Abstract
LodA-like proteins are oxidases with a protein-derived cysteine tryptophylquinone (CTQ) prosthetic group. In Pseudoalteromonas luteoviolacea glycine oxidase (PlGoxA), CTQ biosynthesis requires post-translational modifications catalyzed by a modifying enzyme encoded by goxB. The PlGoxB protein was expressed and shown to possess a flavin cofactor. PlGoxB was unstable in solution as it readily lost the flavin and precipitated. PlGoxB precipitation was significantly reduced by incubation with either excess FAD or an equal concentration of prePlGoxA, the precursor protein that is its substrate. In contrast, the mature CTQ-bearing GoxA had no stabilizing effect. A homology model of PlGoxB, was generated using the structure of Alkylhalidase CmIS. The FAD-binding site of PlGoxB in the model was nearly identical to that of the template structure. The bound FAD in PlGoxB had significant solvent exposure, consistent with the observed tendency to lose FAD. This also suggested that interaction of prePlGoxA with PlGoxB at the exposed FAD-binding site could prevent the observed loss of FAD and subsequent precipitation of PlGoxB. A docking model of the putative PlGoxB-prePlGoxA complex was consistent with these hypotheses. The experimental results and computational analysis implicate structural features of PlGoxB that contribute to its stability and function.
Keywords: Flavin, flavoprotein, quinoprotein, protein-protein interaction
Introduction
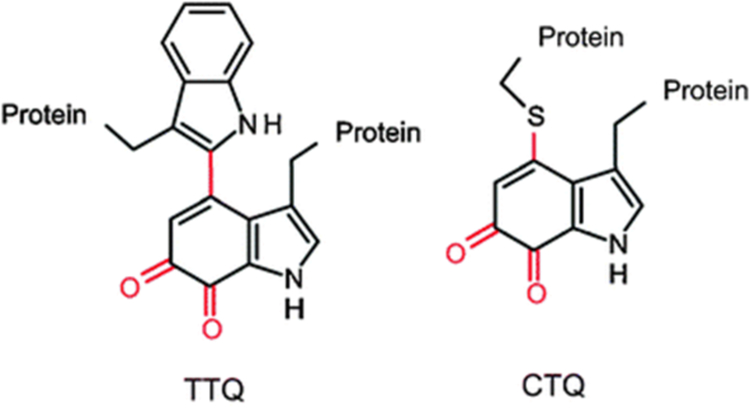
Quinoproteins are enzymes that utilize a quinone cofactor in the active site. The quinone species is either sourced exogenously, as with pyrroloquinoline quinone-dependent enzymes [1, 2], or produced endogenously through post-translational modifications of active site residues [2–4]. These modifications form redox-active catalytic species groups derived from tyrosine or tryptophan residues [5]. The protein-derived tryptophylquinone cofactors described thus far are tryptophan tryptophylquinone (TTQ), which is present in amine dehydrogenases, and cysteine tryptophylquinone (CTQ), which is found in dehydrogenases and oxidases [6] (Figure 1). In each of these tryptophylquinone cofactors, two oxygens have been inserted into a tryptophan side-chain, which is also covalently linked to either another tryptophan or a cysteine side chain. These post-translational modifications are catalyzed by modifying enzymes that are encoded by genes in the same operon as the genes for the precursor proteins. The topic of this study is a modifying enzyme, PlGoxB, which is required for the post-translational modifications to form CTQ on a glycine oxidase from Pseudoalteromonas luteoviolacea CPMOR-2 (PlGoxA).
Figure 1.
Protein-derived tryptophylquinone cofactors. Tryptophan tryptophylquinone (TTQ), cysteine tryptophylquinone (CTQ)
A family of CTQ-dependent oxidases has been designated LodA-like proteins, as the first one to be characterized was an L-lysine-ε-oxidase (LodA) from the marine bacterium Marinomonas mediterranea [7, 8]. The second was a glycine oxidase from the same bacterium (MmGoxA) [9]. These CTQ-bearing oxidases are a class of enzymes distinct from other amino acid oxidases that utilize a flavin cofactor for catalysis. Phylogenetic analysis of LodA-like proteins described five different major groups, I-V, and subgroups [10]. LodA is in Group I and MmGoxA is in Group IIB. The post-translational modifications required for production of the mature active LodA-like proteins requires modifying enzymes specific for each protein, LodB [11, 12] and MmGoxB [13], respectively. Expression of lodA or MmgoxA alone, without the gene for the modifying enzyme, yielded an inactive precursor protein (preLodA or preMmGoxA) that lacks the quinone functional group [14]. Analysis by mass spectrometry revealed that these precursor protein did have a single hydroxyl group present on the tryptophan side chain, but no additional modifications [14]. This initial hydroxylation is thought to be an autocatalytic event requiring copper [15]. In fact, for all of the lodA-like genes that have been identified, a lodB-like gene is present in the same operon, usually downstream of the lodA-like gene [10]. Recombinant expression of only the His-tagged lodB or MmgoxB gene from M. mediterranea did not yield any detectable protein. However, co-expression of lodA or MmgoxA with lodB or MmgoxB, respectively, did yield the mature enzymes [13, 16]. Thus, LodB and MmGoxB must be getting expressed in the system, and the lack of recovery of either LodB or MmGoxB when expressed alone is possibly due to rapid degradation of the protein. Furthermore, the expression of the inactive precursor proteins, preLodA and preMmGoxA, exhibited much lower yields when expressed alone, than the yields of the mature proteins that were expressed in the presence of their respective modifying enzyme. This suggests that the interaction between the proteins and the modifying enzymes may exert stabilizing effects on each other [13].
Recently, another LodA-like protein was characterized. It is a glycine oxidase from Pseudoalteromonas luteoviolacea, PlGoxA [17]. It was assigned to Group IID of the LodA-like proteins. In contrast to M. mediterranea, P. luteoviolacea only possesses this LodA-like protein and does not contain a LodA with L-lysine-ε-oxidase activity. Again, expression of mature active PlGoxA requires co-expression of PlgoxA with PlgoxB. In contrast to what is describe above for LodB and MmGoxB, expression of PlgoxB alone did yield the PlGoxB protein, which possessed a flavin cofactor. While the protein was unstable and prone to precipitation, in this study it was possible to characterize some of its physical properties and identify factors that enhanced the stability of the protein. A homology model was also constructed to provide insight into the structural features of PlGoxB that contribute to its stability and interaction with prePlGoxA.
Materials and Methods
Expression and purification.
Mature PlGoxA [17] and preMmGoxA [13] were expressed in E.coli and purified as previously described. PlGoxB and prePlGoxA were expressed in E. coli Rosetta cells. In each case, cells were transformed with a pET15 plasmid containing the gene to be expressed with a hexa-histidine tag at the N-terminus. Cells were grown overnight at 30° C to an OD of 0.8, at which point they were induced with 1 mM IPTG for four hours at the same temperature. Cells were harvested by centrifugation and lysed through sonication in 50 mM potassium phosphate, pH 7.5, on ice. The cell lysate was centrifuged and the supernatant was applied to Ni-NTA affinity column. The column was washed with an imidazole gradient in the same buffer. PlGoxB and prePLGoxA eluted between 30 and 150 mM imidazole, with the majority of each protein eluting at about 60 mM.
Analytical techniques.
Purity and size of proteins was determined by SDS-PAGE and size exclusion chromatography using a HiPrep 16/60 column packed with Sephacryl S-300 HR collected in an ÄKTA Pure FPLC system (GE Healthcare Life Sciences, Pittsburgh PA, USA). Absorption spectra were recorded using an HP 8452 Diode Array spectrophotometer controlled with Olis Globalworks software (Olis, Bogart, GA). Protein precipitation studies were monitored by measurement of light scattering at 550 nm. These studies were performed 50 mM potassium phosphate, pH 7.5, at 30 °C. Staining to identify the His-tagged peptide of the cleaved prePlGoxA on SDS-PAGE was performed as follows. Post-electrophoresis, the gel was incubated for one h in three 100 mL exchanges of H2O. It was then incubated in 50 mL of 6xHis Protein Tag Stain (Thermo Scientific, Rockford IL, USA) for 5 min followed by incubation in H2O as before. This was followed by incubation in 50 mL of 6xHis Protein Tag Developer (Thermo Scientific) for 15 min. A third water incubation was performed, at which point the gel is exposed to 300 nm light to observe fluorescence from the His tag.
Homology modeling of GoxB.
Homology models of PlGoxB were generated using the Swiss-Model Homology modelling online software (swissmodel.expasy.org) [18]. Template searches with BLAST [19] and HHBlitts [20] were performed against the Swiss-Model template library which includes the PDB. A docking model was constructed with using the ZDOCK software and server (zdock.umassmed.edu) [21]. Figures were prepared using Pymol (https://www.pymol.org).
Results
Physical and spectroscopic properties of PlGoxB.
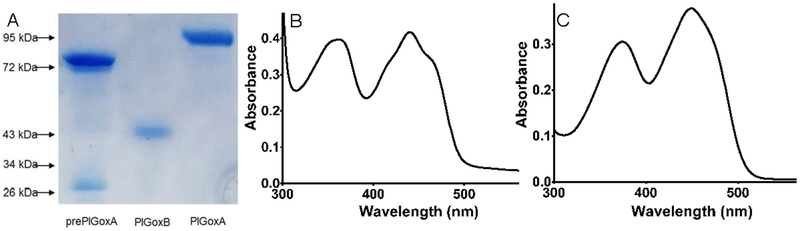
The yield of PlGoxB from the recombinant expression system was approximately 3 mg per liter of cultured cells. However, after isolation the PlGoxB had limited stability and precipitated over several minutes from solution. The pure PlGoxB yielded a single band on SDS-PAGE at a position consistent with the mass of the protein predicted from the sequence of 41.4 kDa (Figure 2A). When subjected to size exclusion chromatography, PlGoxB eluted in the void volume most likely to due to precipitation and aggregation that occurred during the procedure. Thus, the native molecular weight could not be determined. The absorbance spectrum of PlGoxB clearly showed the presence of flavin with two peaks at approximately 360 and 440 nm (Figure 2B). After the spontaneous precipitation of PlGoxB from solution followed by centrifugation, the pellet was white and the spectrum of the supernatant contained the flavin (Figure 2C), consistent with it being non-covalently bound.
Figure 2.
Physical properties of proteins and cofactors. A. SDS-PAGE of prePlGoxA, PlGoxB and PlGoxA. The positions of molecular weight markers are indicated on the left. B. Visible absorbance spectra of PlGoxB. C. Visible absorbance spectra of the flavin released from PlGoxB during precipitation.
The yield of prePlGoxA was approximately 1.5 mg per liter of cultured cells. When the protein was subjected to SDS-PAGE, two bands were observed at approximately 72 and 20 kDa. In contrast, mature PlGoxA migrates as a single band at the predicted mass of 91 kDa (Figure 2A). This result suggests the prePlGoxA may have been cleaved during expression or purification, but that the two segments remained tightly bound since they co-purified. The sum of the molecular weights of the two components of the isolated prePlGoxA are approximately equals the expected size of mature PlGoxA protein. Fluorescent His tag staining of the gel after SDS-PAGE indicated that the 72 kDa peptide was at the N-terminus and the smaller 20 kDa species was the C-terminal peptide. It is noteworthy that in previous studies [14], preLodA and preMmGox were isolated as intact proteins of the expected molecular weight with no evidence of cleavage.
Factors affecting the stability of PlGoxB.
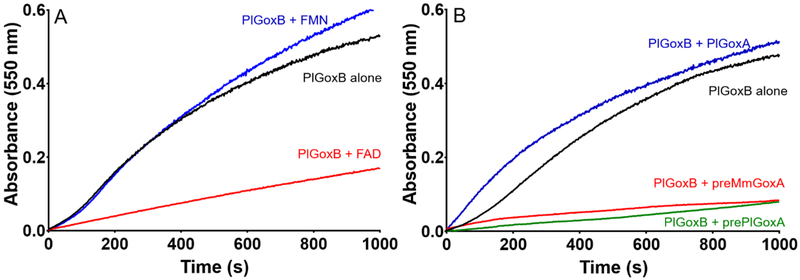
The rate of precipitation of PlGoxB was monitored and quantitated by the rate of increase in absorbance at 550 nm due to light scattering. As stability of flavoproteins is often an issue, several different buffer conditions were tested for enhancement of stability. TRIS and HEPES buffer were used as an alternative to potassium phosphate. Values of pH from 6.5 to 9.0 were tested. Addition of up to 300 mM NaCl or 12% glycerol were examined. None of these modifications to the buffer significantly reduced the rate of precipitation. As the flavin detached from the protein during this process, the effects of added FAD and FMN to the solution on the rate of precipitation were examined (Figure 3A). The presence of FAD significantly retarded the rate of precipitation of PlGoxB. In contrast, addition of FMN had no effect. This is consistent with FAD being the flavin cofactor for PlGoxB. This result also suggests that loss of FAD is the initial step in the precipitation process, and that after loss of FAD the apoPlGoxB molecules then precipitate.
Figure 3.
Factors that influence the stability of PlGoxB in solution. A. The effects of externally added FMN and FAD on the rate of precipitation of PlGoxB as monitored by the rate of increase in absorbance at 550 nm. The concentration of PlGoxB was 5 μM and the concentrations of FMN and FAD were 60 μM. B. The effects of externally added GoxA precursor proteins and mature PlGoxA on the rate of precipitation of PlGoxB as monitored by the rate of increase in absorbance at 550 nm. The concentrations of all proteins were 5 μM.
Since PlGoxB must interact with prePlGoxA in vivo, the effect of addition of prePlGoxA on the rate of precipitation of PlGoxB was examined. The presence of prePlGoxA significantly reduced the rate of precipitation of PlGoxB (Figure 3B). In contrast, addition of mature PlGoxA did not stabilize the PlGoxB. The precursor protein of MmGoxA was also tested and incubation with the preMmGoxA significantly reduced the rate of precipitation. As a control two unrelated proteins, albumin and thioredoxin, were added to the solution and these did not significantly decrease the rate of precipitation of PlGoxB (data not shown). Thus, the stabilizing effect is specific for the protein precursors of GoxA.
Homology modeling.
Homology modelling was performed with the Swiss-Model Homology online software [22]. The software scanned the PDB data base and filtered 3140 templates. Table 1 displays the four templates with the highest QMEAN Z-scores. The QMEAN Z-score [23] provides an estimate of the quality of the structural model. Scores around zero indicate good agreement between the model structure and experimental structures of similar size, and scores below −4.0 are considered low quality. These were the only four models that scored higher than −4.0. The highest scoring template, Alkylhalidase CmIS [24], is a much larger protein, having 566 residues compared to 358 for PlGoxB, so the agreement is actually higher for the overlapping portions than the score indicates.
Table 1.
Highest scoring template structures for modeling the structure of GoxB
| Protein | PDB entry | Coverage | Sequence similaritya | QMEAN Z-scoreb |
|---|---|---|---|---|
| Alkylhalidase CmlS | 3i3l | 0.95 | 0.31 | −2.90 |
| Tryptophan halogenase | 2pyx | 0.92 | 0.28 | −2.99 |
| Halogenase | 3e1t | 0.95 | 0.34 | −3.37 |
| Kynurenine 3-monooxygenase | 5mzc | 0.94 | 0.27 | −3.47 |
The similarity between the target and template sequences is calculated from a normalized BLOSUM62 substitution matrix [44].
The QMEAN Z-score [23] provides an estimate of the quality of the structural model. Scores around zero indicate good agreement between the model structure and experimental structures of similar size and scores below −4.0 indicate models with low quality.
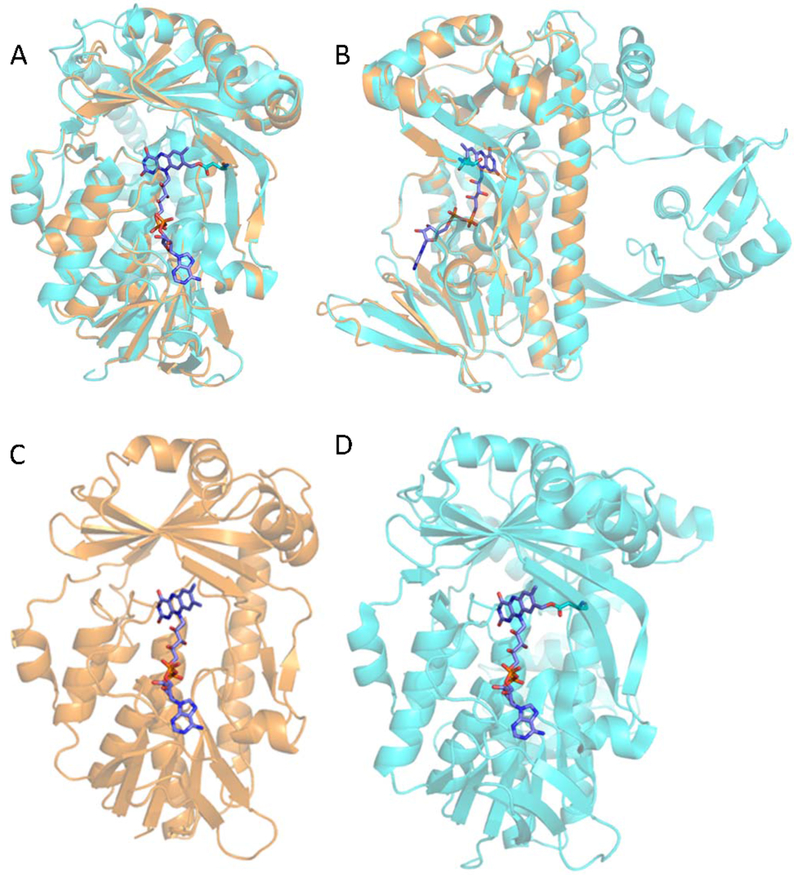
The homology model of GoxB is superimposed with the structure of Alkylhalidase CmlS in Figure 4A. The FAD ligand was not present in the GoxB model. As such, the FAD from the Alkylhalidase CmIS structure was introduced into the GoxB model at the same position. The major distinction in the overall structures of GoxB and Alkylhalidase CmIS is the unusual C-terminal domain of Alkylhalidase CmIS that extends from the domain, which is highly similar to that in PlGoxB (Figure 4B). The homologous domains contain several α-helices and β-sheets that overlay nicely (Figure 4 C and D).
Figure 4.
Homology model of PlGoxB. A. The model of PlGoxB is in orange and the crystal structure of the template, Alkylhalidase CmIS (PDB entry 3i3l) is in light blue. The FAD ligand is represented in stick form. A. A view of the model of PlGoxB superimposed the crystal structure of the template, Alkylhalidase CmIS with the FAD binding site facing forward. B. A view of the superimposed structures from a different angle rotated 90° that highlights the large C-terminal domain, which is present in the template but not in PlGoxB. C. and D. Comparison of PlGoxB model (C) and Alkylhalidase CmIS structure (D) which highlights the conservation of positions of α-helices and β-sheets in the main flavin-binding domain of the proteins.
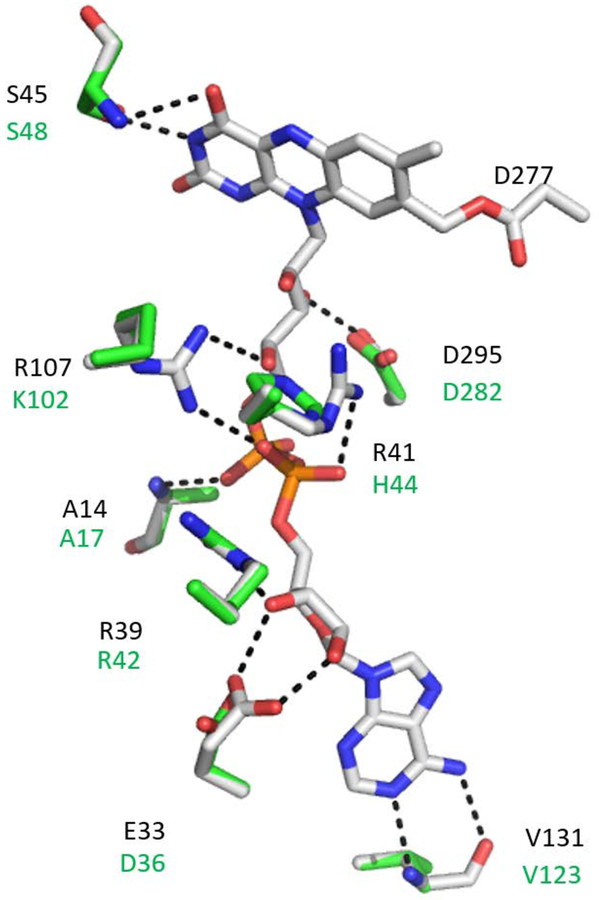
In each structure, the interactions of the host protein are primarily with the tail of the FAD cofactor (Figure 5). There are few stabilizing interactions between the proteins and the isoalloxazine ring of the flavin. However, in each interaction with a Ser residue is common. Several residues in the two proteins that interact with the FAD are conserved or have similar functional groups. In Figure 5 the residues of Alkylhalidase CmIS that are with 3Å of FAD are shown with the corresponding residues in the PlGoxB model superimposed. A major distinction between the Alkylhalidase CmIS structure and the PlGoxB model with regard to interaction with FAD is the unusual type of covalent bond between the flavin and the protein in the former. A covalent bond between the side-chain of Asp277 and a carbon on the flavin ring is present in Alkylhalidase CmIS. There is no Asp in this position in the PlGoxB model, and experimental data indicates that the FAD in PlGoxB is not covalently bound. This may explain why the FAD of PlGoxB readily dissociates in solution, leading to precipitation. The members of the family of flavin-dependent halogenases, which includes Alkylhalidase CmIS, share a structural motif of D(W/Y)SY. The Asp residue in this motif is the one that is covalently bound in Alkylhalidase CmIS. However, other members with this motif do not necessarily have the covalent link to the cofactor [24]. Interestingly, the only members of this family that lacks this motif are tryptophan halogenases, which exhibited the second best QMEAN Z-score (Table 1) during the modelling. That particular structure (PDB entry 2pyx) was not used in the modeling because FAD was not present in its flavin-binding site in that crystal structure.
Figure 5.
Interactions between FAD and amino acid residues in the FAD-binding site. The FAD and residues within 3Å of FAD in the Alkylhalidase CmIS structure are displayed with the corresponding residues in the PlGoxB model superimposed. The interactions with the FAD that are inferred in the Alkylhalidase CmIS structure are shown as dashed lines. The covalent bond between Asp277 of Alkylhalidase CmIS and FAD, which is not present in PlGoxB is also shown. Three additional residues within 3.5 Å, which are not shown for clarity, are V42/L45, S306/S292 and G307/G294. Black lettering indicates the Alkylhalidase CmIS residues and the green lettering indicated the PlGoxA residues. The carbons in FAD and the residues from Alkylhalidase CmIS are grey, and the carbons of the residues of PlGoxB are green. Oxygens are red, Nitrogens are blue and Phosphorus is orange.
Docking model of the prePlGoxA-plGoxB complex
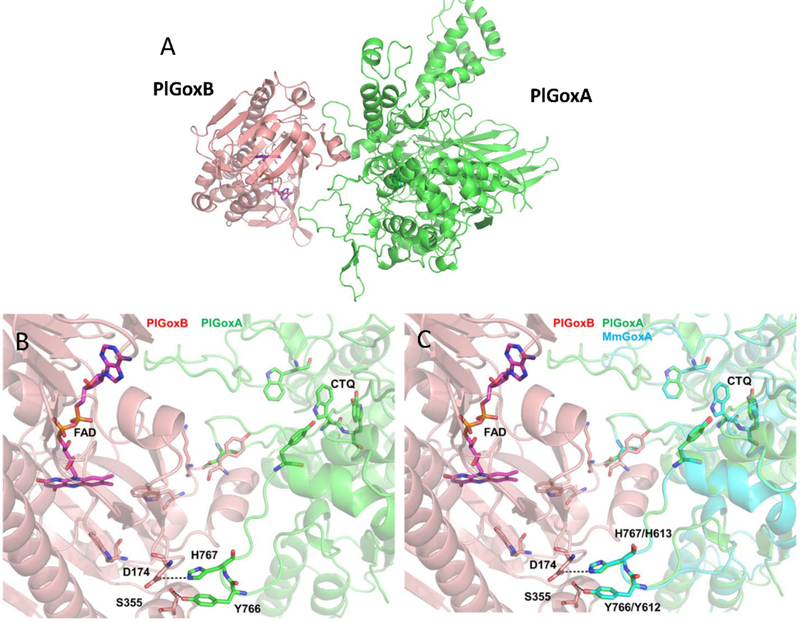
In order to gain insight into the nature of the protein-protein interface within the prePlGoxA-PlGoxB complex, a docking model was constructed with the ZDOCK software and server [21]. The homology model of PlGoxB was used as ligand and the structure of a PlGoxA monomer from the structure of the homotetramer (PDB entry 6byw) [17] was the receptor. The highest ranking docking model is shown in Figure 6A. In this model, the portion of PlGoxB where the FAD resides faces a groove in PlGoxA that leads to the active site tryptophan of CTQ (Figure 6B). This conformation is consistent with the observation that interaction with prePlGoxA prevents the loss of FAD from PlGoxB. Interestingly, the Tyr766 and His767 of PlGoxA interact with Ser155and Asp174, respectively, of PlGoxB via hydrogen and ionic bond. In the mature homotetramer, which does not stabilize PlGoxB, Tyr766 and His767 of PlGoxA stabilize the bound glycine substrate in an unusual manner[17]. The residues on one subunit point towards the active site of an adjacent subunit and interact with the carboxyl group of the glycine substrate. This suggests possible roles for these residues in both interaction of prePlGoxA with PlGoxB and catalysis by mature PlGoxA.
Figure 6.
A. Docking model of the PlGoxA-PlGoxB complex. B. Interface between PlGoxA and PlGoxB in the docking model. C. Docking model of the MmGoxA-PlGoxB complex (blue) superimposed with the model of the PlGoxA-PlGoxB complex (green).
Since the preMmGoxA was also able to stabilize prePlGoxB in solution, it was of interest to determine whether a similar interaction could be inferred for that protein. The crystal structure of MmGoxA has not been obtained. As such, a homology model was generated of the MmGoxA monomer using the MmGoxA sequence and the PlGoxA monomer structure. This MmGoxA model was then docked to PlGoxB. A similar result was obtained (Figure 6c) with the portion of MmGoxA that interacts with PlGoxB aligning very closely with that of PlGoxA. The Tyr and His residues discussed above are in an identical position in the docking model using MmGoxA. Comparison of the sequences of MmGoxA and PlGoxA shows 29% identity. However, much of the difference is due the 20 kDa larger mass of PlGoxA relative to MmGoxA. As seen in the structural model, the core structure of the proteins in which CTQ resides shows extensive homology between the two proteins, especially in the active site.
DISCUSSION
This study describes the first expression and purification of a flavoprotein that is required to catalyze the multiple post-translational modifications that generate CTQ on a LodA-like protein. The limited stability of PlGoxB allowed for only a partial characterization, but the experimental results and determination of a reasonable structural model of the protein provide significant insights into the structure and function of this enzyme. The presence of non-covalently bound flavin was demonstrated. It was observed that the flavin was loosely bound and lost, leading to precipitation of the protein. The presence of external FAD, but not FMN, significantly decreased the rate of precipitation of the protein. Furthermore, it was shown that the stability of both PlGoxB and prePlGoxA are enhanced by the presence of the partner protein. Expression of prePlGoxA in the absence of PlGoxB yields a cleaved protein product. Expression of PlGoxB in the absence of prePlGoxA yields PlGoxB that is unstable and precipitates unless prePlGoxA is also present. It is noteworthy that while prePlGoxA stabilizes PlGoxB, the presence of the mature PlGoxA exerted no stabilization at all, a result replicated with other unrelated proteins. Only preMmGoxA had a similar stabilizing effect. These results suggest that prePlGoxA and preMmGoxA each have a similar site that interacts with PlGoxB, and that is accessible in the precursor but not in the mature protein. The prePlGoxa precursor is a monomer and the mature protein PlGoxA is a tetramer [17], so it is likely that this site is at the subunit-subunit interface in the mature protein, and thus inaccessible.
The homology modeling of PlGoxB yielded several interesting results. It is noteworthy that the three best hits in the search for a template structure are different flavin-dependent halogenases. While there is no indication that PlGoxB exhibits such an activity, the mechanism of action of these halogenase may share common features. During the reaction mechanism of these halogenases [25, 26], it is proposed that Cl− reacts with a FAD-C4a-OOH intermediate to generate a reactive HOCl species. However, the site of halogenation of the substrate is at a site distant from the flavin. For tryptophan 7-halogenase the substrate tryptophan binding site and the FAD are separated by a 20 Å long tunnel [27]. Similarly, for PlGoxB, the FAD must somehow catalyze the oxidation of amino acid residues located inside the prePlGoxA protein substrate, distant from the FAD. The PlGoxB model structure reveals that the FAD is relatively exposed, consistent with the observation that the bound FAD is easily lost in solution. The structural model of PlGoxB also shows that the pocket in which the flavin resides is large enough to allow for FAD binding and still leave room for significant solvent exposure. Although FMN can fit into this pocket, the interactions between protein residues and FAD primarily involve the portion of the FAD tail, which is not present in FMN. This would explain the inability of FMN to stabilize the protein in solution, as the excess FMN is not likely to stay bound to the protein. The stabilization of PlGoxB in solution by prePlGoxA could be a consequence of FAD being present at the PlGoxB-prePlGoxA interface. If so, then the protein-protein interaction could prevent the loss of the flavin. This would also make sense mechanistically, as this would minimize the distance from the FAD to the residues that are modified in in prePlGoxA. It is also possible that the substrate channel in the prePlGox could be positioned so that it serves as a tunnel connecting FAD to the active site. The features of the docking model of PlGoxB with the PlGoxA monomer (Figure 6) are consistent with these ideas.
These results raise the question of why PlGoxB, as well as MmGoxB and LodB, are so unstable. The latter two could not be isolated at all when expressed alone in the recombinant system. One must consider that these enzymes somehow generate a sufficiently high-potential state of FAD to oxidize amino acid residues on the substrate protein. One reason that these enzymes have a short lifetime may be that they have the potential to damage other proteins in the cell if they persist in the absence of their natural substrate. That the operons of LodA-like proteins all contain genes for both LodB-like proteins as well, strongly suggests that the LodB-like proteins are meant to be single-use in a short time frame. A similar observation was made concerning MauG, the diheme enzyme that catalyzes post-translational modifications on a precursor protein to generate the TTQ cofactor of methylamine dehydrogenase. It is present in the same inducible operon as the structural protein. The reaction mechanism of MauG involved generation of a high potential bis-FeIV species [28] that oxidized the residues on the substrate protein by long-range electron transfer via radical intermediates [29, 30]. Although the resting form of MauG was relatively stable, if the bis-FeIV state was generated in the absence of the natural substrate then the protein oxidized residues on itself and became inactive [31, 32].
In the case of most other protein-derived cofactors, the post-translational modifications that form the active site are autocatalytic events that do not require a modifying enzyme. The formation of topaquinone in copper amine oxidases requires the insertion of two oxygen atoms into the phenyl ring of a Tyr residue [33]. The mechanism of biosynthesis is an autocatlytic event that requires copper and O2 [34–38]. The same is true for the autocatalytic biosynthesis of lysine tyrosylquinone, in which one atom of oxygen is incorporated into the phenyl ring of a Tyr, and a covalent bond is formed between Tyr a Lys residue [39, 40] The formation of the reactive carbonyl moieties of the pyruvoyl cofactor [41] and 4-methylideneimidazole-5-one (MIO) cofactor [42, 43] are also autocatalytic events. Thus, the presence of a potentially damaging modifying enzyme is not a concern for the post-translational modifications that form these protein-derived cofactors. In contrast, such modifying enzymes are a central part of the biosynthesis of the tryptophylquinone cofactors.
HIGHLIGHTS.
GoxB, the enzyme required for CTQ biosynthesis in the glycine oxidase (GoxA) is a flavoprotein
GoxB interacts specifically with the GoxA precursor protein that lacks CTQ and not with mature GoxA
Interaction of GoxB with the precursor of GoxA stabilizes both of these unstable proteins
A homology model of GoxB reveals structural features that contribute to stability and function
Acknowledgments
The authors thank Yu Tang for providing technical assistance.
Funding Sources
This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health under awards R37GM41574 and R35GM130173 (V.L.D.).
ABBREVIATIONS
- CTQ
cysteine tryptophylquinone
- PlGoxA
glycine oxidase from Pseudoalteromonas luteoviolacea
- LodA
L-lysine-ε-oxidase
- MmGoxA
glycine oxidase from Marinomonas mediterranea
- TTQ
tryptophan tryptophylquinone
References
- [1].Davidson VL, Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase, Adv. Protein Chem 58 (2001) 95–140. [DOI] [PubMed] [Google Scholar]
- [2].Klinman JP, Bonnot F, Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ, Chem Rev 114 (2014) 4343–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davidson VL, Protein-Derived cofactors. Expanding the scope of post-translational modifications, Biochemistry 46 (2007) 5283–5292. [DOI] [PubMed] [Google Scholar]
- [4].Davidson VL, Protein-derived cofactors revisited: Empowering amino acid residues with new functions, Biochemistry 57 (2018) 3115–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Davidson VL, Generation of protein-derived redox cofactors by posttranslational modification, Mol Biosyst 7 (2011) 29–37. [DOI] [PubMed] [Google Scholar]
- [6].Yukl ET, Davidson VL, Diversity of structures, catalytic mechanisms and processes of cofactor biosynthesis of tryptophylquinone-bearing enzymes, Arch Biochem Biophys 654 (2018) 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gomez D, Lucas-Elio P, Sanchez-Amat A, Solano F, A novel type of lysine oxidase: L-lysine-epsilon-oxidase, Biochim Biophys Acta 1764 (2006) 1577–1585. [DOI] [PubMed] [Google Scholar]
- [8].Okazaki S, Nakano S, Matsui D, Akaji S, Inagaki K, Asano Y, X-ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine epsilon-oxidase from Marinomonas mediterranea, J Biochem 154 (2013) 233–236. [DOI] [PubMed] [Google Scholar]
- [9].Campillo-Brocal JC, Lucas-Elio P, Sanchez-Amat A, Identification in Marinomonas mediterranea of a novel quinoprotein with glycine oxidase activity, Microbiologyopen 2 (2013) 684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Campillo-Brocal JC, Chacon-Verdu MD, Lucas-Elio P, Sanchez-Amat A, Distribution in microbial genomes of genes similar to lodA and goxA which encode a novel family of quinoproteins with amino acid oxidase activity, BMC.Genomics 16 (2015) 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chacón-Verdú MD, Gómez D, Solano F, Lucas-Elío P, Sánchez-Amat A, LodB is required for the recombinant synthesis of the quinoprotein L-lysine-ε-oxidase from Marinomonas mediterranea, Appl Microbiol Biot 98 (2014) 2981–2989. [DOI] [PubMed] [Google Scholar]
- [12].Gomez D, Lucas-Elio P, Solano F, Sanchez-Amat A, Both genes in the Marinomonas mediterranea lodAB operon are required for the expression of the antimicrobial protein lysine oxidase, Mol Microbiol 75 (2010) 462–473. [DOI] [PubMed] [Google Scholar]
- [13].Sehanobish E, Campillo-Brocal JC, Williamson HR, Sanchez-Amat A, Davidson VL, Interaction of GoxA with its modifying enzyme and its subunit assembly are dependent on the extent of cysteine tryptophylquinone biosynthesis, Biochemistry 55 (2016) 2305–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chacón-Verdú MD, Campillo-Brocal JC, Lucas-Elío P, Davidson VL, Sánchez-Amat A, Characterization of recombinant biosynthetic precursors of the cysteine tryptophlquinone cofactors of L-lysine-epsilon-oxidase and glycine oxidase from Marinomonas mediterranea, Biochim Biophys Acta 1854 (2015) 1123–1131. [DOI] [PubMed] [Google Scholar]
- [15].Williamson HR, Sehanobish E, Shiller AM, Sanchez-Amat A, Davidson VL, Roles of copper and a conserved aspartic acid in the autocatalytic hydroxylation of a specific tryptophan residue during cysteine tryptophylquinone biogenesis, Biochemistry 56 (2017) 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sehanobish E, Shin S, Sanchez-Amat A, Davidson VL, Steady-state kinetic mechanism of LodA, a novel cysteine tryptophylquinone-dependent oxidase, FEBS Lett 588 (2014) 752–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Andreo-Vidal A, Mamounis K, Sehanobish E, Avalos D, Campillo-Brocal JC, Sanchez-Amat A, Yukl ET, Davidson VL, Structure and enzymatic properties of an unusual cysteine tryptophylquinone-dependent glycine oxidase from, Biochemistry 57 (2018) 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T, Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology, Sci Rep 7 (2017) 10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos KB, Thomas L Madden, BLAST+: architecture and applications, BMC Bioinformatics 10 (2009) 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Remmert M, Biegert A, Hauser A, Soding J, HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment, Nat Methods 9 (2011) 173–175. [DOI] [PubMed] [Google Scholar]
- [21].Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z, ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers, Bioinformatics 30 (2014) 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res 46 (2018) W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Benkert P, Biasini M, Schwede T, Toward the estimation of the absolute quality of individual protein structure models, Bioinformatics 27 (2010) 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Podzelinska K, Latimer R, Bhattacharya A, Vining LC, Zechel DL, Jia Z, Chloramphenicol biosynthesis: the structure of CmlS, a flavin-dependent halogenase showing a covalent flavin-aspartate bond, J Mol Biol 397 (2010) 316–331. [DOI] [PubMed] [Google Scholar]
- [25].Blasiak LC, Drennan CL, Structural perspective on enzymatic halogenation, Acc Chem Res 42 (2009) 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gkotsi DS, Dhaliwal J, McLachlan MM, Mulholand KR, Goss RJ, Halogenases: powerful tools for biocatalysis (mechanisms applications and scope), Curr Opin Chem Biol 43 (2018) 119–126. [DOI] [PubMed] [Google Scholar]
- [27].Dong C, Flecks S, Unversucht S, Haupt C, van Pée K-H, Naismith JH, Tryptophan 7-Halogenase (PrnA) structure suggests a mechanism for regioselective chlorination, Science 309 (2005) 2216–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li X, Fu R, Lee S, Krebs C, Davidson VL, Liu A, A catalytic di-heme bis-Fe(IV) intermediate, alternative to an Fe(IV)=O porphyrin radical, Proc. Natl. Acad. Sci. USA 105 (2008) 8597–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Abu Tarboush N, Jensen LMR, Yukl ET, Geng J, Liu A, Wilmot CM, Davidson VL, Mutagenesis of tryptophan199 suggets that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 16956–16961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jensen LM, Sanishvili R, Davidson VL, Wilmot CM, In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex, Science 327 (2010) 1392–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ma Z, Williamson HR, Davidson VL, Mechanism of protein oxidative damage that is coupled to long-range electron transfer to high-valent hemes, Biochem J 473 (2016) 1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yukl ET, Williamson HR, Higgins L, Davidson VL, Wilmot CM, Oxidative damage in MauG: implications for the control of high-valent iron species and radical propagation pathways, Biochemistry 52 (2013) 9447–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Janes SM, Mu D, Wemmer D, Smith AJ, Kaur S, Maltby D, Burlingame AL, Klinman JP, A new redox cofactor in eukaryotic enzymes: 6-hydroxydopa at the active site of bovine serum amine oxidase, Science 248 (1990) 981–987. [DOI] [PubMed] [Google Scholar]
- [34].Brazeau BJ, Johnson BJ, Wilmot CM, Copper-containing amine oxidases. Biogenesis and catalysis; a structural perspective, Arch Biochem Biophys 428 (2004) 22–31. [DOI] [PubMed] [Google Scholar]
- [35].Wilce MC, Dooley DM, Freeman HC, Guss JM, Matsunami H, McIntire WS, Ruggiero CE, Tanizawa K, Yamaguchi H, Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone, Biochemistry 36 (1997) 16116–16133. [DOI] [PubMed] [Google Scholar]
- [36].Kim M, Okajima T, Kishishita S, Yoshimura M, Kawamori A, Tanizawa K, Yamaguchi H, X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase, Nat Struct Biol 9 (2002) 591–596. [DOI] [PubMed] [Google Scholar]
- [37].Dove JE, Klinman JP, Trihydroxyphenylalanine quinone (TPQ) from copper amine oxidases and lysyl tyrosylquinone (LTQ) from lysyl oxidase, Adv Protein Chem 58 (2001) 141–174. [DOI] [PubMed] [Google Scholar]
- [38].DuBois JL, Klinman JP, Role of a strictly conserved active site tyrosine in cofactor genesis in the copper amine oxidase from Hansenula polymorpha, Biochemistry 45 (2006) 3178–3188. [DOI] [PubMed] [Google Scholar]
- [39].Moore RH, Spies MA, Culpepper MB, Murakawa T, Hirota S, Okajima T, Tanizawa K, Mure M, Trapping of a dopaquinone intermediate in the TPQ cofactor biogenesis in a copper-containing amine oxidase from Arthrobacter globiformis, J Am Chem Soc 129 (2007) 11524–11534. [DOI] [PubMed] [Google Scholar]
- [40].Wang SX, Mure M, Medzihradszky KF, Burlingame AL, Brown DE, Dooley DM, Smith AJ, Kagan HM, Klinman JP, A crosslinked cofactor in lysyl oxidase: redox function for amino acid side chains, Science 273 (1996) 1078–1084. [DOI] [PubMed] [Google Scholar]
- [41].Ramjee MK, Genschel U, Abell C, Smith AG, Escherichia coli L-aspartate-alpha-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry, Biochem J 323 (1997) 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Retey J, Discovery and role of methylidene imidazolone, a highly electrophilic prosthetic group, Biochim Biophys Acta 1647 (2003) 179–184. [DOI] [PubMed] [Google Scholar]
- [43].Poppe L, Methylidene-imidazolone: a novel electrophile for substrate activation, Curr Opin Chem Biol 5 (2001) 512–524. [DOI] [PubMed] [Google Scholar]
- [44].Henikoff S, Henikoff JG, Amino acid substitution matrices from protein blocks, Proc Natl Acad Sci U.S.A. 89 (1992) 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]