Abstract
Macrophages play an essential role in skeletal muscle regeneration. The phagocytosis of muscle cell debris induces a switch of pro-inflammatory macrophages into an anti-inflammatory phenotype, but the cellular receptors mediating this phagocytosis are still unclear. In this paper, we report novel roles for SRB1 (scavenger receptor class BI) in regulating macrophage phagocytosis and macrophage phenotypic transitions for skeletal muscle regeneration. In a mouse model of cardiotoxin-induced muscle injury/regeneration, infiltrated macrophages expressed a high level of SRB1. Using SRB1 knockout mice, we observed the impairment of muscle regeneration along with decreased myogenin expression and increased matrix deposit. Bone marrow transplantation experiments indicated that SRB1 deficiency in bone marrow cells was responsible for impaired muscle regeneration. Compared with WT mice, SRB1 deficiency increased pro-inflammatory macrophage number and pro-inflammatory gene expression and decreased anti-inflammatory macrophage number and anti-inflammatory gene expression in injured muscle. In vitro, SRB1 deficiency led to a strong decrease in macrophage phagocytic activity on myoblast debris. SRB1-deficient macrophages easily acquired an M1 phenotype and failed to acquire an M2 phenotype in lipopolysaccharide/myoblast debris activation. Furthermore, SRB1 deficiency promoted activation of ERK1/2 MAPK signaling in macrophages stimulated with lipopolysaccharide/myoblast debris. Taken together, SRB1 in macrophages regulates phagocytosis and promotes M1 switch into M2 macrophages, contributing to muscle regeneration.
Keywords: muscle regeneration, scavenger receptor, macrophage, phagocytosis, inflammation, macrophage transition, scavenger receptor class BI
Introduction
Microenvironments play critical roles in regulating muscle regeneration. Interaction between leukocytes and differentiated monocytes, such as different types of macrophages, determines the formation of a coordinated microenvironment that is optimal for muscle regeneration (1). The tissue injuries stimulate infiltration of T cells into injured muscle and induce expression of Th1 cytokines, such as interferon-γ (IFN-γ)4 and TNFα, that drive the classical activation of macrophages to type 1 phenotype (M1) or inflammatory macrophages (2, 3). M1 macrophage has dual impacts on injured muscle. It could induce muscle damage by production of cytotoxic levels of nitric oxide (NO), which is generated by inducible nitric-oxide synthase (4). On the other hand, M1 macrophage could clear up the damaged cellular debris by phagocytosis (5). After injury, the muscle recruits pro-inflammatory CX3CR1lo/Ly-6C+ monocytes/macrophages first, and then these cells switch their phenotype to become anti-inflammatory CX3CR1hi/Ly-6C− monocytes/macrophages through phagocytosis of muscle cell debris (6). M2 macrophages are activated by Th2 cytokines. Interleukin4 (IL4), IL10 and IL13 play particularly well-characterized roles in their activation (7). M2 macrophages promote muscle regeneration and have proved to be associated with M2 macrophage–specific CD antigens, such as CD163 and CD206 (8–10). M2 macrophages increase the expression of anti-inflammatory cytokines and inactivate Th1 cells (11). However, the molecular mechanisms that regulate M2 differentiation of macrophages in injured muscle remain unclear.
Scavenger receptors (SRs) belong to a cell surface transmembrane glycoprotein family with broad ligand-binding abilities (12). SRs may have diverse function, as they have 1) broad ligand binding (13), 2) tissue-specific expression (14–16), and 3) an important role in innate immunity and tissue homeostasis (17). Scavenger receptor class B member 1 (SRB1) was originally identified as a high-density lipoprotein receptor that binds HDL with high affinity and mediates both the selective lipid uptake of cholesterol esters from lipid-rich HDL to cells and the efflux of unesterified cholesterol from cells to HDL and is required for microvillar channel formation and the localization of HDL particles to the surface of adrenocortical cells (18). SRB1 is highly expressed in steroidogenic cells as well as in the liver (19). It is expressed on platelets and plays dual roles in thrombosis and contributes to acute cardiovascular events in vivo in hypercholesterolemia (20). SRB1 expressed in fibroblast-like cells mediates acute-phase apoprotein serum amyloid A–induced pro-inflammatory pathways in rheumatoid arthritis synovial membrane (21). Recent research shows that SRB1 is also expressed in macrophages (22, 23), but its function in the macrophages is still unclear. However, type I and II class A macrophage scavenger receptors (SR-AI/II) are reported as homotrimeric membrane proteins of mononuclear phagocytes mediating phagocytosis of apoptotic thymocytes and endocytosis of modified lipoproteins (24). Reduction of CD36, a class B scavenger in macrophages, leads to a decrease of the phagocytic ability of macrophages in the peritoneum of women with endometriosis (25). SRB1, as a phosphatidylserine receptor, enables Sertoli cells to recognize and phagocytose apoptotic spermatogenic cells at all stages of differentiation (26). Thus, SRB1 may regulate macrophages' phagocytosis.
In this study, we aimed to examine the role of SRB1 in macrophage phagocytosis in injured skeletal muscle. We found that, through regulation of phagocytosis, SRB1 regulated macrophage differentiation, contributing to muscle regeneration.
Results
The abundance of SRB1 protein was increased in injured muscles of WT mice
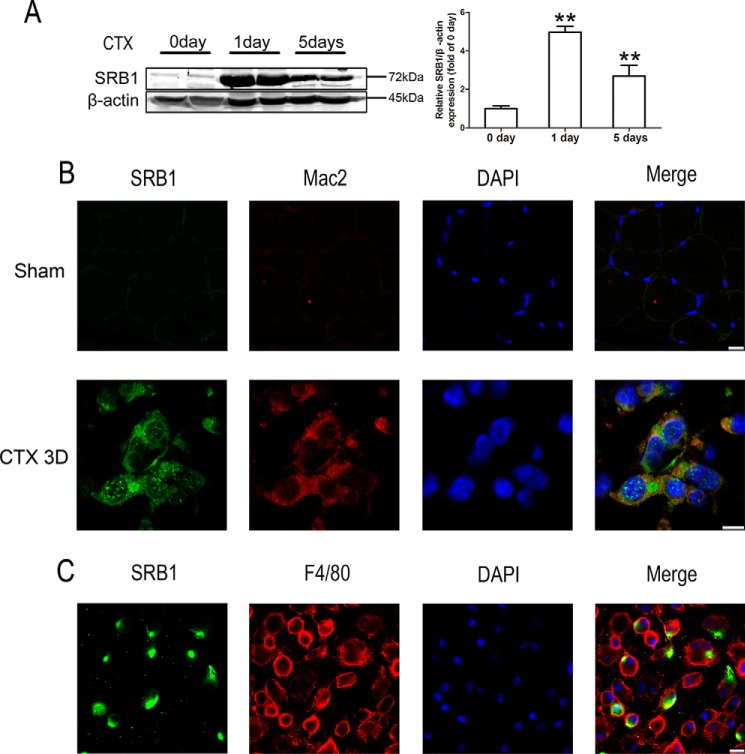
We used a muscle injury/recovery models to study the impact of SRB1 on muscle regeneration in vivo. Cardiotoxin was injected into the tibialis anterior (TA) muscles of WT mice (C57BL/6J background) for inducing muscle injury, and the mice whose TA muscles were injected with PBS served as the sham group. The amount of SRB1 protein was sharply increased 1 day after injury compared with uninjured muscles (Fig. 1A). Immunofluorescence staining showed the presence of SRB1 expression in the injured skeletal muscle tissue at day 3. To identify whether the increased SRB1 was due to infiltration of macrophages, we double-stained Mac2, a marker of phagocytic macrophage, and SRB1 in the muscle 3 days after injury. As shown in Fig. 1B, the staining of Mac2 was co-localized with SRB1, which indicated that SRB1 is primarily expressed in phagocytic macrophages. To confirm that macrophage expresses SRB1, we isolated bone marrow–derived macrophages. By immunostaining with F4/80 (another macrophage marker), we found that F4/80 co-localized with SRB1 (Fig. 1C). These findings indicate that macrophages express SRB1 in injured skeletal muscle.
Figure 1.
SRB1 protein expression is increased in injured muscles of WT mice. A, cardiotoxin was injected into the tibialis anterior muscle of WT mice (C57BL/6J background). Muscles were collected 0, 1, and 5 days after CTX injection, and Western blots were performed using antibody against SRB1. β-Actin was used as an internal control. Densitometric analysis of SRB1 levels is shown in the right histogram. Data represent the results of three independent experiments. **, p < 0.01. B, top panels, immunohistochemical analysis of SRB1 and Mac2 in undamaged skeletal muscle (scale bar, 10 μm); bottom panels, immunohistochemical analysis of SRB1 and Mac2 in skeletal muscle 3 days after CTX-induced injury (scale bar, 7.5 μm). C, immunohistochemical analysis of SRB1 and F4/80 on macrophages derived from the bone marrow of WT mice. DAPI, 4′,6-diamidino-2-phenylindole. Scale bar, 10 μm. Error bars, S.D.
Knockout of SRB1 impairs muscle regeneration capacity
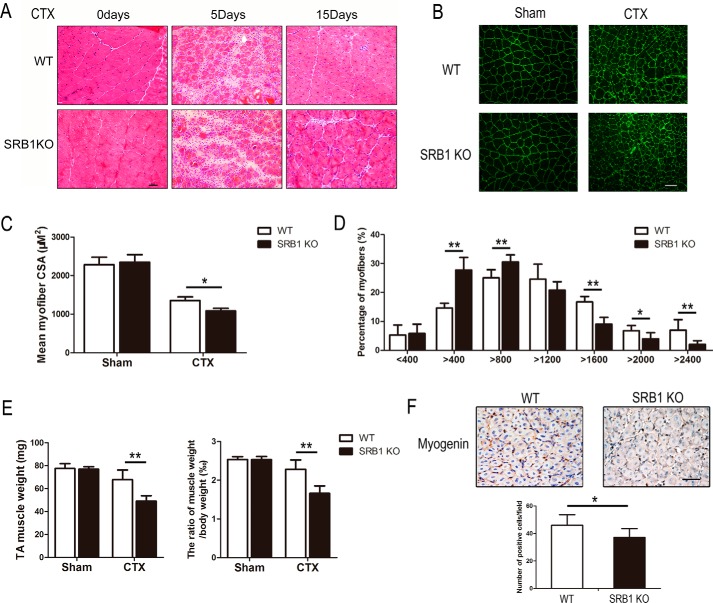
To determine whether SRB1 affects the capacity for muscle regeneration, cardiotoxin (CTX) was injected into TA muscles of both SRB1 KO and WT mice. There was no significant difference between SRB1 KO and WT muscle before injury. Five days after injury, new myofibers (identified by central nuclei) formed in TA muscles of WT mice, but less in TA muscles of SRB1 KO mice (Fig. 2A). At day 15, muscles from SRB1 KO mice had smaller new myofibers compared with those of WT mice (Fig. 2, B and C). The size distribution of myofibers showed that fiber sizes have a significant left shift in SRB1 KO mice from those in WT mice, which indicated that muscle regeneration capacity was decreased in SRB1 KO mice (Fig. 2D). Muscle regeneration was also quantified by TA muscle weight and the ratio of TA muscle weight to body weight at 15 days after injury; the TA muscle weight and ratio of TA muscle weight to body weight was lower in SRB1 KO mice than in WT mice (Fig. 2E). Myogenic regulatory factor myogenin-positive cells were counted to determine muscle regeneration capacity. The number of myogenin-positive cells was significantly lower in the injured muscle of SRB1 KO mice versus WT mice (Fig. 2F). These results indicate that SRB1 is required for muscle progenitor cell differentiation and skeletal muscle regeneration.
Figure 2.
SRB1 deficiency impairs muscle regeneration capacity. A, cardiotoxin was injected into the tibialis anterior muscle of WT and SRB1 KO mice. Muscles were collected and performed with HE staining at 0, 5, and 15 days after injection. Scale bar, 50 μm. B, muscles from WT and SRB1 KO mice were immunostained with laminin at 15 days after injury. Scale bar, 100 μm. C, the graph indicates the mean cross-sectional area (CSA) of muscle myofiber at 15 days after injury (n = 6 in each group). D, the graph indicates the distribution of myofiber size in WT and SRB1 KO mice at 15 days after CTX injections (n = 6 in each group). E, quantitative analysis of the TA muscle weight in each group at 15 days after injury is shown in the left histogram, and quantitative analysis of the ratio of TA muscle weight to body weight in each group at 15 days after injury is shown in the right histogram (n = 8 in each group). F, immunohistochemical staining and quantitative analysis of myogenin+ cells in skeletal muscle of WT and SRB1 KO mice at 5 days after CTX-induced injury. Scale bar, 50 μm (n = 4 in each group). *, p < 0.05; **, p < 0.01. Error bars, S.D.
Knockout of SRB1 promotes interstitial fibrosis in injured muscle
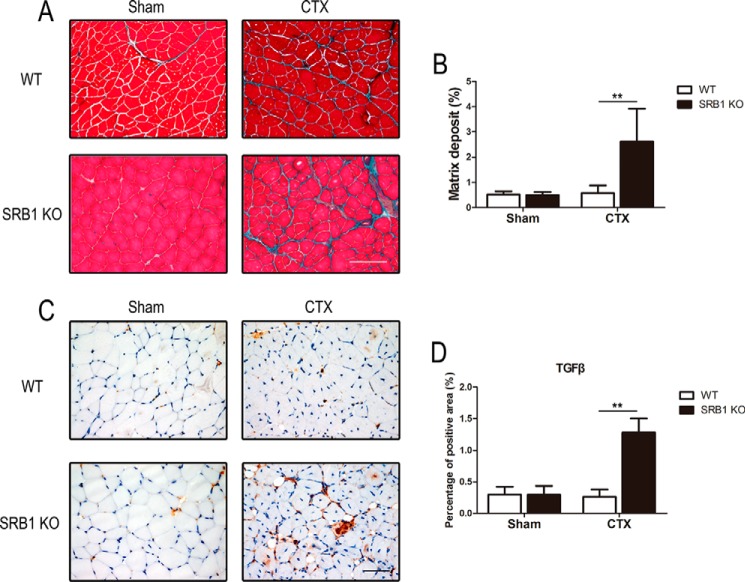
To detect the interstitial fibrosis in injured muscle, the collagen deposition was measured by Masson's trichrome staining at 15 days after muscle damage. There was significantly increased interstitial fibrosis in injured muscle of SRB1 KO mice compared with that of WT mice, whereas there was no significant difference between fibrosis in sham muscle of WT mice and sham muscle of SRB1 KO mice (Fig. 3, A and B). The amount of TGFβ (a pro-fibrotic cytokine) was markedly higher in the muscle of SRB1 KO mice than in WT mice after the injection of CTX for 15 days (Fig. 3, C and D). These data suggest that knockout of SRB1 promotes interstitial fibrosis during skeletal muscle regeneration.
Figure 3.
SRB1 deficiency promotes interstitial fibrosis in injured muscle. A, at 15 days after injury, muscles from WT and SRB1 KO mice were histochemically stained with Masson trichrome. Muscle fiber (red color) and collagen (green) are shown. Scale bar, 100 μm. B, quantitative analysis shows the ratio of interstitial fibrosis to total TA area in each group (n = 6 in each group). C, representative immunohistochemical staining of TGFβ in WT and SRB1 KO muscles at 15 days after CTX-induced injury. Scale bar, 50 μm. D, quantification of immunohistochemical staining of TGFβ in each group (n = 6 in each group). **, p < 0.01. Error bars, S.D.
SRB1 deficiency in bone marrow–derived cells inhibits muscle regeneration
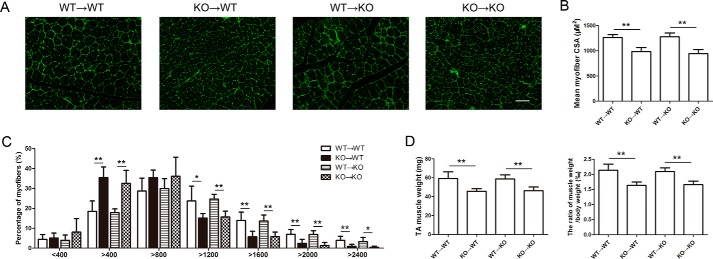
We performed bone marrow transplantation experiments to address whether SRB1 deficiency in skeletal muscle cells or bone marrow–derived cells inhibited muscle regeneration. Bone marrow chimeric mice were reconstituted after transplantation of WT or SRB1-deficient bone marrow cells. 8 weeks after bone marrow transplantation, mice underwent CTX injury. At day 15, mice that received SRB1 knockout bone marrow had smaller new myofibers compared with that received WT bone marrow, regardless of the genotype of the recipient mice (Fig. 4, A and B). The fiber sizes have a left shift in mice that received SRB1 knockout bone marrow compared with those that received WT bone marrow (Fig. 4C). The TA muscle weight and ratio of TA muscle weight to body weight was lower in mice that received SRB1 knockout bone marrow compared with those that received WT bone marrow (Fig. 4D). Thus, SRB1 deficiency in bone marrow cells rather than skeletal muscle cells is responsible for the muscle regeneration defect.
Figure 4.
SRB1 deficiency in bone marrow–derived cells inhibits muscle regeneration. A, four groups of chimeric mice were injured by CTX injection. Muscles were immunostained with laminin at 15 days after injury. Scale bar, 100 μm. B, the graph indicates the mean cross-sectional area (CSA) of muscle myofiber at 15 days after injury (n = 6 in each group). C, the graph indicates the distribution of myofiber size in each group at 15 days after CTX injections (n = 6 in each group). D, quantitative analysis of the TA muscle weight in each group at 15 days after injury is shown in the left histogram, and quantitative analysis of the ratio of TA muscle weight to body weight in each group at 15 days after injury is shown in the right histogram (n = 6 in each group). *, p < 0.05; **, p < 0.01. Error bars, S.D.
The early pro- to anti-inflammatory macrophage phenotypic transition is regulated by SRB1 during skeletal muscle regeneration
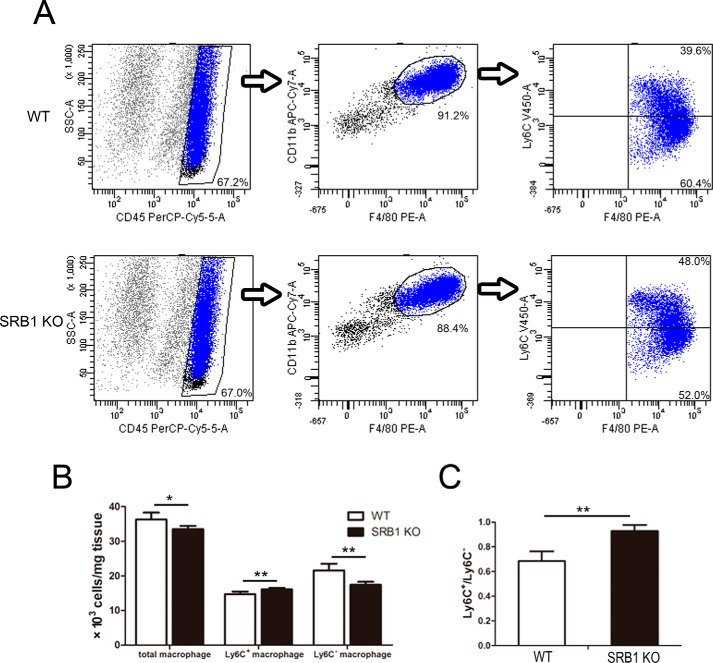
Pro-inflammatory (M1) macrophage to anti-inflammatory (M2) macrophage transition plays a key role during skeletal muscle regeneration. Considering SRB1 expressed on macrophage, we addressed whether macrophage transition was regulated by SRB1 deficiency. We measured muscle macrophage phenotype by flow cytometry. The F4/80 and CD11b antibodies were used to identify macrophages, and the Ly6C antibody was used to identify different macrophage subsets. Ly6C+ macrophages were considered as the pro-inflammatory (M1) macrophages, whereas Ly6C− macrophages were considered as the anti-inflammatory (M2) macrophages (Fig. 5A). 3 days after injury, the total and Ly6C− macrophages were decreased in SRB1 KO mice compared with those in WT mice, and Ly6C+ macrophages were increased in SRB1 KO mice (Fig. 5B). The ratio of Ly6C+ macrophage to Ly6C− macrophage was increased in SRB1 KO mice compared with that in WT mice (Fig. 5C).
Figure 5.
SRB1 deficiency blocks M1 macrophages switching into M2 macrophages during skeletal muscle regeneration. A, flow cytometry analysis of Ly6C+ and Ly6C− macrophages present in skeletal muscle of WT and SRB1 KO mice at 3 days after CTX-induced injury. B, the cell population of total, Ly6C+, and Ly6C− macrophages was analyzed (n = 5 in each group). C, the ratio of Ly6C+ macrophages to Ly6C− macrophages was analyzed (n = 5 in each group). *, p < 0.05; **, p < 0.01. Error bars, S.D.
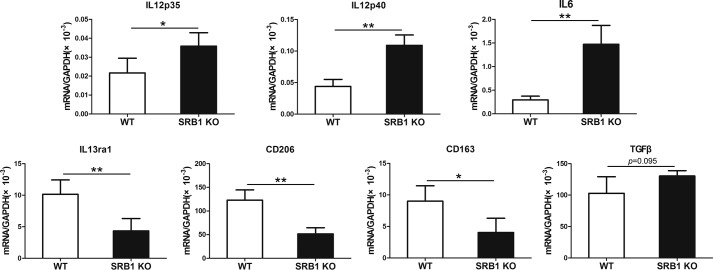
To further investigate the phenotype of the macrophages infiltrating damaged muscle, we isolated WT and SRB1 KO macrophage by cell sorting and detected pro-inflammatory (M1) and anti-inflammatory (M2) marker expression by real-time PCR (Fig. 6). Results showed that macrophages from SRB1 KO mice expressed M1-specific genes (IL12p35, IL12p40, and IL6) more strongly than macrophages from WT mice. Conversely, macrophages from WT mice expressed M2-specific genes (IL13ra1, CD206, and CD163) more strongly than macrophages from SRB1 KO mice. There was no significant difference in TGFβ expression between WT and SRB1 KO macrophages.
Figure 6.
SRB1 regulates M1 to M2 macrophage phenotypic transition during skeletal muscle regeneration. Macrophages were isolated by cell sorting from WT or SRB1 KO skeletal muscle at 3 days after CTX-induced injury. Expression of IL12p35, IL12p40, IL6, IL13ra1, CD206, CD163, and TGFβ was analyzed by real-time PCR in isolated macrophages from WT and SRB1 KO mice. GAPDH was used to normalize the quantitative real-time data (n = 4 in each group). *, p < 0.05; **, p < 0.01. Error bars, S.D.
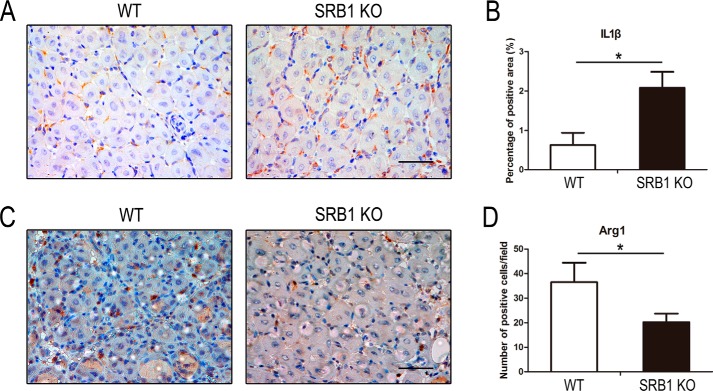
We also measured the levels of M1-specific (IL1β) and M2-specific (arginase 1, Arg1) gene expression in TA muscle by immunohistochemistry analysis. The level of IL1β was increased in the muscle of SRB1 KO mice at 5 days after injury, and the level of Arg1 was significantly decreased in the muscle of SRB1 KO mice compared with that of WT mice (Fig. 7). These results demonstrated that SRB1 deficiency limits pro-inflammatory macrophages switch into anti-inflammatory macrophages in response to acute muscle injury.
Figure 7.
SRB1 deficiency promotes M1-specific gene expression and inhibits M2-specific gene expression during skeletal muscle regeneration. A, injured TA muscles (5 days after CTX injection) were immunostained with IL1β (brown), and nuclei were stained by hematoxylin (blue). Scale bar, 50 μm. B, quantification of IL1β staining in injured skeletal muscle (n = 4 in each group). C, injured TA muscles (5 days after CTX injection) were immunostained with Arg1 (brown), and nuclei were stained by hematoxylin (blue). Scale bar, 50 μm. D, quantification of Arg1 staining in injured skeletal muscle (n = 4 in each group). *, p < 0.05. Error bars, S.D.
SRB1 is essential for macrophage phagocytosis of myoblast debris and macrophage migration
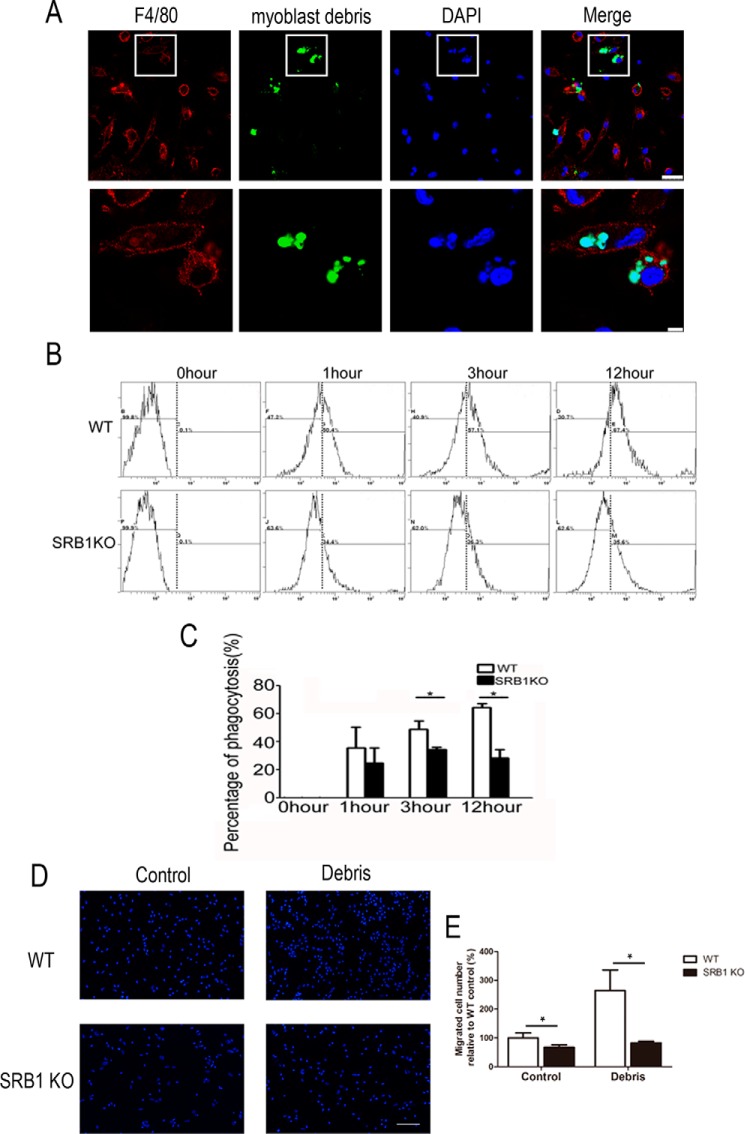
Study has shown that infiltrated macrophage phagocytosis will support myogenesis (6). Because SRB1 was mainly expressed in macrophages, we isolated macrophages from bone marrow of WT and SRB1 KO mice and treated them with C2C12 myoblast debris to measure the phagocytosis of macrophages. Myoblast debris was incubated with macrophages for 1, 3, and 12 h. The myoblast debris could be engulfed by WT macrophages (Fig. 8A). Phagocytosis was quantified by counting fluorescent microspheres inside of macrophage using flow cytometry analysis (Fig. 8B). The rate of phagocytosis was much lower in SRB1 KO mice compared with that in WT mice (Fig. 8C).
Figure 8.
SRB1 is essential for phagocytosis and migration of macrophages. A, macrophages isolated from bone marrow of WT mice were cultured and stimulated with C2C12 myoblast debris, which was labeled by CellTracker Green CMFDA. Immunohistochemical analysis of F4/80 (red) and myoblast debris (green) on WT macrophages. Top panels, low-magnification images of F4/80 (red) and myoblast debris (green). Scale bar, 25 μm. Bottom panels, high-magnification insets from the white boxes of the top panels. Scale bar, 5 μm. B, phagocytosis was quantified by counting fluorescent microspheres inside of the macrophage using flow cytometry analysis at different time points (0, 1, 3, and 12 h). C, the bar graph shows the percentage of debris swallowed by macrophages at different time points. The data are expressed as a percentage of the microspheres relative to the total cell number. D, representative images of the migration of WT or SRB1 KO macrophages stimulated with or without myoblast debris treatment. Scale bar, 100 μm. E, the number of migrated WT or SRB1 KO macrophages was counted. Data represent the results of three independent experiments. *, p < 0.05. Error bars, S.D.
Flow cytometry analysis demonstrated that total macrophages were decreased in SRB1 KO mice compared with that in WT mice 3 days after injury (Fig. 5B). The migration assay was performed to evaluate the chemotaxis of WT and SRB1 KO macrophages to myoblast debris. SRB1 deficiency inhibited migration of macrophages with or without myoblast debris treatment (Fig. 8, D and E). Thus, SRB1 knockout limits macrophage phagocytosis of myoblast debris and limits macrophage migration.
SRB1 promotes macrophage phenotype transition after phagocytosis of myoblast debris in vitro
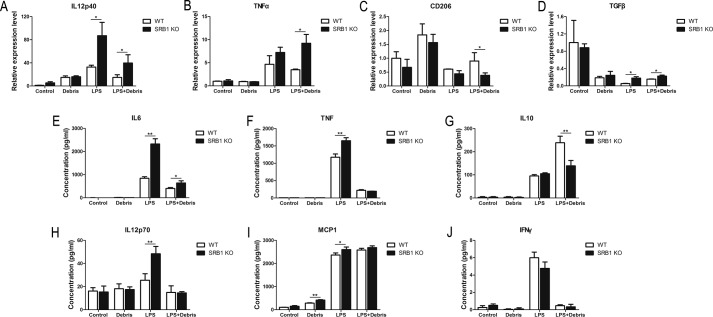
To assess the role of SRB1 in macrophage phenotype transition, we detected the effects of M1-specific or M2-specific gene expression in bone marrow derived macrophages from WT and SRB1 KO mice by adding LPS, myoblast debris, or LPS plus myoblast debris. After activation with these stimuli, we detected the M1-specific and M2-specific gene expression on RNA levels by real-time PCR. Deletion of SRB1 led to increased M1-specific gene expression (IL12p40, TNFα) and decreased M2-specific gene expression (CD206) after LPS plus myoblast debris activation (Fig. 9, A–C). We also collected WT and SRB1 KO macrophage conditioned medium and detected cytokine expression with the BD Biosciences Cytometric Bead Array (CBA) mouse inflammation kit. Deletion of SRB1 led to increased M1-specific gene expression (IL6, TNF, IL12p70, and MCP1) after LPS activation, and deletion of SRB1 led to increased M1-specific gene expression (IL6) and decreased M2-specific gene expression (IL10) after LPS plus myoblast debris activation (Fig. 9, E–J). These results confirmed that SRB1-deficient macrophages easily acquired M1 phenotypes but not M2 phenotypes.
Figure 9.
SRB1 promotes macrophage phenotype transition after phagocytosis of myoblast debris in vitro. Macrophages were isolated from bone marrow of WT and SRB1 KO mice. Expression of IL12p40 (A), TNFα (B), CD206 (C), and TGFβ (D) was analyzed by real-time PCR in isolated macrophages with LPS (50 ng/ml), myoblast debris, or LPS plus myoblast debris activation for 3 days. GAPDH was used to normalize the quantitative real-time data. IL6 (E), TNF (F), IL10 (G), IL12p70 (H), MCP1 (I), and IFN-γ (J) concentrations in WT or SRB1 KO macrophages with LPS/myoblast debris activation were analyzed by a BD Biosciences CBA mouse inflammation kit. Data represent the results of three independent experiments. *, p < 0.05; **, p < 0.01. Error bars, S.D.
SRB1 deficiency promoted ERK1/2 MAPK signaling activation in macrophages stimulated with LPS/myoblast debris
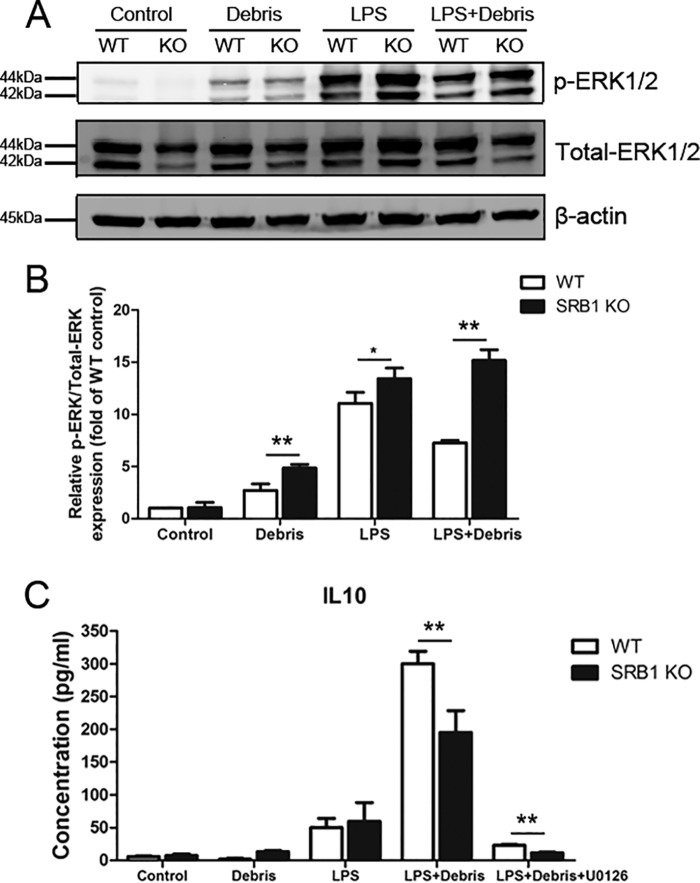
We investigated which downstream signaling pathway was activated by SRB1 in macrophages stimulated with LPS/myoblast debris. SRB1 deficiency increased phosphorylated ERK1/2 in macrophages with myoblast debris/LPS treatment (Fig. 10, A and B). To determine whether activation of the ERK1/2 MAPK pathway was necessary for IL10 expression of macrophages, U0126 was used to block activity of the ERK1/2 MAPK signal pathway. Pretreatment with U0126 significantly suppressed IL10 expression induced by LPS/myoblast debris (Fig. 10C). These results indicated that SRB1 deficiency promotes activation of the ERK1/2 MAPK pathway in macrophages, and the ERK1/2 MAPK pathway induced IL10 expression with treatment of LPS/myoblast debris.
Figure 10.
SRB1 deficiency promoted ERK1/2 MAPK signaling activation. A, both phosphorylated ERK1/2 (p-ERK1/2) and total ERK1/2 (Total-ERK1/2) were measured by immunoblotting. B, densitometric analysis was performed to determine the ratio of phospho-ERK1/2 to total ERK1/2. C, IL10 concentrations in macrophages in the presence of LPS/myoblast debris with or without U0126 (ERK1/2 MAPK inhibitor, 10 μm). Data represent the results of three independent experiments. *, p < 0.05; **, p < 0.01. Error bars, S.D.
Discussion
In this study, we identified the role of SRB1 on macrophage in skeletal muscle regeneration process. Our results indicated that SRB1, a scavenger receptor of macrophage, plays an important role in determining the microenvironment for muscle regeneration. SRB1-positive macrophages were significantly infiltrated into the CTX-injured skeletal muscle. Knockout of SRB1 impaired muscle regeneration capacity by regulating a switch from M1 into M2 macrophages. SRB1 deficiency in bone marrow cells was responsible for impaired muscle regeneration. The SRB1 is involved in phagocytosis and migration of macrophages, and SRB1 promoted the differentiation into M2 phenotype and inhibited differentiation into M1 phenotype in vitro. SRB1 deficiency promoted activation of ERK1/2 MAPK signaling in macrophages stimulated with LPS/myoblast debris. Thus, SRB1 promotes muscle regeneration by mediating the phagocytosis ability of macrophage and promoting M1 switch into M2 macrophage.
It is well-known that inflammation is involved in skeletal muscle regeneration. In response to injury, inflammatory cells are infiltrated into the injury site. After acute muscle injury, the first-response inflammatory cells are neutrophils, which appear within 2 h in injured muscle (9). Following the infiltration of neutrophils, macrophages infiltrate into the injured muscle, with peak numbers at about 3 days. After that, macrophages begin to decline, and only a small number of macrophages maintain in muscle after 7 days (27). As an important part of the inflammatory microenvironment, MP plays a key role in muscle regeneration (6, 28, 29). The inhibition of macrophage infiltration leads to incomplete skeletal muscle regeneration and causes adipogenesis and fibrosis (28), and depletion of the MPs reduces the diameter of myofibers (29). In acute skeletal muscle injury, the peripheral monocyte/macrophage (MP) depletion is associated with impaired regeneration after acute skeletal muscle injury (5, 6). In the CTX injection–induced acute muscle injury/regeneration model, we demonstrated that SRB1 deficiency impaired muscle regeneration (Fig. 2) and aggravated muscle interstitial fibrosis (Fig. 3). Thus, in acute skeletal muscle injury/regeneration, SRB1 is critical for muscle regeneration. The number of total macrophages in injured muscle was suppressed in SRB1 KO mice (Fig. 5, A and B), suggesting that the decrease in total macrophage number in SRB1 KO mice might contribute to impaired muscle regeneration. We also found that SRB1 was expressed in macrophages of injured muscle (Fig. 1), and SRB1 deficiency on bone marrow impaired skeletal muscle regeneration (Fig. 4). Therefore, we thought the phenotypes observed in the global SRB1 KO mice might be attributed to SRB1 deficiency on bone marrow–derived monocytes/macrophages rather than resident skeletal muscle cells.
Different types of macrophage have different functions in the injury/recovery process. The macrophages are classified into pro-inflammatory (M1) and anti-inflammatory (M2) subtypes. At an early stage of skeletal muscle injury, M1 macrophages infiltrate into injured muscle. M1 macrophages are characterized by expressing proinflammatory cytokines TNFα and IL1β and others (2). Then M1 macrophages are replaced by M2 macrophages to attenuate the inflammatory response. M2 macrophages are characterized by expressing M2-specific genes, such as CD206 (7). In our study, we also identified two subsets of macrophages, Ly6C+ and Ly6C−, that were infiltrated into injured skeletal muscle. SRB1 deficiency decreased the number of Ly6C− (M2) macrophages and increased the number of Ly6C+ (M1) macrophages (Fig. 5). SRB1 deficiency promoted macrophage expressing M1-specific genes and inhibited macrophage expressing M2-specific genes (Figs. 6 and 7). Thus, SRB1 regulates the phenotype of macrophages in injured skeletal muscle.
Previous research has demonstrated the M1 macrophages increase myogenic precursor cell growth and proliferation, whereas M2 macrophages promote myogenic precursor cell differentiation and fusion (6, 30). Our previous research also demonstrated that coculture of myoblasts in indirect contact with M1 macrophages rather than M2 macrophages could promote myoblast proliferation (31). M1 macrophage–conditioned medium stimulates myogenic precursor cell proliferation, whereas M2 macrophage–conditioned medium promotes myogenic precursor cell differentiation and fusion into myotubes (30). Because M1 macrophages secrete proinflammatory cytokines, such as TNFα, and M2 macrophages secrete anti-inflammatory cytokines, such as IL10, the effects of M1 or M2 macrophage–conditioned medium on myoblasts are likely to be related with these cytokines. Different cytokines play different roles in skeletal muscle regeneration. IL10 deficiency in mdx mice increased muscle damage in vivo and reduced mouse strength (32). TNFα inhibited myoblast differentiation and fusion (33, 34). It has been reported that IL6 could promote the proliferation of myoblasts (35). Sometimes the synergistic effect of cytokines is powerful. For example, the combined stimulation of IL6 and TNFα greatly promote myoblast proliferation (36). Therefore, the effect of M1 or M2 macrophages on myoblasts is likely to be the synergistic effect of many cytokines. The expression of myogenin reflects the differentiation of satellite cells, and the decrease of myogenin expression in muscle of SRB1 KO mice (Fig. 2F) indicates that satellite cell differentiation is inhibited. In our study, the decrease in differentiation of satellite cells may be related to the decrease in the number of M2 macrophages in SRB1 KO mice.
TGFβ plays a key role in skeletal muscle interstitial fibrosis. TGFβ stimulates the fibroblasts to produce extracellular matrix proteins (37), and TGFβ also induces the transdifferentiation of several resident cell types into myofibroblasts, leading to excessive extracellular matrix deposition and fibrosis (38). In our study, SRB1 deficiency led to aggravated muscle interstitial fibrosis and elevated TGFβ expression after skeletal muscle injury (Fig. 3). Although SRB1 deficiency increased TGFβ expression in macrophages with the treatment of LPS/myoblast debris, the expression of TGFβ in both WT and SRB1 KO macrophages was low in LPS/myoblast debris stimulation (Fig. 9D). Meanwhile, there was no significant difference between TGFβ expression in WT and SRB1 KO macrophage sorting from injured skeletal muscle (Fig. 6). Thus, we speculate that the source of TGFβ expressing in skeletal muscle after injury may be multiple. In fact, TGFβ can be produced by different types of cells, including inflammatory cells, mesenchymal cells, and epithelial cells (39). In skeletal muscle, impaired muscle regeneration is often accompanied by more severe interstitial fibrosis. A variety of cells and various cytokines are involved in the interstitial fibrosis of skeletal muscle (40). In our study, we found that SRB1 deficiency impairs skeletal muscle regeneration by directly inhibiting macrophage phenotype transition from M1 to M2. SRB1 deficiency promoted fibrosis as a concomitant result of impaired regeneration, but the effect of SRB1 on fibrosis might be indirect.
Phagocytosis is an important function of macrophages, which clear cell debris from necrosis or apoptosis to assist inflammation resolution. The phagocytosis plays an important role in skeletal muscle myogenesis because accumulated necrosis debris would cause excessive inflammation, which would impair muscle regeneration. During myogenesis after injury, a decrease in the phagocytosis capacity of macrophage would inhibit the expression of inflammatory factors in macrophages, which would regulate muscle regeneration (6). It is known that molecules, including SRs, are involved in phagocytosis of clear debris during the injury/recovery process. SR-A, a member of the SR superreceptor family in macrophages, has been shown to mediate macrophage adhesion in vitro, and a role for SR-A in the phagocytosis of apoptotic thymocytes has been inferred from the in vitro analysis of SR-A gene knockout animals (41). We showed in this study that lack of SRB1 impairs macrophage phagocytosis of myoblast debris (Fig. 8, B and C). SRB1 is a subtype of SR-B members, and previous work has demonstrated that SRB1 plays an essential role in meditating the uptake of HDL-derived cholesterol and cholesteryl ester in the liver and steroidogenic tissues (42); therefore, our findings add a novel role of SRB1 in regulating macrophage function during skeletal muscle regeneration. We also detected the chemotaxis of WT and SRB1 KO macrophages to myoblast debris and found that SRB1 deficiency inhibited macrophage migration even without myoblast debris treatment (Fig. 8, D and E). Therefore, SRB1 also promotes skeletal muscle regeneration through stimulating macrophage migration.
Because anti-inflammatory effectors are increased and pro-inflammatory effectors are decreased in phagocyting macrophages (43, 44), the transition of macrophages from the M1 to the M2 phenotype is considered to be accomplished by phagocytosis. Injured skeletal muscle recruits monocytes from blood exhibiting pro-inflammatory profiles that operate phagocytosis and rapidly convert to anti-inflammatory macrophages (6). Previous research has shown that AMPKα1 deficiency led to a strong decrease in macrophage phagocytic activity, and the defect in phagocytosis of AMPKα1 deficiency macrophages is associated with decreased phenotypic transition from M1 to M2. Skeletal muscle regeneration is also impaired in AMPKα1 deficiency mice (30). In the present study, we demonstrated that SRB1 deficiency in macrophages prevented the acquisition of an M2 phenotype in vitro, as assessed by the lower expression of M2 markers and the higher expression of M1 markers upon LPS/myoblast debris stimulation (Fig. 9). Thus, muscle injury promotes macrophage switch into M2 type macrophage, and lack of SRB1 significantly impaired this process and stimulated M1 macrophage differentiation and inhibited M2 macrophage differentiation. Our results established a critical role of SRB1 in linking macrophage phagocytosis ability and macrophage transition. It is well-established that the ERK1/2 MAPK signaling pathways participate in cytokine and growth factor actions in macrophages and the transcription of a variety of genes involved in inflammation (45, 46). Our results demonstrated that LPS/myoblast debris activated ERK1/2 MAPK signaling pathway, and the secretion of IL10 in macrophages upon LPS/myoblast stimulation was abrogated by pretreatment with ERK1/2 MAPK inhibitor (Fig. 10). Thus, the ERK1/2 MAPK signaling pathway is essential for the LPS/myoblast-induced secretion of IL10 from macrophages.
In conclusion, we identified a critical role of SRB1 on skeletal muscle myogenesis during muscle injury/recovery. We demonstrated that SRB1 promotes muscle regeneration by mediating the phagocytosis ability of macrophage and promoting M1 macrophage switch into M2 macrophages.
Experimental procedures
Animals
The SRB1 knockout (SRB1 KO) mice were from the Jackson Laboratory (Bar Harbor, ME) and were crossed into C57BL/6J background for 10 generations, and the C57BL/6J WT mice were used as controls. The Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication 85-23, 1996) was followed, and the study was approved by the Animal Care and Use Committee of Capital Medical University.
Muscle injury/regeneration model
WT (C57BL/6J) and SRB1-KO mice were kept in a 12-h light/12-h dark cycle and studied at 8–12 weeks old. For muscle regeneration models, the anesthetized mice were injected with 40 μl of 10 μm CTX (C9759, Sigma-Aldrich) into the TA muscle using a 27-gauge needle as described (47). The sham muscle was injected with the same volume of PBS.
Histological and immunohistochemical analyses
Serial, transverse cryosections (7 μm thick) of the mid-belly region of frozen TA muscles were stained with hematoxylin and eosin, Masson trichrome. Standard immunohistochemical techniques were used to detect changes in muscle regeneration. Collagen in the interstitial of muscle was stained by Gomori trichrome, and the green colors of collagen were selected to calculate the percentage of fibrosis relative to muscle area. To calculate the cross-sectional area of individual myofibers, the muscle sections were incubated with antibody against laminin (1:100; ZSGB-BIO, Beijing, China) at 4 °C overnight and then incubated with goat anti-mouse IgG labeled with Alexa Fluor 488 (1:500; Invitrogen). Some muscle tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned (5 μm thick). These muscle sections were incubated with antibody against myogenin (1:100; Abcam, Cambridge, MA), TGFβ1 (1:200; Santa Cruz Biotechnology, Inc.), arginase 1 (Arg1, 1:200; Santa Cruz Biotechnology), and IL1β (1:200; Santa Cruz Biotechnology) and then incubated with secondary antibody and detected with 3,3-diaminobenzidine for immunohistochemistry. Images were captured by a Nikon Eclipse 90i microscope (Nikon, Tokyo, Japan) and analyzed by a person blinded to treatment with the use of NIS-Elements BR 3.1 (Nikon).
To detect the expression of SRB1, the muscle sections were incubated with antibody against galectin-3 (Mac2; 1:200; ZSGB-BIO), SRB1 (1:100; Novus Biologicals LLC, Littleton, CO) at 4 °C overnight and then incubated with Alexa Fluor 488 secondary antibodies (1:500; Invitrogen) or Alexa Fluor 555 secondary antibodies (1:500; Invitrogen) at room temperature for 1 h. To detect the expression of SRB1 on macrophage, the bone marrow–derived macrophages from WT mice were incubated with antibody against F4/80 (1:100; Abcam), SRB1 (1:100; Novus Biologicals LLC) at 4 °C overnight and incubated with secondary antibodies at room temperature for 1 h. Images were captured by a Leica TSC-SP5 laser-scanning confocal microscope (Leica, Wetzlar, Germany).
Generation of bone marrow chimeric mice
Bone marrow cells were collected from femurs and tibias of WT or SRB1 KO mice and resuspended in RPMI 1640. 4 h after irradiation, recipient mice were intravenously injected with 1 × 107 bone marrow cells. The mice were then kept in a specific pathogen-free environment for another 8 weeks to reconstitute their bone marrow with sterilized water and food. Four groups of chimeric mice were generated: WT to WT, SRB1 KO to WT, WT to SRB1 KO, and SRB1 KO to SRB1 KO.
Flow cytometry
Flow cytometry was performed using muscle single-cell suspension, which was prepared as described with minor modifications (31). In brief, TA muscles were dissected and then gently torn with tissue forceps. The tissue was digested by collagenase type II (2.5 units/ml; Sigma-Aldrich) at 37 °C for 30 min. After washing, the second round of enzymatic digestion was performed with collagenase D (1.5 units/ml; Roche Diagnostics GmbH, Mannheim, Germany) and Dispase II (2.4 units/ml; Roche Diagnostics) at 37 °C for 30 min, and cells were collected at 300 × g for 10 min. Flow cytometry was carried out using the following antibodies: PerCP-Cy5.5 anti-mouse CD45.2 (BD Biosciences), PE anti-mouse F4/80 (Biolegend, San Diego, CA), allophycocyanin-Cy7 anti-mouse CD11b (BD Biosciences), and V450 anti-mouse Ly6C (BD Biosciences). Flow cytometric analysis was performed on LSRFortessa (BD Biosciences), and flow cytometric sorting was performed on a FACSAria III instrument (BD Biosciences) and analyzed using BD FACSDiva software (BD Biosciences).
Quantitative real-time PCR
Total RNA was extracted with TRIzol (Invitrogen). Equal amounts of RNA (2 μg) were added to the reverse transcriptase reaction mix with oligo(dT) primers (Promega, Southampton, UK). SYBR Premix Ex Taq (TaKaRa, Shiga, Japan) was used to perform quantitative real-time PCRs with an IQ5 multicolor real-time PCR detection system (Bio-Rad). The following primers were used: IL12p35 (5′-AGTTTGGCCAGGGTCATTCC-3′ (forward) and 5′-TCTCTGGCCGTCTTCACCAT-3′ (reverse)), IL12p40 (5′-AGACCCTGCCCATTGAACTG-3′ (forward) and 5′-CGGGTCTGGTTTGATGATGTC-3′ (reverse)), IL6 (5′-TTCCATCCAGTTGCCTTCTTG-3′ (forward) and 5′-TTGGGAGTGGTATCCTCTGTGA-3′ (reverse)), IL13ra1 (5′-CAGCTGGGATACAGGCATCT-3′ (forward) and 5′-TGGTTTCCACAGCATTTCAA-3′ (reverse)), CD206 (5′-CCCAAGGGCTCTTCTAAAGCA-3′ (forward) and 5′-CGCCGGCACCTATCACA-3′ (reverse)), CD163 (5′-TCCCAGACACTATTGCCATGTAGT-3′ (forward) and 5′-CCTTTGGAATTGTAGAGCTTGTTG-3′ (reverse)), TNFα (5′-CACAAGATGCTGGGACAGTGA-3′ (forward) and 5′-TCCTTGATGGTGGTGCATGA-3′ (reverse)), TGFβ (5′-TCAGACATTCGGGAAGCAGT-3′ (forward) and 5′-TCGAAAGCCCTGTATTCCGT-3′ (reverse)), and GAPDH (5′-CATGGCCTTCCGTGTTCCTA-3′ (forward) and 5′-GCGGCACGTCAGATCCA-3′ (reverse)).
Cell culture and bone marrow–derived macrophage isolation
These cells were isolated from tibia and femur bone marrow of WT (C57B/L6) mice and SRB1 KO mice as described by phagocytosis (48). Briefly, unaggregated cells were passed through a 100-μm filter and subjected to density centrifugation. Culture medium was 1640 supplemented with 10% horse serum and 30 ng/ml macrophage colony–stimulating factor. Cells were cultured at 37 °C in a 5% CO2 incubator. Cells were identified as macrophages by staining with antibodies against F4/80. C2C12 myoblasts were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37 °C in a 5% CO2 incubator.
Phagocytosis
The phagocytosis was performed as described previously (6). C2C12 myoblasts were labeled with CellTracker Green CMFDA (C7025, Invitrogen) and then induced by H2O treatment for 1 h at 37 °C. 100% of cells were propidium iodide–positive. Myoblast debris was incubated with macrophages for 1, 3, and 12 h at 37 °C. The number of CellTracker–positive cells was quantified under a flow test and expressed as a percentage of total cells. Phagocytosis was quantified by counting fluorescent microspheres inside of the macrophage using flow cytometry analysis (Epics XL, MCL, Beckman Coulter, Miami, FL).
Cell migration assay
Cell migration was quantitated by the use of 24-well Transwell inserts with a polycarbonate membrane filter (8-mm pore size; Corning, Inc.). The bone marrow–derived macrophages were seeded onto the upper chamber of the insert. The bottom chambers were treated with or without myoblast debris. These macrophages were allowed to migrate for 12 h at 37 °C with 5% CO2.
Analysis of cytokine concentrations
WT and SRB1 KO macrophage conditioned medium were analyzed for cytokine expression by the BD Biosciences CBA mouse inflammation kit according to the manufacturer's protocol. The fluorescence produced by the beads was measured on a FACSAria III instrument (BD Biosciences), and the latter results were analyzed with FCAP Array version 3.0 (Soft Flow, Duesseldorf, Germany).
Western blotting
The proteins were extracted from muscle collected 0, 1, and 5 days after CTX injection or cultured macrophages stimulated with LPS/myoblast debris by T-PER reagent (Thermo Fisher Scientific). A protease inhibitor mixture and phosphatase inhibitors (Roche Diagnostics) were added according to the manufacturer's instructions. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. After blocking, the nitrocellulose membranes were incubated with SRB1 (Novus Biologicals LLC), phosphorylated ERK1/2 (Cell Signaling Technology, Beverly, MA), total ERK1/2 (Cell Signaling Technology), or β-actin (Cell Signaling Technology) overnight at 4 °C and then incubated with IRDye 800CW secondary antibodies (LI-COR Biosciences, Lincoln, NE) for 1 h at room temperature. Quantification was done using the Odyssey IR imaging system (LI-COR Biosciences).
Statistical analysis
Values are presented as means ± S.D. Results were analyzed using Student's t test when results from two experimental groups were compared. One-way analysis of variance was conducted followed by Bonferroni post hoc test for comparisons between more than two groups. Statistical analysis was performed using SPSS 13.0 software (SPSS Inc.).
Author contributions
J. Z. data curation; J. Z. and C. Q. investigation; J. Z., T. L., and W. C. methodology; J. Z. writing-original draft; C. Q. validation; C. Q. visualization; C. Q. and X. W. writing-review and editing; T. L. and W. C. software; X. W. and J. D. supervision; X. W. and J. D. project administration; J. D. conceptualization; J. D. funding acquisition.
This work was supported by National Natural Science Foundation of China Grants 81430050 and 81672151, Ministry of Science and Technology of China (Key Projects of the Precision Medicine Program) Grant 2016YFC0903000, and Beijing Municipal Science and Technology Commission Grant Z171100000417002. The authors declare that they have no conflicts of interest with the contents of this article.
- IFN
- interferon
- TNF
- tumor necrosis factor
- IL
- interleukin
- SR
- scavenger receptor
- HDL
- high-density lipoprotein
- TA
- tibialis anterior
- CTX
- cardiotoxin
- KO
- knockout
- CBA
- Cytometric Bead Array
- ERK
- extracellular signal–regulated kinase
- MAPK
- mitogen-activated protein kinase
- MP
- macrophage
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- LPS
- lipopolysaccharide
- TGF
- transforming growth factor.
References
- 1. Kharraz Y., Guerra J., Mann C. J., Serrano A. L., and Muñoz-Cánoves P. (2013) Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediat. Inflamm. 2013, 491497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gordon S. (2003) Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 10.1038/nri978 [DOI] [PubMed] [Google Scholar]
- 3. Biswas S. K., and Mantovani A. (2010) Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- 4. Villalta S. A., Nguyen H. X., Deng B., Gotoh T., and Tidball J. G. (2009) Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 18, 482–496 10.1093/hmg/ddn376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Summan M., Warren G. L., Mercer R. R., Chapman R., Hulderman T., Van Rooijen N., and Simeonova P. P. (2006) Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1488–R1495 10.1152/ajpregu.00465.2005 [DOI] [PubMed] [Google Scholar]
- 6. Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R. K., and Chazaud B. (2007) Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 10.1084/jem.20070075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weisser S. B., McLarren K. W., Kuroda E., and Sly L. M. (2013) Generation and characterization of murine alternatively activated macrophages. Methods Mol. Biol. 946, 225–239 10.1007/978-1-62703-128-8_14 [DOI] [PubMed] [Google Scholar]
- 8. Deng B., Wehling-Henricks M., Villalta S. A., Wang Y., and Tidball J. G. (2012) IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J. Immunol. 189, 3669–3680 10.4049/jimmunol.1103180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tidball J. G., and Villalta S. A. (2010) Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1173–R1187 10.1152/ajpregu.00735.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Massimino M. L., Rapizzi E., Cantini M., Libera L. D., Mazzoleni F., Arslan P., and Carraro U. (1997) ED2+ macrophages increase selectively myoblast proliferation in muscle cultures. Biochem. Biophys. Res. Commun. 235, 754–759 10.1006/bbrc.1997.6823 [DOI] [PubMed] [Google Scholar]
- 11. Hao N. B., Lü M. H., Fan Y. H., Cao Y. L., Zhang Z. R., and Yang S. M. (2012) Macrophages in tumor microenvironments and the progression of tumors. Clin. Dev. Immunol. 2012, 948098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schäfer G., Guler R., Murray G., Brombacher F., and Brown G. D. (2009) The role of scavenger receptor B1 in infection with Mycobacterium tuberculosis in a murine model. PLoS One 4, e8448 10.1371/journal.pone.0008448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peiser L., and Gordon S. (2001) The function of scavenger receptors expressed by macrophages and their role in the regulation of inflammation. Microbes Infect. 3, 149–159 10.1016/S1286-4579(00)01362-9 [DOI] [PubMed] [Google Scholar]
- 14. Greaves D. R., Gough P. J., and Gordon S. (1998) Recent progress in defining the role of scavenger receptors in lipid transport, atherosclerosis and host defence. Curr. Opin. Lipidol. 9, 425–432 10.1097/00041433-199810000-00006 [DOI] [PubMed] [Google Scholar]
- 15. Hughes D. A., Fraser I. P., and Gordon S. (1995) Murine macrophage scavenger receptor: in vivo expression and function as receptor for macrophage adhesion in lymphoid and non-lymphoid organs. Eur. J. Immunol. 25, 466–473 10.1002/eji.1830250224 [DOI] [PubMed] [Google Scholar]
- 16. Krieger M. (1998) The “best” of cholesterols, the “worst” of cholesterols: a tale of two receptors. Proc. Natl. Acad. Sci. U.S.A. 95, 4077–4080 10.1073/pnas.95.8.4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Canton J., Neculai D., and Grinstein S. (2013) Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634 10.1038/nri3515 [DOI] [PubMed] [Google Scholar]
- 18. Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., and Krieger M. (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271, 518–520 10.1126/science.271.5248.518 [DOI] [PubMed] [Google Scholar]
- 19. Rigotti A., Miettinen H. E., and Krieger M. (2003) The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24, 357–387 10.1210/er.2001-0037 [DOI] [PubMed] [Google Scholar]
- 20. Ma Y., Ashraf M. Z., and Podrez E. A. (2010) Scavenger receptor BI modulates platelet reactivity and thrombosis in dyslipidemia. Blood 116, 1932–1941 10.1182/blood-2010-02-268508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mullan R. H., McCormick J., Connolly M., Bresnihan B., Veale D. J., and Fearon U. (2010) A role for the high-density lipoprotein receptor SR-B1 in synovial inflammation via serum amyloid-A. Am. J. Pathol. 176, 1999–2008 10.2353/ajpath.2010.090014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fluiter K., van der Westhuijzen D. R., and van Berkel T. J. (1998) In vivo regulation of scavenger receptor BI and the selective uptake of high density lipoprotein cholesteryl esters in rat liver parenchymal and Kupffer cells. J. Biol. Chem. 273, 8434–8438 10.1074/jbc.273.14.8434 [DOI] [PubMed] [Google Scholar]
- 23. Chinetti G., Gbaguidi F. G., Griglio S., Mallat Z., Antonucci M., Poulain P., Chapman J., Fruchart J. C., Tedgui A., Najib-Fruchart J., and Staels B. (2000) CLA-1/SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome proliferator-activated receptors. Circulation 101, 2411–2417 10.1161/01.CIR.101.20.2411 [DOI] [PubMed] [Google Scholar]
- 24. Thomas C. A., Li Y., Kodama T., Suzuki H., Silverstein S. C., and El Khoury J. (2000) Protection from lethal Gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. J. Exp. Med. 191, 147–156 10.1084/jem.191.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chuang P. C., Wu M. H., Shoji Y., and Tsai S. J. (2009) Downregulation of CD36 results in reduced phagocytic ability of peritoneal macrophages of women with endometriosis. J. Pathol. 219, 232–241 10.1002/path.2588 [DOI] [PubMed] [Google Scholar]
- 26. Nakagawa A., Shiratsuchi A., Tsuda K., and Nakanishi Y. (2005) In vivo analysis of phagocytosis of apoptotic cells by testicular Sertoli cells. Mol. Reprod. Dev. 71, 166–177 10.1002/mrd.20278 [DOI] [PubMed] [Google Scholar]
- 27. Kohno S., Yamashita Y., Abe T., Hirasaka K., Oarada M., Ohno A., Teshima-Kondo S., Higashibata A., Choi I., Mills E. M., Okumura Y., Terao J., and Nikawa T. (2012) Unloading stress disturbs muscle regeneration through perturbed recruitment and function of macrophages. J. Appl. Physiol. (1985) 112, 1773–1782 10.1152/japplphysiol.00103.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Segawa M., Fukada S., Yamamoto Y., Yahagi H., Kanematsu M., Sato M., Ito T., Uezumi A., Hayashi S., Miyagoe-Suzuki Y., Takeda S., Tsujikawa K., and Yamamoto H. (2008) Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis. Exp. Cell Res. 314, 3232–3244 10.1016/j.yexcr.2008.08.008 [DOI] [PubMed] [Google Scholar]
- 29. Chazaud B., Brigitte M., Yacoub-Youssef H., Arnold L., Gherardi R., Sonnet C., Lafuste P., and Chretien F. (2009) Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc. Sport Sci. Rev. 37, 18–22 10.1097/JES.0b013e318190ebdb [DOI] [PubMed] [Google Scholar]
- 30. Mounier R., Théret M., Arnold L., Cuvellier S., Bultot L., Göransson O., Sanz N., Ferry A., Sakamoto K., Foretz M., Viollet B., and Chazaud B. (2013) AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 18, 251–264 10.1016/j.cmet.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 31. Zhang J., Xiao Z., Qu C., Cui W., Wang X., and Du J. (2014) CD8 T cells are involved in skeletal muscle regeneration through facilitating MCP-1 secretion and Gr1(high) macrophage infiltration. J. Immunol. 193, 5149–5160 10.4049/jimmunol.1303486 [DOI] [PubMed] [Google Scholar]
- 32. Villalta S. A., Rinaldi C., Deng B., Liu G., Fedor B., and Tidball J. G. (2011) Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum. Mol. Genet. 20, 790–805 10.1093/hmg/ddq523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langen R. C., Schols A. M., Kelders M. C., Wouters E. F., and Janssen-Heininger Y. M. (2001) Inflammatory cytokines inhibit myogenic differentiation through activation of nuclear factor-κB. FASEB J. 15, 1169–1180 10.1096/fj.00-0463 [DOI] [PubMed] [Google Scholar]
- 34. Dargelos E., Renaud V., Decossas M., Bure C., Lambert O., and Poussard S. (2018) Caveolae-mediated effects of TNF-α on human skeletal muscle cells. Exp. Cell Res. 370, 623–631 10.1016/j.yexcr.2018.07.027 [DOI] [PubMed] [Google Scholar]
- 35. Wang X., Wu H., Zhang Z., Liu S., Yang J., Chen X., Fan M., and Wang X. (2008) Effects of interleukin-6, leukemia inhibitory factor, and ciliary neurotrophic factor on the proliferation and differentiation of adult human myoblasts. Cell Mol. Neurobiol. 28, 113–124 10.1007/s10571-007-9247-9 [DOI] [PubMed] [Google Scholar]
- 36. Al-Shanti N., Saini A., Faulkner S. H., and Stewart C. E. (2008) Beneficial synergistic interactions of TNF-α and IL-6 in C2 skeletal myoblasts–potential cross-talk with IGF system. Growth Factors 26, 61–73 10.1080/08977190802025024 [DOI] [PubMed] [Google Scholar]
- 37. Kim J., and Lee J. (2017) Role of transforming growth factor-β in muscle damage and regeneration: focused on eccentric muscle contraction. J. Exerc. Rehabil. 13, 621–626 10.12965/jer.1735072.536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Darby I. A., Zakuan N., Billet F., and Desmoulière A. (2016) The myofibroblast, a key cell in normal and pathological tissue repair. Cell Mol. Life Sci. 73, 1145–1157 10.1007/s00018-015-2110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bersini S., Gilardi M., Mora M., Krol S., Arrigoni C., Candrian C., Zanotti S., and Moretti M. (2018) Tackling muscle fibrosis: from molecular mechanisms to next generation engineered models to predict drug delivery. Adv. Drug Deliv. Rev. 129, 64–77 10.1016/j.addr.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 40. Mahdy M. A. A. (2019) Skeletal muscle fibrosis: an overview. Cell Tissue Res. 375, 575–588 10.1007/s00441-018-2955-2 [DOI] [PubMed] [Google Scholar]
- 41. Greaves D. R., and Gordon S. (2005) Thematic review series: the immune system and atherogenesis: recent insights into the biology of macrophage scavenger receptors. J. Lipid Res. 46, 11–20 10.1194/jlr.R400011-JLR200 [DOI] [PubMed] [Google Scholar]
- 42. Valacchi G., Sticozzi C., Lim Y., and Pecorelli A. (2011) Scavenger receptor class B type I: a multifunctional receptor. Ann. N.Y. Acad. Sci. 1229, E1–E7 10.1111/j.1749-6632.2011.06205.x [DOI] [PubMed] [Google Scholar]
- 43. Freire-de-Lima C. G., Xiao Y. Q., Gardai S. J., Bratton D. L., Schiemann W. P., and Henson P. M. (2006) Apoptotic cells, through transforming growth factor-β, coordinately induce anti-inflammatory and suppress pro-inflammatory eicosanoid and NO synthesis in murine macrophages. J. Biol. Chem. 281, 38376–38384 10.1074/jbc.M605146200 [DOI] [PubMed] [Google Scholar]
- 44. Johann A. M., von Knethen A., Lindemann D., and Brüne B. (2006) Recognition of apoptotic cells by macrophages activates the peroxisome proliferator-activated receptor-γ and attenuates the oxidative burst. Cell Death Differ. 13, 1533–1540 10.1038/sj.cdd.4401832 [DOI] [PubMed] [Google Scholar]
- 45. Arya S. B., Kumar G., Kaur H., Kaur A., and Tuli A. (2018) ARL11 regulates lipopolysaccharide-stimulated macrophage activation by promoting mitogen-activated protein kinase (MAPK) signaling. J. Biol. Chem. 293, 9892–9909 10.1074/jbc.RA117.000727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Noh K. T., Son K. H., Jung I. D., Kang H. K., Hwang S. A., Lee W. S., You J. C., and Park Y. M. (2012) Protein kinase Cδ (PKCδ)-extracellular signal-regulated kinase 1/2 (ERK1/2) signaling cascade regulates glycogen synthase kinase-3 (GSK-3) inhibition-mediated interleukin-10 (IL-10) expression in lipopolysaccharide (LPS)-induced endotoxemia. J. Biol. Chem. 287, 14226–14233 10.1074/jbc.M111.308841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang L., Wang X. H., Wang H., Du J., and Mitch W. E. (2010) Satellite cell dysfunction and impaired IGF-1 signaling cause CKD-induced muscle atrophy. J. Am. Soc. Nephrol. 21, 419–427 10.1681/ASN.2009060571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brouckaert G., Kalai M., Krysko D. V., Saelens X., Vercammen D., Ndlovu M., Haegeman G., D'Herde K., and Vandenabeele P. (2004) Phagocytosis of necrotic cells by macrophages is phosphatidylserine dependent and does not induce inflammatory cytokine production. Mol. Biol. Cell 15, 1089–1100 10.1091/mbc.e03-09-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]