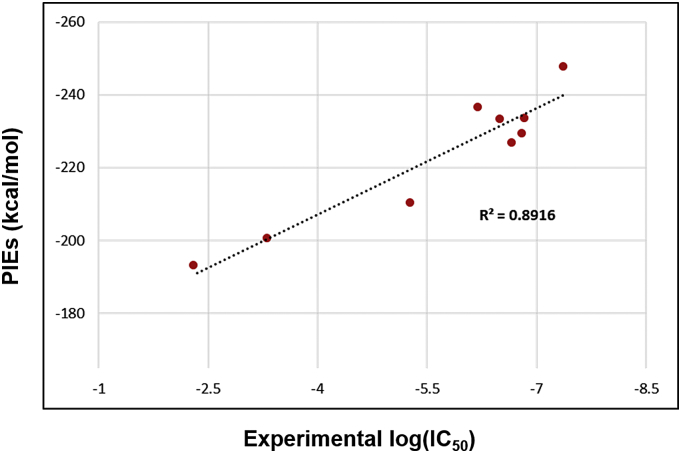
Arulmozhiraja et al. [18] showed that the FMO-PIE represented the strength of the interactions between the ligand and protein residues in the complex and the total PIEs (sum of PIEs of each residue in XIAP-BIR3), but not the exact binding affinity. Despite the fact that PIEs cannot describe all factors in the binding event contributed by different energy terms like enthalpy, entropy, and solvation, we expected to see some correlation between measured and calculated binding affinity. Here, we compared the calculated PIEs with the experimentally measured log(IC50) values to assess how accurately the total PIEs described changes in the inhibitory activities of ligands 1–8. Notably, the calculated PIEs of the ligands with XIAP-BIR3 correlate well with their log(IC50) values, with an R2 of 0.8916 in Fig. 4. This observation was consistent with the trend obtained by a FBDD study, showing that calculated PIEs can quantitate the interactions between XIAP-BIR3 and its ligands and can be used to determine the structure-activity relationship for ligands 1–8.
Fig. 4.
Correlation plot between experimentally measured binding affinity log(IC50) and total PIEs calculated using the FMO method.
To reveal the important interactions in the XIAP-BIR3-ligand complexes, we evaluated the change in PIEs from fragment 1 to lead 8 and compared the PIEs with AVPI (Supporting Information, Table S1–S9). Comparing the PIEs for each XIAP-BIR3 subpocket (Table 2) revealed that the interactions in the P1 pocket were much stronger than in the other subpockets, indicating that AVPI and the ligands preferentially interact with the P1 pocket. These FMO results agree with the experimental data published by Tamanini et al. [21], suggesting that the P1 pocket of XIAP-BIR3 plays a significant role in inhibitor binding affinity. Pearson correlation coefficients between log(IC50) and PIEs in each subpocket were 0.71 for P1, −0.22 for P2, −0.32 for P3, and 0.85 for P4, indicating that the P1 and P4 pockets exhibit stronger binding affinity for 1 to 8.
Table 2.
Total PIEs of each XIAP-BIR3 subpocket.
| AVPI | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | PCCa | |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | −208.60 | −186.27 | −209.10 | −202.28 | −230.72 | −211.72 | −212.64 | −209.00 | −222.38 | 0.71 |
| P2 | −26.68 | −12.54 | −9.71 | −6.51 | −8.18 | −11.08 | −11.11 | −10.53 | −9.49 | −0.22 |
| P3 | −6.37 | −3.54 | −3.66 | −8.65 | −8.51 | −5.28 | −8.14 | −7.47 | −2.05 | −0.32 |
| P4 | −4.44 | −2.44 | 4.61 | −3.45 | −9.66 | −13.67 | −15.54 | −19.41 | −12.99 | 0.85 |
| PIEsb | −233.517 | −193.37 | −200.701 | −210.366 | −236.598 | −233.517 | −226.997 | −233.636 | −247.822 | 0.944 |
Pearson correlation coefficient between log(IC50) and sum of PIEs in each subpocket.
PIEs is sum of pair interaction energies of all residues.
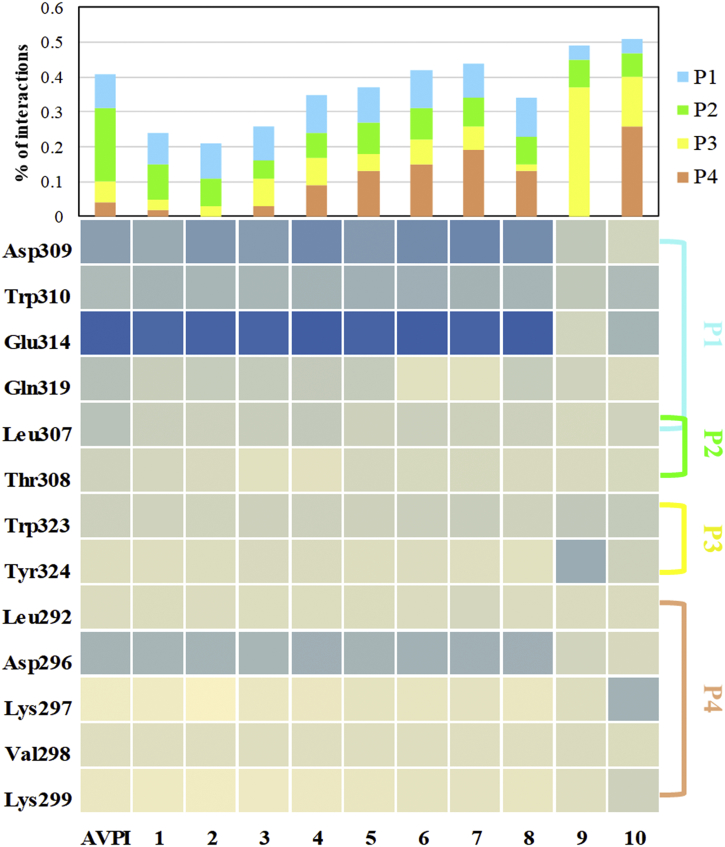
We used the FMO calculations to investigate the PIEs between each residue of XIAP-BIR3 and ligands 1–8. As shown in Fig. 5, P1–P4 pocket residues Asp296, Leu307, Asp309, Trp310, Glu314, and Trp323 interacted with the ligands, indicating the importance of these residues for ligand binding in XIAP-BIR3 (Supporting Information, Fig. S1–S8, Table S2–S9). The calculated PIEs of Asp296, Asp309, and Glu314 in the P1 and P4 pockets increased with increasing binding affinity of the ligands for XIAP-BIR3, which may derive from previously described charge-induced dipole interactions [13].
Fig. 5.
Comparison of residues involved in ligand binding. Protein-ligand information is shown in the columns, and the numbers of residues are shown in the rows. In the matrix, contact between the ligand and the residue is shown in blue, and the absence of a contact is shown in light-yellow. Boxes are colored from dark blue (PIE < −100 kcal/mol) to light yellow (PIE >0 kcal/mol). The level of interaction consensus is shown above, with colors indicating the fraction of the PIE values for each of the four XIAP-BIR3 subpockets: P1 - light blue; P2 - green; P3 - yellow; and P4 - orange.
Water molecules were involved in the binding of ligands 4–8 to XIAP-BIR3 (Supporting Information, Fig. S4–S8). The PIE between lead 8 and the P4 pocket residues (−12.993 kcal/mol) was smaller than the PIEs of 5–7 (Table 2), however, one water molecule was found to form a hydrogen bond between 8 and the side chain of Lys299 in the P4 pocket (−20.762 kcal/mol). Therefore, the interactions mediated by water molecules increased the binding affinity of ligands with XIAP-BIR3. The FMO analysis of water molecules in the binding pocket can help to distinguish between energetically favorable and unfavorable water molecules and design analogs that can interact or displace certain water molecules [31].
3.3. Specific Interactions between Ligands and XIAP-BIR3
The FMO analysis revealed that the calculated PIEs of Asp296, Leu307, Asp309, Trp310, Glu314, Gln319, and Trp323 with all of the ligands were similar to the PIEs of AVPI (Supporting Information, Table S1–S9). This result can be explained by the fact that these residues are important for the binding modes between the inhibitors and XIAP-BIR3. We used PIEDA between the inhibitors and XIAP-BIR3 residues to elucidate the main reason for the difference in binding affinity for fragment optimization [27].
PIEDA of the XIAP-BIR3 residues showed that the interaction energies were mainly the result of the electrostatic term. The dominant contribution of electrostatic interactions supports the assumption that these interactions derive from charge-induced dipole interactions. A major difference in the PIEDA components of the interactions between 1 and 8 and the residues of XIAP-BIR3 was the dispersion term, particularly in the P4 pocket, indicating that differences in the interactions in the XIAP-BIR3-ligand complexes were attributed to dispersion energies. Consequently, we found that increased dispersion energies involving Lys297 and Lys299 contributed to increases in binding affinity observed for 1–8. In addition, differences between the dispersion energies of 8 and AVPI in Lys297 (−4.712 vs. −2.98 kcal/mol) and Lys299 (−2.446 vs. −1.193 kcal/mol) agreed with the experimental data published by Tamanini et al. [21], demonstrating that the P4 binding pocket is a promising target. These findings indicate that these P4 pocket residues provide scope for further fragment optimization.
Ichihara etal. [14] described the application of PIEDA for the FBDD process, and proposed that increased ligand binding affinity can be explained by the electrostatic, charge transfer, and dispersion terms. Accordingly, we investigated the ratio of the sum of the dispersion and charge transfer terms over the electrostatic term ((∆Ect+∆Edi)/∆Ees) for 1–8. The values were 0.30, 0.36, 0.35, 0.36, 0.41, 0.43, and 0.44 for fragments 1 to 7, respectively, whereas the values for lead 8 and AVPI were 0.39 (Table 3). The values for 1–7 increased according to their binding affinity, but not for lead 8, suggesting that the charge transfer and dispersion terms may be the dominant factors in fragment optimization; however, the ligands could also be optimized by converting the component of the energy term [13]. Overall, the chemical nature of the interactions computed by PIEDA could highlight the most appropriate types of modification for improving binding affinity, whether it be modifying dispersive, electrostatic, or charge–charge interactions [30].
Table 3.
Calculated PIEDA, IC50 values, and attraction energy ratios of XIAP-BIR3-ligand complexes.
| Ligand | XIAP-BIR3 IC50 (μM) | ∆Ees | ∆Eex | ∆Ect+mix | ∆Edi | |
|---|---|---|---|---|---|---|
| AVPI | 0.32 | −231.525 | 64.683 | −30.563 | −62.044 | 0.39 |
| 1 | >5000 | −205.073 | 34.466 | −19.708 | −42.212 | 0.30 |
| 2 | >495 | −215.027 | 45.393 | −23.288 | −54.857 | 0.36 |
| 3 | 5.5 | −224.581 | 47.522 | −22.017 | −57.309 | 0.35 |
| 4 | 0.64 | −237.385 | 52.817 | −24.415 | −60.467 | 0.36 |
| 5 | 0.22 | −232.429 | 57.841 | −26.326 | −68.943 | 0.41 |
| 6 | 0.16 | −233.483 | 64.355 | −27.424 | −73.141 | 0.43 |
| 7 | 0.15 | −246.712 | 69.402 | −30.024 | −79.102 | 0.44 |
| 8 | 0.044 | −258.229 | 59.614 | −27.575 | −73.148 | 0.39 |
The FMO analysis demonstrated that the residues of the P1 and P4 pockets of XIAP-BIR3 play an important role in the binding of XIAP-BIR3 inhibitors. The FMO/PIEDA results provided evidence that varying the substituents of the P4 pocket could provide scope for further fragment optimization. It is difficult to detect small changes in the residue interactions in these four subpockets by visual inspection and to provide a good explanation for binding affinity; however, the FMO method can reveal key interactions between ligands and XIAP-BIR3.
3.4. A Novel Ligand Targeting XIAP-BIR3
We investigated a natural product, epicatechin gallate 9, which directly binds XIAP-BIR3 with a KD value of 107 μM [22] and is known to regulate XIAP expression [49,50]. Comparing the PIEs of the XIAP-BIR3 inhibitors provided a list of important interactions which allowed us to analyze the structure of the inhibitors for strong, weak, missing, or new interactions with XIAP-BIR3. In order to develop XIAP-BIR3 inhibitors with a higher potency than 9, we used the FMO method to investigate the interactions between 9 and XIAP-BIR3.
Firstly, the PIEs obtained from the XIAP-BIR3-9 complex were analyzed to identify the XIAP-BIR3 residues that interacted with 9. The FMO method detected seven strong interactions between seven residues of 9 and XIAP-BIR3: Asp296, Asp309, Trp310, Glu314, Gln319, Trp323, and Tyr324 (Supporting Information, Fig. S9). As expected, FMO confirmed previously identified interactions in the XIAP-BIR3-ligand complexes (1–8, Asp296, Asp309, Trp310, Glu314, and Trp323) and identified a new strong interaction with Tyr324. The interaction energy between 9 and the P3 pocket residues was −54.961 kcal/mol, higher than that of other inhibitors, indicating that 9 can bind more strongly with the P3 pocket (Supporting Information, Fig. S9 and Table S10). PIEDA showed that 9 lacks a side chain to penetrate the P4 pocket (Supporting Information, Fig. S9). The FMO method provided information to guide the structural-based virtual screening of future ligands to target XIAP-BIR3.
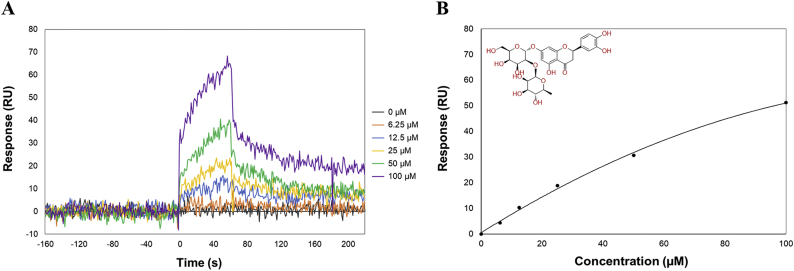
Based on the FMO results, we performed docking-based virtual screening on our in-house natural product database (3000), selecting 45 virtual hits (XP Gscore >3) that matched the hot-spot features of XIAP-BIR3 well (Supplementary Information). SPR analysis was performed to determine whether the selected compounds bind XIAP-BIR3 (data not shown). One novel natural product, neoeriocitrin 10, altered the refractive index and mass concentration when injected into the XIAP-BIR3-immobilized sensor surface. The compound also exhibited a dose-dependent change with a KD value of 13.3 μM, indicating that 10 can bind more efficiently to XIAP-BIR3 than 9 (KD=107 μM) or the XIAP inhibitor, embelin (KD = 37.6 μM) [22] (Fig. 6).
Fig. 6.
SPR analysis of 10 binding to XIAP-BIR3-10. A. Sensorgrams of 10 binding to XIAP-BIR3. B. Equilibrium binding analysis and chemical structure of 10. For SPR analysis, XIAP-BIR3 was immobilized onto the vertical channels of a NeutrAvidin-precoated ProteOn GLH sensor chip and 10 was injected into the horizontal channels of the sensor chip at concentrations of 0, 6.25, 12.5, 25, 50, and 100 μM.
To elucidate the reason for the different binding affinities of these two natural products, we compared the PIEs of the XIAP-BIR3-10 and XIAP-BIR3-9 complexes. The structure of the XIAP-BIR3-10 complex was obtained by molecular docking, the MM-GBSA method, and MD simulations, as for the XIAP-BIR3-9 complex [22]. The structure and initial conformation of XIAP-BIR3-10 were generated using CDOCKER in Discovery Studio 4.1. The top-hit docking pose of 10 was identified using the MM-GBSA method with a ∆Gbind value of −49.89 kcal/mol. To evaluate the dynamic stability of the proposed binding mode of the XIAP-BIR3-10 complex, we performed MD simulations twice using the backbone atom root-mean-square deviation (RMSD) of XIAP-BIR3 and the heavy atom RMSD of 10. The trajectory of the MD simulations confirmed the docking pose and revealed the stability and interactions of the protein-ligand complex (Supporting Information, Fig. S10). The structure of the XIAP-BIR3–10 complex was optimized and the binding energies between 10 and XIAP-BIR3 were evaluated using the FMO results.
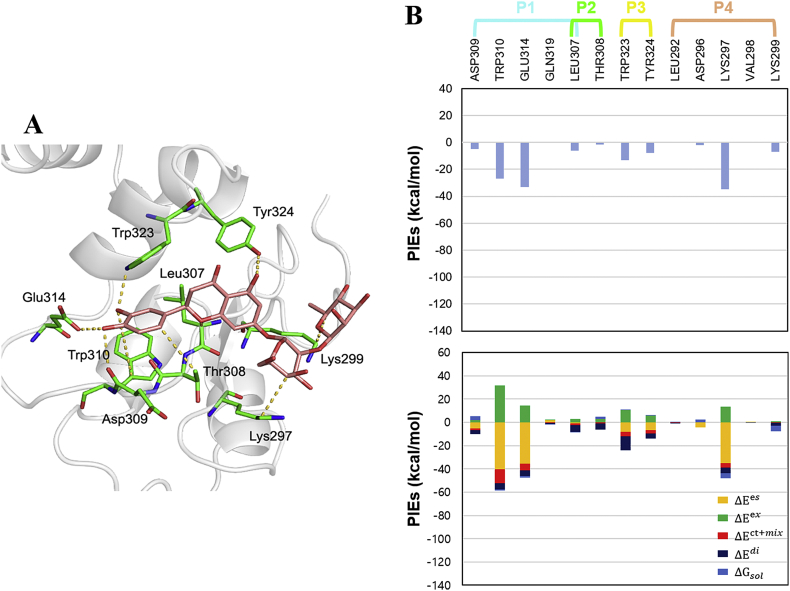
The FMO method detected seven strong interactions between eight residues of 10 and XIAP-BIR3: Lys297, Lys299, Leu307, Asp309, Trp310, Glu314, Trp323, and Tyr324 (Fig. 7A). As expected, these results confirmed previously identified interactions in the XIAP-BIR3-9 complex (Asp309, Trp310, Glu314, Trp323, and Tyr324) and identified two new strong interactions (Lys297 and Lys299). The total PIEs of the residues in the P1–P3 pockets of 9 were stronger than the total PIEs between 10 and XIAP-BIR3 (Supporting Information, Table S10–S11); however, the total PIEs of the residues in the P4 pocket (−43.385 kcal/mol) of 10 were much stronger than the total PIEs between 9 and XIAP-BIR3 (0.570 kcal/mol), indicating that strong interactions in the P4 pocket strongly affect binding affinity. PIEDA revealed that the sugar moiety of 10 interacts with Lys297 and Lys299 in the P4 pocket, supporting the assumption that the interactions formed by these residues are important for improving XIAP-BIR3 binding affinity (Fig. 7B). Therefore, the FMO method is a useful tool for exploring key interactions in ligand binding and for validating the structural optimization process.
Fig. 7.
FMO results for 10 in complex with XIAP-BIR3-10. A. The structure of 10 binding to the XIAP-BIR3 subpockets. The ligand is shown in light pink, the protein residues are shown in green, and the nitrogen and oxygen atoms are shown in blue and red, respectively. Key interactions according to the FMO results are shown as yellow dashed lines. B. The upper bar plot describes the PIEs of the significant residues in the four XIAP-BIR3 subpockets, while the lower bar plot describes the PIEDA of these key interactions. The electrostatic, exchange repulsion, charge transfer with higher order mixed term, dispersion, and solvation energy terms are shown in yellow, green, red, dark blue, and light blue, respectively.
4. Conclusions
In this study, the interactions in XIAP-BIR3-ligand complexes, including AVPI, fragment hit-to-lead 1–8, the previously reported 9, and the novel proposed 10, were quantitatively studied using the FMO method at a correlated MP2/6-31G**/PCM level. The FMO method provides comprehensive structural information on both protein-protein and protein-ligand systems. The outcomes of this study can be used to generate novel XIAP-BIR3 inhibitors and perform detailed analyses on the binding affinities of XIAP-BIR3 inhibitors. We have shown how the FMO method can be applied to the rational design of natural products for binding XIAP-BIR3. By applying the information provided by our FMO results, we can develop novel synthetic approaches to extend diverse analogs and improve their potency. The FMO/PIEDA method allowed us to identify the contribution of different energy components from PIEs, while the FMO method provided significant information on hydrogen bonds, salt bridges, CH-π interactions, and induced charges at the molecular level. Therefore, the FMO method is a good tool for analyzing the molecular interactions between ligands and proteins for drug discovery.
Author Contributions
‡H.L. and ‡X.J. contributed equally. All authors contributed to writing the manuscript and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare no conflicting interests.
Acknowledgements
This work was supported by the Ministry of Knowledge Economy through Korea Research Institute of Chemical Technology (SI-1505, SI-1605, SI-1705), Brain Korea 21 (BK21) PLUS program, and the National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2018M3A9G2062552).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.08.004.
Appendix A. Supplementary Data
Supplementary material 1
Supplementary material 2
References
- 1.Scott D.E., Bayly A.R., Abell C., Skidmore J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat Rev Drug Discov. 2016;15(8):533. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson J.C., Cepero E., Boise L.H., Duckett C.S. Upstream regulatory role for XIAP in receptor-mediated apoptosis. Mol Cell Biol. 2004;24(16):7003–7014. doi: 10.1128/MCB.24.16.7003-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulathila R., Vash B., Sage D., Cornell-Kennon S., Wright K., Koehn J. The structure of the BIR3 domain of cIAP1 in complex with the N-terminal peptides of SMAC and caspase-9. Acta Crystallogr D Biol Crystallogr. 2009;65(1):58–66. doi: 10.1107/S0907444908039243. [DOI] [PubMed] [Google Scholar]
- 4.Fesik S.W., Shi Y. Controlling the caspases. Science. 2001;294(5546):1477–1478. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- 5.Huang J.-W., Zhang Z., Wu B., Cellitti J.F., Zhang X., Dahl R. Fragment-based design of small molecule X-linked inhibitor of apoptosis protein inhibitors. J Med Chem. 2008;51(22):7111–7118. doi: 10.1021/jm8006992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., Sun C., Olejniczak E.T., Meadows R.P., Betz S.F., Oost T. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408(6815):1004. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 7.Mannhold R., Fulda S., Carosati E. IAP antagonists: promising candidates for cancer therapy. Drug Discov Today. 2010;15(5–6):210–219. doi: 10.1016/j.drudis.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Vucic D., Deshayes K., Ackerly H., Pisabarro M.T., Kadkhodayan S., Fairbrother W.J. SMAC negatively regulates the anti-apoptotic activity of melanoma inhibitor of apoptosis (ML-IAP) J Biol Chem. 2002;277(14):12275–12279. doi: 10.1074/jbc.M112045200. [DOI] [PubMed] [Google Scholar]
- 9.Wu G., Chai J., Suber T.L., Wu J.-W., Du C., Wang X. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408(6815):1008. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- 10.Sheng C., Dong G., Miao Z., Zhang W., Wang W. State-of-the-art strategies for targeting protein–protein interactions by small-molecule inhibitors. Chem Soc Rev. 2015;44(22):8238–8259. doi: 10.1039/c5cs00252d. [DOI] [PubMed] [Google Scholar]
- 11.Clackson T., Wells J.A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 12.Smith M.C., Gestwicki J.E. Features of protein–protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev Mol Med. 2012;14 doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozawa M., Ozawa T., Ueda K. Application of the fragment molecular orbital method analysis to fragment-based drug discovery of BET (bromodomain and extra-terminal proteins) inhibitors. J Mol Graph Model. 2017;74:73–82. doi: 10.1016/j.jmgm.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Ichihara O., Barker J., Law R.J., Whittaker M. Compound design by fragment-linking. Molecular Informatics. 2011;30(4):298–306. doi: 10.1002/minf.201000174. [DOI] [PubMed] [Google Scholar]
- 15.Kitaura K., Ikeo E., Asada T., Nakano T., Uebayasi M. Fragment molecular orbital method: an approximate computational method for large molecules. Chemical Physics Letters. 1999;313(3–4):701–706. [Google Scholar]
- 16.Fedorov D.G., Nagata T., Kitaura K. Exploring chemistry with the fragment molecular orbital method. Phys Chem Chem Phys. 2012;14(21):7562–7577. doi: 10.1039/c2cp23784a. [DOI] [PubMed] [Google Scholar]
- 17.Fedorov D.G., Kitaura K. Extending the power of quantum chemistry to large systems with the fragment molecular orbital method. Chem A Eur J. 2007;111(30):6904–6914. doi: 10.1021/jp0716740. [DOI] [PubMed] [Google Scholar]
- 18.Arulmozhiraja S., Matsuo N., Ishitsubo E., Okazaki S., Shimano H., Tokiwa H. Comparative binding analysis of dipeptidyl peptidase IV (DPP-4) with antidiabetic drugs–an ab initio fragment molecular orbital study. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M., Fukuzawa K., Ishikawa T., Mochizuki Y., Nakano T., Tanaka S. Ab initio fragment molecular orbital study of molecular interactions in liganded retinoid X receptor: specification of residues associated with ligand inducible information transmission. J Phys Chem B. 2008;112(38):12081–12094. doi: 10.1021/jp803369x. [DOI] [PubMed] [Google Scholar]
- 20.Chessari G., Buck I.M., Day J.E., Day P.J., Iqbal A., Johnson C.N. Fragment-based drug discovery targeting inhibitor of apoptosis proteins: discovery of a non-alanine lead series with dual activity against cIAP1 and XIAP. J Med Chem. 2015;58(16):6574–6588. doi: 10.1021/acs.jmedchem.5b00706. [DOI] [PubMed] [Google Scholar]
- 21.Tamanini E., Buck I.M., Chessari G., Chiarparin E., Day J.E., Frederickson M. Discovery of a potent nonpeptidomimetic, small-molecule antagonist of cellular inhibitor of apoptosis protein 1 (cIAP1) and X-linked inhibitor of apoptosis protein (XIAP) J Med Chem. 2017;60(11):4611–4625. doi: 10.1021/acs.jmedchem.6b01877. [DOI] [PubMed] [Google Scholar]
- 22.Jin X., Lee K., Kim N.H., Kim H.S., Yook J.I., Choi J. Natural products used as a chemical library for protein–protein interaction targeted drug discovery. J Mol Graph Model. 2018;79:46–58. doi: 10.1016/j.jmgm.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat Struct Mol Biol. 2002;9(9):646. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 24.Neumann T., Junker H., Schmidt K., Sekul R. SPR-based fragment screening: advantages and applications. Curr Top Med Chem. 2007;7(16):1630–1642. doi: 10.2174/156802607782341073. [DOI] [PubMed] [Google Scholar]
- 25.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 26.Nakano T., Kaminuma T., Sato T., Akiyama Y., Uebayasi M., Kitaura K. Fragment molecular orbital method: application to polypeptides. Chemical Physics Letters. 2000;318(6):614–618. [Google Scholar]
- 27.Fedorov D.G., Kitaura K. Pair interaction energy decomposition analysis. J Comput Chem. 2007;28(1):222–237. doi: 10.1002/jcc.20496. [DOI] [PubMed] [Google Scholar]
- 28.Heifetz A., Chudyk E.I., Gleave L., Aldeghi M., Cherezov V., Fedorov D.G. The fragment molecular orbital method reveals new insight into the chemical nature of GPCR–ligand interactions. J Chem Inf Model. 2015;56(1):159–172. doi: 10.1021/acs.jcim.5b00644. [DOI] [PubMed] [Google Scholar]
- 29.Fedorov D.G., Kitaura K. Subsystem analysis for the fragment molecular orbital method and its application to protein–ligand binding in solution. Chem A Eur J. 2016;120(14):2218–2231. doi: 10.1021/acs.jpca.6b00163. [DOI] [PubMed] [Google Scholar]
- 30.Heifetz A., Trani G., Aldeghi M., MacKinnon C.H., McEwan P.A., Brookfield F.A. Fragment molecular orbital method applied to lead optimization of novel interleukin-2 inducible T-cell kinase (ITK) inhibitors. J Med Chem. 2016;59(9):4352–4363. doi: 10.1021/acs.jmedchem.6b00045. [DOI] [PubMed] [Google Scholar]
- 31.Heifetz A., Aldeghi M., Chudyk E.I., Fedorov D.G., Bodkin M.J., Biggin P.C. Using the fragment molecular orbital method to investigate agonist–orexin-2 receptor interactions. Biochem Soc Trans. 2016;44(2):574–581. doi: 10.1042/BST20150250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexeev Y., Mazanetz M.P., Ichihara O., Fedorov D.G. GAMESS as a free quantum-mechanical platform for drug research. Curr Top Med Chem. 2012;12(18):2013–2033. doi: 10.2174/156802612804910269. [DOI] [PubMed] [Google Scholar]
- 33.Fedorov D.G., Kitaura K. Second order Møller-Plesset perturbation theory based upon the fragment molecular orbital method. J Chem Phys. 2004;121(6):2483–2490. doi: 10.1063/1.1769362. [DOI] [PubMed] [Google Scholar]
- 34.Fedorov D.G., Kitaura K., Li H., Jensen J.H., Gordon M.S. The polarizable continuum model (PCM) interfaced with the fragment molecular orbital method (FMO) J Comput Chem. 2006;27(8):976–985. doi: 10.1002/jcc.20406. [DOI] [PubMed] [Google Scholar]
- 35.Fedorov D.G., Alexeev Y., Kitaura K. Geometry optimization of the active site of a large system with the fragment molecular orbital method. J Phys Chem Lett. 2011;2(4):282–288. [Google Scholar]
- 36.Hayes J.M., Archontis G. Molecular dynamics-studies of synthetic and biological macromolecules. InTech; 2012. MM-GB (PB) SA calculations of protein-ligand binding free energies. [Google Scholar]
- 37.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discovery. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munnaluri R., Sivan S.K., Manga V. Molecular docking and MM/GBSA integrated protocol for designing small molecule inhibitors against HIV-1 gp41. Med Chem Res. 2015;24(2):829–841. [Google Scholar]
- 39.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 40.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 41.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 42.Humphreys D.D., Friesner R.A., Berne B.J. A multiple-time-step molecular dynamics algorithm for macromolecules. J Phys Chem. 1994;98(27):6885–6892. [Google Scholar]
- 43.Sun C., Cai M., Meadows R.P., Xu N., Gunasekera A.H., Herrmann J. NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP. J Biol Chem. 2000;275(43):33777–33781. doi: 10.1074/jbc.M006226200. [DOI] [PubMed] [Google Scholar]
- 44.Nikolovska-Coleska Z., Meagher J.L., Jiang S., Kawamoto S.A., Gao W., Yi H. Design and characterization of bivalent Smac-based peptides as antagonists of XIAP and development and validation of a fluorescence polarization assay for XIAP containing both BIR2 and BIR3 domains. Anal Biochem. 2008;374(1):87–98. doi: 10.1016/j.ab.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 45.Sun H., Stuckey J.A., Nikolovska-Coleska Z., Qin D., Meagher J.L., Qiu S. Structure-based design, synthesis, evaluation, and crystallographic studies of conformationally constrained Smac mimetics as inhibitors of the X-linked inhibitor of apoptosis protein (XIAP) J Med Chem. 2008;51(22):7169–7180. doi: 10.1021/jm8006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cossu F., Malvezzi F., Canevari G., Mastrangelo E., Lecis D., Delia D. Recognition of Smac-mimetic compounds by the BIR domain of cIAP1. Protein Sci. 2010;19(12):2418–2429. doi: 10.1002/pro.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzoni L., Belvisi L., Bianchi A., Conti A., Drago C., De Matteo M. Homo-and heterodimeric Smac mimetics/IAP inhibitors as in vivo-active pro-apoptotic agents. Part I: synthesis. Bioorg Med Chem. 2012;20(22):6687–6708. doi: 10.1016/j.bmc.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 48.Hashimoto K., Saito B., Miyamoto N., Oguro Y., Tomita D., Shiokawa Z. Design and synthesis of potent inhibitor of apoptosis (IAP) proteins antagonists bearing an octahydropyrrolo [1, 2-a] pyrazine scaffold as a novel proline mimetic. J Med Chem. 2013;56(3):1228–1246. doi: 10.1021/jm301674z. [DOI] [PubMed] [Google Scholar]
- 49.Wu P.-P., Kuo S.-C., Huang W.-W., Yang J.-S., Lai K.-C., Chen H.-J. (-)-Epigallocatechin gallate induced apoptosis in human adrenal cancer NCI-H295 cells through caspase-dependent and caspase-independent pathway. Anticancer Res. 2009;29(4):1435–1442. [PubMed] [Google Scholar]
- 50.Ly B.T.K., Chi H.T., Yamagishi M., Kano Y., Hara Y., Nakano K. Inhibition of FLT3 expression by green tea catechins in FLT3 mutated-AML cells. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2