Abstract
Clinical trials, whether industry, cooperative group sponsored, or investigator initiated, have an unacceptable rate of failure as a result of the inability to recruit sufficient numbers of patients. Even those trials that are completed often require time-consuming protocol amendments to achieve accrual goals. These inefficiencies in clinical trial research result in increasing costs and prolong the time needed to bring improved treatments to cancer clinical practice. TriNetX has developed a clinical research collaboration platform—deployed by a federated network of health care organizations (HCOs), pharmaceutical firms (Pharma), and contract research organizations (CROs)—to enable data-driven clinical research study design to reduce accrual failure and protocol amendment. Currently, the network extends to 55 HCOs and covers 84 million patients, mostly within the United States, but with a growing international presence. (Many of the HCOs in United States are Clinical and Translational Science Awardees and/or National Cancer Institute–designated cancer centers.) The TriNetX business model includes Pharma and the CROs as sponsors whose subscriptions financially support the network, including the software and hardware costs of the HCOs. Furthermore, as each HCO network member has their data harmonized with the TriNetX model upon joining, data sharing among them does not require any technical processes to establish connectivity. To date, on the basis of the data on the network, HCOs have been presented approximately 757 studies by Pharma and CROs, and four data-sharing subnetworks have been formed among member HCOs.
INTRODUCTION
Despite recent improvements in clinical trial management, there remains a need for better design and patient recruitment strategies to reduce the significant number of trials that still fail to reach completion because of a lack of participation.1,2 Clearly, clinical research could benefit from leveraging the capabilities of new health informatics tools and data analytic approaches.3,4 Many clinical trialists and data scientists have focused on data sharing as a key pathway to improving clinical research efficiency and maximizing its value to researchers and patients.5
The number of participants who are enrolled directly correlates with the success of a study, and, consequently, low accrual for studies is a major barrier confronting diagnostic and therapeutic developments. It is estimated that up to 50% of trials are not completed because of insufficient enrollment.6,7 In addition, protocol amendments often cause delays and dramatically increase the costs of developing new therapies.8
The current approach to site selection is often an art. Decisions are frequently based on incorrect guestimates of patient availability or the site’s prior history with other projects. From the perspective of the participating site, the time that is required to obtain approval and activation of a study, on average, is 6 months to 1 year.9 Between 2008 and 2013, study activation costs for study sites increased to $50,000 per trial,10 a steady increase of 88%.11 Activation delays also result in the diminished relevance of these studies.12 To reduce study initiation costs and accelerate the opening of studies, many academic medical centers, pharmaceutical firms (Pharma), and contract research organizations (CROs) have identified process improvements to increase the efficiency of study activation.13
Whereas these efficiency improvement efforts are beneficial, consideration should also be given to the likelihood of successful patient accrual on the basis of site-specific real-world data. Using a site’s real-world data to analyze the size of the local patient population that satisfies a clinical trial’s eligibility criteria has been shown to accurately predict whether a trial could conceivably attain its accrual goals.14 Prospectively eliminating studies that lack the required patient cohort size via a data-driven prescreening analysis would reduce the financial loss associated with unsuccessfully accruing trials. Enterprise research repositories, such as those that are built on the Informatics for Integrating Biology and the Bedside (i2b2)15 platform, have been widely used in cohort identification for prospective studies, and those that have regular refreshes of clinical data can provide a reliable indication of prospective trial accrual.16
There are several data networks in existence across the globe—for example, Swedish Integrated Electronic Health Records17 and the United Kingdom’s National Cancer Data Repository18 —in which real-world electronic medical record data are used in clinical research for specific disease studies (asthma and cancer, respectively). The TriNetX Research Network expands on the use of real-world data. The goal of the TriNetX Research Network is to improve the efficiency of the clinical trial process by utilizing analyses of real-world data—for example, electronic health records (EHRs) and cancer registries—from network member sites in the trial design and site selection processes to reduce the risk of accrual failure and decrease the need for protocol amendments.
TRINETX FEDERATED DATA NETWORK
TriNetX was initially motivated by the desire of Pharma clinical trialists to make collaborative industry–academia clinical trial research more efficient—to use real-world data to design trials that have the potential to reach their accrual requirements, and to rationally identify performance sites that should be invited to open a trial. Beginning in 2015, TriNetX first approached health care organizations (HCOs) with well-established i2b2 research repositories to participate in the fledgling network by becoming data providers. Over the past 2 years, data harmonization processes within TriNetX have matured such that an i2b2 data source is no longer a requirement for HCO network data providers.
The TriNetX business model relies on industry sponsors—Pharma and CROs—who pay a subscription fee to query for aggregate counts (by HCO) across a hub-and-spoke network of research repositories populated by HCOs with deidentified patient data. It has been successful thus far, with 14 leading Pharma and CRO sponsors subscribed and 55 HCO data providers in the network. In less than 2 years, the network expanded to seven countries with approximately 84 million patients and 8.1 billion observation facts.
DATA-DRIVEN APPROACH TO INDUSTRY TRIAL STUDY DESIGN AND SITE SELECTION
Pharma and CROs can make site selection on the basis of the size of the relevant patient cohort at a given site, as well as other information, such as the rate at which patients of interest appear at the institution. These site-specific cohort sizes are the result of queries that correspond to the trial inclusion/exclusion rules for deidentified patient data provided by HCOs. Only aggregate counts are provided by each site, which minimizes the risk of patient reidentification.
If the Pharma and/or CRO trial sponsor determines that sufficient patient populations exist for the trial as designed, the company relies on TriNetX to establish whether there is interest from the HCO in the study. If there is, TriNetX connects the sponsor and the site so that the trial can be activated. For some trials, upon a decision to pursue participation in the trial by the HCO, the patients in the TriNetX cohort for that site may be reidentified on the basis of synthetic patient IDs. This process is mediated by the institution’s honest broker process, assuming all the necessary regulatory permissions are in place.
HCO BENEFITS OF TRINETX NETWORK MEMBERSHIP
The current 55 HCO TriNetX network members are mainly in the United States, with an additional six HCOs in other countries that include the United Kingdom, Germany, Italy, Israel, and Singapore. In return for providing deidentified clinical data, which includes patient demographics, diagnoses, procedures, medications, and laboratory and genomic test results, the HCO members receive benefits in three areas: data analytical tools for their own researchers, facilitated collaboration with industry trials, and facilitated data sharing with their HCO peers via the creation of peer networks that operate isolated from the main network to support clinical research, clinical trial design, the initiation of clinical trials, and other relevant operations.
First, HCOs obtain the TriNetX research data analytics software at no cost. This data visualization platform allows HCO researchers to use an English-language query tool with their institution’s clinical data to visualize data subsets for cohort definition and hypothesis generation. Deployment of the platform requires little institutional effort, as data harmonization to the TriNetX data model for each site is performed by the TriNetX engineering team. The platform complies with institutional review board requirements, as it consumes only deidentified data, and the industry sponsors of the network only obtain aggregate site counts. Furthermore, TriNetX provides an appliance hardware in the HCO’s secure data center for use with the software. (The motivation for TriNetX to provide the hardware is to assure the necessary responsiveness to sponsor queries.)
Second, with the increased number of targeted-therapy industry trials, multisite clinical trials are increasingly necessary. Membership in the TriNetX network allows Pharma and CROs to be aware of when an HCO has a desired patient subpopulation, thereby facilitating an industry–academic clinical research collaboration. Both parties benefit from these opportunities to work together.
Third, local investigator-initiated studies benefit from having access to data sets from other institutions to increase the available patient population for these studies. Network membership greatly expedites data sharing among HCO members. The process of establishing network membership requires the harmonization of the HCO’s data model with that of the TriNetX data model. As noted above, this work is performed by the TriNetX engineering team, which maps any institutional data source, with i2b2 no longer required. Because all members are harmonized with the TriNetX data model, members themselves also have harmonized data models; therefore, data sharing among network members does not require any technical effort, but does have legal and compliance requirements that must be met. TriNetX requires each participating HCO to establish a data use agreement among participants as well as to obtain institutional review board approval for human participant research-designated efforts. To date, several collaborative networks are already functioning, and multiple HCOs are in the process of creating large and small collaborative networks with peer institutions.
NETWORK INFRASTRUCTURE
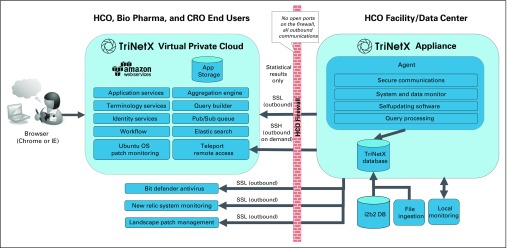
TriNetX has been established as multitenant software-as-a-service platform on Amazon Web Services (AWS), implementing the architecture depicted in Figure 1. HCO data that are accessible through the TriNetX network resides on the appliance located at each HCO data center. During the onboarding process, these data are loaded onto the appliance with a simplified extract-transform-load process that leverages the existing capabilities and scripting of the TriNetX agent. In addition to i2b2, TriNetX supports the loading of data from other source systems with a combination of product and service capabilities.
Fig 1.
TriNetX network architecture and modules. CRO, contract research organization; HCO, health care organization; i2b2 DB, Informatics for Integrating Biology and the Bedside database; IE, Internet Explorer; OS, operating system; Pharma, pharmaceutical firm; SSH, secure shell ; SSL, secure socket layer.
SECURITY
TriNetX is deployed in a secure Health Insurance Portability and Accountability Act–compliant virtual private cloud hosted by AWS, which supports the Federal Risk and Authorization Management Program, NIST 800-53, and other industry-standard security certifications. Access to TriNetX is over secure transport layer security with a 2,048-bit security certificate. TriNetX services that are hosted behind an AWS elastic load balancer are configured to use the AWS Elastic Load Balancer Security Policy 2015-05.19
The TriNetX appliance is highly secure (expert attestation by Brad Malin, PhD; full documentation is available to the members) and locked down for any extraneous system processes, and no processes listen for inbound connections; the appliance only initiates outbound communication. On a regular basis, TriNetX runs penetration testing against its hosted application environment as well as Nessus vulnerability scanning.
DATA OBFUSCATION
TriNetX has implemented several safeguards to minimize the risk of patient reidentification and to prevent exposing the data of a single HCO. First, for Pharma-initiated query results, all HCO patient counts are rounded to the next 10 so that the exact number of patients per site is obfuscated. Second, to avoid the risk that a series of individual queries could identify small subsets of cohorts, total patient counts greater than 10 are rounded up to the nearest 10. Lastly, when a query returns a patient count on a term and the patient count is ≤ 10, results shows the count as 10.
CLINICAL DATA, CONTROLLED TERMINOLOGIES, AND SEMANTIC MAPPING
Semantic frameworks are instrumental in minimizing potential data misinterpretations and discovery challenges that can arise during data integration and analyses. Ultimately, standardizing clinical—and other—data to publicly available coding systems will foster semantic integration and interoperability among the HCOs.
TriNetX began by supporting demographics, diagnosis, procedures, medications, and laboratories. As part of the ongoing data expansion initiative, tumor registry and molecular genomic data are now available on the network. Soon to be added are vital signs and additional observations that are pertinent to such selected therapeutic areas as oncology and pulmonology.
The data taken into the TriNetX appliance varies in provenance from institution to institution. Some HCOs extract data directly from their EHRs, whereas others have data warehouses with varying common data models 20, such as i2b2 and observational health data sciences and informatics. A typical commercial EHR uses a plethora of proprietary code system standards or terminology standards that may also vary by country—for example, the United States uses the Clinical Modification version of the International Classification of Diseases (ICD), Tenth Revision. In the United States, procedures are coded using ICD-10-PCS and Current Procedural Terminology (CPT), but there is no accepted standard for procedures in other countries. Many EHRs incorporate proprietary drug data, including First DataBank, Wolters Kluwer’s Medi-Span, and Cerner’s Multum, each of which has a different identifier for the same drug. Medications may also be coded to national drug codes or to anatomic therapeutic chemical—used in many European countries—or local codes. Many laboratory information systems at the HCOs and commercial laboratories rarely use standard codes—that is, Logical Observation Identifiers Names and Codes (LOINCs)—for test results.
To enable queries for such heterogeneous semantics across a federated network, data must be mapped to agreed-upon terminologies. TriNetX assumes responsibility for generating and maintaining such mappings for HCOs. In the United States, diagnoses and procedures are consistently coded to ICD and CPT and require no additional mapping. Medications are mapped to the RxNorm ingredients via the RxNorm Application Programming Interface,21 which allows code-to-code mapping and mapping on the basis of the text of the drug description. Laboratory test results are mapped to LOINC with the Regenstrief LOINC Mapping Assistant.
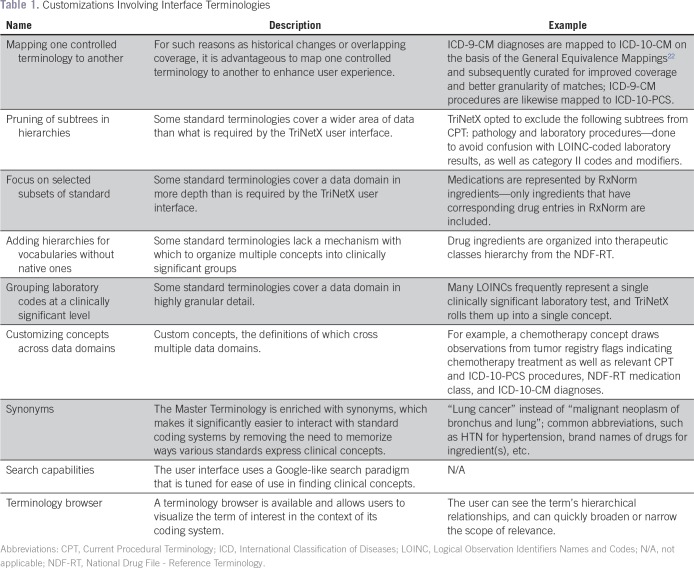
Users interact with TriNetX Master Terminology to enumerate eligibility criteria that describe patient cohorts of interest. The Master Terminology consists of 300,000 terms and standard vocabularies that cover data domains that are supported by the TriNetX application—demographics, such as patient sex, are drawn from Health Level 7 administrative standards; diagnoses use ICD22; procedures (ICD and CPT), medications (RxNorm), laboratory results (LOINC), and some oncology-specific data elements are coded to ICD-O; gene names are from the Human Genome Organization Gene Nomenclature Committee; and variants use codes from ClinVar, dbSNP, and dbVar, and are expressed using Human Genome Variation Society syntax. The Master Terminology undergoes extensive curation and customization to ensure high usability. Table 1 lists various types of customization.
Table 1.
Customizations Involving Interface Terminologies
ANALYTICS
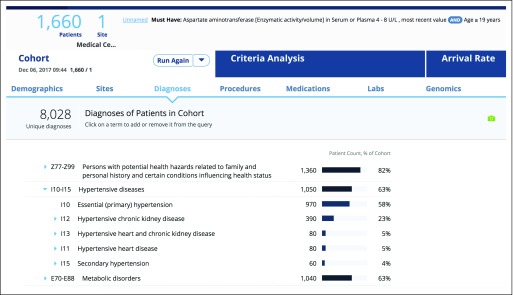
Once the user enters the eligibility criteria into the TriNetX query builder, the first action is to count the patients who conform to these criteria. Explore Cohort begins with the query criteria and requests clinical data about patients in the cohort. Results include a graphic summary of demographics, with age, sex, race, and ethnicity breakdowns. Age breakdown is presented in an interactive histogram, and diagnoses, procedures, and medications are presented as hierarchical lists of observations ordered by frequency. The user can get an understanding of which comorbidities are present in their patient cohort (Fig 2). The list of diagnoses can be filtered by time and chronic diseases on the basis of the Chronic Condition Indicator.23
Fig 2.
TriNetX query builder user interface. Shown are the results of cohort exploration analysis focusing on comorbidities in a patient cohort of interest.
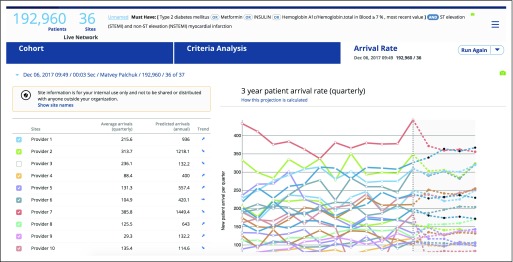
A logical consideration when characterizing a cohort of patients is how often patients like these appear at a given institution. Arrival rate analysis goes back in time and establishes a 3-year-long baseline by calculating the size of the cohort of interest on a quarterly basis in the past, then uses multiple logistic regression to project the size of the cohort into the future (Fig 3). The user gains an understanding of the relative velocity at which new qualifying patients appear at an HCO.
Fig 3.
Arrival rate graph representation on the basis of the historical data analyses for each health care organization.
Researchers who are responsible for study design and feasibility analyses are concerned with the relative impact of individual criteria and groupings of criteria on the overall size of the patient cohort, and are focused on optimizing the size of the cohort. We designed an analysis that precalculates cohort sizes for each permutation of eligibility criteria, which requires 2n total queries, where n is the number of criteria being analyzed, and presents the results as a funnel image for easy visualization of the impact of individual criteria on cohort size. The graph is interactive, with the user being able to remove one or more criteria from the funnel and see in real time the effect that that will have on the overall size of the cohort of interest.
The TriNetX application has extensive features that allow users to collaborate on studies by working in teams. A study can be shared among colleagues, and the application provides a comprehensive history of individual queries that shows the user their query logic and other pertinent information. The TriNetX application supports various modes of collaboration, including the ability to securely share necessary documents, as well as enabling collaboration among colleagues on individual studies and providing other workflow-related functionality.
DATA QUALITY
Varying data quality is among the major hurdles to the proper use of research data, and, in some cases, may compromise the validity of the research results.24 Although the adoption of EHRs has exploded as a result of federal incentives and meaningful use requirements, the quality of the data contained in EHRs and, correspondingly, that used in research is slowly improving. Many EHRs were designed and operate with billing and patient care functions in mind, which limits the quality of data for research purposes.16 Such potential limitations in observational data warrant a comprehensive data-quality framework and approach.25 There is limited literature on the topic of data quality, and the majority of work described has focused on assessing the quality of data in a single system or a single institution, as expected.26 Furthermore, efforts to quantify the goodness of data typically focus on whether the data are good enough for the primary purpose, which is providing clinical care to patients.
The data that underpin all analytic functions in TriNetX originates in the EHR systems and other systems used for treatment, payment, and operations. The data in these systems are collected mainly to provide clinical care and to satisfy the applicable regulatory requirements. For use by TriNetX, data are extracted from the source systems and undergo transformation, clean-up, deduplication, deidentification, optional obfuscation, and semantic mapping, etc. Despite not having direct control over the primary data, TriNetX nonetheless attempts to assess its quality. A comprehensive methodology has been developed to do so that consists of four Cs—cleanliness, consistency, correctness, and completeness.
Cleanliness is a high-level assessment performed upon data intake. These analyses are largely automatic and evaluate the requirements for basic formatting (dates as strings, codes with dots removed, etc), ensure the presence of required fields, check the referential integrity (eg, encounter IDs present in fact and dimension tables), and assess temporal variability across refreshes (the volume of facts over time).
Consistency begins with standard data profiling and proceeds to analyze the data by using deep domain-specific assessments. Typical data profiling collects the following metrics and basic statistics: percent of nulls, minimum, maximum, average, and standard deviation for fields with continuous variables. The volume of observations is tracked over time, including changes by data type, to ensure that the volume of observations remains steady over time. Semantic interoperability, or data mapping, is necessary for subsequent analyses of data against the codes in the TriNetX Master Terminology. Domain-specific analysis includes generating observation signatures, distribution of observations across a coding system for a given data domain, etc.
Correctness focuses on the evaluation of data that come from individual HCOs from a clinical perspective. Such evaluation is important for correct site selection on the basis of protocol criteria as well as for correct patient reidentification at the site level. At this level, structured validations are performed—that is, logic, context, and temporality—on the data.
Completeness establishes an agreement of network insights with external information via cross-network analysis. The importance of completeness evaluation lies in correct cohort analysis during protocol design and correct criteria analysis to evaluate the feasibility of eligibility criteria. A typical metric ascertains whether a result makes medical sense—for example, there is a higher risk of stroke for patients with atrial fibrillation and diabetes compared with that of patients with atrial fibrillation alone.
MEMBERSHIP AND CONTRACTING PROCESS
TriNetX is open to HCOs willing to participate in industry-sponsored trials and have operational patient data repositories. Because paid subscription is only required for CROs and Pharma, there is no associated cost with HCO participation. TriNetX desires to establish a business associate agreement with HCOs, which is required for those HCOs with limited data sets and optional for those joining with deidentified data sets. HCOs are expected to complete deidentification and perform iterative data quality improvement activities.
Joining a federated network and thus retaining data within institutional boundaries has had a positive influence on the time and effort required to obtain necessary legal, compliance, and other approvals, all of which took approximately 3 months. Upon delivery of the appliance, a biweekly call is established, and mapping and the intake of data are completed within 2 months.
DISCUSSION
Several large-scale, real-world data networking efforts have been made in the past,27 and such efforts continue today. Past attempts provide learned lessons for strategies to avoid. The existing data networks all have different focuses, and TriNetX has taken a nonexclusionary approach of coexistence with them given its specific goal of improving clinical research efficiency. With its focus on building a large federated network of HCOs, TriNetX relies on its business model of monetizing HCO data by providing, at no cost, data visualization software and hardware for institutional use, the ease of intermember data sharing, and facilitated access to industry–academia research collaboration. This strategy has quickly gained interest in the community, with a rapidly growing international network of HCOs and industry sponsors. The business model does not rely on government funding and will be sustainable if the underlying premise—data-driven clinical trial design and trial site selection as implemented by TriNetX—proves to be a more efficient pathway to successfully completing clinical trials.
FUTURE WORK
TriNetX aims to increase HCO, CRO, and Pharma participation. With regard to data security, TriNetX plans to obtain ISO/IEC 27001 certification. For better data analytics, a beta project has recently opened to test a natural language processing application to allow for the incorporation of text-based patient data into the TriNetX data set.
ACKNOWLEDGMENT
We thank Brian Ostasiewski and Michael Horvath for their help.
Footnotes
The work is partially supported by the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197). The authors acknowledge use of the services and facilities, funded by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (UL1TR001420).
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Collection and assembly of data: All authors
Data analysis and interpretation: Matvey B. Palchuk
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Umit Topaloglu
No relationship to disclose
Matvey Palchuk
Employment: TriNetX
REFERENCES
- 1.Topaloglu U, Niedner H: Advances in clinical research towards the data-driven economy. Clin Res 30:58-62, 2016 [Google Scholar]
- 2.Chow S-C, Chang M: Adaptive design methods in clinical trials—A review. Orphanet J Rare Dis 3:11, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinze D, Kahn S, McOwen P, et al. : Building a richly connected and highly analyzed genotype/phenotype ecosystem in a world of data silos. 2014 AMIA Joint Summits on Translational Science, San Francisco, CA, April 7-11, 2014 [Google Scholar]
- 4.US Food and Drug Administration : Guidance for industry: Use of electronic health record data in clinical investigations. https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM501068.pdf
- 5.Rosenbaum L: Bridging the data-sharing divide: Seeing the devil in the details, not the other camp. N Engl J Med 376:2201-2203, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Cheng SK, Dietrich MS, Dilts DM: A sense of urgency: Evaluating the link between clinical trial development time and the accrual performance of cancer therapy evaluation program (NCI-CTEP) sponsored studies. Clin Cancer Res 16:5557-5563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korn EL, Freidlin B, Mooney M, et al. : Accrual experience of National Cancer Institute Cooperative Group phase III trials activated from 2000 to 2007. J Clin Oncol 28:5197-5201, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Applied Clinical Trials : Protocol amendments: A costly solution. http://www.appliedclinicaltrialsonline.com/protocol-amendments-costly-solution
- 9.Turgon JL, Welter TL: The nuts and bolts of clinical research billing. Oncol Iss 23:36-39, 2017 [Google Scholar]
- 10.Handelsman D; SAS : Optimizing clinical research operations with business analytics. http://support.sas.com/resources/papers/proceedings11/204-2011.pdf
- 11.Goldfarb NM: Clinical operations: Benchmarking per-patient trial costs, staffing and adaptive design. J Clin Res Best Pract 7:1-174, 2011 [Google Scholar]
- 12.Rosas SR, Schouten JT, Dixon D, et al. : Evaluating protocol lifecycle time intervals in HIV/AIDS clinical trials. Clin Trials 11:553-559, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez DA, Tsalatsanis A, Yalcin A, et al. : Activating clinical trials: A process improvement approach. Trials 17:106, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.London JW, Balestrucci L, Chatterjee D, et al. : Design-phase prediction of potential cancer clinical trial accrual success using a research data mart. J Am Med Inform Assoc 20:e260-e266, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.i2b2 Foundation : Informatics for integrating biology & the bedside. https://www.i2b2.org/
- 16.Psaty BM, Breckenridge AM: Mini-Sentinel and regulatory science—Big data rendered fit and functional. N Engl J Med 370:2165-2167, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Franzén S, Janson C, Larsson K, et al. : Evaluation of the use of Swedish integrated electronic health records and register health care data as support clinical trials in severe asthma: The PACEHR study. Respir Res 17:152, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Cancer Registration and Analysis Service : National cancer data repository. http://www.ncin.org.uk/collecting_and_using_data/national_cancer_data_repository
- 19.Amazon.com : AWS cloud security. https://aws.amazon.com/security/
- 20.MacKenzie SL, Wyatt MC, Schuff R, et al. : Practices and perspectives on building integrated data repositories: Results from a 2010 CTSA survey. J Am Med Inform Assoc 19:e119-e124, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Library of Medicine : RxNorm API https://rxnav.nlm.nih.gov/RxNormAPIs.html [DOI] [PubMed]
- 22.Centers for Medicare & Medicaid Services : General equivalence mappings. https://www.cms.gov/Medicare/Coding/ICD10/2017-ICD-10-CM-and-GEMs.html
- 23.Agency for Healthcare Research and Quality : Beta chronic condition indicator: https://www.hcup-us.ahrq.gov/toolssoftware/chronic_icd10/chronic_icd10.jsp
- 24.Hudson C, Topaloglu U, Bian J, et al. : Automated tools for clinical research data quality control using NCI common data elements. 2014 AMIA Joint Summits on Translational Science, San Francisco, CA, April 7-11, 2014 [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn MG, Brown JS, Chun AT, et al. : Transparent reporting of data quality in distributed data networks. EGEMS (Wash DC) 3:1052, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiskopf NG, Hripcsak G, Swaminathan S, et al. : Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform 46:830-836, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greater Plains Collaborative : Home. www.gpcnetwork.org