Abstract
The nematodes of genus Oscheius are insect parasites with a potential role as biological control agents. The composition of gut microbiota and its potential assistant role in the complex pathogenic mechanism of nematodes have been poorly illustrated. In this study, the intestinal bacteria associated with dauer juveniles of the nematode Oscheius chongmingensis Tumian were classified by 16S rDNA high‐throughput sequencing. The raw reads were assigned to 845 operational taxonomic units (OTUs) after quality filtering. The results showed that the genus Ochrobactrum, with a proportion of 59.82%, was the most abundant genus, followed by 7.13% Bacillus, 4.7% Albidiferax, 4.26% Acinetobacter, and 3.09% Rhodococcus. The two dominant bacteria, Ochrobactrum and Bacillus, were further isolated by culturing on NBTA and LB medium respectively, and then identified as Ochrobactrum tritici and Bacillus cereus by morphological and 16S rDNA sequence analysis. Furthermore, the entomopathogenicity of these two bacterial species was studied. The results showed that O. tritici caused 93.33% mortality within 144 hr in the 4th‐instar larvae of Galleria mellonella treated with 2 × 109 CFU/ml, whereas B. cereus showed 100% mortality at a concentration of 3.3 × 107 CFU/ml within 48 hr. These findings, especially the presence of O. tritici, which had not been found in other nematode species in the genus Oscheius, indicate that the associated nematode O. chongmingensis may have particular utility as a biocontrol agent.
Keywords: Bacillus cereus, entomopathogenicity, high‐throughput sequencing, Ochrobactrum tritici, Oscheius chongmingensis Tumian
The nematodes of genus Oscheius are insect parasites with a potential role as biological control agents. We identified and analyzed the gut bacterial community of the dauer juveniles of O. chongmingensis Tumian.
1. INTRODUCTION
Nematode species in the genus Rhabditis (Oscheius) have been characterized as entomopathogenic nematodes (EPNs) (Poinar,1979; Schulte, 1989; Smart & Nguyen, 1994). Recent studies have reported that some rhabditid nematodes could act as potential biocontrol agents to control various invertebrate pests, such as the burrower bug (Stock, Caicedo, & Calatayud, 2005), Formosan subterranean termite (Carta & Osbrink, 2005), areca nut spindle bug (Mohandas, Sheeba, Firoza, & Rajamma, 2007), rice yellow stem borer (Padmakumari, Prasad, Katti, & Sankar, 2007) and burying beetle (Sangeetha, Rajitha, Shyni, & Mohandas, 2016).
The dauer juvenile of entomopathogenic nematodes is the only infective juvenile stage (IJs) that invades insect hosts, and releases bacteria to produce toxins (Dillman et al., 2012). Furthermore, entomopathogenic nematodes are highly virulent due to the symbiotic bacteria found in IJs. In addition, entomopathogenic bacteria are promising sources of antimicrobial, insecticidal and nematicidal compounds, which might become potential biopesticides. Some nematodes of Oscheius sp. have been found to associate with insect pathogenic bacteria. However, the diversity of bacteria associated with nematodes of Oschieus sp. differs from the specific bacterial symbionts of the genera Heterorhabditis and Steinernema. Some reports have shown that the some species of intestinal bacteria associated with rhabditid nematodes could serve to promote nematode biological control effects. Nineteen bacterial genera have been found to be mutually associated with Rhabditis (Oscheius) nematodes: Bacillus (Mohandas et al., 2007), Klebsiella, Acinetobacter, Comamonas, Brucellaceae, Achromobacter (Deepa, Mohandas, & Siji, 2011), Alcaligenes, Flavobacterium, Providencia (Park, Kim, Ha, & Youn, 2011), Stenotrophomonas, Enterobacter, Proteus, Pseudomonas, Enterococcus, Lysinibacillus (Padmakumari et al., 2007; Sangeetha et al., 2016), Microbacterium, Serratia, Rheinheimera and Staphylococcus (Tambong, 2013). Among them, Bacillus cereus isolate 03BB102 (Mohandas et al., 2007), Flavobacterium sp., Providencia vermicola (Deepa et al., 2011), Serratia marcescens (Tambong, 2013) and Serratia nematodiphila (Zhang, Yang, Xu, & Sun, 2009) exhibit virulence against insects and help improve nematode reproduction.
Notably, S. nematodiphila was associated with the nematode species O. chongmingensis, which was collected from the soil of Chongming Island in the southeastern area of Shanghai, China (Zhang et al., 2008). In the present work, the same nematode species, the Oscheius chongmingensis Tumian strain, isolated from an alfalfa field in the city of Hailar, Inner Mongolia, China (Liu, Mráček, Zhang, & Půža, 2012), the presence of the same or different associated bacterial genera in its intestine was assessed. In previous studies, dauer juveniles of O. chongmingensis Tumian showed pathogenicity to Galleria mellonella and Tenebrio molitor (Cao, Liu, Xie, Cao, & Li, 2007), and this nematode strain also led to 90% mortality of the longhorn beetle, Batocera lineolata (Coleoptera: Cerambycidae), which attacks walnut trees in walnut fields (Liu & Wei, 2015). Hence, O. chongmingensis Tumian has been identified as a potential biocontrol agent.
Therefore, in the present study, the intestinal bacteria associated with the dauer juveniles of O. chongmingensis Tumian were investigated and the effects of two associated bacteria on pathogenicity were analyzed. The results will benefit further study of the infection mechanism of this nematode against insect pests.
2. MATERIALS AND METHODS
2.1. Nematode culture and DNA extraction
Galleria mellonella larvae were reared in the Entomology and Nematology Laboratory, Department of Entomology, China Agricultural University (CAU), using artificial diets (Pu & Liu, 2009). The nematode O. chongmingensis Tumian was originally isolated from a soil sample collected from an alfalfa field in the city of Hailar, Inner Mongolia, using the Galleria bait method. The nematode has been subsequently maintained through reinfections of fourth‐instar larvae of G. mellonella (Lepidoptera: Galleridae). To produce fresh nematodes for the study, G. mellonella larvae were infected with O. chongmingensis Tumian at room temperature and then transferred to White traps to facilitate the harvest of dauer juveniles. The nematodes were suspended in distilled water and counted. Only newly emerged IJs (<48 hr) were utilized for the inoculation test in each experiment.
The freeze‐thaw lysis protocol was adopted to extract genomic DNA from 100 dauer juveniles of O. chongmingensis Tumian. The nematodes were surface‐sterilized by immersion in 1% (w/v) NaClO (sodium hypochlorite solution) for 30 min and then in 75% alcohol for 1 min, followed by washing three times with sterile water. To avoid contamination by foreign bacteria, 200 μl of the treated nematode suspension was dropped onto Luria‐Bertani (LB) plates and incubated for 24 hr. Only nematode samples with no bacterial growth on LB were used for further analysis. Total DNA from dauer juvenile nematodes was extracted using a Power Soil®DNA Isolation Kit (MoBio) according to the manufacturer's instructions. All operations were carried out under a sterile laminar flow hood. Purified DNA extracts were stored at −20°C until PCR amplification.
2.2. PCR amplification and high‐throughput sequencing
DNA fragments of the bacterial 16S rDNA gene, targeting the hypervariable region V3–V4, were amplified using the primer pairs 336F (5′‐GTACTCCTACGGGAGGCAGCA‐3′) and 806R (5′‐GTGGACTACHVGGGTWTCTAAT‐3′). PCR amplification was carried out in triplicate 50 μl reactions containing 30 ng of genomic DNA, 0.3 μl of Pyrobest DNA Polymerase (2.5 U/μl, TaKaRa), 2 μl of each barcoded fusion primer (10 μM), and 4 μl of dNTPs (2.5 mM), in the appropriate 10 × Pyrobest Buffer and ddH2O. Negative control samples were treated similarly with the exclusion of template DNA. The following thermal program was used for amplification: an initial denaturation at 95°C for 5 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 40 s, with a final extension step at 72°C for 10 min. The triplicate PCR products were pooled and purified from 2.0% agarose gels. The amplicon products were quantified by the Qubit fluorescence quantitative system and then combined in an equimolar ratio in a single tube. Paired‐end sequencing was performed on the Illumina platform (MiSeq) (Allwegene Technology Co., Ltd., Beijing, China).
2.3. Analysis of data processing and bacterial diversity
The data were processed by removing low‐quality reads using Trimmomatic software and Readfq (version 6.0) and then splicing the paired reads into a sequence based on the PE data by FLASH (version 1.2.10) and Pear software. Sequences with lengths shorter than 200 bp or with maxhomop >10 were removed using the Mothur pipeline (Schloss, Westcott, Ryabin, & Hall, 2009). Sequences with ambiguous bases, primer mismatches, errors in barcodes, and chimerism were filtered using Usearch (version 8.0.1623) (Edgar, Haas, Clemente, & Quince, 2011). The remaining high‐quality sequences were aligned to a reference alignment derived from the 16S rDNA gene database (DeSantis, Hugenholtz, Larsen, & Rojas, 2006). Then, the spliced and filtered clean tags were clustered into OTUs (Operational Taxonomic Units) using a 3% distance cutoff by using the QIIME pipeline (v.1.8.0) (Caporaso, Kuczynski, Stombaugh, & Bittinger, 2010; Schloss & Westcott, 2011).
To improve the accuracy of the annotation, sequences were also searched against the NCBI nt database using BLASTn (Altschul, Gish, Miller, & Myers, 1990). Taxonomic information was assigned by the NCBI database according to the highest scoring sequence. The diversity index and species richness estimate were calculated using the Mothur pipeline. The α‐diversity was represented by rarefaction curves plotting the cumulative number of OTUs at a 3% distance level. Diversity was measured by counting the number of observed OTUs using the Shannon index, Chao1 index, phylogenetic diversity and observed number of species as described previously (Magurran, 2004; Chao, 1984). The estimators were calculated by subsampling the smallest number of sequences from each sample. Statistical analysis was performed in R (version 3.11) (R Development Core Team, 2014). The dominant taxonomic unit, species richness and relative abundances in each sample were determined from rank‐abundance curves. Beta diversity and species composition analysis were constructed by using the QIIME pipeline and R software.
2.4. Isolation of dominant bacteria from nematodes
Associated bacteria were obtained from the infective stages of O. chongmingensis Tumian by two methods. Dauer juveniles of nematodes were surface‐sterilized by immersion in 1% (w/v) NaClO (sodium hypochlorite solution) for 30 min and then in 75% alcohol for 1 min; after washing three times in sterile water, they were streaked onto nutrient agar NBTA (peptone, 10 g; beef extract, 5 g; NaCl, 5 g; agar, 18 g; water, 1 L, 0.0025% bromothymol blue, 0.004% triphenyltetrazolium chloride; pH 7.0–7.2) and LB (tryptone, 10 g; NaCl, 10 g; yeast extract, 5 g; water, 1 L; pH 7.0–7.2). In addition, associated bacteria were isolated from the hemolymph of dead G. mellonella larvae infected with the nematode species. The dead larvae were surface‐sterilized by dipping into 75% ethanol for 1 min and placed in a sterile petri dish to dry. Sterile scissors were used to dissect the 3rd segment from the head of each larva. A sterile loop was used to touch the hemolymph and streak it on the nutrient agar media. The plates were sealed and incubated in the dark at 28°C for 48 hr. Developing bacterial colonies that differed in morphology or in color were transferred to NBTA or LB plates (Akhurst, 1980). Before and after each test, cultures were streaked onto NBTA and LB to confirm that there had been no changes from one form to the other.
These colonies were added to LB nutrient broth and shaken for 48 hr (150 rpm) at 28°C in the dark. Subsequently, the bacterial suspension was centrifuged at 4,000 g for 8 min and the supernatant decanted. Sterile water was added to the pellets and mixed thoroughly to obtain a concentrated suspension of the bacterial symbionts. The total number of bacteria in the suspension was measured with a spectrophotometer with a wavelength at 600 nm. The cell number in the suspension was estimated by counting colonies on culture plates of different concentration gradients and measuring the OD600 value. Some of the colonies were sampled for molecular analysis.
2.5. Morphology and identification of bacterial isolates
Cultural properties such as colony size, shape, and color were observed after 48 hr incubation at 28°C on nutrient agars (NBTA and LB), and then, the bacteria were gram stained.
Pure isolates of two strains, NMA‐1 and NMA‐2, were used for identification following 16S rDNA gene sequencing. For DNA extraction, bacterial isolates were grown individually in 5 ml of LB broth at 28°C for 24 hr. DNA was extracted following the procedure described previously (Krsek & Wellington, 1999) with some modifications. The sample was suspended in 100 μl 1 × TE (pH 8.0) and 100 μl of lysozyme solution (50 mg·mL−1) was added. The tube was placed in a 37oC water bath for 20 min. Subsequently, 100 μl of 10% SDS solution (10% w/v SDS) and 5 μl of proteinase K (20 mg·mL−1) were added, and the contents of the tube were gently mixed and placed in a 37oC water bath for 30 min. Then, 100 μl of NaClO solution (5 M) was added, and the mixture was gently whirled for 1 min; the supernatant was treated with phenol:chloroform:isoamyl alcohol (25:24:1) and chloroform:isoamyl alcohol (24:1) successively to remove proteins and other impurities. Thereafter, the DNA was precipitated with 90 μl of isopropanol for 10 min at room temperature and then pelleted by centrifugation at 12,000 g and 4°C for 10 min. The precipitate was washed with 75% ethanol and dried in a laminar flow cabinet for 1 hr prior to resuspension with 50 μl of sterile water. DNA extracts were stored at −20°C until further analysis.
The gene sequences were amplified using three different primer sets: two targeting the bacterial 16S rDNA gene and a second pair targeting a different gene, recA (Table 1). The primer sets for bacteria were 27f/1492r and S1/A1 (Yang, 2008) (Table 1), while the primer set for the Brucella spp. recA gene was recA‐f/recA‐r (Scholz, Pfeffer, Witte, & Neubauer, 2008). PCR amplification was performed in a thermal cycler (Bio‐Gener) using approximately 1.0 μl of bacterial DNA, 2.5 μl of 10 × PCR buffer, 2.0 μl of 2.5 mM dNTPs, 1.0 μl of each primer (10 μM), 0.3 μl of Taq DNA polymerase, and 17.2 μl of ddH2O. Thermal cycling conditions were as follows: 3 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C for 27f/1492r and recA‐f/recA‐r or 1 min at 63°C for S1/A1, 1.5 min at 72°C, and a final step at 72°C for 10 min. The PCR products were sequenced by BGI (Beijing, China).
Table 1.
Primer pairs used for identification in this study
| Gene | Primer | Sequence (5′ → 3′) | Tm (°C) |
|---|---|---|---|
| 16S rDNA | 27F | AGAGTTTGATCCTGGCTCAG | 55 |
| 1492R | GGTTACCTTGTTACGACTT | ||
| S1 | GAAGAGTTTGATCATGGCTC | 63 | |
| A1 | AAGGAGGTGATCCAGCCGCA | ||
| recA gene | recA‐f | ATGTCTCAAAATTCATTGCGAC | 54 |
| recA‐r | AGCATCTTCTTCCGGTCCGC |
2.6. Sequence and phylogenetic relationship analysis of bacteria
For phylogenetic analysis, all nucleotide sequences obtained were compared with NCBI database sequences using BLASTn. The selected sequences were aligned using ClustalW, and neighbor‐joining trees of the homologous sequences were constructed using the maximum likelihood method with 1,000 bootstrap replications in the MEGA 7.0 program (Kumar, Steche, & Tamura, 2016).
2.7. Physiological and biochemical characteristics of bacterial isolates
Physiological and biochemical tests were performed at 28°C according to Bergey's Manual of Determinative Bacteriology (Buchanan & Gibbons, 1986) and Common Bacterial System Identification Manual (Dong, Cai, Lu, Xie, & Liu, 2001).
2.8. Evaluation of potential pathogenicity of bacterial isolates to G. mellonella larvae
The two species of bacteria were cultured separately for 48 hr, concentrated, and rinsed three times with sterile water as previously described (Delalibera, Handelsman, & Raffa, 2005). Serial dilutions were performed in sterile water to obtain dilutions from 1 × 10 to 1 × 106, which corresponded to suspensions of 2.0 × 105–2.0 × 1010 CFU/ml for the strain NMA‐1 and 3.3 × 102–3.3 × 107 CFU/ml for the strain NMA‐2. In addition, 100 μl of serial dilutions was streaked onto LB agar plates to count the number of colonies, and the OD600 values of gradient dilutions were measured by an ultraviolet‐visible light detector (LabTech) to make a standard curve. G. mellonella larvae were used as the test insect for assessing the pathogenicity of the two dominant bacteria associated with O. chongmingensis Tumian. A cell suspension (50 μl) was injected into 4th‐instar G. mellonella larvae using a 50 μl Anting microsyringe (Shanghai, China). Sterilized water was used as a control. Thirty larvae were used per concentration, and three replications were performed. Insect mortality was observed every 24 hr after injection.
3. RESULTS
3.1. Basic data processing and statistics
16S rRNA gene sequencing of the dauer juvenile nematode samples was performed by paired‐end sequencing. After the quality check, unqualified sequences shorter than 200 bp were removed. According to the quality criteria, approximately 97.47% of the raw sequences were used for subsequent analysis (Table 2). Next, filtering processes were performed to remove edundant, chimeras and undesirable sequences, generating 51,651 clean reads (91.69%) for dauer juveniles. After rarefying the reads to the smallest number, 51,651 bacterial sequences from nematodes were retained.
Table 2.
Sequence statistics
| Sample | Raw tags | High‐quality tags | Clean tags | ||
|---|---|---|---|---|---|
| Number | %a | Number | %a | ||
| Dauer Juvenile | 56,334 | 54,907 | 97.47 | 51,651 | 91.69 |
Percent of raw reads.
3.2. Diversity and richness of bacterial species
The Chao1 index was used to analyze OTU richness and species richness at the 0.03 dissimilarity level. The shannon index, phylogenetic diversity (PD, whole tree) and observed number of species were used to assess the diversity of the bacterial communities from nematodes. Similar results were obtained (Table 3). The Shannon index of the nematode sample was 3.60, and the value of Good's coverage was 0.99. In addition, the value of the PD whole tree was 65.2 (Table 3). These results suggested that dauer juveniles of O. chongmingensis Tumian had high bacterial diversity.
Table 3.
Species richness estimator and diversity index of bacterial community
| Sample | OTUs | Chao1 | Goods_coverage | Observed_species | PD_whole_tree | Shannon |
|---|---|---|---|---|---|---|
| Dauer juvenile | 845 | 924.61 | 0.99 | 766 | 65.20 | 3.60 |
3.3. Analysis of bacterial community structure and predominant species
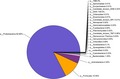
The 845 OTUs from the nematode sample were assigned to corresponding taxonomic groups based on the combined search results from the Greengenes and NCBI databases. The relative abundances of different phyla in the sample are shown in Figure 1. The results showed that Proteobacteria was the most heavily sequenced phylum associated with the dauer juvenile nematodes (82.66%). Firmicutes (10.45%), Actinobacteria (4.34%) and Bacteroidetes (1.25%) were also present in the juvenile communities.
Figure 1.
The composition and relative abundances of bacterial phyla associated with the dauer juveniles of Oscheius chongmingensis Tumian. Only OTUs with at least 20 sequences are represented[Colour figure can be viewed at wileyonlinelibrary.com]
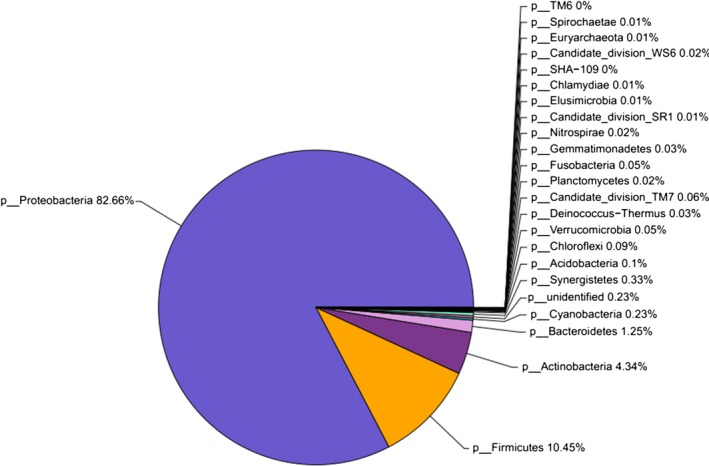
Under the 97% similarity level, the trimmed clean tags were clustered into OTUs for species classification by QIIME (v1.8.0) software. To describe the majority of bacterial genera, OTUs of the nematode samples were assigned to corresponding taxonomic groups based on the combined search results from the NCBI databases. Ochrobactrum was the most abundant bacterial genus detected in dauer juveniles of O. chongmingensis Tumian, with an abundance of 59.82%, followed by Bacillus (7.13%), Albidiferax (4.7%), Acinetobacter (4.26%), Rhodococcus (3.09%), Pseudomonas (2.69%), Delftia (1.96%), Stenotrophomonas (1.94%), and Citrobacter (1.39%). All other genera associated with dauer juveniles are shown in Figure 2.
Figure 2.
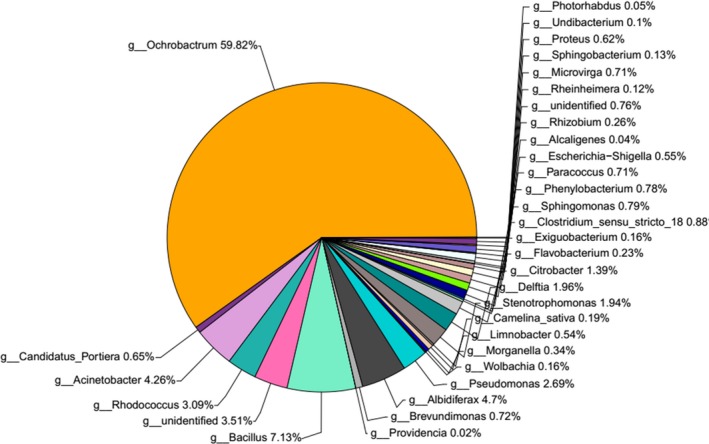
The composition and relative abundances of bacterial genera associated with the dauer juveniles of Oscheius chongmingensis Tumian
3.4. Morphological characteristics of bacteria
Two bacterial species were isolated from surface‐sterilized nematodes and from the hemolymph of G. mellonella larvae infected by the nematodes. After growth on NBTA plates for 48 hr, the NMA‐1 strain was obtained and distinguished as gram‐negative (Figure 3a). During incubation on NBTA, the pink colonies of NMA‐1 were opaque, mucoid, smooth, protuberant and rapidly confluent. The gram‐positive NMA‐2 strain was obtained on LB nutrient agar after 24 hr (Figure 3b). The colonies of the NMA‐2 strain were large (approximately 15 mm in diameter), and white waxy with a rough surface and a flat, irregular shape.
Figure 3.
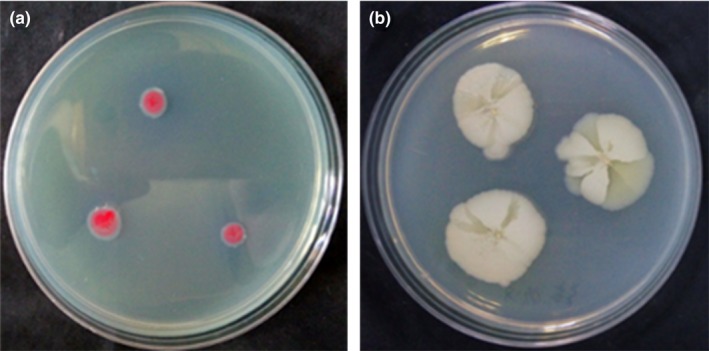
Colony morphology of the two main bacteria species isolated from Oscheius chongmingensis Tumian on nutrient agar culture. (a) NMA‐1 strain on NBTA. (b) NMA‐2 strain on LB
3.5. 16S rDNA sequence and phylogeny of two dominant bacteria
The associated bacteria were confirmed by molecular analysis based on the comparison of their 16S rDNA. Strain NMA‐1 was 99% homologous to O. tritici, and strain NMA‐2 was B. cereus with 100% identity. The analysis of morphological characteristics and 16S rDNA sequencing showed conclusively that strain NMA‐1 belonged to O. tritici and strain NMA‐2 was close to B. cereus. The phylogenetic relationship analysis (Figure 4) demonstrated that the genetic distance between the amplified fragment and O. tritici (GenBank accession no. AM087901) was the smallest, and they were in the same clade (with a confidence level of 88%). Alternatively, NJ trees (Figure 5) based on the S1/A1 sequence showed that all 17 B. cereus isolates belonged to a single cluster, including strain NMA‐2 (with 86% bootstrap support).
Figure 4.
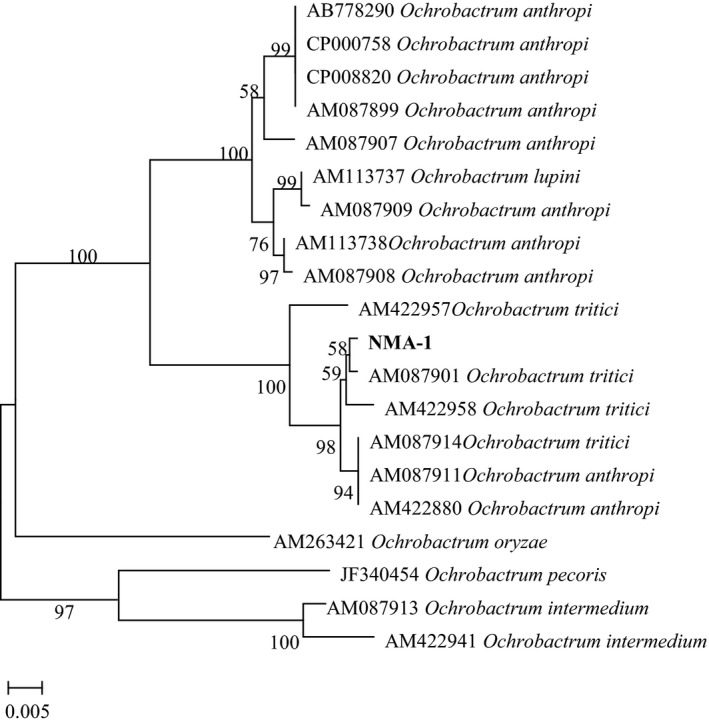
Phylogenetic relationships of the strain NMA‐1 and other closely related Ochrobactrum species in a neighbor‐joining tree based on analysis of the recA sequence. Bootstrap values are 1,000 replications and above 50% are shown at the branch points by MEGA 7.0
Figure 5.

Phylogenetic tree of strain NMA‐2 and other related Bacillus species in a neighbor‐joining tree based on analysis of the S1/A1 sequence data. Bootstrap values are 1,000 replications and above 50% are shown at the branch points by MEGA 7.0
3.6. Physiological characteristics of two dominant bacteria
Phenotypic characteristics are shown in Table 4. Both strains assimilated D‐glucose, sucrose, D‐fructose, and D‐trehalose but not mannitol or lactose, and they produced urease, lecithinase and amylase. Furthermore, both strains hydrolyzed starch. Strain NMA‐1 produced indole, but did not hydrolyze gelatin, while strain NMA‐2 did both.
Table 4.
Physiological and biochemical characteristics of strains NMA‐1 and NMA‐2
| Characteristics | NMA‐1 | NMA‐2 |
|---|---|---|
| Glucose | + | + |
| Sucrose | + | + |
| Fructose | + | + |
| Trehalose | + | + |
| Mannitol | − | − |
| Lactose | − | − |
| Starch | + | + |
| Voges–Proskauer | − | − |
| Methyl red | + | + |
| Mycoderm | + | + |
| Gelatinase | − | + |
| Urease | + | + |
| Catalase | + | − |
| Arginine dihydrolase | − | − |
| Lecithinase | + | + |
| Aesculin | − | + |
| Tween 80 | − | − |
| Malonate | − | − |
| Phenylalanine deaminase | − | − |
| Citrate | + | + |
| H2S | − | − |
| Indole | + | + |
| Litmus milk | + | + |
| Protease | − | + |
+, Positive;−, Negative.
3.7. Pathogenicity of associated bacteria to G. mellonella larvae
To assess the ability of the nematode symbiotic bacteria to grow and survive within the host larval hemocoel and to test their pathogenicity, 50 μl of different bacterial suspensions was injected into G. mellonella larvae. Over time, the body color of the G. mellonella larvae became dark brown as the infected larvae died, while no changes were observed in the control (H2O) (Figure 6).
Figure 6.
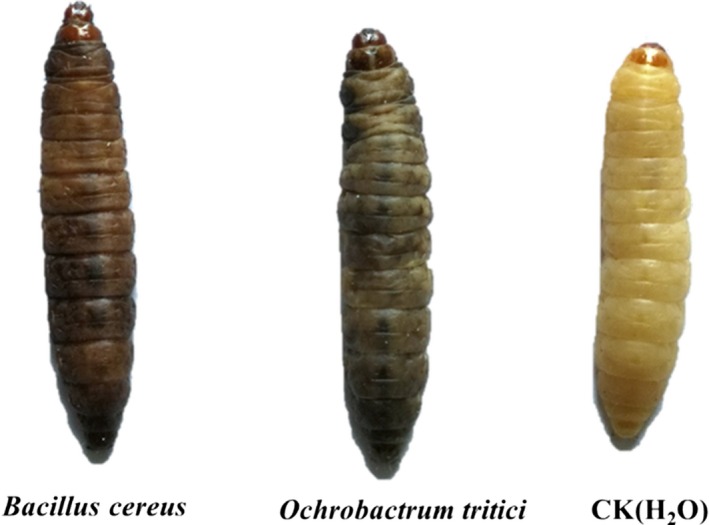
Death symptoms of Galleria mellonella larvae caused by injection of cell suspensions of Ochrobactrum tritici NMA‐1 and Bacillus cereus NMA‐2, sterile water as the control
The pathogenicity of serial dilutions of O. tritici and B. cereus at 48 hr after injection into the hemocoel of G. mellonella larvae is shown in Figure 7; a significant difference in insect mortality (p < 0.05) was observed between bacterial species. After 24 hr treatment, low insect mortalities of 0%–6.67% were observed for O. tritici, while 90% mortality caused by B. cereus was detected. Up to 120 hr after injection, larvae injected with the highest concentration (2.0 × 1010 CFU/ml) showed 90% mortality (Figure 7a). However, the highest mortality of larvae was 100% at 48 hr after injection of B. cereus NMA‐2 (3.3 × 107 CFU/ml) (Figure 7b). Since B. cereus NMA‐2 was more pathogenic than O. tritici NMA‐1 after culture for 48 hr, it was possible that these bacteria had different characteristics and different effects on nematode reproduction and pathogenicity.
Figure 7.
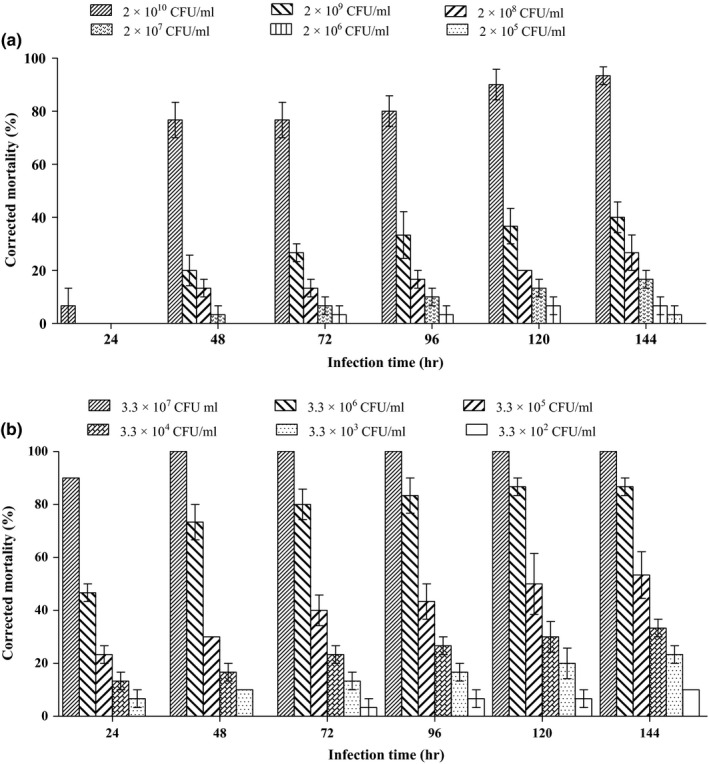
Corrected mortality of Galleria mellonella larvae injected with cell suspensions of (a) Ochrobactrum tritici NMA‐1 and (b) Bacillus cereus NMA‐2. Each data represents the mean of three replicates with 10 larvae per replicate and error bars standard errors of the means (p < 0.05; One‐way ANOVA, post hoc LSD)
4. DISCUSSION
In the present work, we identified and analyzed the gut bacterial community of dauer juveniles of O. chongmingensis Tumian. We found gut microbiota with high diversity and two dominant bacterial OTUs. Comparison of the microbial composition revealed that the most abundant OTUs of the sequences belonged to the phyla (Proteobacteria (82.66%), Firmicutes (10.45%) and Actinobacteria (4.34%). Among them, the phylum Proteobacteria contains a variety of species, that are adapted to diverse environments (Delalibera et al., 2005). At least 33 bacterial genera were found to be associated with the dauer juveniles of O. chongmingensis Tumian, which was initially isolated from an alfalfa field in the city of Hailar, Inner Mongolia, China. The genus Serratia, as a biological control agent, has been found to associate with the nematode species O. chongmingensis, as well as some other nematodes, such as O. carolinensis, O. myriophila and O. rugaoensis (Zhang et al., 2009; Liu et al., 2012; Ye, Torres‐Barragan, & Cardoza, 2010; Torres‐Barragan, Suazo, Buhler, & Cardoza, 2011; Liu, Zhang, & Zeng, 2016). However, no Serratia sp. was discovered in the O. chongmingensis Tumian strain in this report. Thus, differences in the dominant bacteria associated with rhabditid nematodes may be due to differences in the species of nematodes, isolation methods and bacterial species in the rhizosphere soil of different plants from different regions. Hence, different nematode habitats may affect their associated bacterial communities and, consequently, nematode lifestyle plasticity and pathogenicity.
The two highly abundant genera, Ochrobactrum and Bacillus, with high frequencies of separation, were further isolated by culture methods and identified as O. tritici and B. cereus, respectively. Both caused high mortality of G. mellonella larvae. In the genus Ochrobactrum, most members occur in the environment (soil, water, plant, human) (Jelveh & Cunha, 1999). Some species of Ochrobactrum can biodegrade polycyclic aromatic hydrocarbons (Arulazhagan & Vasudevan, 2011). Some endophytic bacteria belonging to Ochrobactrum sp. have been isolated from plants, and these strains exhibit maximum antagonistic activity and possess higher chitinase and glucanase activity (Zhao, Xu, Chang, & Li, 2016; Zang, Zhao, Liu, & Liang, 2014; Sowndhararajan, Marimuthu, & Manian, 2013). Metabolites produced by bacterial species of Ochrobactrum inhibit the growth of a wide range of bacteria and fungi (Han, Peng, & Yi, 2011; Lebuhn, Achouak, Schloter, & Berge, 2000). Only one report on one species in the genus, O. anthropi, has previously associated this genus with a nematode, Steinernema scapterisci (Aguillera, Hodge, Stall, & Smart, 1993). Nematodes in monoxenic culture with O. anthropi could result in 81% mortality to the southern mole cricket, Scapteriscus borellii. Until now, there had been no report on Ochrobactrum sp. in association with Rhabditis (Oscheius) sp. However, the strain O. tritici NMA‐1 was discovered and isolated from dauer juveniles of O. chongmingensis Tumian for the first time and showed high efficacy for controing G. mellonella larvae in this study.
The second most abundant genus investigated in this study was Bacillus. Previous studies had demonstrated that the bacterial species B. cereus was closely associated with different rhabditid nematodes (Deepa et al., 2011). The insecticidal proteins produced by B. cereus were significantly pathogenic to insects from the orders Lepidoptera and Diptera (Kaaya & Darji, 1989; Warren, Koziel, Mullins, & Nye, 1996). Moreover, some studies have revealed that bioactive metabolites produced by the strain of B. cereus associated with rhabditid nematodes cause insect mortality (Anju, Archana, Mohandas, & Nambisan, 2015a; Kumar, Nambisan, Sundaresan, & Mohandas, 2014; Nishanth, Nath, Pratap, & Nambisan, 2014). Similarly, the strain B. cereus NMA‐2 isolated from dauer juveniles of O. chongmingensis Tumian in the present study may have high potential insecticidal bioactivity.
The other most abundant genus detected in this study was Acinetobacter. One species of Acinetobacter, A. baumannii, is an opportunistic pathogen of great concern in nosocomial pneumonias and especially as an invader of burn wounds (Livermore & Woodford, 2006). Acinetobacter calcoaceticus, associated with the nematode Steinernema sp., produces compounds bioactive against Candida albicans (Reghunath, Siji, Mohandas, & Nambisan, 2017). In addition, the genus Rhodococcus has been found in the intestine and thrives in a broad range of environments, including soil, water, and eukaryotic cells. Two of the most important species were the plant pathogen R. fascians which causes leafy gall disease of gymnosperm plants and angiosperm (Goethals, Vereecke, Jaziri, & Montagu, 2001), and the human and animal pathogen R. equi, which infects foal pneumonia, cattle and immunocompromised humans (Muscatello, Leadon, Klayt, & Ocampo‐Sosa, 2007). Nevertheless, it was rare to isolate bacteria in the genera of Acinetobacter and Rhodococcus using nutrient agar in this study, although isolation was performed more than 10 times. These results suggest that O. chongmingensis Tumian could be a potential biocontrol agent, because it carries different pathogenic bacteria. Further studies are needed to understand the interactions between nematodes and associated bacteria in the natural environment.
Similar to O. chongmingensis Tumian, the nematode Rhabditis blumi also has more than one species of associated bacteria. The bacterium Providencia vermicola promotes was one of the bacteria that contributed to promoting nematode culture, in addition to Flavobacterium sp. Another species, Alcaligenes faecalis, however, is unfavorable for nematode reproduction and pathogenicity (Park et al., 2011). In addition, a slug‐parasitic nematode, Phasmarhabditis hermaphrodita, has been associated with multiple bacteria as well. Finally, the core bacterium Moraxella osloensis is the optimum species that can improve nematode production and pathogenicity (Wilson, Glen, George, & Pearce, 1995; Wilson, Glen, Pearce, & Rodgers, 1995).
Further studies are needed to analyze the function of O. tritici, B. cereus or other intestinal bacteria associated with O. chongmingensis Tumian to explore the mechanism of nematode infection pathogenicity to invertebrate pests.
CONFLICT OF INTERESTS
The authors declare no conflicts of interest.
AUTHORS CONTRIBUTION
Qi‐zhi Liu conceived and designed the experiments, confirmed the analyzed data and revised manuscript; Jun‐rui Fu performed the experiments, analyzed the data, drafted the manuscript and amended the revised manuscript according to the language company’s comments
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
This research was supported by the China National Key Technology R & D Program (No. 2014BAD23B01 and No. 2014BAD16B07).
Fu J‐R, Liu Q‐Z. Evaluation and entomopathogenicity of gut bacteria associated with dauer juveniles of Oscheius chongmingensis (Nematoda: Rhabditidae). MicrobiologyOpen. 2019;8:e823 10.1002/mbo3.823
Funding information
China National Key Technology R & D Program, Grant number: 2014BAD23B01; China National Key Technology R & D Program, Grant number: 2014BAD16B07.
DATA ACCESSIBILITY
All data are provided in full in the results section of this paper apart from the DNA sequences of Ochrobactrum tritici NMA‐1 and Bacillus cereus NMA‐2 genes which are available at https://submit.ncbi.nlm.nih.gov/subs/genbank/ under accession number MK434214 and MK434215, respectively.
REFERENCES
- Aguillera, M. M. , Hodge, N. C. , Stall, R. E. , & Smart, G. C. (1993). Development, reproduction, and pathogenicity of Steinernema scapterisci in monoxenic culture with different species of bacteria. Journal of Invertebrate Pathology, 62, 289–294. 10.1006/jipa.1993.1115 [DOI] [Google Scholar]
- Akhurst, R. J. (1980). Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis . Microbiology, 121, 303–309. 10.1099/00221287-121-2-303 [DOI] [Google Scholar]
- Altschul, S. F. , Gish, W. , Miller, W. , & Myers, E. W. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Anju, K. M. , Archana, M. M. , Mohandas, C. , & Nambisan, B. (2015). An antimicrobial phthalate derivative from Bacillus cereus, the symbiotic bacterium associated with a novel entomopathogenic nematode, Rhabditis (Oscheius) sp. International Journal of Pharmacy & Pharmaceutical Sciences, 7, 238–242. [Google Scholar]
- Arulazhagan, P. , & Vasudevan, N. (2011). Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain ochrobactrum sp. VA1. Marine Pollution Bulletin, 62, 388–394. 10.1016/j.marpolbul.2010.09.020 [DOI] [PubMed] [Google Scholar]
- Buchanan, R. E. , & Gibbons, N. E. (1986). Bergey's manual of determinative bacteriology, 8th edition Philadelphia, PA: The Williams & Wilkins Company. [Google Scholar]
- Cao, H. F. , Liu, Q. Z. , Xie, W. W. , Cao, J. , & Li, L. (2007). Suppression of Rhabditis sp. on plant parasitic nematodes of cucumber in greenhouse. Acta Phytopathologica Sinica, 37, 210–213. 10.13926/j.cnki.apps.2007.02.016 [DOI] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , & Bittinger, K. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta, L. K. , & Osbrink, W. (2005). Rhabditis rainain. sp. (Nematoda: Rhabditida) associated with the Formosan subterranean termite, Coptotermes formosanus (Isoptera: Rhinotermitidae). Nematology, 7, 863–879. 10.1163/156854105776186299 [DOI] [Google Scholar]
- Chao, A. (1984). Non‐parametric estimation of the number of classes in a population. Scandinavian Journal of Statistics, 11, 265–270. 10.1080/02664760220136221 [DOI] [Google Scholar]
- Deepa, I. , Mohandas, C. , & Siji, J. V. (2011). Molecular characterization of novel symbiotic bacteria from entomopathogenic nematodes. Nature Seminar on Climate Change and Food Security: challenges and opportunities. Tuber Crops , 24, 399–404. [Google Scholar]
- Delalibera, I. , Handelsman, J. , & Raffa, K. F. (2005). Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environmental Entomology, 34, 541–547. 10.1603/0046-225x-34.3.541 [DOI] [Google Scholar]
- DeSantis, T. Z. , Hugenholtz, P. , Larsen, N. , & Rojas, M. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied & Environmental Microbiology, 72, 5069–5072. 10.1128/aem.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman, A. R. , Chaston, J. M. , Adams, B. J. , Ciche, T. A. , Goodrich‐Blair, H. , & Sternberg, P. W. (2012). An entomopathogenic nematode by any other name. PLoS Pathogens, 8, e1002527 10.1371/journal.ppat.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. Z. , Cai, M. Y. , Lu, Y. Y. , Xie, J. Y. , & Liu, X. L. (2001). Identification methods of common bacteria. In: Dong X. Z. & Cai M. Y. (Eds.), Handbook of common bacteria systematic identify (pp. 349–398). Beijing, China: Beijing Science Press. [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , & Quince, C. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals, K. , Vereecke, D. , Jaziri, M. , & Montagu, M. V. (2001). Leafy gall formation by Rhodococcus fascians . Annual Review of Phytopathology, 39, 27–52. 10.1146/annurev.phyto.39.1.27 [DOI] [PubMed] [Google Scholar]
- Han, M. , Peng, S. , & Yi, Y. L. (2011). Identification of the antagonistic bacterium YB01 against Bipolaris maydis and its antagonism. Plant Protection, 37, 151–154. 10.3969/j.issn.0529-1542.2011.05.029 [DOI] [Google Scholar]
- Jelveh, N. , & Cunha, B. A. (1999). Ochrobactrum anthropi bacteremia. Heart & Lung the Journal of Critical Care, 28, 145–146. 10.1053/hl.1999.v28.a94602 [DOI] [PubMed] [Google Scholar]
- Kaaya, G. P. , & Darji, N. (1989). Mortality in adult tsetse, Glossina morsitans, caused by entomopathogenic bacteria. Journal of Invertebrate Pathology, 54, 32–38. 10.1016/0022-2011(89)90136-5 [DOI] [PubMed] [Google Scholar]
- Krsek, M. , & Wellington, E. M. (1999). Comparison of different methods for the isolation and purification of total community DNA from soil. Journal of Microbiological Methods, 39, 1‐16. 10.1016/S0167-7012(99)00093-7 [DOI] [PubMed] [Google Scholar]
- Kumar, S. N. , Nambisan, B. , Sundaresan, A. , & Mohandas, C. (2014). Isolation and identification of antimicrobial secondary metabolites from Bacillus cereus associated with a rhabditid entomopathogenic nematode. Annals of Microbiology, 64, 209–218. 10.1007/s13213-013-0653-6 [DOI] [Google Scholar]
- Kumar, S. , Steche, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebuhn, M. , Achouak, W. , Schloter, M. , & Berge, O. (2000). Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. International Journal of Systematic and Evolutionary Microbiology, 50, 2207–2223. 10.1099/00207713-50-6-2207 [DOI] [PubMed] [Google Scholar]
- Liu, Q. Z. , Mráček, Z. , Zhang, L. , & Půža, V. (2012). Re‐description of Oscheius chongmingensis (Zhang et al., 2008) (Nematoda: Rhabditidae) and its entomopathogenicity. Nematology, 14, 139–149. 10.1163/138855411X580777 [DOI] [Google Scholar]
- Liu, Q. Z. , & Wei, T. Y . (2015). Application of Oscheius chongmingensis Tumian in the control of walnut cloudling beetles. Patent. CN201510275635.7.
- Liu, J. , Zhang, K. H. , & Zeng, Y. S. (2016). Isolation and identification of a symbiotic bacterial strain (B1) from an entomopathogenic nematode, Oscheius myriophila . Guangdong Agricultural Sciences, 3, 111–115. 10.16768/j.issn.1004-874x.2016.03.022 [DOI] [Google Scholar]
- Livermore, D. M. , & Woodford, N. (2006). The beta‐lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter . Trends in Microbiology, 2006(14),413–420. 10.1016/j.tim.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Magurran, A. E. (2004). Measuring biological diversity. Oxford: Blackwell. [Google Scholar]
- Mohandas, C. , Sheeba, M. , Firoza, A. J. , & Rajamma, P . (2007). Bacteria associated with Rhabditis (Oscheius) spp. (Rhabditidae: Nematoda) for the biocontrol of insect pests. Proceeding of the National Seminar on Achievements and Opportunities in Post harvest Management and Value Addition in Root and Tuber Crops (NSRTC–2) (Trivandrum), 195–198.
- Muscatello, G. , Leadon, D. P. , Klayt, M. , & Ocampo‐Sosa, A. (2007). Rhodococcus equi infection in foals: The science of ‘rattles’. Equine Veterinary Journal, 39, 470–478. 10.2746/042516407X209217 [DOI] [PubMed] [Google Scholar]
- Nishanth, K. S. , Nath, V. S. , Pratap, C. R. , & Nambisan, B. (2014). Cyclic dipeptides from rhabditid entomopathogenic nematode‐associated Bacillus cereus have antimicrobial activities. World Journal of Microbiology and Biotechnology, 30, 439–449. 10.1007/s11274-013-1461-7 [DOI] [PubMed] [Google Scholar]
- Padmakumari, A. P. , Prasad, J. S. , Katti, G. , & Sankar, M. (2007). Rhabditis sp (Oscheius sp), a biocontrol agent against rice yellow stem borer, Scirpophaga incertulas . Indian Journal of Plant Protection, 35, 255–258. [Google Scholar]
- Park, H. W. , Kim, Y. O. , Ha, J. S. , & Youn, S. H. (2011). Effects of associated bacteria on the pathogenicity and reproduction of the insect‐parasitic nematode Rhabditis blumi (Nematoda: Rhabditida). Canadian Journal of Microbiology, 57, 750–758. 10.1139/w11-067 [DOI] [PubMed] [Google Scholar]
- Poinar, G. O. J . (1979). Nematodes for biological control of insects. Boca Raton, FL: CRC Press, 0–289. [Google Scholar]
- Pu, H. H. , & Liu, Q. Z. (2009). Evaluation on the effect of different feed formulas on the growth of Galleria mellonella L. Journal of Anhui Agricultural Sciences, 37, 13647–13668. 10.13989/j.cnki.0517-6611.2009.28.142 [DOI] [Google Scholar]
- R Development Core . (2014). R: A language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing. [Google Scholar]
- Reghunath, S. R. , Siji, J. V. , Mohandas, C. , & Nambisan, B. (2017). Isolation and identification of bioactive molecules produced by entomopathogenic bacteria, Acinetobacter calcoaceticus . Applied Microbiology Open Access, 3, 1000134 10.4172/2471-9315.1000134 [DOI] [Google Scholar]
- Sangeetha, B. G. , Rajitha, M. , Shyni, B. , & Mohandas, C. (2016). Molecular characterization and amplified ribosomal DNA restriction analysis of entomopathogenic bacteria associated with Rhabditis (Oscheius) spp. Biotech, 6, 32–44. 10.1007/s13205-015-0326-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , & Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit‐based approaches for 16S rRNA gene sequence analysis. Applied and Environmental Microbiology, 77, 3219–3226. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , Westcott, S. L. , Ryabin, T. , & Hall, J. R. (2009). Introducing mothur: Open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz, H. C. , Pfeffer, M. , Witte, A. , & Neubauer, H. (2008). Specific detection and differentiation of Ochrobactrum anthropi, Ochrobactrum intermedium and Brucella spp. by a multi‐primer PCR that targets the recA gene. Journal of Medical Microbiology, 57, 64–71. 10.1099/jmm.0.47507-0 [DOI] [PubMed] [Google Scholar]
- Schulte, F. (1989). The association between Rhabditis Necromena Sudhaus & Schulte, 1989 (Nematoda: Rhabditidae) and native and introduced millipedes in South Australia. Nematologica, 35, 82–89. 10.1163/002825989X00089 [DOI] [Google Scholar]
- Smart, G. C. , & Nguyen, K. B. (1994). Rhabditis (Oscheius) pheropsophi n. sp. (Rhabditida: Rhabditidae). Journal of Nematology, 26, 19–24. 10.1006/jipa.1994.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowndhararajan, K. , Marimuthu, S. , & Manian, S. (2013). Biocontrol potential of phylloplane bacterium Ochrobactrum anthropi BMO‐111 against blister blight disease of tea. Journal of Applied Microbiology, 114, 209–218. 10.1111/jam.12026 [DOI] [PubMed] [Google Scholar]
- Stock, S. P. , Caicedo, A. M. , & Calatayud, P. A. (2005). Rhabditis (Oscheius) colombiana n. sp. (Nematoda: Rhabditidae), a necromenic associate of the subterranean burrower bug Cyrtomenus bergi (Hemiptera: Cydnidae) from the Cauca Valley, Colombia. Nematology, 7, 363–373. 10.1163/156854105774355590 [DOI] [Google Scholar]
- Tambong, J. T. (2013). Phylogeny of bacteria isolated from Rhabditis sp. (Nematoda) and identification of novel entomopathogenic Serratia marcescens strains. Current Microbiology, 66, 138–144. 10.1007/s00284-012-0250-0 [DOI] [PubMed] [Google Scholar]
- Torres‐Barragan, A. , Suazo, A. , Buhler, W. G. , & Cardoza, Y. J. (2011). Studies on the entomopathogenicity and bacterial associates of the nematode Oscheius carolinensis . Biological Control, 59, 123–129. 10.1016/j.biocontrol.2011.05.020 [DOI] [Google Scholar]
- Warren, G. W. , Koziel, M. G. , Mullins, M. A. , & Nye, G. J. (1996). Novel pesticidal proteins and strains. Patent WO, 96, 10083. [Google Scholar]
- Wilson, M. J. , Glen, D. M. , George, S. K. , & Pearce, J. D. (1995). Selection of a bacterium for the mass production of Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) as a biocontrol agent for slugs. Fundamental and Applied Nematology, 18, 419–425. [Google Scholar]
- Wilson, M. J. , Glen, D. M. , Pearce, J. D. , & Rodgers, P. B. (1995). Monoxenic culture of the slug parasite Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) with different bacteria in liquid and solid phase. Fundamental and Applied Nematology, 18, 159–166. [Google Scholar]
- Yang, X. F. (2008). Isolation and identification of seven symbiotic bacteria from local entomopathogenic nematodes. Microbiology, 35, 225–229. 10.1163/156939308783122788 [DOI] [Google Scholar]
- Ye, W. , Torres‐Barragan, A. , & Cardoza, Y. J. (2010). Oscheius carolinensis n.sp. (Nematoda: Rhabditidae), a potential entomopathogenic nematode from vermicompost. Nematology, 12, 121–135. 10.1163/156854109X458464 [DOI] [Google Scholar]
- Zang, C. , Zhao, K. , Liu, C. , & Liang, C. (2014). Screening of antagonistic bacteria SY286 and its effect on grapevine downy mildew. Chinese Journal of Biology Control, 30, 402–407. 10.16409/j.cnki.2095-039x.2014.03.015 [DOI] [Google Scholar]
- Zhang, C. , Liu, J. , Xu, M. , Sun, J. , Yang, S. , An, X. , … Wu, Y. (2008). Heterorhabditidoides chongmingensis gen. nov., sp. nov (Rhabditida: Rhabditidae), a novel member of the entomopathogenic nematodes. Journal of Invertebrate Pathology, 98, 0–168. 10.1016/j.jip.2008.02.011 [DOI] [PubMed] [Google Scholar]
- Zhang, C. X. , Yang, S. Y. , Xu, M. X. , & Sun, J. (2009). Serratia nematodiphila sp. nov., symbiotically associated with entomopathogenic nematode Heterorhabditidoides chongmingensis (Rhabditida: Rhabditidae). International Journal of Systematic and Evolutionary Microbiology, 59, 1603–1608. 10.1099/ijs.0.65718-0 [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Xu, Y. , Chang, J. , & Li, M. (2016). Screening, resistance and growth‐promoting effect of endophytic bacteria with ACC deaminase activity isolated from soybean nodules. Acta Microbiologica Sinica, 56, 1009–1021. 10.13343/j.cnki.wsxb.20150434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in full in the results section of this paper apart from the DNA sequences of Ochrobactrum tritici NMA‐1 and Bacillus cereus NMA‐2 genes which are available at https://submit.ncbi.nlm.nih.gov/subs/genbank/ under accession number MK434214 and MK434215, respectively.


