Abstract
The reductive pyrimidine catabolic pathway is the most widespread pathway for pyrimidine degradation in bacteria, enabling assimilation of nitrogen for growth. This pathway, which has been studied in several bacteria including Escherichia coli B, releases only one utilizable nitrogen atom from each molecule of uracil, whereas the other nitrogen atom remains trapped in the end product β-alanine. Here, we report the biochemical characterization of a β-alanine:2-oxoglutarate aminotransferase (PydD) and an NAD(P)H-dependent malonic semialdehyde reductase (PydE) from a pyrimidine degradation gene cluster in the bacterium Lysinibacillus massiliensis. Together, these two enzymes converted β-alanine into 3-hydroxypropionate (3-HP) and generated glutamate, thereby making the second nitrogen from the pyrimidine ring available for assimilation. Using bioinformatics analyses, we found that PydDE homologs are associated with reductive pyrimidine pathway genes in many Gram-positive bacteria in the classes Bacilli and Clostridia. We demonstrate that Bacillus smithii grows in a defined medium with uracil or uridine as its sole nitrogen source and detected the accumulation of 3-HP as a waste product. Our findings extend the reductive pyrimidine catabolic pathway and expand the diversity of enzymes involved in bacterial pyrimidine degradation.
Keywords: pyrimidine, reductase, nitrogen metabolism, pyridoxal phosphate, enzyme kinetics, beta-alanine, 2-oxoglutarate, 3-hydroxypropionate, aminotransferase, enzyme phylogeny, reductive pathway, nitrogen assimilation, malonic semialdehyde reductase
Introduction
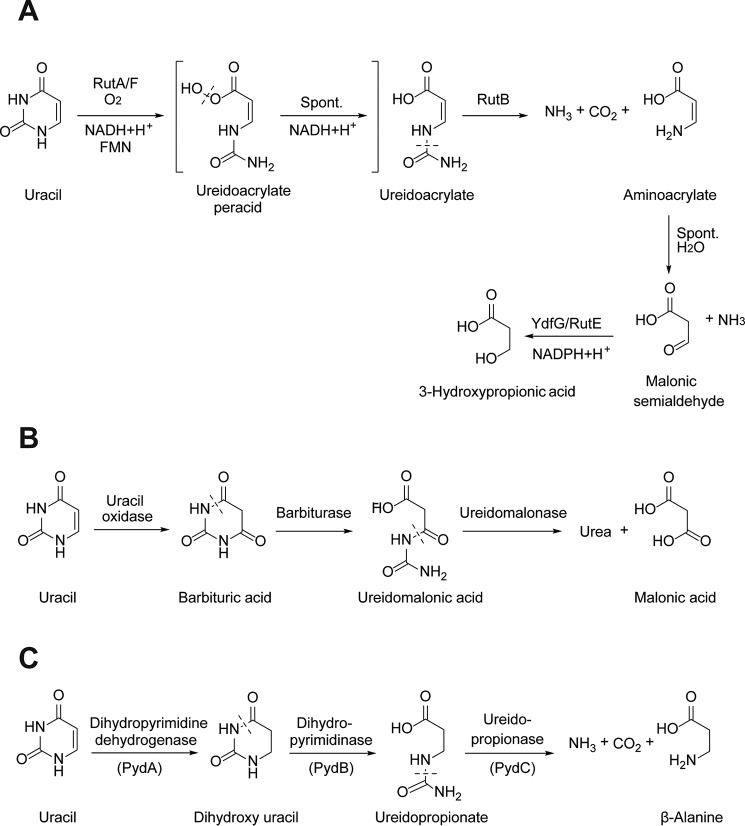
Pyrimidines are a rich source of nitrogen available for assimilation by bacteria (1). Three bacterial pathways are known for the degradation of the stable aromatic pyrimidine ring, as exemplified by the degradation of uracil (Fig. 1). These are known as the pyrimidine utilization (RUT),4 oxidative, and reductive pathways. The RUT pathway is present in certain Proteobacteria, including Escherichia coli K-12, and utilizes a C4a-flavin peroxide for pyrimidine ring opening, releasing two utilizable nitrogen atoms in the form of ammonia (Fig. 1A) (2). The oxidative pathway has only been reported in a limited number of bacteria, including Rhodococcus erythropolis and Enterobacter aerogenes, and proceeds through the oxidation of uracil to barbiturate, yielding two utilizable nitrogen atoms subsequent to hydrolysis of urea (Fig. 1B) (3, 4).
Figure 1.
Three pathways for uracil degradation in bacteria. A, RUT pathway; B, oxidative pathway; C, reductive (Pyd) pathway.
The reductive pathway (Pyd) is the most well-studied and widespread pyrimidine degradation pathway and, in addition to bacteria, also occurs in archaea and eukaryotes. It proceeds through the reduction of uracil to dihydrouracil by dihydropyrimidine dehydrogenase (PydA), followed by two hydrolytic steps catalyzed by dihydropyrimidinase (PydB) and ureidopropionase (PydC) (5). In many bacteria, including E. coli B, the reductive pathway releases only one utilizable nitrogen atom per uracil, the other being trapped in the end product β-alanine (Fig. 1C) (6).
Pathways for the degradation of β-alanine are known in eukaryotes (7) and certain bacteria (8) and involve β-alanine aminotransferase, generating the toxic intermediate malonic semialdehyde. There are several known mechanisms for the degradation of malonic semialdehyde generated in various biochemical pathways. These include reduction to 3-hydroxypropionate catalyzed by the short-chain alcohol dehydrogenase/reductase YdfG in the RUT pathway (Fig. 1A) (2), oxidation to malonyl-CoA catalyzed by malonic semialdehyde dehydrogenase (9), and decarboxylation to acetaldehyde catalyzed by malonic semialdehyde decarboxylase (10). Therefore, it is conceivable that, in some bacteria, the Pyd pathway may include additional enzymatic steps for the acquisition of nitrogen from β-alanine by transamination followed by degradation of malonic semialdehyde. However, the association of β-alanine degradation enzymes with the Pyd pathway in bacteria has not been systematically investigated.
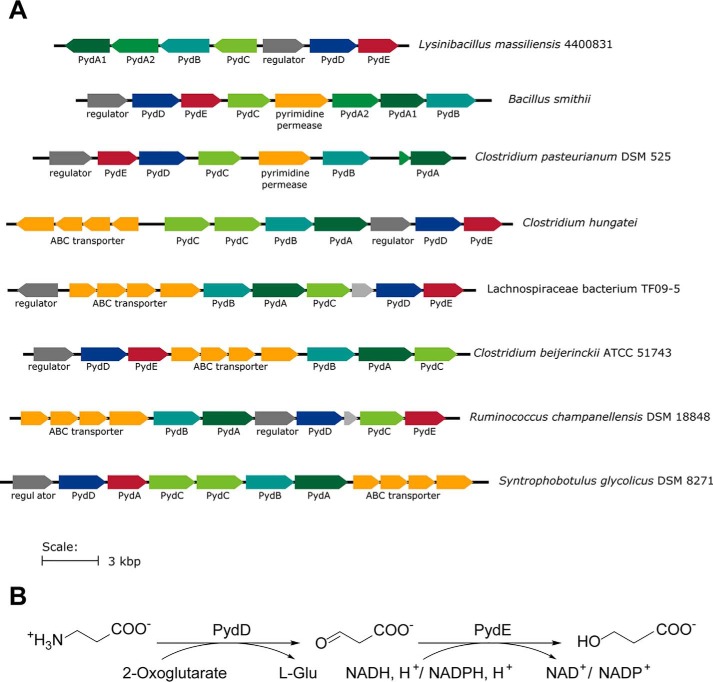
The study of bacterial pyrimidine degradation has relied on the isolation and culturing of bacteria for their ability to utilize pyrimidines as a nitrogen source (1). However, the increasing volume of publicly available genetic sequences presents the opportunity to systematically investigate bacterial pyrimidine degradation gene clusters using bioinformatics. While studying the distribution of aminoethylsulfonate (taurine) degradation enzymes, we observed the occurrence of close homologs of some of these enzymes in Pyd gene clusters. For example, the Lysinibacillus massiliensis Pyd gene cluster contains a putative aminotransferase and a metal-dependent alcohol dehydrogenase related to Bifidobacterium kashiwanohense taurine:oxoglutarate aminotransferase (BkToa) (11) and sulfoacetaldehyde reductase (BkTauF) (12), respectively (Fig. 2A). Because of the structural similarity between taurine and β-alanine, we hypothesized that these two enzymes could play a role in nitrogen acquisition from β-alanine produced in the Pyd pathway (Fig. 2B). Here, we report the biochemical characterization of these two enzymes, a β-alanine:2-oxoglutarate aminotransferase (PydD) and a malonic semialdehyde reductase (PydE), which extend the Pyd pathway by converting β-alanine into 3-HP, generating glutamate for nitrogen assimilation. We further explored the prevalence of this extended pathway in bacteria through bioinformatics. The physiological relevance of this extended pathway was supported by the growth of Bacillus smithii with uracil or uridine as the sole nitrogen source, accompanied by the secretion of 3-HP.
Figure 2.
Association of PydD and PydE with reductive pyrimidine catabolism. A, genome neighborhoods of close homologs of PydA and PydB in different bacteria with some containing PydD and PydE. B, proposed pathway for β-alanine degradation by PydDE.
Results
Recombinant production of PydD and PydE
L. massiliensis PydD and PydE with an N-terminal His6 tag were recombinantly produced in E. coli. Both proteins were purified to near homogeneity with a TALON Co2+-affinity column, with an additional anion-exchange chromatography employed for the purification of PydE (Fig. S1). The yields of the proteins are 52 and 75 mg/liter of cell culture for PydD and PydE, respectively. The UV-visible spectrum of the purified recombinant PydD contains characteristic absorbance values at 330 and 410 nm, which correlate with the enolimine and ketoenamine forms of the pyridoxal phosphate (PLP) cofactor, respectively (Fig. S2).
β-Alanine:2-oxoglutarate aminotransferase activity of PydD
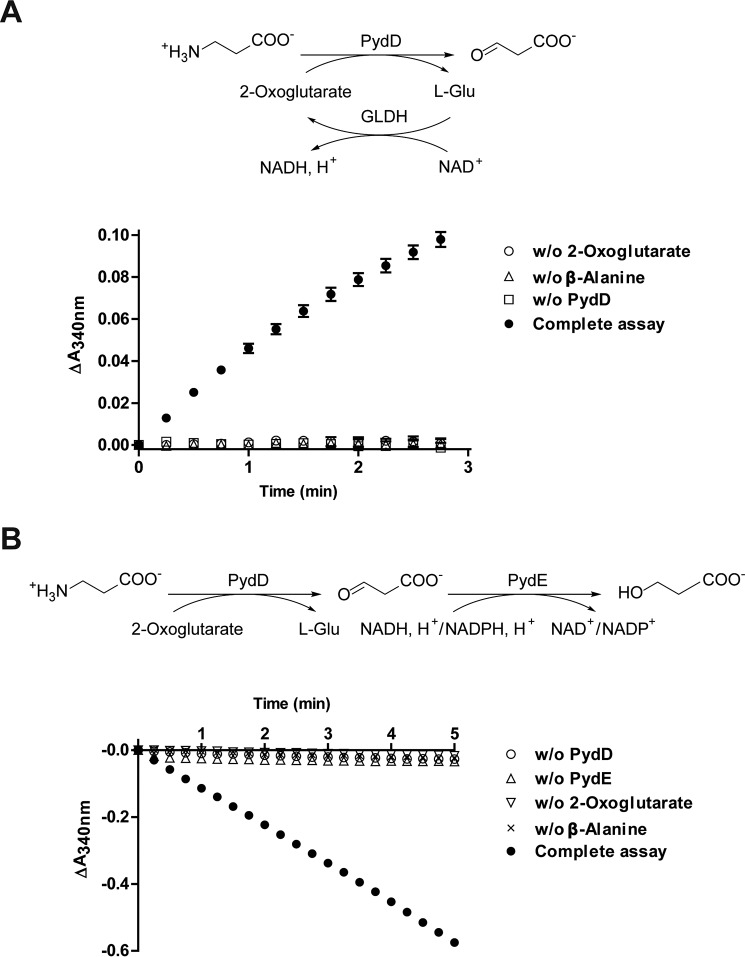
PydD catalyzed the formation of l-glutamate from β-alanine and 2-oxoglutarate, as detected by an assay with glutamate dehydrogenase (GLDH) (Fig. S3 and Fig. 3A). PydD also catalyzed the formation of l-alanine under similar reaction conditions when pyruvate was used as the amine acceptor, although with a lower conversion (Fig. S3). Incubation of PydD together with PydE, β-alanine, 2-oxoglutarate, and NADPH led to the time-dependent consumption of NADPH (Fig. 3B), suggesting that following the reversible conversion of β-alanine to malonic semialdehyde by PydD, the reaction is pulled forward by the NADPH-dependent reduction of malonic semialdehyde by PydE. Further characterization of PydD was carried out using this PydE-coupled spectrophotometric assay, at the optimal pH of 8.0 (Fig. S4).
Figure 3.
PydD activity assays. A, detection of l-glutamate as a reaction product of PydD-catalyzed β-alanine:2-oxoglutarate transamination. Assays monitor NADH formation accompanying l-glutamate oxidation by GLDH. B, coupled activity assays for PydD, detecting malonic semialdehyde generated by PydD-catalyzed β-alanine:2-oxoglutarate transamination. Assays monitor NADPH consumption accompanying malonic semialdehyde reduction by PydE. Error bars, S.E.
The Michaelis–Menten kinetic parameters of PydD were determined for different amine donor (β-alanine, taurine) and acceptor (2-oxoglutarate, pyruvate, oxaloacetate) substrates (Fig. S5) and summarized in Table 1. With β-alanine as the amine donor, the highest activity was obtained with 2-oxoglutarate as the amine acceptor (Figs. S5 and S6), suggesting that 2-oxoglutarate is the physiological substrate for PydD. The apparent kcat/Km for pyruvate was 63-fold lower than that for 2-oxoglutarate (Table 1). Under the same reaction conditions, the activity was the lowest for oxaloacetate (Fig. S6), and its apparent kcat/Km was not determined. The sulfonate-containing analog taurine also served as a substrate by PydD with 2-oxoglutarate as the amine acceptor (Fig. S3). Its apparent kcat/Km was ∼60-fold lower than that for β-alanine, consistent with our hypothesis that β-alanine is the physiological substrate of PydD.
Table 1.
Michaelis–Menten kinetic parameters of PydD (error values reported are the S.E. for the fits)
| Enzyme | Substrate | kcat | Km | kcat/Km |
|---|---|---|---|---|
| s−1 | mm | m−1 s−1 | ||
| PydD | β-Alanine | 5.6 ± 0.2 | 2.3 ± 0.4 | 2400 ± 500 |
| 2-Oxoglutarate | 5.6 ± 0.2 | 3.3 ± 0.3 | 1700 ± 200 | |
| PydD | β-Alanine; pyruvate | 1.25 ± 0.06 | 46 ± 5 (pyruvate) | 27 ± 3 |
| PydD | Taurine; 2-oxoglutarate | 0.51 ± 0.02 | 12 ± 2 (taurine) | 41 ± 7 |
Malonic semialdehyde reductase activity of PydE
PydE catalyzed the NADPH-dependent reduction of malonic semialdehyde, generated in situ in the presence of excess PydD, β-alanine, and 2-oxoglutarate (Fig. S7). PydE also catalyzed the reverse reaction, NADP+-dependent oxidation of 3-HP, with an optimum pH of 10.0 (Fig. S8). The Michaelis–Menten kinetic parameters of PydE were determined for different alcohol and nucleotide (NAD+ and NADP+) substrates (Fig. S9 and Table 2). The apparent kcat/Km for the sulfonate-containing analog 2-hydroxyethylsulfonate (isethionate) was ∼10-fold lower than that for 3-HP. PydE exhibited comparable activities with both NAD+ and NADP+ (Fig. S9 and Table 2).
Table 2.
Michaelis–Menten kinetic parameters of PydE (error values reported are the S.E. for the fits)
| Enzyme | Substrate | kcat | Km | kcat/Km |
|---|---|---|---|---|
| s−1 | mm | m−1 s−1 | ||
| PydE | NADP+; 3-HP | 4.23 ± 0.07 | 7.4 ± 0.4 (3-HP) | 570 ± 30 |
| PydE | NADP+; isethionate | 3.07 ± 0.09 | 55 ± 3 (isethionate) | 56 ± 4 |
| PydE | NAD+; 3-HP | 3.4 ± 0.2 | 4.0 ± 0.7 (NAD+) | 680 ± 160 |
| PydE | NADP+; 3-HP | 2.9 ± 0.1 | 3.4 ± 0.5 (NADP+) | 840 ± 130 |
LC/MS analyses of the PydD and PydE-catalyzed reactions
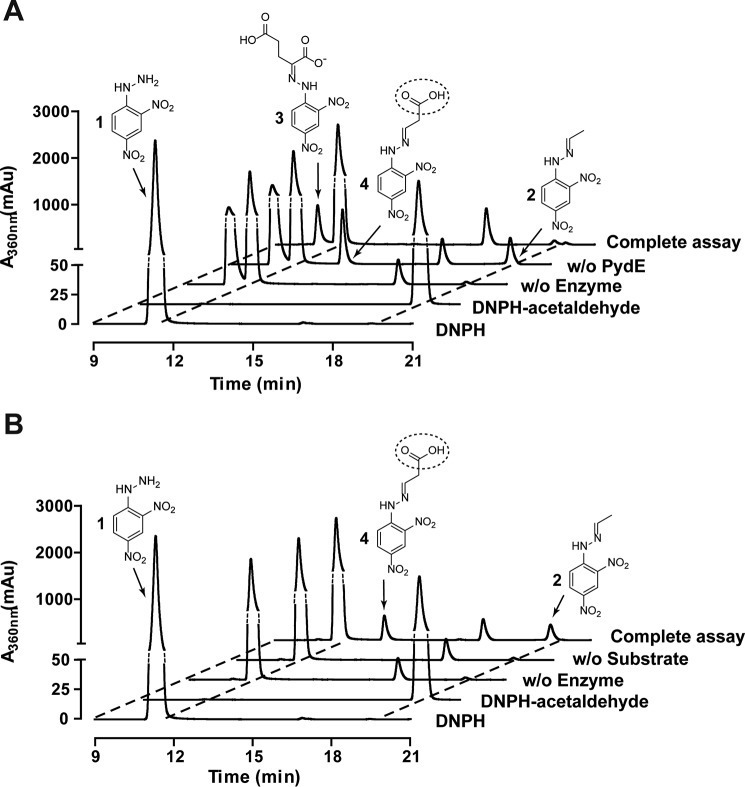
The PydD- and PydE-catalyzed reactions were analyzed by derivatization with 2,4-dinitrophenylhydrazine (DNPH) followed by LC/MS. The DNPH (compound 1) and acetaldehyde-DNPH (compound 2) standards eluted at 11.3 and 19.5 min (Fig. 4A), and their identities were confirmed by electrospray ionization-MS (m/z) (Fig. S10, A and B). 2-Oxoglutarate-DNPH (compound 3) eluted at 10.6 min, was prominent in the assay mixture without enzymes (Fig. 4A and Fig. S10C). In the assay containing PydD but not PydE, the 2-oxoglutarate-DNPH peak was diminished, accompanied by the appearance of a peak eluting at 13.2 min, corresponding to malonic semialdehyde-DNPH (compound 4, Fig. 4A). The negative ion mass spectrum of this peak suggested decarboxylation accompanying the ionization process (Fig. S10D). A peak corresponding to acetaldehyde-DNPH was also present in this assay, suggesting spontaneous decarboxylation of malonic semialdehyde during the reaction or workup (Fig. 4A). In the complete assay containing both PydD and PydE, the malonic semialdehyde-DNPH peak was eliminated, consistent with PydE-catalyzed malonic semialdehyde reduction. The 2-oxoglutarate-DNPH peak was also further diminished, suggesting that PydE drives the reaction toward completion.
Figure 4.
LC/MS analyses of product formation in PydD- and PydE-catalyzed reactions. A, elution profiles of the LC/MS assays for the PydDE-catalyzed β-alanine degradation, monitoring the absorbance at 360 nm. The complete assay contains both PydD and PydE, and controls omitting both enzymes and omitting PydE are included. B, elution profiles of the LC/MS assays for PydE-catalyzed 3-HP oxidation, monitoring absorbance at 360 nm. Reaction products were derivatized with DNPH prior to LC/MS analyses. The break in the vertical axis of the plot is indicated in dotted lines.
The PydE-catalyzed NADP+-dependent oxidation of 3-HP was also analyzed. A peak corresponding to malonic semialdehyde-DNPH, eluting at 13.2 min, was present in the complete reaction mixture (Fig. 4B) and confirmed by electrospray ionization-MS (m/z) (Fig. S11B). Similar to the PydD assay, a peak corresponding to acetaldehyde-DNPH was also observed (Fig. 4B and Fig. S11C). The malonic semialdehyde-DNPH and acetaldehyde-DNPH peaks were absent in negative controls omitting either PydE or 3-HP.
Taken together, these observations are consistent with our hypothesis that PydD catalyzes the transamination reaction converting β-alanine and 2-oxoglutarate to glutamate and malonic semialdehyde, which is then reduced by PydE to 3-HP.
Use of pyrimidines as the sole nitrogen source supports growth of B. smithii
Next, we investigated the physiological relevance of the extended Pyd gene cluster in pyrimidine nitrogen assimilation in bacteria. Despite our attempts, we were unsuccessful in growing L. massiliensis using defined medium with uracil or uridine as the sole nitrogen source. However, we were able to grow B. smithii, which contains an extended Pyd gene cluster similar to that in L. massiliensis (66.3 and 43.1% identity between B. smithii and L. massiliensis PydD and PydE, respectively). This enabled us to assay the spent medium for 3-HP, the predicted end product of the extended Pyd pathway. Using an enzymatic assay with PydE, 3-HP was detected in the spent medium with uracil or uridine as the sole nitrogen source (Table S2 and Fig. 5), providing evidence for our proposed pathway. Conversely, 3-HP was not detected in the spent media with ammonium or cytidine as the nitrogen source (Table S2 and Fig. 5). This is consistent with the fact that utilization of cytidine/cytosine as a nitrogen source only requires hydrolysis of the 4-amino group (e.g. by cytidine/cytosine deaminase) and does not necessarily require the Pyd pathway.
Figure 5.
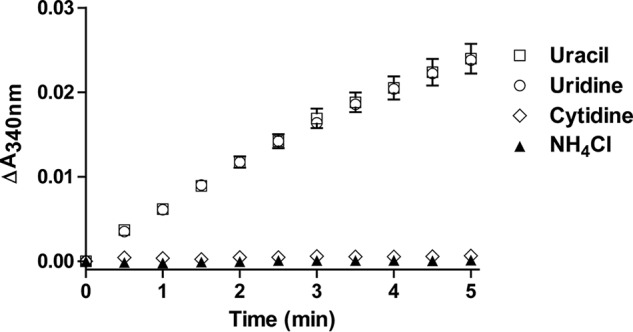
Enzymatic detection of the presence of 3-HP in bacterial culture media. All assays were performed with PydE monitoring the increase of A340 representing the formation of NADPH. The sole nitrogen source used in each of the culture media was as labeled. Error bars, S.E.
PydDE is present in the Pyd gene clusters of many bacteria in the phylum Firmicutes
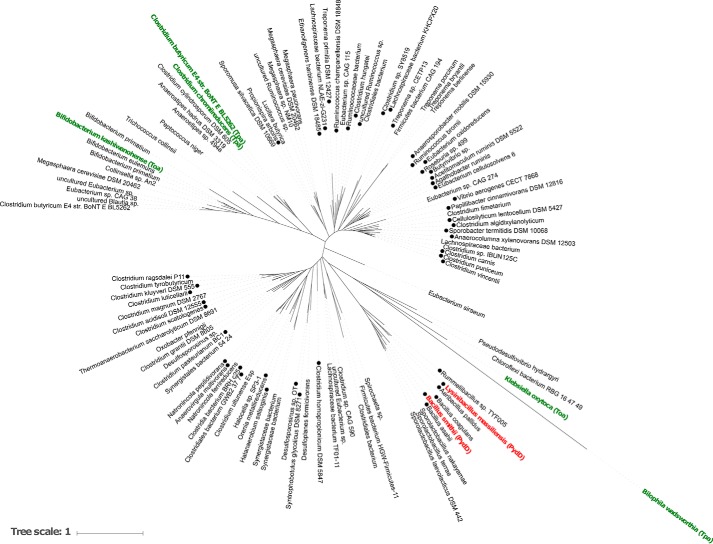
Having established that PydDE form a viable pathway for nitrogen acquisition from β-alanine, we next investigated the occurrence of PydDE in Pyd gene clusters in different bacteria. We examined the 236 PydD homologs in the UniRef cluster UniRef50_A0A0H4P1N9 (where each member shares ≥50% sequence identity and ≥80% overlap with the seed sequence of the cluster (13)). A maximum likelihood phylogenetic tree of representative PydD homologs is given in Fig. 6. Of these sequences, 93 fall within gene clusters containing homologs of PydA, PydB, and PydC (Table S3 and Fig. 6).
Figure 6.
Maximum likelihood phylogenetic tree of representative PydD homologs belonging to UniRef50_A0A0H4P1N9. Klebsiella oxytoca taurine:2-oxoglutarate aminotransferase (Toa) and Bilophila wadsworthia taurine:pyruvate aminotransferase (Tpa) were included as an outgroup. Labels of known enzymes are colored green (taurine aminotransferases) and red (β-alanine aminotransferases). ●, sequences containing PydABC homologs in their genome neighborhood, within a 10-ORF window.
Of the 93 PydABCD clusters, 88 contain a metal-dependent alcohol dehydrogenase in the same family of PydE. 90 belong to bacteria in the phylum Firmicutes, with 82 in the class Clostridia and 7 in the class Bacilli (Table S3). This provides evidence for the prevalence of the extended Pyd pathway in Firmicutes bacteria.
Discussion
The biochemical characterization of the β-alanine:2-oxoglutarate aminotransferase PydD and the malonic semialdehyde reductase PydE, associated with the PydABC gene cluster in L. massiliensis, provides evidence for an extended Pyd pathway that allows the acquisition of both nitrogens from the pyrimidine ring. The physiological relevance of this pathway is supported by the production of 3-HP by B. smithii when uracil or uridine is used as a nitrogen source. Our inability to grow L. massiliensis in defined medium with uracil or uridine as the sole nitrogen source might be due to an unknown growth requirement of this bacterium or a different function of the Pyd pathway in this bacterium (e.g. recycling of intracellular pyrimidines).
The occurrence of the standard and extended variants of the Pyd pathway is unlike the other pyrimidine degradation pathways, where degradation is concomitant with the release of both nitrogen atoms of the pyrimidine ring. In the oxidative pathway, two equivalents of ammonia are released upon hydrolysis of urea. In the RUT pathway, ammonia is released on the hydrolysis of ureidoacrylate and on hydrolysis of the unstable intermediate aminoacrylate. For the Pyd pathway, the presence of the standard or extended variant in different bacteria might reflect the balance between cellular requirement for nitrogen and the costs of producing two additional enzymes PydD and PydE. The extended pathway might be advantageous under conditions of extreme nitrogen limitation.
Our study also deepened the link between enzymes involved in the metabolism of sulfonic acids and structurally similar carboxylic acid substrates. PydD possesses taurine:2-oxoglutarate aminotransferase activity, and the PydD UniRef50_A0A0H4P1N9 cluster contains the previously reported B. kashiwanohense taurine:2-oxoglutarate aminotransferase (11), thought to be involved in taurine nitrogen assimilation. Many of the members of UniRef50_A0A0H4P1N9 that are not associated with Pyd appear to be associated instead with taurine degradation genes. Similar promiscuity has been observed with sulfoacetaldehyde reductases (12, 14), and this observation will aid future bioinformatics-based enzyme function prediction.
Compared with PydD, the sequences of candidate PydE in the Pyd clusters are much more divergent. Malonic semialdehyde as an intermediate in pyrimidine degradation is also produced by the E. coli RUT pathway and yeast URC pathway, where it is reduced to 3-HP by YdfG (2) and URC8 (15), respectively. Both YdfG and URC8 belonging to the short-chain dehydrogenase/reductase (SDR) family are structurally unrelated to PydE. 3-hydroxyacid dehydrogenases belonging to the SDR family are widespread in bacteria and are specific to using NADPH as a reductant. Malonic semialdehyde reductases belonging to the metal-dependent alcohol dehydrogenase family have been reported in the archaea, such as Metallosphaera sedula (16) and Nitrosopumilus maritimus (16), involved in CO2 fixation through the 3-HP pathway, with a preference for NADPH as a reductant. To our knowledge, PydE is the only bacterial malonic semialdehyde reductase belonging to this enzyme family reported to date. Its ability to use both NADH and NADPH might find applications in the biotechnological production of 3-HP, an industrially important chemical.
Materials and methods
General
L. massiliensis (DSMZ 27596) was purchased from DSMZ (Deutsche Sammlung von mikroorganismen und Zellkulturen GmbH). B. smithii (CGMCC 1.3752) was purchased from CGMCC (China General Microbiological Culture Collection Center). Lysogeny broth (DifcoTM) was purchased from BD Biosciences. CASO medium for the growth of L. massiliensis was prepared by dissolving 15 g of peptone from casein, 5 g of peptone from soymeal, and 5 g of NaCl in 1 liter of distilled water and adjusting the pH to 7.3. Nutrient broth for the growth of B. smithii was prepared by dissolving 5 g of peptone (BactoTM, BD Biosciences), 3 g of meat extract (AoBox, Beijing), 0.1 g of agar, and 10 mg of MnSO4·H2O in 1 liter of distilled water and adjusting the pH to 7.0. Defined medium was prepared as specified in the supporting Methods.
Acetonitrile and methanol were purchased from TJConcord Technology (Tianjin, China). Formic acid was purchased from Merck. TALON resin was purchased from CLONTECH. All protein purification chromatographic experiments were performed on an ÄKTA pure or ÄKTA prime plus FPLC machine equipped with appropriate columns (GE Healthcare). Protein concentrations were calculated from their absorption at 280 nm measured using a Nanodrop One (Thermo Fisher Scientific). NAD(P)H-coupled activity assays were carried out by monitoring the absorbance at 340 nm using a plate reader (Tecan M200).
Cloning, expression, and purification of PydD and PydE
L. massiliensis cells (DSMZ 27596) were streaked on a CASO agar plate. Colony PCRs were performed to amplify the pydD and pydE genes using the primer pairs 1F/1R and 2F/2R (Table S1), respectively. The amplified DNA fragments were inserted by Gibson assembly into the SspI site of a modified pET-28a(+) vector (HT), containing an N-terminal His6 tag followed by a tobacco etch virus protease cleavage site (17). The resulting plasmids HT-PydD and HT-PydE were confirmed by sequencing.
For the recombinant production of PydD and PydE, E. coli BL21 (DE3) cells were transformed with the plasmids HT-PydD and HT-PydE, respectively. The transformants were grown in lysogeny broth supplemented with 50 μg/ml kanamycin (1 liter in a 2.6-liter flask) at 37 °C while being shaken at 220 rpm. When A600 reached ∼0.8, the temperature was decreased to 16 °C, and isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.3 mm to induce the production of the proteins. After 18 h, cells were harvested by centrifugation (8000 × g for 15 min at 4 °C).
The cell paste was resuspended in lysis buffer (5 ml/g of cell paste) containing 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethanesulfonyl fluoride, 0.4 mg/ml lysozyme, 0.03% Triton X-100, and 0.03 mg/ml DNase I (Roche Applied Science). The cell suspension was frozen in a −80 °C freezer and then thawed and incubated at 25 °C for 40 min to allow for lysis. A 6% solution of streptomycin sulfate in water was added to a final concentration of 1% to precipitate the DNA. The precipitate was removed by centrifugation (12,000 × g for 10 min at 4 °C). The supernatant was then filtered through a 0.45-μm filter. β-Mercaptoethanol (BME) was added to a final concentration of 5 mm, and the sample was loaded onto a 10-ml TALON Co2+-affinity column, pre-equilibrated with buffer A (20 mm Tris-HCl, pH 7.5, 5 mm BME, and 0.2 m KCl). The column was washed with 10 column volumes (CV) of buffer A, and then the protein was eluted with 5 CV of buffer A containing 150 mm imidazole. The eluted protein (∼20 ml) was dialyzed against 2 liters of buffer A to remove imidazole (3 h at 4 °C). The dialyzed PydD was frozen in aliquots with liquid nitrogen and stored at −80 °C until further use. The dialyzed PydE was diluted 4-fold with buffer B (20 mm Tris-HCl, pH 7.5, 5 mm BME) and loaded onto a 10-ml Q column, pre-equilibrated with buffer B. The column was washed with 10 CV of buffer B and eluted with 10 CV of buffer B containing a linear gradient of 50–500 mm KCl. PydE eluted at ∼270 mm KCl. PydE (∼30 ml) was then dialyzed against 2 liters of buffer A for 3 h at 4 °C. The purified PydD and PydE were quantified using their calculated extinction coefficients (ϵ280 = 46,760 and 29,340 m−1 cm−1, respectively). The purity of both proteins was assessed by SDS-PAGE.
Determination of the amine acceptor of PydD
GLDH was purchased from Sigma (catalogue no. 2626). The procedure for production of recombinant Bacillus subtilis alanine dehydrogenase (18) is described in the supporting Methods.
To carry out the transamination reaction, a 200-μl reaction mixture containing 100 mm HEPES, pH 8.0, 20 μm PLP, 5 mm 2-oxoglutarate or pyruvate, 5 mm β-alanine, and 1 μm PydD was incubated in a 35 °C water bath for 30 min. The enzyme was then heat-inactivated in a boiling water bath for 2 min. To detect the amino acid product, 100 μl of the reaction mixture was mixed with a 100-μl solution containing 10 mm NAD+ and 72 milliunits of GLDH or alanine dehydrogenase in the same reaction buffer. Absorbance at 340 nm was monitored over 10 min.
Activity assays for PydD
A PydE-coupled spectrophotometric assay was used to assay the β-alanine:2-oxoglutarate aminotransferase activity of PydD. In a typical assay, a 200-μl reaction mixture containing a limiting amount (0.1 μm) of PydD and excess (0.5 μm) PydE, 10 μm PLP, 25 mm 2-oxoglutarate, 25 mm β-alanine, and 0.4 mm NADPH in 100 mm HEPES, pH 8.0, was prepared, and the decrease in absorbance at 340 nm was monitored over 3 min.
The optimal pH of PydD was determined by carrying out the PydD activity assay in a 100 mm concentration of different buffers: MES, pH 6.0; HEPES, pH 7.0, 7.5, and 8.0; Tricine, pH 8.5; Tris/HCl, pH 9.0. The ability of PydD to use different amine acceptors was determined with pyruvate or oxaloacetate in place of 2-oxoglutarate. The taurine:2-oxoglutarate aminotransferase activity of PydD was determined with taurine in place of β-alanine. Higher concentrations of enzymes (0.5 or 1 μm PydD and 2.5 μm PydE) were used for these experiments.
To obtain the Michaelis–Menten kinetic parameters of PydD, the concentration of one substrate (β-alanine/taurine or 2-oxoglutarate/pyruvate) was fixed at 25 mm while varying the concentration of the other substrate. The β-alanine:2-oxoglutarate aminotransferase assay was conducted with 0.1 μm PydD. The β-alanine:pyruvate and taurine:2-oxoglutarate aminotransferase assays were conducted with 0.5 μm PydD. Data analysis was conducted with GraphPad Prism version 6 (GraphPad Software, La Jolla, CA).
Activity assays for PydE
Malonic semialdehyde is chemically unstable and was generated in situ by PydD to assay the malonic semialdehyde reductase activity of PydE. The PydE assay conditions were identical to that of PydD, except that a limiting amount (0.05, 0.1, and 0.2 μm) of PydE and excess (10 μm) PydD were used.
To assay the 3-HP dehydrogenase activity of PydE, the 200-μl reaction mixture contained 0.25 μm PydE, 100 mm 3-HP, 15 mm NADP+, 0.2 m KCl, and 40 mm different buffers (HEPES, pH 7.0 and 8.0; Tris/HCl, pH 9.0; CAPSO, pH 10.0 and 11.0). To obtain the Michaelis–Menten kinetic parameters of PydE for 3-HP oxidation, the assay was conducted with 0.25 μm PydE at the optimal pH of 10.0, and the concentration of one substrate was varied while the concentration of the other substrate was fixed (3-HP at 50 mm or NADP+ at 15 mm). Kinetic parameters for other substrates, including isethionate and NAD+, were also measured.
LC/MS detection of malonic semialdehyde
To detect malonic semialdehyde produced as an intermediate in the PydD-PydE reaction, 200-μl reaction mixtures containing 100 mm HEPES, pH 8.0, 10 μm PLP, 5 mm 2-oxoglutarate, 5 mm β-alanine, and either 1) no enzyme, 2) 2 μm PydD, or 3) 2 μm PydD, 10 μm PydE, and 5 mm NADPH were incubated at 35 °C for 30 min. To detect malonic semialdehyde produced by oxidation of 3-HP by PydE, a 200-μl reaction mixture containing 40 mm CAPSO, pH 10.0, 2 mm NADP+, 50 mm 3-HP, and 2 μm PydE was incubated at room temperature for 15 min.
The product was derivatized with DNPH (J&K) (19) prior to LC/MS analysis. After the enzyme reaction, 100 μl of reaction solution was mixed with 1.1 ml of 0.73 m sodium acetate buffer, pH 5.0, followed by 800 μl of freshly prepared DNPH solution (40 mg dissolved in 100 ml of methanol), and the mixture was incubated at 50 °C for 1 h and then filtered. A commercial acetaldehyde-DNPH standard (Sigma) (40 mg in 100 ml of methanol) was also prepared. LC/MS analysis was performed on an Agilent 6420 Triple Quadrupole LC/MS instrument (Agilent Technologies). The drying gas temperature was maintained at 350 °C with a flow rate of 12 liters min−1 and a nebulizer pressure of 25 p.s.i. LC/MS analysis was carried out on an Agilent ZORBAX SB-C18 column (4.6 × 250 mm, product number 880975-902). A linear gradient of acetonitrile (25–85% in H2O) containing 0.1% formic acid and a flow rate of 1.0 ml/min for 30 min were used to elute. UV detection was set at 360 nm.
Bacterial culture using pyrimidine as the sole nitrogen source
B. smithii was grown in nutrient broth at 55 °C overnight. A 50-μl portion of the culture was then transferred into defined medium with different nitrogen sources (20 mm ammonium chloride, 10 mm uracil/uridine, or 6.7 mm cytidine). Cells were incubated for a further 3–5 days.
To detect 3-HP produced, the bacterial cultures were centrifuged at 20,000 × g, and 20 μl of supernatant was added to an assay mixture, with a final volume of 200 μl, containing 40 mm CAPSO, pH 10.0, 2 mm NADP+, and 2 μm PydE. The absorbance at 340 nm was monitored over 5 min.
Bioinformatics
The genome neighborhoods of several PydD homologs were examined by using the web-based Enzyme Function Initiative Genome Neighborhood Tool (20) to identify homologs of PydA (belonging to the “dihydroorotate dehydrogenase” PF01180 and 4Fe-4S dicluster PF14697 Pfam families), PydB (amidohydrolase family PF01979), PydC (peptidase family M20 PF01546), and PydE (metal-containing alcohol dehydrogenase family PF00465), within a 10-ORF window.
Phylogenetic tree construction
A maximum-likelihood phylogenetic tree was constructed using PhyML (21) with the WAG substitution model (22). The unequal rate of variation among amino acid sites was modeled with a γ distribution with a shape parameter (23) of 0.932. The level of confidence for the branches was determined based on 100 bootstrap replicates (24). The resulting consensus tree (Fig. 6) was rendered using the web-based program iTOL (25).
Author contributions
J. Y., Y. W., and Y. Z. conceptualization; J. Y., Y. W., D. L., Y. H., and Y. Z. data curation; J. Y., Y. W., D. L., Y. H., Q. L., and Y. Z. formal analysis; J. Y., Y. W., D. L., Y. H., and Y. Z. validation; J. Y., Y. W., D. L., H. Z., and Y. Z. visualization; J. Y., Y. W., and Y. Z. methodology; J. Y., Y. W., E. L. A., H. Z., and Y. Z. writing-original draft; Y. W., E. L. A., H. Z., and Y. Z. supervision; Y. W. and Y. Z. investigation; Y. W., E. L. A., H. Z., and Y. Z. writing-review and editing; Q. L., E. L. A., H. Z., and Y. Z. funding acquisition; E. L. A., H. Z., and Y. Z. resources; E. L. A., H. Z., and Y. Z. project administration.
Supplementary Material
Acknowledgments
We thank the instrument analytical center of the School of Pharmaceutical Science and Technology at Tianjin University for providing the LC/MS analysis and Zhi Li, Dr. Xinghua Jin, and Prof. Xiangyang Zhang for helpful discussion.
This work was supported by National Science Foundation of China Grants 31870049 and 31570060 (to Y. Z.) and by the Agency for Science, Technology, and Research of Singapore Visiting Investigator Program Grant 1535j00137 (to H. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting Methods, Tables S1–S3, and Figs. S1–S11.
- RUT
- pyrimidine utilization
- CAPSO
- 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid
- BME
- β-mercaptoethanol
- CV
- column volumes
- GLDH
- glutamate dehydrogenase
- PLP
- pyridoxal phosphate
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- 3-HP
- 3-hydroxypropionate
- DNPH
- 2,4-dinitrophenylhydrazine.
References
- 1. Vogels G. D., and Van der Drift C. (1976) Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40, 403–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim K. S., Pelton J. G., Inwood W. B., Andersen U., Kustu S., and Wemmer D. E. (2010) The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J. Bacteriol. 192, 4089–4102 10.1128/JB.00201-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel B. N., and West T. P. (1987) Oxidative catabolism of uracil by Enterobacter aerogenes. FEMS Microbiol. Lett. 40, 33–36 10.1111/j.1574-6968.1987.tb01977.x [DOI] [Google Scholar]
- 4. Soong C. L., Ogawa J., Sakuradani E., and Shimizu S. (2002) Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 277, 7051–7058 10.1074/jbc.M110784200 [DOI] [PubMed] [Google Scholar]
- 5. Traut T. W., and Loechel S. (1984) Pyrimidine catabolism: individual characterization of the three sequential enzymes with a new assay. Biochemistry 23, 2533–2539 10.1021/bi00306a033 [DOI] [PubMed] [Google Scholar]
- 6. West T. P. (1998) Isolation and characterization of an Escherichia coli B mutant strain defective in uracil catabolism. Can. J. Microbiol. 44, 1106–1109 10.1139/cjm-44-11-1106 [DOI] [PubMed] [Google Scholar]
- 7. Schnackerz K. D., Andersen G., Dobritzsch D., and Piskur J. (2008) Degradation of pyrimidines in Saccharomyces kluyveri: transamination of β-alanine, Nucleosides Nucleotides Nucleic Acids 27, 794–799 10.1080/15257770802145983 [DOI] [PubMed] [Google Scholar]
- 8. Wilding M., Peat T. S., Newman J., and Scott C. (2016) A β-alanine catabolism pathway containing a highly promiscuous ω-transaminase in the 12-aminododecanate-degrading Pseudomonas sp. strain AAC. Appl. Environ. Microbiol. 82, 3846–3856 10.1128/AEM.00665-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakamura K., and Bernheim F. (1961) Studies on malonic semialdehyde dehydrogenase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 50, 147–152 10.1016/0006-3002(61)91071-X [DOI] [PubMed] [Google Scholar]
- 10. Poelarends G. J., Johnson W. H. Jr., Murzin A. G., and Whitman C. P. (2003) Mechanistic characterization of a bacterial malonate semialdehyde decarboxylase: identification of a new activity on the tautomerase superfamily. J. Biol. Chem. 278, 48674–48683 10.1074/jbc.M306706200 [DOI] [PubMed] [Google Scholar]
- 11. Li M., Wei Y., Yin J., Lin L., Zhou Y., Hua G., Cao P., Ang E. L., Zhao H., Yuchi Z., and Zhang Y. (2019) Biochemical and structural investigation of taurine:2-oxoglutarate aminotransferase from Bifidobacterium kashiwanohense. Biochem. J. 476, 1605–1619 10.1042/BCJ20190206 [DOI] [PubMed] [Google Scholar]
- 12. Zhou Y., Wei Y., Nanjaraj Urs A. N., Lin L., Xu T., Hu Y., Ang E. L., Zhao H., Yuchi Z., and Zhang Y. (2019) Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacterium Bifidobacterium kashiwanohense. Biosci. Rep. 39, BSR20190715 10.1042/BSR20190715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H., and UniProt Consortium (2015) UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 10.1093/bioinformatics/btu739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Y., Wei Y., Lin L., Xu T., Ang E. L., Zhao H., Yuchi Z., and Zhang Y. (2019) Biochemical and structural investigation of sulfoacetaldehyde reductase from Klebsiella oxytoca. Biochem. J. 476, 733–746 10.1042/BCJ20190005 [DOI] [PubMed] [Google Scholar]
- 15. Andersson Rasmussen A., Kandasamy D., Beck H., Crosby S. D., Björnberg O., Schnackerz K. D., and Piškur J. (2014) Global expression analysis of the yeast Lachancea (Saccharomyces) kluyveri reveals new URC genes involved in pyrimidine catabolism. Eukaryot. Cell 13, 31–42 10.1128/EC.00202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Otte J., Mall A., Schubert D. M., Könneke M., and Berg I. A. (2015) Malonic semialdehyde reductase from the archaeon Nitrosopumilus maritimus is involved in the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl. Environ. Microbiol. 81, 1700–1707 10.1128/AEM.03390-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nallamsetty S., and Waugh D. S. (2007) A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat. Protoc. 2, 383–391 10.1038/nprot.2007.50 [DOI] [PubMed] [Google Scholar]
- 18. Grimshaw C. E., and Cleland W. W. (1981) Kinetic mechanism of Bacillus subtilis l-alanine dehydrogenase. Biochemistry 20, 5650–5655 10.1021/bi00523a002 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Q., Li Y., Chen D., Yu Y., Duan L., Shen B., and Liu W. (2011) Radical-mediated enzymatic carbon chain fragmentation-recombination. Nat. Chem. Biol. 7, 154–160 10.1038/nchembio.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao S., Sakai A., Zhang X., Vetting M. W., Kumar R., Hillerich B., San Francisco B., Solbiati J., Steves A., Brown S., Akiva E., Barber A., Seidel R. D., Babbitt P. C., Almo S. C., et al. (2014) Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. Elife 3 10.7554/eLife.03275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S., and Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 22. Whelan S., and Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- 23. Eyre-Walker A., and Keightley P. D. (2007) The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8, 610–618 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- 24. Felsenstein J. (1978) Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 10.2307/2412923 [DOI] [Google Scholar]
- 25. Letunic I., and Bork P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.