Abstract
Defects in the Fanconi anemia (FA) DNA damage–response pathway result in genomic instability, developmental defects, hematopoietic failure, cancer predisposition, and metabolic disorders. The endogenous sources of damage contributing to FA phenotypes and the links between FA and metabolic disease remain poorly understood. Here, using mice lacking the Fancd2 gene, encoding a central FA pathway component, we investigated whether the FA pathway protects against metabolic challenges. Fancd2−/− and wildtype (WT) mice were fed a standard diet (SD), a diet enriched in fat, cholesterol, and cholic acid (Paigen diet), or a diet enriched in lipid alone (high-fat diet (HFD)). Fancd2−/− mice developed hepatobiliary disease and exhibited decreased survival when fed a Paigen diet but not a HFD. Male Paigen diet–fed mice lacking Fancd2 had significant biliary hyperplasia, increased serum bile acid concentration, and increased hepatic pathology. In contrast, female mice were similarly impacted by Paigen diet feeding regardless of Fancd2 status. Upon Paigen diet challenge, male Fancd2−/− mice had altered expression of genes encoding hepatic bile acid transporters and cholesterol and fatty acid metabolism proteins, including Scp2/x, Abcg5/8, Abca1, Ldlr, Srebf1, and Scd-1. Untargeted lipidomic profiling in liver tissue revealed 132 lipid species, including sphingolipids, glycerophospholipids, and glycerolipids, that differed significantly in abundance depending on Fancd2 status in male mice. We conclude that the FA pathway has sex-specific impacts on hepatic lipid and bile acid metabolism, findings that expand the known functions of the FA pathway and may provide mechanistic insight into the metabolic disease predisposition in individuals with FA.
Keywords: cholesterol metabolism, metabolic syndrome, DNA damage response, fatty acid metabolism, lipid metabolism, liver metabolism, cholestasis, Fanconi anemia complementation group D2 (FANCD2), Fanconi anemia, nonalcoholic fatty liver disease
Introduction
Fanconi anemia (FA)2 is a human genetic disorder characterized by developmental defects, sterility, hematopoietic failure, cancer predisposition, and metabolic disease. FA is caused by biallelic mutation of any of the 22 genes encoding components of the FA pathway. Canonically, the FA pathway responds to replicative stress, particularly to DNA interstrand cross-links. FA-deficient cells are hypersensitive to genotoxins, such as DNA cross-linking agents, irradiation, alkylating agents, and oxidative stress. Endocrine and metabolic abnormalities are also components of the FA phenotype (1–3). Close to 80% of FA patients have at least one endocrine abnormality (2). Dyslipidemia has been reported in 55% of FA patients (3) and impaired glucose tolerance in 27–68% of FA patients (2–5). The endogenous agents contributing to DNA damage and the etiologic connection between FA deficiency and the development of metabolic disease remain incompletely characterized.
FA phenotypes may be the direct result of DNA damage arising from endogenous sources normally counteracted by the FA pathway's DNA repair functions. This is thought to be the mechanism underlying the hypersensitivity of FA-deficient cells to aldehydes and formaldehyde by-products generated as a result of cellular metabolism (6–8). Alternatively, the increased risk of metabolic disease might be attributable to a direct connection between the FA pathway and metabolic homeostasis. Cells have numerous mechanisms to coordinate a DNA damage response (DDR) with metabolic regulation, with several factors having dual roles, including p53, ataxia telangiectasia-mutated (ATM), and others (9–11).
Evidence for a direct connection between the FA pathway and metabolic homeostasis is accumulating. The FA pathway modulates cellular antioxidant defenses (12, 13). FA-deficient cells have altered energy metabolism and increased reliance on glycolysis (14), and the FA pathway regulates energy metabolism via ATP synthesis (15). The FA pathway also influences lipid metabolism (16). FA deficiency has been reported to impact the abundance of lipids and other metabolites in multiple cancer types, as well as FA cell lines and lymphoblasts (17–20). Abnormal production of glycerophospholipids by FA bone marrow mesenchymal stromal cells may contribute to altered hematopoietic cell physiology (21). MicroRNAs that regulate cholesterol and lipid metabolism, miR-122 and miR-206, have decreased abundance in FA bone marrow mononuclear cells and lymphoblast cell lines (22). Thus, regulation of cellular metabolism, including lipid metabolism, may be an important component of the tumor suppression roles of the FA pathway and may relate to the metabolic phenotypes seen in FA patients.
We evaluated how FA deficiency alters the metabolic impacts of challenge with a high-fat diet (HFD) or the high-fat/high-cholesterol Paigen diet in vivo. HFD is used routinely in obesity and insulin-resistance models, whereas the Paigen high-fat diet with cholesterol and cholic acid induces steatohepatitis and hepatobiliary disease without obesity (23). We predicted that FA mutant mice would be hypersensitive to challenge with these diets through either increased DNA damage, increased metabolic derangements, or both. The damaging impacts of excessive lipid metabolism on hepatic physiology include oxidative stress, lipid peroxidation, mitochondrial damage, endoplasmic reticulum stress, cytokine imbalances, and inflammation, factors in nonalcoholic fatty liver disease and steatohepatitis (24). Our data indicate the FA pathway is essential for protection against hepatobiliary disease in the face of Paigen diet challenge and impacts hepatic lipid and BA homeostasis in male mice.
Results
Male Fancd2−/− mice have increased susceptibility to hepatobiliary disease and hepatic damage when fed Paigen diet
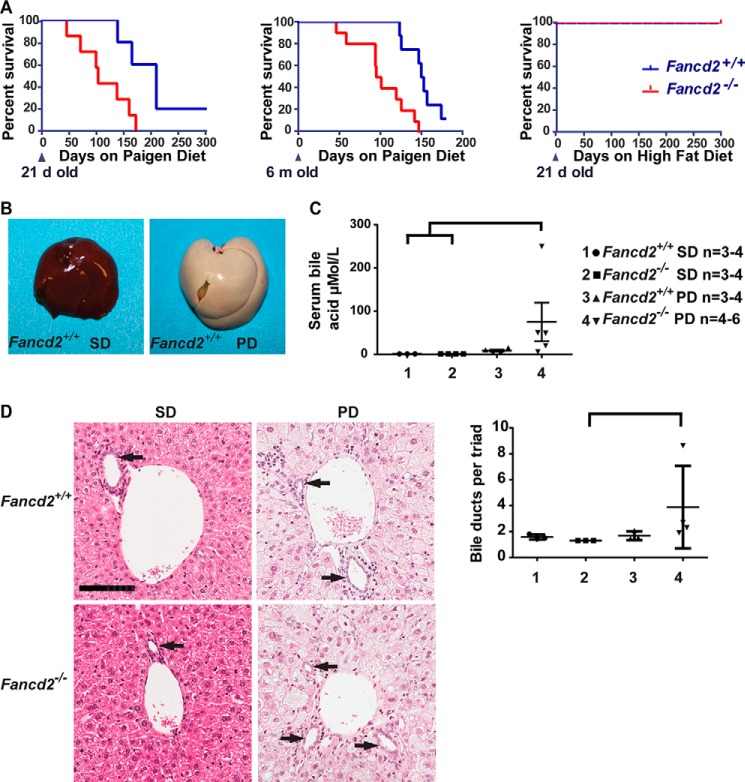
We aimed to test whether deficiency for FANCD2, a central component of the FA pathway (25), would be associated with increased sensitivity to hepatic metabolic challenge by feeding mice a high-fat, high-cholesterol diet with cholic acid (Paigen diet) or a diet enriched in lipid alone (HFD). Paigen diet feeding led to increased morbidity and mortality in Fancd2−/− mice compared with WT controls when the Paigen diet was fed from weaning or beginning at 6 months of age (Fig. 1A, left and middle panels). No morbidity or mortality was observed in Fancd2−/− or WT mice fed HFD (Fig. 1A, right panel) or fed normal mouse chow (standard diet, SD). These data demonstrate that Fancd2−/− mice were hypersensitive to Paigen diet feeding but not HFD feeding.
Figure 1.
Male Fancd2−/− mice had increased susceptibility to hepatobiliary disease when fed a Paigen diet. A, Kaplan-Meier survival curves showing increased mortality in Fancd2−/− mice fed a Paigen diet beginning at 21 days (left; p = 0.014; Fancd2+/+ = 5 and Fancd2−/− = 8) or 6 months (middle; p = 0.001; Fancd2+/+ = 8 and Fancd2−/− = 10) of age relative to WT mice, and a lack of sensitivity to a high-fat diet (right; p = 1.0; Fancd2+/+ = 8, Fancd2−/− = 5). B, livers were collected from mice fed SD or Paigen diet (PD) for 50–55 days, demonstrating Paigen diet-induced fatty liver disease. There were no grossly detectable differences between genotypes on either diet. C, serum was collected from mice at 50–55 days for blood chemistry analysis using an Abaxis VetScan VS2 chemistry analyzer. Bile acid concentration was significantly increased in male Fancd2−/− mice fed a Paigen diet relative to WT SD–fed mice (p = 0.012) and Fancd2−/− SD-fed mice (p = 0.010). D, hematoxylin and eosin (H&E)-stained liver sections focusing on portal triads. Bile ducts are lined by cuboidal epithelium (arrows). Scale bar equals 100 μm. Quantification of the average number of bile ducts per hepatic portal triad. Fancd2−/− Paigen diet–fed mice had significantly more bile duct profiles than Fancd2−/− SD–fed mice (p = 0.011) indicating Paigen diet feeding induced biliary hyperplasia in Fancd2−/− mice. Error bars represent S.E.
High-fat, high-cholesterol diets enriched with cholic acid are lithogenic and cause severe hepatic pathology (26). At necropsy, Paigen diet–fed mice of both genotypes had significant hepatomegaly characterized by firm, pale livers with rounded margins, whereas SD-fed mice of both genotypes had grossly normal livers (Fig. 1B). Morbidity in both genotypes fed Paigen diet was consistent with liver failure and/or cholestasis. Icterus was an experimental end point, and choleliths and distended gallbladders were found at necropsy. Observed hepatobiliary phenotypes were sex-dependent. In male mice, serum BA concentration increased upon Paigen diet feeding in Fancd2−/− mice by roughly 9-fold more than in WT mice (Fig. 1C), although in females, there was no genotype-dependent difference in serum BA concentration upon Paigen diet feeding (Fig. S1A). Additionally, male Fancd2−/− mice fed the Paigen diet had hepatic biliary hyperplasia, with an increase in bile duct profiles (Fig. 1D). Female mice of both genotypes had a nonstatistically significant trend toward increased bile ducts upon Paigen diet feeding (Fig. S1B). Biliary hyperplasia was not observed in mice of either genotype fed HFD (WT average 1.15 ± 0.09 ducts/triad; Fancd2−/− average 1.15 ± 0.074 ducts/triad on HFD) or another DDR-deficient mouse model (Atm−/−) on a FVB/N background fed Paigen diet (Fig. S1C) (27). These data indicate that the increased morbidity and mortality observed upon Paigen diet feeding in Fancd2−/− mice were associated with hepatobiliary disease consistent with biliary outflow obstruction.
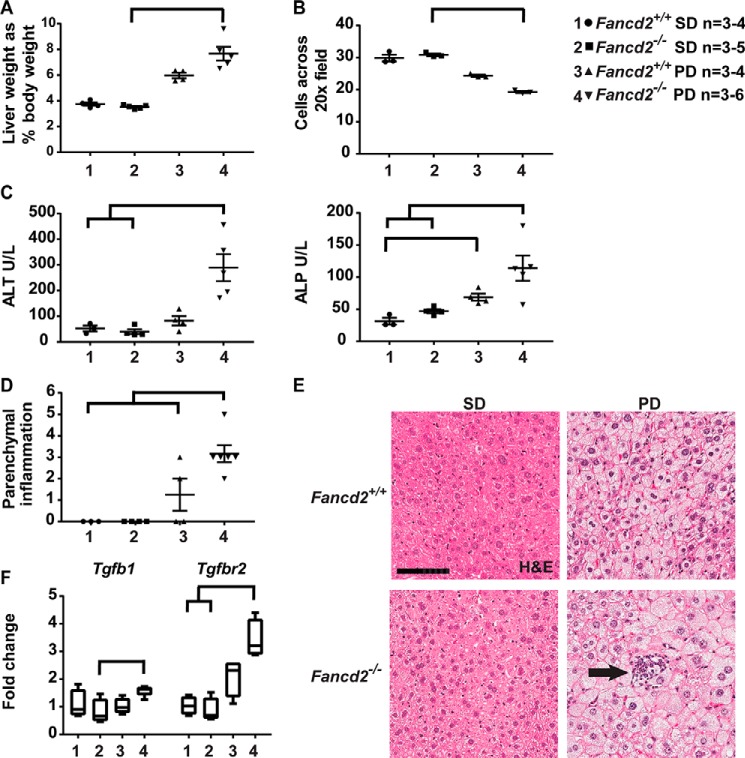
Concurrent with the biliary phenotype, male Fancd2−/− mice fed the Paigen diet had increased indices of hepatic disease. Male Fancd2−/− mice fed the Paigen diet had an increased liver weight relative to body weight versus Fancd2−/− SD-fed controls (Fig. 2A), whereas Paigen diet-fed female mice of both genotypes had a similar increase in liver weight (Fig. S2A). No difference by genotype was seen in mean liver weight for mice fed HFD (WT 2.78 ± 0.3%; Fancd2−/− 2.68 ± 0.3%, p = 0.5476); we therefore focused subsequent analyses on the Paigen diet-fed cohort. Male Paigen diet-fed Fancd2−/− mice had significantly fewer hepatocytes per ×20 field than SD fed Fancd2−/− mice (Fig. 2B), whereas Paigen diet–fed females of both genotypes had a similar number of hepatocytes per field (Fig. S2B). These data indicate that Paigen diet feeding resulted in greater hepatomegaly and hepatocellular swelling in male Fancd2−/− mice.
Figure 2.
Paigen diet feeding induced greater hepatomegaly, hepatic damage, and hepatic inflammation in male Fancd2−/− mice. Mice were fed PD or SD for 50–55 days and then sacrificed. A, male Fancd2−/− mice had increased liver weight relative to body weight on Paigen diet versus SD (p = 0.002). B, Paigen diet feeding increased hepatocyte size, decreasing the average number of cells across ten 20× fields. Male Fancd2−/− mice had marginally significantly larger hepatocytes than male WT mice fed SD (p = 0.052) and significantly larger hepatocytes than male Fancd2−/− mice fed SD (p = 0.039). C, serum chemistry analysis was performed after 50–55 days of diet feeding using an Abaxis VetScan VS2 chemistry analyzer. Male Fancd2−/− mice fed the Paigen diet had increased serum alanine aminotransferase (ALT) (left panel), a marker of hepatocellular damage, versus WT mice fed SD (p = 0.0496), and Fancd2−/− mice fed SD (p = 0.006). Male Fancd2−/− mice fed the Paigen diet had increased alkaline phosphatase (ALP) (right panel), a marker of hepatobiliary damage, versus WT mice fed SD (p = 0.011) and Fancd2−/− mice fed SD (p = 0.038). WT Paigen diet–fed mice also had a significant increase in serum alkaline phosphatase versus WT SD–fed mice (p = 0.049). D and E, H&E-stained liver sections were scored for hepatic parenchymal polymorphonuclear cells (inflammation, arrow). Scale bar equals 100 μm. Paigen diet feeding increased hepatic inflammation in male Fancd2−/− mice versus all three other groups (p < 0.005 for all pairwise comparisons). F, Paigen diet feeding was associated with increased expression of the gene encoding the inflammatory marker TGFβ (Tgfb1) in Fancd2−/− mice versus Fancd2−/− mice fed SD (p = 0.047). Expression of the gene encoding the TGFβ receptor 2 (Tgfbr2) increased in Fancd2−/− mice fed the Paigen diet versus Fancd2−/− mice fed SD (p = 0.005) and WT mice fed SD (p = 0.014). Expression presented as fold change relative to the WT SD group. Error bars in dot plots represent S.E. Box and whisker plots show 25th to 75th percentiles (box) and minimum and maximum (whiskers), with the median indicated by the horizontal bar. p < 0.05 for all pairwise comparison indicated.
The sex specificity of the phenotypes observed raised the possibility that sex hormones influence the impact of dietary challenge. Gonadal dysfunction is common in FA (3), and Fancd2−/− mice show testis degeneration and germ cell loss (28). With the exception of one individual, male Fancd2−/− mice in our study tended to have lower serum testosterone than WT males, although individual mice varied significantly (Fig. S3). Thus, male Fancd2−/− mice may lack potential protective effects of testosterone, leading to the Paigen diet-induced hepatic pathologies observed, although we cannot rule out roles for other sex-specific mechanisms influencing hepatic metabolism (29–32).
Plasma was used for liver profile chemistry analysis. The increase in alanine aminotransferase, a marker of hepatocellular injury, was 10.9-fold greater in male Fancd2−/− mice than WT mice upon Paigen diet challenge (Fig. 2C, left panel). Hepatobiliary disease is also typically associated with elevated alkaline phosphatase. Male Fancd2−/− mice fed the Paigen diet had higher alkaline phosphatase, although it was increased in both WT and Fancd2−/− male mice fed a Paigen diet (Fig. 2C, right panel). In contrast, Paigen diet feeding resulted in similar increases in these serologic markers of hepatic pathology in female mice of both genotypes (Fig. S2, C and D). These data indicate that Fancd2 deficiency was associated with a greater severity of liver damage upon Paigen diet feeding in male mice.
Hepatic disease can be associated with altered hepatic insulin and glucose homeostasis. Although we did not observe differences in fasting glucose between diets or genotypes in either sex, gluconeogenesis enzymes encoded by G6pc and Pck1 trended toward lower expression in Fancd2−/− male mice fed a Paigen diet (Fig. S4). We found no evidence that Paigen diet feeding leads to altered bone marrow or blood cell counts, suggesting that metabolic by-products resulting from lipid metabolism did not overtly impact hematopoietic cells in Fancd2−/− mice (supporting Results and Fig. S5).
However, male Fancd2−/− mice fed the Paigen diet had a significantly higher hepatic parenchymal polymorphonuclear cell (inflammation) score versus WTs (Fig. 2, D and E). Inflammatory cell presence was accompanied by increased gene expression for transforming growth factor β (Tgfb) and TGFB receptor 2 (Tgfbr2) in male Fancd2−/− mice upon Paigen diet feeding (Fig. 2F). In contrast, female mice of both genotypes had similar increases in hepatic inflammation score and gene expression of Tgfb and Tgfbr2 (Fig. S2, E and F). Taken together, these data demonstrate that Paigen diet feeding elicited more severe biliary hyperplasia, hepatic injury, and inflammation in male Fancd2-deficient mice as compared with Fancd2-proficient controls. We therefore focused subsequent mechanistic analyses on male mice.
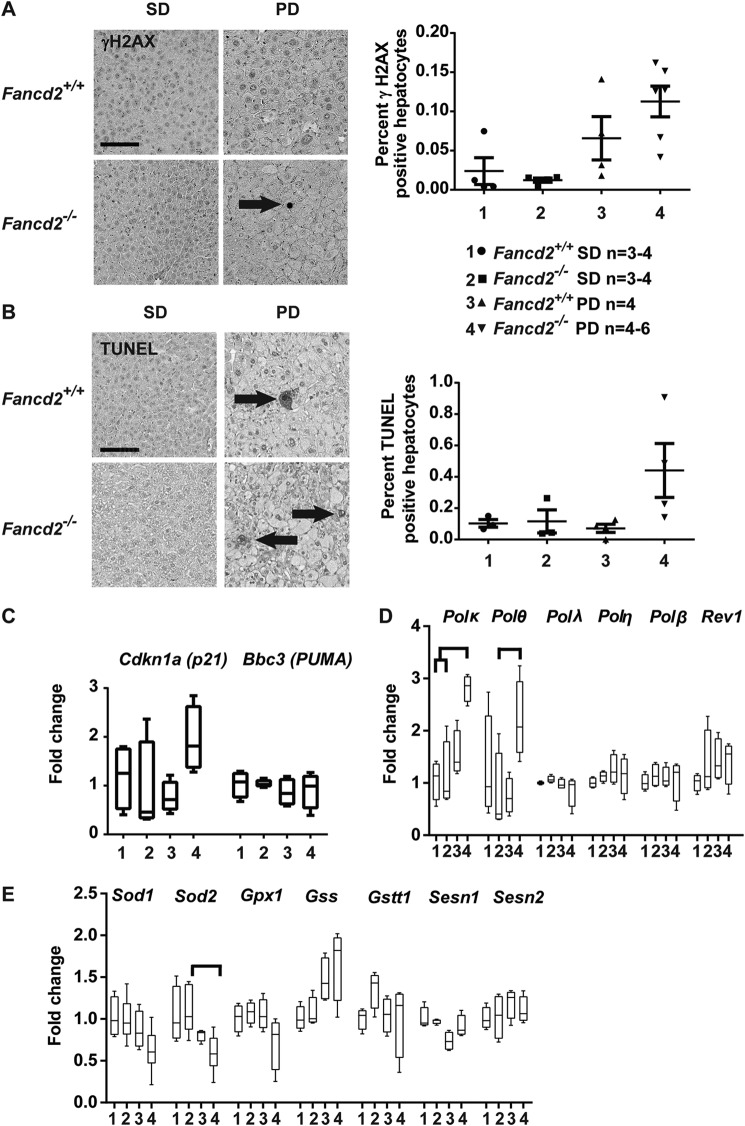
Hepatic DNA damage was only modestly increased by Paigen diet feeding in Fancd2 deficient mice relative to controls
As the canonical function of the FA pathway is in the DDR, we evaluated whether the more severe hepatic phenotypes in Paigen diet-fed Fancd2−/− mice were associated with elevated hepatic DNA damage. There was a trend toward increased staining for the DNA damage marker γH2AX (Fig. 3A) in hepatocytes from male Fancd2−/− mice fed the Paigen diet. Apoptosis was also increased as measured by terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining (Fig. 3B). A similar trend toward increased expression of p21, which encodes a cell cycle regulator and indicator of DDR activation, was observed upon Paigen diet feeding in Fancd2−/− mice, whereas expression of Bbc3 (PUMA), encoding a pro-apoptotic Bcl-2 family member, did not differ by genotype or diet (Fig. 3C). Although the trends toward increased DNA damage and apoptosis seen in the Fancd2−/− Paigen diet–fed mice were consistent with hepatobiliary pathology from accumulated BA and/or lipids, the magnitude of the changes in the IHC and DDR gene expression data suggest that Paigen diet feeding did not elicit a robust DDR in either genotype.
Figure 3.
Male Fancd2−/− mice showed modestly increased hepatocellular DNA damage when fed Paigen diet. IHC or quantitative PCR was performed on livers collected after 50–55 days of PD or SD feeding. Comparisons were expressed as fold change relative to the wildtype (WT) SD group. A, IHC on liver sections for γH2AX, a DNA damage marker, and quantification of γH2AX-positive hepatocytes (arrow) as a percentage of total hepatocytes. Male Fancd2−/− mice fed the Paigen diet had a trend toward increased γH2AX-positive hepatocytes (p = 0.08 versus WT SD–fed mice). B, IHC of liver sections by TUNEL, which labels DNA nicks associated with apoptosis, and quantification of TUNEL-positive apoptotic hepatocytes (arrow) quantified as the percent of total hepatocytes. C, expression of DNA damage-response genes Cdkn1a (p21) and Bbc3 (PUMA). D, expression of genes encoding translesion synthesis DNA polymerases. Male Fancd2−/− Paigen diet–fed mice had increased expression of Polκ relative to SD-fed WT (p = 0.043) and Fancd2−/− (p = 0.036) mice. Male Fancd2−/− Paigen diet–fed mice had increased expression of Polθ relative to FancdD2−/− SD-fed mice (p = 0.014). E, expression of genes encoding enzymes involved in detoxifying superoxide radicals and GSH metabolism. Error bars in dot plots represent S.E. Box and whisker plots show 25th to 75th percentiles (box) and minimum and maximum (whiskers), with the median indicated by the horizontal bar. p < 0.05 for all pairwise comparison indicated. Scale bars are 100 μm.
Lipid peroxidation can result in DNA interstrand cross-links, and FA pathway-directed repair of such lesions involves coordination of translesion synthesis (TLS) DNA polymerases (25, 33). Only in Fancd2−/− male mice, Paigen diet feeding was associated with a significant increase in expression of TLS polymerase encoding genes Polθ and Polκ, but not Polλ, Polη, Polβ, or Rev1 (Fig. 3D). Elevated Polκ and Polθ expression in Fancd2−/− mice suggests increased reliance on potentially error-prone DNA damage tolerance mechanisms following Paigen diet-induced DNA damage.
Oxidative stress is characteristic of FA-deficient cells, and mitochondrial β-oxidation of free fatty acids generates reactive oxygen species (34). We therefore quantified expression of genes associated with GSH metabolism and superoxide detoxification. There were no significant differences by genotype or diet in gene expression levels of GSH peroxidase 1 (Gpx1), glutathione S-transferase θ (Gstt), GSH synthetase (Gss), sestrin 1 (Sesn1), or sestrin 2 (Sesn2) (Fig. 3E). However, male Fancd2−/− mice fed the Paigen diet had significantly lower superoxide dismutase 2 (Sod2) gene expression than male Fancd2−/− mice fed a SD. A similar trend was seen in Sod1 expression. This suggests Fancd2−/− mice fed the Paigen diet may be less able to detoxify superoxide due to lower Sod1/2 expression; however, up-regulation of antioxidant defenses was not observed in either genotype upon Paigen diet feeding. Female mice of both genotypes had similar indices of DNA damage and DDR gene expression levels (Fig. S6). Together, these observations that Paigen diet sensitivity in Fancd2−/− mice was not associated with a robust DDR or antioxidant defense response suggested the possibility of a noncanonical metabolic role for the FA pathway.
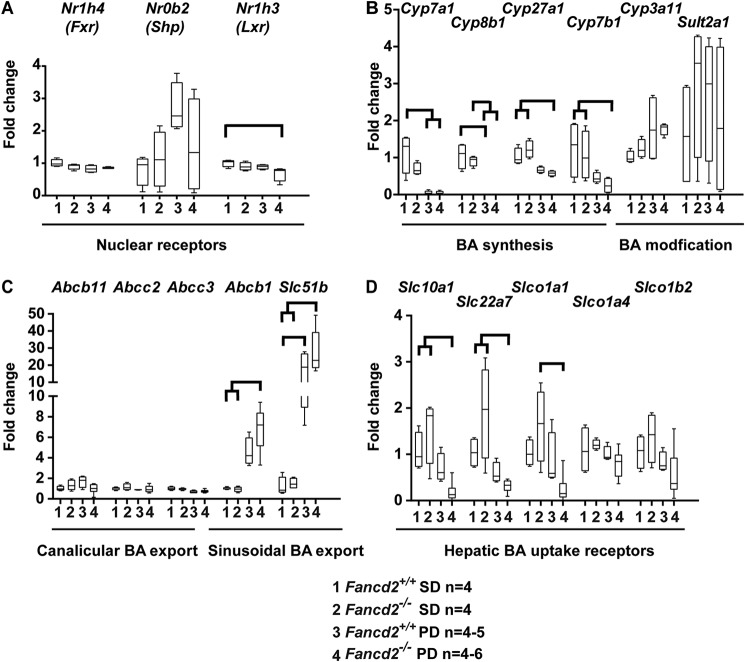
Sensitivity to hepatobiliary damage in male Fancd2−/− mice fed the Paigen diet is not due to gene expression differences for rate-limiting enzymes in hepatic bile acid synthesis
As male Fancd2−/− mice fed the Paigen diet had increased susceptibility to hepatobiliary disease, increased serum BA, and biliary hyperplasia, we probed expression of genes involved in hepatic BA, cholesterol, and lipid homeostasis. Expression of the genes encoding the nuclear BA receptor Fxr and its downstream mediator Shp did not differ by genotype or diet (Fig. 4A). There was a slight but statistically significant reduction in expression of Lxr, which encodes a nuclear receptor and cholesterol and lipid regulator, in Fancd2−/− Paigen diet–fed male mice. As expected with a cholic acid–enriched diet, expression levels of genes encoding BA synthesis enzymes Cyp7a1, Cyp8b1, Cyp27a1, and Cyp7b1 were down-regulated in WT and Fancd2−/− male mice upon Paigen diet feeding (Fig. 4B). Expression of Cyp3A11 and Sult2a1, which encode enzymes that modify BA, making them less cytotoxic, was not significantly different by genotype or diet. Expression of canalicular and sinusoidal hepatic BA exporters Abcb11 (Bsep), Abcc2 (Mrp2), and Mrp3 (Abcc3) did not differ significantly by diet or genotype in male mice, although expression of Abcb1 (Mdr1) and Ostβ (Slc51b) increased upon Paigen diet feeding in both genotypes (Fig. 4C). Thus, it appears that Fancd2−/− mice were capable of modulating gene expression following Paigen diet challenge to decrease de novo BA synthesis and detoxify and export BA.
Figure 4.
Differential expression of genes encoding proteins involved in BA metabolism was unlikely to be the cause of hepatobiliary disease in Fancd2−/− mice fed the Paigen diet. Mice were sacrificed after feeding PD or SD for 50–55 days, and RNA was extracted from liver, reverse-transcribed, and used for quantitative PCR. All comparisons are expressed as fold change relative to the WT SD group. A, expression of genes encoding the nuclear receptor transcription factors regulating BA (Fxr and Shp) and cholesterol and lipid (Lxr). There was a small but significant reduction in expression of Nrlh3 (Lxr) in Fancd2−/− Paigen diet–fed mice relative to WT SD–fed mice (p = 0.036). B, expression of genes encoding BA synthesis and modification enzymes. WT and Fancd2−/− mice down-regulated BA synthesis enzyme gene expression when fed Paigen diet (p < 0.05 for all pairwise comparisons). C, expression of hepatic canalicular and sinusoidal BA transporter-encoding genes. The expression of the sinusoidal BA exporter encoded by Abcb1 (Mdr1) increased upon Paigen diet feeding in both genotypes, but the increase was larger and statistically significant only for Fancd2−/− Paigen diet–fed mice versus SD-fed mice (p < 0.05 for both pairwise comparisons). The expression of the sinusoidal exporter encoded by Slc51b increased upon Paigen diet feeding in both genotypes, but to a larger degree in Fancd2−/− mice (p < 0.05 for all pairwise comparisons indicated). D, expression of genes encoding hepatic BA uptake receptors. Fancd2−/− Paigen diet–fed mice had a significant reduction in the expression of three hepatic BA uptake receptor-encoding genes (Slc10a1 (Ntcp), Slc22a7 (Oat2), and Slc1a1), which is consistent with bile acid accumulation and cytotoxicity. Box and whisker plots show 25th to 75th percentiles (box) and minimum and maximum (whiskers), with the median indicated by the horizontal bar. p < 0.05 for all pairwise comparisons indicated.
We also quantified expression of genes encoding hepatic BA importers. Male Fancd2−/− mice fed the Paigen diet had a greater decrease in expression of BA importer encoding genes Slc10a1 (NTCP; 2.59-fold greater decrease), Slc22a7 (Oat2; 1.95-fold greater decrease), and Slco1a1 (Oatp1a1; 4.92-fold greater decrease) than WT Paigen diet–fed mice (Fig. 4D). Reduction in these BA import receptors can be interpreted as a compensatory hepatoprotective response to increased intracellular BA accumulation. In addition to being consistent with the observed morbidity, the decreased expression of hepatic BA importer genes in Paigen diet–fed Fancd2−/− mice likely contributed to the observed increase in serum BA. In contrast to males, female mice of both genotypes had similar expression of genes encoding BA metabolism and export/import receptors (Fig. S7).
Male Fancd2−/− mice have impaired cholesterol and lipid regulation upon Paigen diet feeding
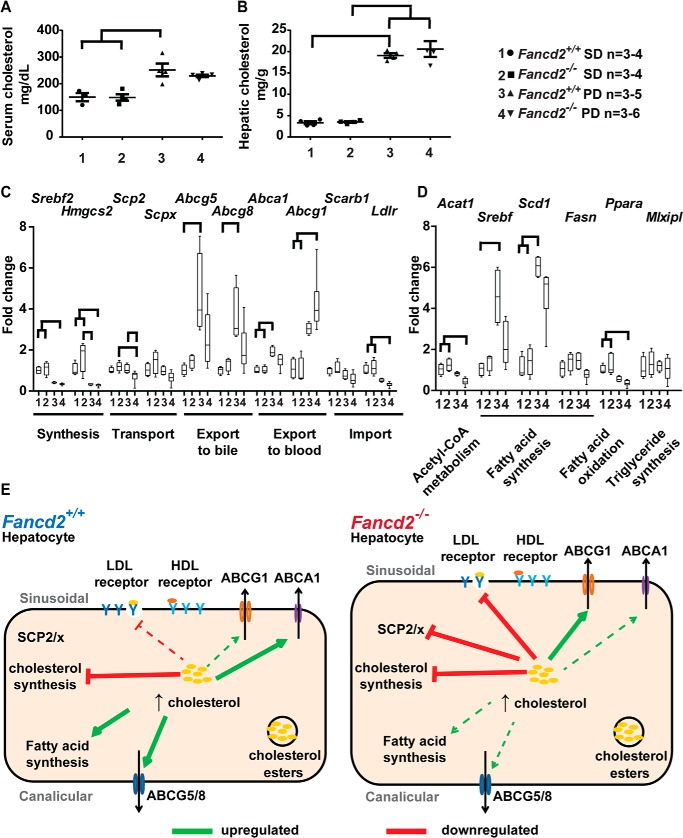
As we did not observe differential BA metabolism gene expression that appeared causal for the hepatobiliary pathology in Fancd2−/− mice upon Paigen diet feeding, we evaluated whether the phenotype was associated with impaired cholesterol metabolism. Serum and hepatic total cholesterol increased in a genotype-independent manner upon Paigen diet feeding (Fig. 5, A and B), and there was no detectable difference in the proportion of serum cholesterol HDL and non-HDL subtypes (Fig. S8). Expression of Scarb1 (HDL receptor) did not vary significantly. Expression of Ldlr (LDL receptor) decreased in both genotypes on the Paigen diet; however, the decrease was 1.5-fold greater and statistically significant only in male Fancd2−/− mice (Fig. 5C). These data indicate that the hepatobiliary phenotype observed in male Fancd2−/− mice was not associated with gene expression changes predictive of increased serum or hepatic cholesterol; however, factors regulating hepatic uptake of cholesterol may differ by Fancd2 status.
Figure 5.
Male Fancd2−/− mice had differential cholesterol and lipid metabolism gene expression upon Paigen diet feeding. After feeding PD or SD for 50–55 days, livers were collected for RNA extraction and used for quantitative PCR. All comparisons are expressed as fold change relative to the WT SD group. Total serum (A) and hepatic (B) cholesterol increased on Paigen diet feeding in both genotypes in male mice (p < 0.05 for indicated comparisons). C, expression of genes encoding proteins involved in cholesterol metabolism and transport. Both genotypes decreased expression of Srebf2 (Srebp2), encoding the major transcription factor regulating cholesterol synthesis and Hmgcs2, encoding a cholesterol synthesis enzyme, upon Paigen diet feeding. Fancd2−/− mice had decreased expression of the cholesterol transporters encoded by Scp2/x upon Paigen diet feeding and failed to up-regulate expression of the cholesterol exporters encoded by Abcg5/8 and Abca1 upon Paigen diet feeding, as was observed in WT Paigen diet–fed mice. Fancd2−/− but not WT mice had a significantly increased expression of the cholesterol exporter Abcg1 upon Paigen diet feeding. LDL receptor (Ldlr) expression decreased in both genotypes upon Paigen diet feeding but decreased significantly only in Fancd2−/− mice. D, expression of genes involved in acetyl-CoA, fatty acid, and triglyceride metabolism. Fancd2−/− mice had decreased expression of Acat1, which converts two acetyl-CoAs to acetoacetyl-CoA as an early step in lipid, ketone, and amino acid synthesis, whereas Paigen diet feeding did not significantly impact expression of Acat1 in WT mice. Upon Paigen diet feeding, WT but not Fancd2−/− mice increased expression of Srebf1 (Srebp1c), the major transcription factor regulating fatty acid synthesis. A trend toward lower expression of Fasn (fatty-acid synthase), a transcriptional target of SREPB1c, was seen in Fancd2−/− mice fed the Paigen diet. Ppara, a gene regulating fatty acid oxidation, decreased upon Paigen diet feeding in both genotypes, although the decrease was significant only for Fancd2−/− mice fed the Paigen diet. E, summary of differential expression of genes encoding proteins involved in cholesterol and lipid metabolism by FANCD2 status upon Paigen diet feeding in male WT (left) and Fancd2−/− mice (right). Solid lines indicate statistically significant up/down-regulation. Dashed lines indicate nonsignificant trends. Error bars in dot plots represent S.E. Box and whisker plots show 25th to 75th percentiles (box) and minimum and maximum (whiskers), with the median indicated by the horizontal bar. p < 0.05 for all pairwise comparisons indicated.
We subsequently uncovered differential regulation of genes encoding proteins involved in cholesterol transport, export, import, and fatty acid synthesis in male Fancd2−/− mice fed the Paigen diet. The expression level of genes encoding cholesterol synthesis enzymes Srebf2 (SREBP2) and Hmgcs2 (hydroxymethylglutaryl-CoA synthase) decreased in both genotypes upon Paigen diet feeding, suggesting cholesterol synthesis decreased irrespective of genotype. Expression of genes encoding sterol carrier protein 2/sterol carrier protein-x (SCP-2/SCP-x), which are generated via use of alternative transcriptional start sites and involved in cholesterol trafficking and BA synthesis (35), were differentially regulated by Fancd2 status. Paigen diet feeding was associated with significantly reduced Scp2 expression and a 10-fold greater reduction in Scpx in male Fancd2−/− mice (Fig. 5C), but it did not significantly impact expression of Scp2/x in WT mice, indicating that Fancd2 loss impairs hepatic expression of cholesterol transport and enzymatic conversion genes.
Cholesterol is excreted from hepatocytes into bile via ABC family members ABCG5/8 (36). Unlike male WT mice, male Fancd2−/− mice did not demonstrate increased Abcg5/8 expression upon Paigen diet feeding (Fig. 5C). Cholesterol is also exported to lipoprotein particles in blood. ABCA1 promotes the efflux of cholesterol and phospholipids to lipid-poor apoA-I as part of the early formation of HDL particles (37). Paigen diet feeding was associated with a significant increase in Abca1 expression in WT mice only. ABCG1 exports phospholipids and cholesterol to HDL, LDL, and phospholipid particles and is involved in the intracellular movement of cholesterol from the endoplasmic reticulum to the plasma membrane (37, 38). On the Paigen diet, only Fancd2−/− mice significantly up-regulated expression of Abcg1. These data suggest that cholesterol storage and export dynamics may have been altered in male Fancd2−/− mice upon Paigen diet challenge.
In addition to assessing genes regulating hepatic BA and cholesterol metabolism, we probed expression of genes encoding proteins involved in fatty acid and triglyceride metabolism. Acetyl-CoA acetyltransferase (Acat1) generates acetoacetyl-CoA from acetyl-CoA and is involved in the metabolism of fatty acids, ketones, amino acids, and energy metabolism (39–41). Acat1 gene expression was significantly decreased in Fancd2−/− mice fed the Paigen diet, and the decrease was 3.2-fold greater than the nonsignificant decrease in Acat1 expression in WT mice fed the Paigen diet (Fig. 5D). SREBP-1c (Srebf1) is a transcription factor that regulates hepatic lipogenesis, glucose metabolism, and the expression of genes needed to produce the fatty acid chains esterified to cholesterol (42). Paigen diet feeding was associated with increased expression of Srebf1 in WT but not Fancd2−/− male mice (Fig. 5D). This alteration in the expression of a fatty acid regulating transcription factor in livers of Fancd2−/− mice fed the Paigen diet is consistent with previously reported altered fatty acid metabolism in FA lymphoblasts and lymphocytes (20). Stearoyl-CoA desaturase is a rate-limiting enzyme that catalyzes the synthesis of monounsaturated fatty acids that are important for hepatic VLDL export and cholesterol esterification (44). On the Paigen diet, WT mice had a significant increase in expression of Scd1, whereas the nonsignificant increase in Scd1 expression in male Fancd2−/− mice was only one-half that seen in the WTs (Fig. 5D). Fancd2 status did not affect expression of Mlxipl, encoding a transcription factor (Carbohydrate Response Element Binding Protein) regulating triglyceride synthesis, or Pparα, which encodes a transcription factor that regulates fatty acid, triglyceride, and glucose metabolism. These data suggest the supply of fatty acids for cholesterol esterification and VLDL formation could have been deficient in Fancd2−/− mice, further perturbing cholesterol homeostasis. Female mice of both genotypes had similar expression of cholesterol metabolism genes (Fig. S9).
We also fed an additional cohort of male mice the Paigen diet for a longer 10-week period, initiated at 21 days of age, in an attempt to potentially exaggerate the hepatobiliary phenotype. Similar genotype-dependent differences in expression of cholesterol metabolism genes were observed in this cohort as in mice aged to 6 months and fed the Paigen diet for 50–55 days (Fig. S10). By contrast, transcriptional responses to HFD feeding did not generally differ by Fancd2−/− status, indicating that the phenotypes are not universal to high-fat diets and likely are dependent on the inclusion of cholesterol and/or cholic acid in Paigen diet (Fig. S11). Together, these data suggest that male Fancd2−/− mice had alterations in multiple genes involved in cholesterol metabolism in response to Paigen diet feeding which may impact hepatic cholesterol storage, intracellular transport and export, ultimately increasing hepatoxicity (Fig. 5E).
Several of the cholesterol and lipid metabolism genes with increased expression in WT but not Fancd2−/− mice fed the Paigen diet were transcriptional targets positively regulated by the nuclear receptor LXR, including Abcg5/8, Srebf, Scd-1, Acat1, and Lxr itself (44–46), suggesting some LXR-dependent transcriptional pathways were differentially regulated in Fancd2−/− mice. LXR also regulates Cyp7a1 (45), which was decreased in Paigen diet–fed mice of both genotypes, although in response to HFD feeding Cyp7a1 had roughly half the expression in Fancd2−/− versus WT mice. However, when treated with the synthetic LXR agonist T0901317, male WT and Fancd2−/− mice had similar up-regulation in LXR target gene expression, indicating that Fancd2−/− mice were capable of appropriate LXR transcriptional activity when LXR was activated by this agonist (Fig. S12).
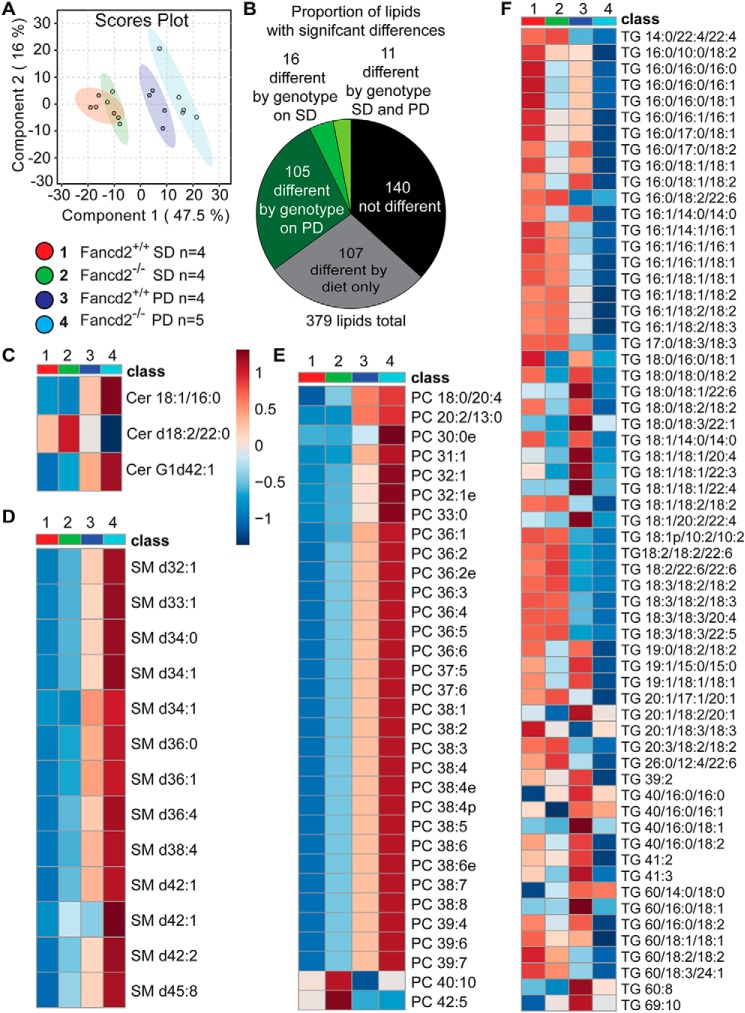
To further analyze hepatic lipid metabolism in Fancd2−/− mice, we performed untargeted lipidomic profiling in positive mode. Partial least-squares discriminate analysis revealed distinct clustering of Paigen diet–fed WT samples relative to Fancd2−/− samples (Fig. 6A). A total of 379 lipid species were detected (Fig. S13A). Approximately 28% (107) of lipids had a significant diet effect only, and notably, nearly 35% (132) of lipids detected differed between WT and Fancd2−/− groups, with the vast majority of genotype differences (105) in lipid abundance occurring on the Paigen diet (Fig. 6B). The top lipids contributing to the component scores are shown in Fig. S13, B and C. Fancd2−/− samples had numerous differences in membrane, energy storage, and signaling lipids.
Figure 6.
Male Fancd2−/− mice differed in hepatic sphingolipid, glycerophospholipid, and glycerolipid species abundance, particularly upon Paigen diet feeding. After feeding PD or SD for 50–55 days, livers were collected and lipids extracted for untargeted lipidomics. A, partial least-squares discriminant analysis revealing partial separation of WT and Fancd2−/− mice on SD and separation of the genotypes on Paigen diet. B, proportions of the 379 lipid species detected that differed significantly by diet or genotype. Heat maps of lipid species with significantly different abundances between WT and Fancd2−/− mice on Paigen diet: ceramides (Cer) (C); sphingomyelins (SM) (D); phosphatidylcholines (PC) (E); and triacylglycerols (TG) (F).
Fancd2−/− mice fed the Paigen diet had significantly different and largely elevated relative abundances of several species of ceramides (Cer), including Cer 18:1/16:0, and sphingomyelins, classes of sphingolipids that are components of cellular membranes and involved in many cellular functions (Fig. 6, C and D, and Fig. S14, A–C). Numerous genotype-dependent differences were detected in the abundance of glycerophospholipids, the major class of lipids in cell membranes. Phosphatidylcholine (PC) species that differed by genotype on Paigen diet, which were generally markedly elevated in Fancd2−/− mice, are shown in Fig. 6E, with all PC species shown in Fig. S15A. Lysophosphatidylcholines (LPC) and phosphatidylethanolamines (PE) also differed by diet and genotype (Fig. S15, B–E). Fancd2−/− mice fed the SD or Paigen diet had higher hepatic concentrations of LPC (e.g. LPC 16:0 or 18:0).
Glycerolipids differed by Fancd2 status as well. Triacylglycerols (TG) were the most frequently detected lipids, and many differed in abundance between genotypes on the Paigen diet (Fig. 6F and Fig. S16A). Increased TGs are associated with lipotoxicity and insulin resistance (47), although interestingly, many TG species were reduced in livers from Fancd2−/− mice fed the Paigen diet. Total hepatic TG was not impacted by diet or genotype (Fig. S16B). Several diacylglycerols (DG), which can be reacylated and converted to TG, as well as function as signaling molecules (48), were differentially impacted by diet and genotype (Fig. S17, A and B). In addition to ceramides, DG contributes to increased hepatic insulin resistance (49, 50). Collectively, it would appear that fatty acids were partitioned away from neutral lipid synthesis and toward sphingolipid and glycerophospholipid production in Fancd2−/− mice.
Cholesterols can exist as free or esterified forms. Esterified forms are generated by acyl-CoA cholesterol acyltransferase and are considered an inert storage form that protects cells from free cholesterol accumulation (51). Seven cholesterol esters were detected in our lipidomic screen, two of which (ChE 20:4NH4 and ChE 22:6NH4) were significantly different between WT mice fed the Paigen diet, in which they increased, and Fancd2−/− mice fed the Paigen diet, in which they decreased (Fig. S17C), suggesting that cholesterol esterification and storage were altered in Fancd2−/− hepatocytes upon Paigen diet feeding. Sphingolipid, glycerophospholipid, and glycerolipid metabolism are interconnected (48), and these data indicate that altered lipid metabolism was characteristic of FA hepatocytes, particularly when faced with dietary metabolic challenge.
Discussion
The etiologic connection between a FA DDR pathway defect and the metabolic abnormalities observed in FA patients and cells is not understood. Furthermore, the specific DNA lesions and cellular aberrations that arise and contribute to disease phenotypes in the absence of an intact FA pathway are incompletely characterized. Recent work suggests the FA pathway regulates multiple components of cellular metabolism, including energy and lipid metabolism, in addition to its canonical functions in DNA repair (14, 15, 17–21). In this study, we tested the in vivo sensitivity of FA-deficient mice to challenge with a high-lipid HFD or the high-fat, high-cholesterol cholic acid–enriched Paigen diet to probe whether FA deficiency would result in increased DNA damage accumulation and/or altered cellular metabolism. Male Fancd2−/− mice were highly sensitive to Paigen diet-induced hepatobiliary pathology relative to WT mice in a sex-dependent manner, having increased serum BA and markers of hepatobiliary damage, biliary hyperplasia, greater hepatomegaly and hepatocellular swelling, increased hepatic inflammation, and differing in hepatic lipid metabolism. Although it is possible that some pathologies were common to both WT and Fancd2−/− mice but differed in severity, several of the Paigen diet-induced pathologies were apparent only in Fancd2−/− mice.
Two independent cohorts fed the Paigen diet for 55 days or 10 weeks showed highly-similar genotype-dependent differences in cholesterol gene expression (Fig. S10), suggesting these cholesterol metabolism differences are intrinsic phenotypes in Fancd2−/− mice upon Paigen diet challenge. Genotype-dependent expression differences in TLS polymerases and inflammation markers were not seen in the 10-week cohort, which may be due to increased damage in both genotypes after the longer 10-week challenge. The transcriptional differences were specific to Paigen diet feeding, as a distinct high-lipid diet, HFD, did not elicit the same genotype-dependent cholesterol gene expression differences (Fig. S11). The lack of transcriptional differences or hepatic sensitivity to HFD feeding in Fancd2−/− mice suggests the cholesterol and/or cholic acid in the Paigen diet were key to the observed hepatic pathology.
We initially evaluated evidence that canonical FA pathway functions in DNA damage and oxidative stress responses protected against hepatic pathology after Paigen diet feeding. It has been proposed that redox imbalances and oxidative stress contribute to cellular and organismal FA phenotypes, potentially including the metabolic phenotypes seen in FA (2, 52, 53). However, we found only mildly increased DNA damage in the Paigen diet-fed Fancd2−/− mice and no genotype-specific Paigen diet-induced changes in antioxidant gene expression. We also report the novel finding of increased expression of genes encoding TLS polymerases Polθ and Polκ upon Paigen diet feeding in male Fancd2−/− mice. It has been reported that Fancd2 and Polθ expressions are correlated in ovarian carcinoma and that homologous recombination-deficient tumors are hypersensitive to inhibition of Polθ-mediated repair (54, 55). Our results are consistent with increased reliance on lower fidelity repair pathways, such as alternative end-joining, in Fancd2−/− mice upon Paigen diet feeding. However, the severity of the hepatobiliary phenotype led us to explore additional roles for FANCD2 in hepatic metabolism.
As Paigen diet–fed male Fancd2−/− mice were hypersensitive to hepatic and biliary pathology, we quantified expression of genes involved in hepatic BA and cholesterol metabolism. We found no BA gene expression differences that appeared causal of the phenotype. More likely to contribute to the Paigen diet sensitivity, we report multiple cholesterol and lipid metabolism gene expression differences in Fancd2−/− mice fed the Paigen diet, including decreased hepatic expression of Ldlr (LDL receptor) and Scpx/2 (cholesterol transporters) and a lack of a significant increase in Abcg5/8 (cholesterol exporters) as well as Srebf and Scd1 (fatty acid synthesis regulators).
Although LXR-target gene expression in Fancd2−/− and WT mice responded similarly to synthetic LXR agonist administration, our finding of altered LXR target gene expression is interesting in light of reports that miR-206, which represses LXR activity, is reduced in FA cells (16, 22). Multiple transcriptional corepressors and coactivators have ligand-dependent interactions with LXR (56); thus differential expression of LXR targets in Fancd2−/− mice could arise from differential context-dependent activity of these cofactors. Additionally, as we have shown many lipid species are present at altered levels in Paigen diet–fed Fancd2−/− livers, the endogenous ligands of LXR may be present at different levels in Fancd2−/− mice.
Although we did not detect differences in serum or hepatic bulk cholesterol or differential proportions of serum cholesterol species, untargeted lipidomic profiling revealed differences across sphingolipid, glycerophospholipid, and glycerolipid classes. As components of lipid rafts, sphingolipids influence cholesterol trafficking and metabolism (57), promote apoptosis, and impact cell–cell connections (58, 59). Elevated ceramides are associated with insulin resistance and lipotoxicity (60, 61). Recent work has demonstrated that C16:0-linked ceramide is considered the principal sphingolipid antagonist of insulin sensitivity (62). Future work should determine whether insulin-stimulated glucose utilization is impaired by ceramide in Fancd2−/− mice. As saturated and unsaturated PC were increased in Paigen diet-fed Fancd2−/− mice, we expect that the CDP-choline and phosphatidylethanolamine N-methyltransferase pathways were fully functional (63). Considering that PC is required for VLDL assembly and secretion (64), the ability of Fancd2−/− mice to modulate VLDL export deserves further investigation. The finding of elevated LPCs in Fancd2−/− mice is of potential significance when we consider the ability of long-chain LPC to regulate innate immunity and inflammatory processes (65) and the observed elevation in hepatic inflammation in male Fancd2−/− mice. Changes in glycerophospholipid metabolism can alter membrane dynamics (48, 66), and our finding of differential abundance of sphingolipids and glycerophospholipids in livers of Fancd2−/− mice is consistent with alterations in these lipid classes reported in other cell types deficient in the FA pathway (17, 21). It should be noted that euthanasia or anesthesia can impact some tissue metabolites, and thus the carbon dioxide method of euthanasia used throughout this study may have impacted some liver metabolites measured (67).
Altered lipid abundances could both contribute to the hepatobiliary phenotype observed in Fancd2−/− mice upon Paigen diet feeding, as well as result from hepatoprotective compensatory responses. Altered lipid accumulation may have led to the increase in hepatocyte size observed in Fancd2−/− mice upon Paigen diet challenge. Components of cholesterol–glycosphingolipid rafts are protective against the cytotoxic effects of bile acids (68, 69), and the ratio of PC and PE may be important in hepatocyte protection from cholestatic liver injury (70).
Many FA patients have abnormalities in insulin and glucose metabolism, dyslipidemia, and metabolic syndrome (2). Our data revealing important impacts of FANCD2 in lipid and BA metabolism may be mechanistically related to these FA patient phenotypes and suggest that FA patients may respond to conventional therapy for metabolic syndrome differently from the general population. It is not yet clear what components of the metabolic alterations in FA cells are due to primary roles of the FA pathway or secondary to other defects, and the mechanism of how the FA pathway regulates lipid metabolism is unknown. The transcriptional differences in cholesterol and lipid metabolism genes indicate that at least some of the differences arise at the gene expression level. Although we did not detect transcriptional differences in TG metabolism genes, lipidomics revealed many differences in the relative abundance of triglyceride species. Likely, a combination of alterations in gene expression and enzymatic activities perturbs hepatic lipid metabolism in FA.
Metabolic derangements are a hallmark of cancer cells, and it is possible some of the pre-existing metabolic phenotypes of FA cells may be relevant to their genomic instability and tumor predisposition. Potentially targeting these metabolic perturbations as part of managing FA patients could reduce their risk of cancer development. It is also possible that combining metabolism-altering therapies with genotoxic chemotherapy may improve treatment for FA cancer patients, who are particularly sensitive to the toxic effects of common chemotherapeutic agents (71). Further exploration of the role the FA pathway plays in hepatic and systemic metabolism may reveal therapeutic targets for treatment and continue to expand the known roles of the FA pathway beyond its function in DNA repair and our understanding of the connection between the DDR and cellular metabolism.
Experimental procedures
Animal care
Fancd2−/− mice on the 129S4 background (28) were crossed for a minimum of 10 generations onto the 129S6 background. Fancd2−/− and WT littermates were generated from Fancd2 heterozygote crosses. The Cornell University Institutional Animal Care and Use Committee approved all animal procedures, and mice were cared for in compliance with the Guide for the Care and Use of Laboratory Animals. Mice were fed one of three diets: a SD (0% cholesterol, 0% cholic acid, 5.8% triglyceride; 7012 Harlan Teklad LM-485 Mouse/Rat Sterilizable Diet); a high-fat diet enriched in cholesterol with cholic acid (Paigen diet) (1.25% cholesterol, 0.5% cholic acid, 15.8% triglyceride; TD.880511, Harlan, Teklad Lab Animal Diets); or a diet high in lipid alone (HFD) (∼0.028% cholesterol, 0% cholic, 34.9% triglyceride; D12492 Research Diets Inc.). For detailed diet composition, housing conditions, and pathogen status, see the supporting information. Male and female mice were fed diets for 50–55 days or 10 weeks (if the diet was started at 6 months of age or weaning, respectively) and fasted 12 h during the dark cycle before euthanasia, which occurred during the light cycle.
Tissue analysis
Mice were euthanized with CO2 and weighed. Blood was collected via cardiac puncture for glucometer reading (Accu-Chek Aviva, Roche Applied Science) or into microtubes containing heparin for serum chemistry or EDTA for serum lipid analysis. The liver was removed aseptically and weighed. A portion of the median lobe was frozen in liquid nitrogen for RNA isolation. Portions of the remaining liver tissue were fixed in paraformaldehyde, processed, and stained with hematoxylin and eosin or IHC. Tissues were scored for inflammation by a boarded veterinary pathologist blinded to sample identity. To assess hepatocellular swelling, the number of hepatocytes across ten 20× fields was counted. To avoid confounding differences in hepatocyte size between liver zones, fields were chosen to center around a central vein of 60–70 μm in diameter. For additional details, see supporting information.
Serum chemistry
Plasma was separated by centrifugation, flash-frozen in liquid N2, and stored at −80 °C until analysis. An Abaxis VetScan VS2 chemistry analyzer and the VetScan VS2 mammalian liver profile kit was used to measure alkaline phosphatase, alanine aminotransferase, total BA concentration, and cholesterol in 100 μl of plasma.
Serum testosterone quantification
Testosterone was measured in heparinized plasma via radioimmunoassay using ImmuChemTM double antibody testosterone 125 RIA kit (MP Biomedicals) following the manufacturer's protocol.
Bone marrow and blood analysis
Heparinized blood was collected for hematocrit quantification. Fresh blood smear and bone marrow slides were stained with Wright's stain. The opposite femur and sternum were fixed in paraformaldehyde for bone marrow histopathology. Blood and bone marrow slides were scored by a boarded veterinary pathologist blinded to sample identity. See the supporting information for the indices scored.
Immunohistochemistry
IHC was performed on paraformaldehyde-fixed, paraffin-embedded 5-μm liver sections. See supporting information for details. DNA damage was assessed by γH2AX staining at 1:200 (Millipore). Secondary biotinylated anti-rabbit antibody (Invitrogen, Histostain) incubation was followed by staining with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Invitrogen) to detect positive cells. Hepatocyte apoptosis was quantified by terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick-end labeling per manufacturers recommendation (TUNEL; ApopTag Kit; Millipore).
Quantitative PCR
RNA was extracted from liver samples using RNA Stat-60 (Tel-Test, Inc.). Complementary DNA (high-capacity cDNA reverse transcription kit; Applied Biosystems) was synthesized from total RNA and used for qPCR (C1000 Touch Thermal Cycler, CFX96 Real-Time System, Bio Rad). Gene expression was normalized to Rplp0 (Arbp) and/or Tbp expression. Quantification was determined via the ΔΔCT method using the Fancd2+/+ SD group as the comparative Δ value. See supporting information for additional details and Table S1 for primer sequences.
Serum lipoprotein agarose electrophoresis
Lipoproteins were quantified as described previously (72). See supporting information for details.
Hepatic cholesterol and triglyceride quantification
Lipid extraction was modified from Ref. 73 (see supporting information). Cholesterol was measured using InfinityTM cholesterol liquid stable reagent (Thermo Fisher Scientific), and triglyceride was measured with InfinityTM triglyceride kit (Thermo Fisher Scientific) with Data-CalTM chemistry calibrator (Thermo Fisher Scientific) for calibration. Absorbance was read at 500 nm on a SpectraMax190 (Molecular Devices) 96-well plate reader.
LXR agonist administration
T0901317 (Cayman Chemical) was dissolved in DMSO to 50 mg/ml, which was then diluted 2:1 with sterile PBS on the day of injection. Drug or DMSO only was administered at 50 mg/kg via intraperitoneal injection to adult male mice of both genotypes on the morning of days 1 and 2, and then the mice were euthanized, and liver tissue was collected on the evening of day 2 following an 8-h fast.
Lipidomic profiling
Lipid extraction was performed as follows. Snap-frozen liver tissues (10 mg) were homogenized in 300 μl of cold 50% methanol, followed by addition of 50 μl of internal standard (25 μg/ml each of TG (15:0)3, PC (17:0)2, phosphatidylglycerol (14:0)2, LPC (20:0), phosphatidylserine (16:0)2, fatty acid (18:1), C17-ceramide (d18:1/17:0) and ChE (17:0). Then 600 μl of dichloromethane was added, and the mixture was vortexed for 10 s. 300 μl of LC-MS–grade water was then added and vortexed again for 10 s. The tissue homogenates were centrifuged at 13,000 × g for 15 min at 4 °C. A total of 370 μl of the lower lipid-rich dichloromethane layer was then collected into silica tubes, and the solvent was evaporated to dryness under vacuum. Samples were reconstituted in 150 μl of acetonitrile/isopropyl alcohol/H2O (65:30:5 v/v/v) prior to injection. Two μl was injected for LC-MS, which was performed on a Vanquish ultra-HPLC system with an Accucore C30, 2.6-μm column (2.1 mm inner diameter × 150 mm) coupled to a Q ExactiveTM hybrid quadrupole–Orbitrap high-resolution mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Lipidomics filter criteria are listed in Table S2. Lipidomics raw data are available in Table S3. Data analysis was conducted with LipidSearchTM (Thermo Fisher Scientific) and Metaboanalyst 4.0 software (43, 74). For additional details, see the supporting information.
Statistical analyses
Data were analyzed nonparametrically with Kruskal-Wallis tests followed by Dunn post hoc tests for multiple comparisons with p values adjusted with the Benjamini-Hochberg method. For histology and IHC data, an average number per mouse was used with the exception of bile duct quantification, in which case all counts were included in the analysis and a random mouse effect was included in the linear mixed effects model. Kaplan Meier survival curves were compared by log rank test. qPCR comparisons were performed on ΔCT values per mouse. p values of ≤0.05 were considered statistically significant. Data are presented as mean ± S.E. of the mean or as box and whisker plots showing 25th to 75th percentiles (box) and minimum and maximum (whiskers), with the median indicated by the horizontal bar. All analyses were made with R-3.4.1 (R Core Team (2015)) and GraphPad Prism version 7.03 for Windows (GraphPad Software). For lipidomics, analyses of normalized data collected in positive ion mode were summed within lipid species. Data were then generalized, log-transformed, auto-scaled, and subjected to partial least-squares discriminant analysis and analysis of variance with an adjusted p value (false discovery rate) cutoff of 0.05 and post hoc analyses by Fisher's least significant difference test. Heat maps were generated using Euclidean distance measure and Ward clustering algorithm. All lipidomics analyses were performed in Metaboanalyst 4.0 software (43, 74).
Author contributions
E. S. M., E. K. D., B. P. C., E. B.-K., and R. S. W. conceptualization; E. S. M., D. M. S., and J. W. M. formal analysis; E. S. M., E. K. D., D. I. K., E. B.-K., D. M. S., and T. L. S. investigation; E. S. M., E. B.-K., D. M. S., and J. W. M. methodology; E. S. M. writing-original draft; E. K. D., B. P. C., E. B.-K., D. M. S., T. L. S., J. W. M., and R. S. W. writing-review and editing; B. P. C., J. W. M., and R. S. W. resources; B. P. C., J. W. M., and R. S. W. supervision; R. S. W. project administration.
Supplementary Material
Acknowledgments
We thank Markus Grompe for Fancd2 knockout mice; Kirk Mauer for helpful discussions; Andrew Dannenberg for comments on the manuscript; Lynn Johnson for statistical assistance; Rebecca Haeusler for select qPCR primer sequences; Maria Elena Diaz Rubio and Jorge Eduardo Rico-Navarrete for assistance with lipidomics; and Timothy Pierpont for assistance with sample collection.
This work was supported in part by a Cornell University College of Veterinary Medicine resident research grant and National Institutes of Health Training Grant Award T32 ODO011000 (to E. S. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S17, Tables S1–S3, and supporting Refs. 1–16.
- FA
- Fanconi anemia
- PD
- Paigen diet
- SD
- standard diet
- IHC
- immunohistochemistry
- BA
- bile acid
- DDR
- DNA damage response
- HFD
- high-fat diet
- TG
- triacylglycerol
- DG
- diacylglycerol
- LPC
- lysophosphatidylcholine
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- Cer
- ceramide
- TUNEL
- terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick-end labeling
- TLS
- translesion synthesis
- LXR
- liver X receptor
- ChE
- cholesterol ester
- HDL
- high-density lipoprotein
- LDL
- low-density lipoprotein
- VLDL
- very-low-density lipoprotein
- H&E
- hematoxylin and eosin.
References
- 1. Wajnrajch M. P., Gertner J. M., Huma Z., Popovic J., Lin K., Verlander P. C., Batish S. D., Giampietro P. F., Davis J. G., New M. I., and Auerbach A. D. (2001) Evaluation of growth and hormonal status in patients referred to the International Fanconi Anemia Registry. Pediatrics 107, 744–754 10.1542/peds.107.4.744 [DOI] [PubMed] [Google Scholar]
- 2. Petryk A., Kanakatti Shankar R., Giri N., Hollenberg A. N., Rutter M. M., Nathan B., Lodish M., Alter B. P., Stratakis C. A., and Rose S. R. (2015) Endocrine disorders in Fanconi anemia: recommendations for screening and treatment. J. Clin. Endocrinol. Metab. 100, 803–811 10.1210/jc.2014-4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giri N., Batista D. L., Alter B. P., and Stratakis C. A. (2007) Endocrine abnormalities in patients with Fanconi anemia. J. Clin. Endocrinol. Metab. 92, 2624–2631 10.1210/jc.2007-0135 [DOI] [PubMed] [Google Scholar]
- 4. Elder D. A., D'Alessio D. A., Eyal O., Mueller R., Smith F. O., Kansra A. R., and Rose S. R. (2008) Abnormalities in glucose tolerance are common in children with Fanconi anemia and associated with impaired insulin secretion. Pediatr. Blood Cancer 51, 256–260 10.1002/pbc.21589 [DOI] [PubMed] [Google Scholar]
- 5. Rose S. R., Myers K. C., Rutter M. M., Mueller R., Khoury J. C., Mehta P. A., Harris R. E., and Davies S. M. (2012) Endocrine phenotype of children and adults with Fanconi anemia. Pediatr. Blood Cancer 59, 690–696 10.1002/pbc.24095 [DOI] [PubMed] [Google Scholar]
- 6. Ridpath J. R., Nakamura A., Tano K., Luke A. M., Sonoda E., Arakawa H., Buerstedde J.-M., Gillespie D. A., Sale J. E., Yamazoe M., Bishop D. K., Takata M., Takeda S., Watanabe M., Swenberg J. A., and Nakamura J. (2007) Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 67, 11117–11122 10.1158/0008-5472.CAN-07-3028 [DOI] [PubMed] [Google Scholar]
- 7. Langevin F., Crossan G. P., Rosado I. V., Arends M. J., and Patel K. J. (2011) Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 10.1038/nature10192 [DOI] [PubMed] [Google Scholar]
- 8. Oberbeck N., Langevin F., King G., de Wind N., Crossan G. P., and Patel K. J. (2014) Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol. Cell 55, 807–817 10.1016/j.molcel.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puzio-Kuter A. M. (2011) The role of p53 in metabolic regulation. Genes Cancer 2, 385–391 10.1177/1947601911409738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D. H., and Lee J. W. (2011) Tumor suppressor p53 regulates bile acid homeostasis via small heterodimer partner. Proc. Natl. Acad. Sci. U.S.A. 108, 12266–12270 10.1073/pnas.1019678108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S. H., Trinh A. T., Larsen M. C., Mastrocola A. S., Jefcoate C. R., Bushel P. R., and Tibbetts R. S. (2016) Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism. Nucleic Acids Res. 44, 9667–9680 10.1093/nar/gkw643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du W., Rani R., Sipple J., Schick J., Myers K. C., Mehta P., Andreassen P. R., Davies S. M., and Pang Q. (2012) The FA pathway counteracts oxidative stress through selective protection of antioxidant defense gene promoters. Blood 119, 4142–4151 10.1182/blood-2011-09-381970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pagano G., Talamanca A. A., Castello G., d'Ischia M., Pallardó F. V., Petrovíc S., Porto B., Tiano L., and Zatterale A. (2013) From clinical description, to in vitro and animal studies, and backward to patients: oxidative stress and mitochondrial dysfunction in Fanconi anemia. Free Radic. Biol. Med. 58, 118–125 10.1016/j.freeradbiomed.2013.01.015 [DOI] [PubMed] [Google Scholar]
- 14. Cappelli E., Cuccarolo P., Stroppiana G., Miano M., Bottega R., Cossu V., Degan P., and Ravera S. (2017) Defects in mitochondrial energetic function compels Fanconi anaemia cells to glycolytic metabolism. Biochim. Biophys. Acta 1863, 1214–1221 10.1016/j.bbadis.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 15. Jayabal P., Ma C., Nepal M., Shen Y., Che R., Turkson J., and Fei P. (2017) Involvement of FANCD2 in energy metabolism via ATP5α. Sci. Rep. 7, 4921 10.1038/s41598-017-05150-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Degan P., Cappelli E., Regis S., and Ravera S. (2019) New insights and perspectives in Fanconi anemia research. Trends Mol. Med. 25, 167–170 10.1016/j.molmed.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 17. Zhao X., Brusadelli M. G., Sauter S., Butsch Kovacic M., Zhang W., Romick-Rosendale L. E., Lambert P. F., Setchell K. D. R., and Wells S. I. (2018) Lipidomic profiling links the Fanconi anemia pathway to glycosphingolipid metabolism in head and neck cancer cells. Clin. Cancer Res. 24, 2700–2709 10.1158/1078-0432.CCR-17-3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nepal M., Ma C., Xie G., Jia W., and Fei P. (2018) Fanconi anemia complementation group C protein in metabolic disorders. Aging 10, 1506–1522 10.18632/aging.101487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panneerselvam J., Xie G., Che R., Su M., Zhang J., Jia W., and Fei P. (2016) Distinct metabolic signature of human bladder cancer cells carrying an impaired Fanconi anemia tumor-suppressor signaling pathway. J. Proteome Res. 15, 1333–1341 10.1021/acs.jproteome.6b00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ravera S., Degan P., Sabatini F., Columbaro M., Dufour C., and Cappelli E. (2019) Altered lipid metabolism could drive the bone marrow failure in Fanconi anaemia. Br. J. Haematol. 184, 693–696 10.1111/bjh.15171 [DOI] [PubMed] [Google Scholar]
- 21. Amarachintha S., Sertorio M., Wilson A., Li X., and Pang Q. (2015) Fanconi anemia mesenchymal stromal cells-derived glycerophospholipids skew hematopoietic stem cell differentiation through Toll-like receptor signaling. Stem Cells 33, 3382–3396 10.1002/stem.2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Degan P., Cappelli E., Longobardi M., Pulliero A., Cuccarolo P., Dufour C., Ravera S., Calzia D., and Izzotti A. (2019) A global microRNA profile in Fanconi anemia: a pilot study. Metab. Syndr. Relat. Disord. 17, 53–59 10.1089/met.2018.0085 [DOI] [PubMed] [Google Scholar]
- 23. Paigen B., Morrow A., Brandon C., Mitchell D., and Holmes P. (1985) Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 57, 65–73 10.1016/0021-9150(85)90138-8 [DOI] [PubMed] [Google Scholar]
- 24. Reccia I., Kumar J., Akladios C., Virdis F., Pai M., Habib N., and Spalding D. (2017) Non-alcoholic fatty liver disease: a sign of systemic disease. Metabolism 72, 94–108 10.1016/j.metabol.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 25. Ceccaldi R., Sarangi P., and D'Andrea A. D. (2016) The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- 26. Wang D. Q., Paigen B., and Carey M. C. (1997) Phenotypic characterization of Lith genes that determine susceptibility to cholesterol cholelithiasis in inbred mice: physical-chemistry of gallbladder bile. J. Lipid Res. 38, 1395–1411 [PubMed] [Google Scholar]
- 27. Daugherity E. K., Balmus G., Al Saei A., Moore E. S., Abi Abdallah D., Rogers A. B., Weiss R. S., and Maurer K. J. (2012) The DNA damage checkpoint protein ATM promotes hepatocellular apoptosis and fibrosis in a mouse model of non-alcoholic fatty liver disease. Cell Cycle 11, 1918–1928 10.4161/cc.20259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houghtaling S., Timmers C., Noll M., Finegold M. J., Jones S. N., Meyn M. S., and Grompe M. (2003) Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17, 2021–2035 10.1101/gad.1103403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turley S. D., Schwarz M., Spady D. K., and Dietschy J. M. (1998) Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets. Hepatology 28, 1088–1094 10.1002/hep.510280425 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., and Klaassen C. D. (2010) Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242 10.1194/jlr.M007641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maxwell K. N., Soccio R. E., Duncan E. M., Sehayek E., and Breslow J. L. (2003) Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 44, 2109–2119 10.1194/jlr.M300203-JLR200 [DOI] [PubMed] [Google Scholar]
- 32. Lorbek G., Perše M., Horvat S., Björkhem I., and Rozman D. (2013) Sex differences in the hepatic cholesterol sensing mechanisms in mice. Molecules 18, 11067–11085 10.3390/molecules180911067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grillari J., Katinger H., and Voglauer R. (2007) Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 35, 7566–7576 10.1093/nar/gkm1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Begriche K., Igoudjil A., Pessayre D., and Fromenty B. (2006) Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion 6, 1–28 10.1016/j.mito.2005.10.004 [DOI] [PubMed] [Google Scholar]
- 35. He H., Wang J., Yannie P. J., Kakiyama G., Korzun W. J., and Ghosh S. (2018) Sterol carrier protein-2 deficiency attenuates diet-induced dyslipidemia and atherosclerosis in mice. J. Biol. Chem. 293, 9223–9231 10.1074/jbc.RA118.002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quazi F., and Molday R. S. (2011) Lipid transport by mammalian ABC proteins. Essays Biochem. 50, 265–290 10.1042/bse0500265 [DOI] [PubMed] [Google Scholar]
- 37. Terasaka N. (2010) in The HDL Handbook (Komoda T., ed) pp. 199–214, Academic Press, Boston [Google Scholar]
- 38. Wang N., Lan D., Chen W., Matsuura F., and Tall A. R. (2004) ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 9774–9779 10.1073/pnas.0403506101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukao T., Mitchell G., Sass J. O., Hori T., Orii K., and Aoyama Y. (2014) Ketone body metabolism and its defects. J. Inherit. Metab. Dis. 37, 541–551 10.1007/s10545-014-9704-9 [DOI] [PubMed] [Google Scholar]
- 40. Garcia-Bermudez J., and Birsoy K. (2016) Drugging ACAT1 for cancer therapy. Mol. Cell 64, 856–857 10.1016/j.molcel.2016.11.023 [DOI] [PubMed] [Google Scholar]
- 41. Fan J., Lin R., Xia S., Chen D., Elf S. E., Liu S., Pan Y., Xu H., Qian Z., Wang M., Shan C., Zhou L., Lei Q. Y., Li Y., Mao H., et al. (2016) Tetrameric acetyl-CoA acetyltransferase 1 is important for tumor growth. Mol. Cell 64, 859–874 10.1016/j.molcel.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldstein J. L., DeBose-Boyd R. A., and Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- 43. Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., and Xia J. (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ntambi J. M. (1999) Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40, 1549–1558 [PubMed] [Google Scholar]
- 45. Maqdasy S., Trousson A., Tauveron I., Volle D. H., Baron S., and Lobaccaro J. M. (2016) Once and for all, LXRα and LXRβ are gatekeepers of the endocrine system. Mol. Aspects Med. 49, 31–46 10.1016/j.mam.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 46. Laffitte B. A., Joseph S. B., Walczak R., Pei L., Wilpitz D. C., Collins J. L., and Tontonoz P. (2001) Autoregulation of the human liver X receptor α promoter. Mol. Cell. Biol. 21, 7558–7568 10.1128/MCB.21.22.7558-7568.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Unger R. H., and Scherer P. E. (2010) Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol. Metab. 21, 345–352 10.1016/j.tem.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han X. (2016) Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 12, 668–679 10.1038/nrendo.2016.98 [DOI] [PubMed] [Google Scholar]
- 49. Jornayvaz F. R., and Shulman G. I. (2012) Diacylglycerol activation of protein kinase Cϵ and hepatic insulin resistance. Cell Metab. 15, 574–584 10.1016/j.cmet.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Samuel V. T., Petersen K. F., and Shulman G. I. (2010) Lipid-induced insulin resistance: unravelling the mechanism. Lancet 375, 2267–2277 10.1016/S0140-6736(10)60408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spector A. A., Mathur S. N., and Kaduce T. L. (1979) Role of acylcoenzyme A:cholesterol o-acyltransferase in cholesterol metabolism. Prog. Lipid Res. 18, 31–53 10.1016/0163-7827(79)90003-1 [DOI] [PubMed] [Google Scholar]
- 52. Pagano G., Degan P., d'Ischia M., Kelly F. J., Pallardó F. V., Zatterale A., Anak S. S., Akisik E. E., Beneduce G., Calzone R., De Nicola E., Dunster C., Lloret A., Manini P., Nobili B., et al. (2004) Gender- and age-related distinctions for the in vivo prooxidant state in Fanconi anaemia patients. Carcinogenesis 25, 1899–1909 10.1093/carcin/bgh194 [DOI] [PubMed] [Google Scholar]
- 53. Li J., Sipple J., Maynard S., Mehta P. A., Rose S. R., Davies S. M., and Pang Q. (2012) Fanconi anemia links reactive oxygen species to insulin resistance and obesity. Antioxid. Redox Signal. 17, 1083–1098 10.1089/ars.2011.4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kais Z., Rondinelli B., Holmes A., O'Leary C., Kozono D., D'Andrea A. D., and Ceccaldi R. (2016) FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 15, 2488–2499 10.1016/j.celrep.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ceccaldi R., Liu J. C., Amunugama R., Hajdu I., Primack B., Petalcorin M. I., O'Connor K. W., Konstantinopoulos P. A., Elledge S. J., Boulton S. J., Yusufzai T., and D'Andrea A. D. (2015) Homologous recombination-deficient tumors are hyper-dependent on POLQ-mediated repair. Nature 518, 258–262 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schulman I. G. (2017) Liver X receptors link lipid metabolism and inflammation. FEBS Lett. 591, 2978–2991 10.1002/1873-3468.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Róg T., and Vattulainen I. (2014) Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes? Chem. Phys. Lipids 184, 82–104 10.1016/j.chemphyslip.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 58. D'Angelo G., Capasso S., Sticco L., and Russo D. (2013) Glycosphingolipids: synthesis and functions. FEBS J. 280, 6338–6353 10.1111/febs.12559 [DOI] [PubMed] [Google Scholar]
- 59. Hakomori S., and Igarashi Y. (1995) Functional role of glycosphingolipids in cell recognition and signaling. J. Biochem. 118, 1091–1103 10.1093/oxfordjournals.jbchem.a124992 [DOI] [PubMed] [Google Scholar]
- 60. Summers S. A. (2006) Ceramides in insulin resistance and lipotoxicity. Prog. Lipid Res. 45, 42–72 10.1016/j.plipres.2005.11.002 [DOI] [PubMed] [Google Scholar]
- 61. Chaurasia B., and Summers S. A. (2015) Ceramides- lipotoxic inducers of metabolic disorders. Trends Endocrinol. Metab. 26, 538–550 10.1016/j.tem.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 62. Hla T., and Kolesnick R. (2014) C16:0-ceramide signals insulin resistance. Cell Metab. 20, 703–705 10.1016/j.cmet.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DeLong C. J., Shen Y. J., Thomas M. J., and Cui Z. (1999) Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J. Biol. Chem. 274, 29683–29688 10.1074/jbc.274.42.29683 [DOI] [PubMed] [Google Scholar]
- 64. Fast D. G., and Vance D. E. (1995) Nascent VLDL phospholipid composition is altered when phosphatidylcholine biosynthesis is inhibited: evidence for a novel mechanism that regulates VLDL secretion. Biochim. Biophys. Acta 1258, 159–168 10.1016/0005-2760(95)00116-T [DOI] [PubMed] [Google Scholar]
- 65. Kabarowski J. H., Xu Y., and Witte O. N. (2002) Lysophosphatidylcholine as a ligand for immunoregulation. Biochem. Pharmacol. 64, 161–167 10.1016/S0006-2952(02)01179-6 [DOI] [PubMed] [Google Scholar]
- 66. McMahon H. T., and Boucrot E. (2015) Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Overmyer K. A., Thonusin C., Qi N. R., Burant C. F., and Evans C. R. (2015) Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One 10, e0117232 10.1371/journal.pone.0117232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smid V., Petr T., Vanova K., Jasprova J., Suk J., Vitek L., Smid F., and Muchova L. (2016) Changes in liver ganglioside metabolism in obstructive cholestasis- the role of oxidative stress. Folia Biol. 62, 148–159 [PubMed] [Google Scholar]
- 69. Guyot C., and Stieger B. (2011) Interaction of bile salts with rat canalicular membrane vesicles: evidence for bile salt resistant microdomains. J. Hepatol. 55, 1368–1376 10.1016/j.jhep.2011.04.014 [DOI] [PubMed] [Google Scholar]
- 70. Morita S. Y., Ikeda N., Horikami M., Soda K., Ishihara K., Teraoka R., Terada T., and Kitagawa S. (2011) Effects of phosphatidylethanolamine N-methyltransferase on phospholipid composition, microvillus formation and bile salt resistance in LLC-PK1 cells. FEBS J. 278, 4768–4781 10.1111/j.1742-4658.2011.08377.x [DOI] [PubMed] [Google Scholar]
- 71. Mehta P. A., and Tolar J. (2002) in GeneReviews (Adam M. P., Ardinger H. H., Pagon R. A., Wallace S. E., Bean L. J. H., Stephens K., and Amemiya A., eds), University of Washington, Seattle University of Washington, Seattle [Google Scholar]
- 72. Behling-Kelly E., and Collins-Cronkright R. (2014) Increases in β-lipoproteins in hyperlipidemic and dyslipidemic dogs are associated with increased erythrocyte osmotic fragility. Vet. Clin. Pathol. 43, 405–415 10.1111/vcp.12155 [DOI] [PubMed] [Google Scholar]
- 73. Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 74. Chong J., and Xia J. (2018) MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34, 4313–4314 10.1093/bioinformatics/bty528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.