Abstract
Temporal lobe epilepsy (TLE) is a prevalent neurological disorder with many patients experiencing poor seizure control with existing anti-epileptic drugs. Thus, novel insights into the mechanisms of epileptogenesis and identification of new drug targets can be transformative. Changes in ion channel function have been shown to play a role in generating the aberrant neuronal activity observed in TLE. Previous work demonstrates that hyperpolarization-activated cyclic nucleotide-gated (HCN) channels regulate neuronal excitability and are mislocalized within CA1 pyramidal cells in a rodent model of TLE. The subcellular distribution of HCN channels is regulated by an auxiliary subunit, tetratricopeptide repeat–containing Rab8b-interacting protein (TRIP8b), and disruption of this interaction correlates with channel mislocalization. However, the molecular mechanisms responsible for HCN channel dysregulation in TLE are unclear. Here we investigated whether changes in TRIP8b phosphorylation are sufficient to alter HCN channel function. We identified a phosphorylation site at residue Ser237 of TRIP8b that enhances binding to HCN channels and influences channel gating by altering the affinity of TRIP8b for the HCN cytoplasmic domain. Using a phosphospecific antibody, we demonstrate that TRIP8b phosphorylated at Ser237 is enriched in CA1 distal dendrites and that phosphorylation is reduced in the kainic acid model of TLE. Overall, our findings indicate that the TRIP8b–HCN interaction can be modulated by changes in phosphorylation and suggest that loss of TRIP8b phosphorylation may affect HCN channel properties during epileptogenesis. These results highlight the potential of drugs targeting posttranslational modifications to restore TRIP8b phosphorylation to reduce excitability in TLE.
Keywords: ion channel, epilepsy, phosphorylation, protein–protein interaction, Ca2+/calmodulin-dependent protein kinase II (CaMKII), channelopathy, HCN, neuronal excitability, TLE, TRIP8b
Introduction
An estimated 3.4 million patients suffer from epilepsy in the United States, and temporal lobe epilepsy (TLE)2 represents one of the most common drug-resistant forms of the disorder (1–3). Although many patients with TLE respond to anti-epileptic drugs, one-third of individuals experience seizures that are refractory to medical treatment (4). Changes in ion channel function that contribute to the epileptic state are observed in individuals suffering from inherited epilepsies and in rodent models of TLE (5–10). Therefore, understanding ion channelopathy in the context of epilepsy may lead to new targets for drugs to treat this disorder.
Hyperpolarization-activated cyclic nucleotide-gatedtk; (HCN1–HCN4) channels are nonspecific cation channels that mediate the Ih current and are highly expressed in the hippocampus (11). HCN1 and HCN2 channels are enriched in the distal dendrites of CA1 pyramidal cells, where they limit neuronal excitability and regulate temporal summation and dendritic integration (12–15). The intracellular region of the HCN channel consists of a cyclic nucleotide-binding domain (CNBD) that binds cAMP and a C-terminal tail consisting of the amino acids serine, asparagine, and leucine (SNL), which are also involved in protein binding. Binding of cAMP to the CNBD speeds channel activation kinetics and depolarizes the activation voltage (16–19).
Within CA1 pyramidal neurons, the auxiliary subunit tetratricopeptide repeat–containing Rab8b-interacting protein (TRIP8b) regulates the subcellular location and function of HCN channels (20–25). TRIP8b is a cytosolic protein that directly binds to HCN channels at two distinct locations to hyperpolarize channel gating and facilitate dendritic trafficking (21, 26). Within the HCN channel intracellular C-terminus, TRIP8b binds to the CNBD, where it acts as a competitive antagonist to inhibit cAMP binding (16, 21, 27–29). The second TRIP8b–HCN interaction occurs between the TRIP8b tetratricopeptide repeat (TPR) domains and the HCN channel C-terminal tail (26, 30). Disrupting the CNBD binding interaction prevents TRIP8b-mediated hyperpolarization of channel gating (21, 26). Both binding sites are necessary for proper dendritic trafficking in vivo, and disruption of either the CNBD or C-terminal tail interaction precludes dendritic enrichment of HCN channels by TRIP8b (22). TRIP8b contains an alternatively spliced N terminus, and the isoforms are named according to included exons. Isoform 1a-4 enhances channel surface expression, dendritic trafficking, and current density (20, 22). In TRIP8b knockout (TKO) mice, there is significant loss of dendritic HCN channels with a concomitant reduction in Ih, highlighting the importance of TRIP8b for channel function (31).
Genetic screens of patients with epilepsy have identified several mutations in HCN channels and their auxiliary subunits (32–38). In rodents, reduced HCN1 expression results in enhanced neuronal excitability as well as increased seizure susceptibility and seizure-related death (15, 39–41). In animal models of TLE, there is loss of Ih, reduced surface expression, and hyperpolarization of channel activation in chronic epilepsy (42–46). Prior work demonstrated mislocalization of the channels throughout CA1 pyramidal cell dendrites that correlated with disruption of the TRIP8b–HCN interaction (47). These results raise the possibility that loss of TRIP8b function is responsible for HCN channel dysregulation in TLE. However, it has been unclear what molecular mechanism might be responsible for regulating the TRIP8b–HCN interaction in vivo. We hypothesized that posttranslational modifications of TRIP8b would limit binding to HCN channels and affect channel function.
HCN channel trafficking and gating can be modulated by changes in kinase or phosphatase activity, including calcium/calmodulin-dependent protein kinase II (CaMKII), p38 mitogen-activated kinase and PKC activity (48–50). During epileptogenesis, several alterations in kinase and phosphatase activity have been observed (43, 51, 52). Thus, we sought to determine whether changes in TRIP8b phosphorylation would affect HCN channel function and whether TRIP8b phosphorylation is dysregulated in TLE.
Through phosphoproteomics analysis, we and others identified a phosphorylation site on TRIP8b at residue Ser237 that is located within the TRIP8b domain critical for CNBD binding (53). We generated a phosphospecific antibody targeting residue Ser237 on TRIP8b and determined that TRIP8b is phosphorylated at Ser237 in areas of dendritic HCN channel enrichment. The phosphorylation of this site enhances TRIP8b binding to the HCN channel CNBD and is necessary for TRIP8b-mediated hyperpolarization of channel gating. Additionally, we determined that both phosphorylation of this residue and CaMKIIα activity are significantly reduced during epileptogenesis in the kainic acid model of TLE. This work demonstrates that restoring TRIP8b phosphorylation may enhance HCN channel function in the distal dendrites of the CA1 and reduce neuronal excitability in epilepsy.
Results
Identification of phosphorylation site Ser237 on TRIP8b
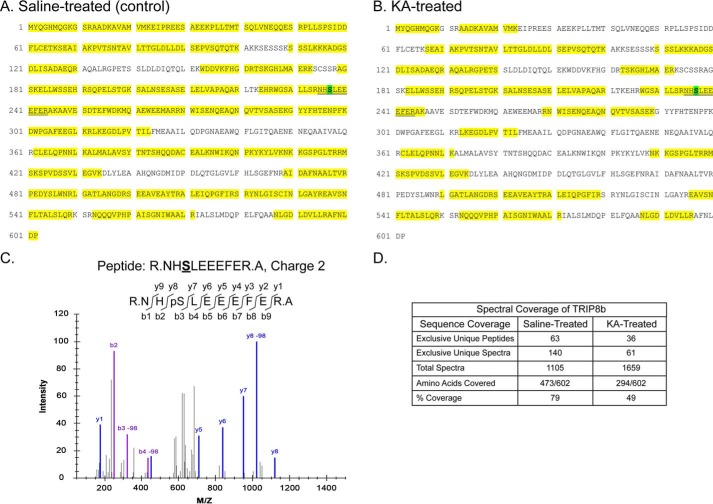
A previous global phosphoproteome analysis identified several possible phosphorylation sites on TRIP8b, including on residue Ser237 (53). We immunopurified TRIP8b from hippocampal lysates prepared from both a control rat and a rat subjected to kainic acid (KA)–induced seizures. We then subjected the immunopurified and gel-purified TRIP8b protein to LC-MS/MS. As shown in Fig. 1, we obtained extensive sequence coverage of the rat TRIP8b protein isoform 1a-4 (UniProt Q925N3) from both samples (473/602 aa or 79% from control and 294/602 or 49% from KA-treated). Fig. 1C shows a representative MS/MS spectrum of the peptide NHSLEEEFER (aa 235–244) that demonstrates unambiguous identification of in vivo phosphorylation of TRIP8b at Ser237. Phosphorylation at Ser237 was identified on TRIP8b purified from both control and KA-treated rats. The normalized spectral counts for the phosphopeptide containing Ser237 were present at a lower level in the KA-induced seizure sample than in the control sample (data not shown), suggesting that phosphorylation at this site may be affected by acute seizures.
Figure 1.
Identification of in vivo TRIP8b phosphorylation at Ser237. A and B, TRIP8b was immunopurified from hippocampal lysates prepared from saline (control) or KA-seized (Racine scale level 5) rats. TRIP8b immunoprecipitation products were analyzed by MS/MS. Spectral coverage of TRIP8b immunopurified from hippocampal lysates from saline-treated (A) and KA-treated (B) rats is highlighted in yellow. pSer237 is highlighted in green within the recovered tryptic peptide fragment, as indicated by the double underline. C, deconvolution of a representative pSer237-containing peptide MS/MS spectrum of the phosphopeptide NHpSLEEEFER derived from TRIP8b immunopurified from a hippocampal lysate prepared from a saline-treated rat. The fragmentation pattern of the peptide is shown, highlighting that the y8 peptide includes the phosphorylated Ser237 residue yielding a 98 m/z shift on the x axis. An m/z shift of +2 for Ser237 was detected with a confidence score of 10−7.6, indicating a low probability of mistaken identification of Ser237 phosphorylation. D, summary of the overall spectral coverage of TRIP8b from saline-treated and KA-treated rats, including the overall sequence coverage as shown in A and B.
Ser237 is located within a conserved “nano” domain of TRIP8b (residues 235–275) that is highly conserved across vertebrate species (Fig. 2, A and B). This previously identified nano domain directly binds to the CNBD of HCN channels and competes with cAMP binding (54). Additionally, removing a segment of TRIP8b that contains the nano domain prevented distal dendritic enrichment of HCN1 in vivo (22). Given that the Ser237 site is included within this functional domain, we hypothesized that Ser237 phosphorylation could play a key role in regulating the TRIP8b–HCN interaction.
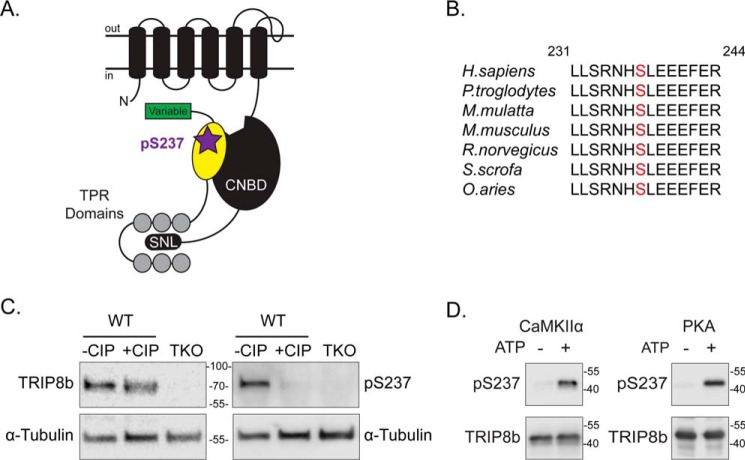
Figure 2.
Location of Ser237 on TRIP8b and phosphospecific antibody validation. A, schematic of one subunit of an HCN channel and TRIP8b. The intracellular CNBD of the HCN subunit and the C-terminal residues SNL are labeled. The unbound cAMP binding site is depicted on the CNBD. On TRIP8b, the N-terminal variable domain, the nano domain (yellow shape), and the C-terminal TPR domains are indicated. Serine 237 is indicated (purple star) within the nano domain of TRIP8b that interacts with the HCN CNBD. B, Ser237 (red text) is conserved in select vertebrates: Homo sapiens (human), Pan troglodytes (chimpanzee), Macaca mulatta (rhesus monkey), Mus musculus (mouse), Rattus norvegicus (rat), Sus scrofa (pig), and Ovis aries (sheep). TRIP8b residue numbers are based on isoform 1a-4 (UniProt Q925N3) as a reference. C, WT and TKO mouse hippocampal lysates incubated with or without CIP for 2 h at 37 °C, n = 4. Samples were immunoblotted with antibodies against TRIP8b, pSer237, and α-tubulin. D, purified TRIP8b(219–602) protein incubated with CaMKIIα or PKA with or without ATP at 30 °C for 30 min; n = 3. Molecular mass markers are shown in kilodaltons.
Generation and validation of the phosphospecific antibody pSer237
An antibody targeted against pSer237 (the phosphorylated TRIP8b residue Ser237) was generated using a 15-amino acid sequence that included the phosphorylated serine in the TRIP8b nano domain as the antigen (see “Experimental procedures”). The specificity of this antibody for the phosphorylated serine was confirmed in Western blots with native and dephosphorylated TRIP8b. Hippocampi from WT mice were incubated with or without the calf intestinal phosphatase (CIP), and the lysates were probed with antibodies targeting total-TRIP8b, pSer237, and α-tubulin (Fig. 2C). Although total TRIP8b was expressed in both samples, the signal from the pSer237 antibody was largely reduced with addition of the phosphatase. Additionally, the pSer237 antibody did not detect any protein in hippocampi from TKO mice (Fig. 2C). Thus, the pSer237 antibody is specific for the phosphorylated serine residue on TRIP8b.
Based on the TRIP8b sequence and known protein kinase motifs, we predicted that CaMKIIα and PKA would phosphorylate TRIP8b(Ser237) (55). We performed in vitro protein kinase assays to investigate this possibility by incubating purified TRIP8b protein with CaMKIIα or PKA, with or without ATP, at 30 °C for 30 min. The reaction mixtures were immunoblotted using antibodies against pSer237 and TRIP8b (Fig. 2D). Treatment of TRIP8b with CaMKIIα or PKA in the presence of ATP enhanced the pSer237 signal compared with the nonphosphorylated protein without ATP. These data demonstrate that both CaMKIIα and PKA can phosphorylate the Ser237 residue of TRIP8b in vitro.
Localization of phosphorylated TRIP8b in the brain
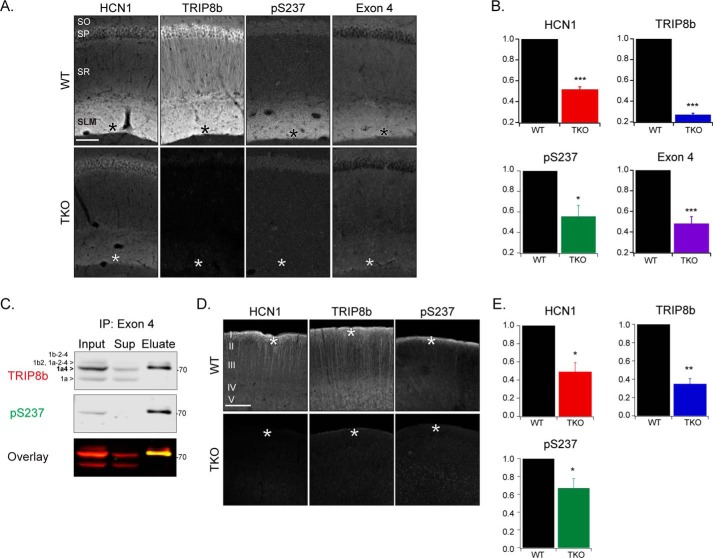
To determine the subcellular distribution of the phosphorylated TRIP8b protein, we performed immunohistochemistry. In the mouse hippocampus, we found that phosphorylated TRIP8b is enriched in the CA1 in a pattern similar to HCN1 and TRIP8b. The data show that HCN1, total TRIP8b, and phosphorylated TRIP8b are expressed in distal dendrites of CA1 pyramidal cells in the stratum lacunosum molecular layer (Fig. 3, A and B, asterisks) (HCN1 (WT: 1, TKO: 0.52 ± 0.03, Student's t test, p < 0.001, n = 5), TRIP8b (WT: 1, TKO: 0.27 ± 0.02, p < 0.001, n = 5), pSer237 (WT: 1, TKO: 0.56 ± 0.1, p = 0.01, n = 5), mean ± S.E.). TRIP8b isoform 1a-4, which enhances HCN channel dendritic trafficking in the hippocampus, has also been shown to be enriched in this location through use of an antibody targeting TRIP8b exon 4 (24) (Fig. 3, A and B) (exon 4 (WT: 1, TKO: 0.5 ± 0.1, p < 0.001, n = 6). Therefore, consistent with our LC-MS/MS results, we hypothesized that isoform 1a-4 in particular is phosphorylated at Ser237. We used an α-exon 4 antibody to immunoprecipitate TRIP8b isoforms containing exon 4 (the majority of which is isoform 1a-4 (23)) from mouse hippocampal lysate. The total TRIP8b antibody detected several TRIP8b isoforms that were present in the hippocampal input sample (Fig. 3C). However, the pSer237 antibody primarily detected one band that was enriched following immunoprecipitation with the α-exon 4 antibody. The pSer237 band corresponds to the isoform 1a-4 band and confirms that, indeed, isoform 1a-4 is significantly phosphorylated in the hippocampus (Fig. 3C).
Figure 3.
Immunohistochemical localization of pSer237 in the CA1 and neocortical distal dendrites. A, the CA1 region of WT and TKO mice probed with antibodies against HCN1, TRIP8b, pSer237, and TRIP8b exon 4. B, the distal dendritic region was quantified in each stain: HCN1 (WT: 1, TKO: 0.52 ± 0.03, Student's t test, p < 0.001, n = 5), TRIP8b (WT: 1, TKO: 0.27 ± 0.02, p < 0.001, n = 5), pSer237 (WT: 1, TKO: 0.56 ± 0.1, p = 0.01, n = 5), and exon 4 (WT: 1, TKO: 0.5 ± 0.1, p < 0.001, n = 6). C, mouse hippocampal extracts before (input) and after (eluate) immunoprecipitation (IP) with an α-TRIP8b exon 4 antibody. Lysates were immunoblotted with antibodies against TRIP8b and pSer237 (n = 3). D, neocortex region of WT and TKO mice probed with antibodies against HCN1, TRIP8b, and pSer237. E, the distal dendritic region was quantified in each stain: HCN1 (WT: 1, TKO: 0.5 ± 0.1, Student's t test, p = 0.01, n = 4), TRIP8b (WT: 1, TKO: 0.35 ± 0.06, p = 0.002, n = 4), and pSer237 (WT: 1, TKO: 0.7 ± 0.1, p = 0.04, n = 5). Scale bars = 100 μm; asterisks, distal dendritic region; SO, stratum oriens; SP, stratum pyramidale; SR, stratum radiatum; SLM, stratum lacunosum moleculare. Molecular mass markers are shown in kilodaltons. All error bars represent mean ± S.E.
Because HCN1 is also trafficked to the distal dendrites of the neocortex in a TRIP8b-dependent manner (31), we examined the retrosplenial and parietal association cortices (Fig. 3, D and E). We observed a similar pattern of pSer237 staining with greater expression in the distal dendrites in layer I (HCN1 (WT: 1, TKO: 0.5 ± 0.1, Student's t test, p = 0.01, n = 4), TRIP8b (WT: 1, TKO: 0.35 ± 0.06, p = 0.002, n = 4), pSer237 (WT: 1, TKO: 0.7 ± 0.1, p = 0.04, n = 5)). These results indicate that both the CA1 and neocortex express HCN1, TRIP8b, and pSer237 enrichment in distal dendrites.
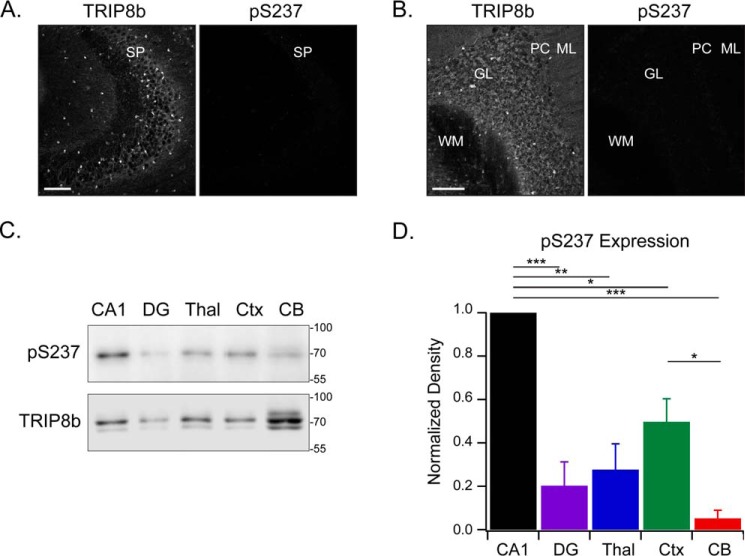
We then examined other areas with TRIP8b expression to determine whether the protein is phosphorylated at Ser237 throughout the brain. TRIP8b is expressed in oligodendrocytes of hippocampal area CA3 (24) and the granular layer of the cerebellum. TRIP8b Ser237 was not phosphorylated to a detectable degree in either region (Fig. 4, A and B). We noted that nonphosphorylated TRIP8b specifically colocalizes with HCN2 in cerebellar oligodendrocytes but not with HCN1 in the Purkinje cell layer (Fig. S1). The data suggest that TRIP8b phosphorylated at Ser237 is neuron-specific in brain regions that exhibit distal dendritic enrichment of HCN channels.
Figure 4.
Phosphorylated TRIP8b is absent from the CA3 and cerebellum but highly expressed in the CA1 region of the hippocampus. A and B, CA3 (A) and the cerebellum region (B) stained with antibodies against TRIP8b and pSer237 (n = 3). SP, stratum pyramidale; WM, white matter; GL, granular layer; PC, Purkinje cells; ML, molecular layer. C, protein lysate was loaded onto the gel from various mouse brain regions: 6 μg total lysate from the cornu ammonis 1 (CA1), dentate gyrus (DG), thalamus (Thal), and neocortex (Ctx) and 30 μg of total lysate from the cerebellum (CB). The brain regions were immunoblotted with the indicated antibodies. Note that different quantities of protein had to be loaded to obtain any pSer237 signal, and this led to predictable differences in the total TRIP8b loading control. D, quantification of C, with the pSer237 level normalized to total TRIP8b; one-way analysis of variance with Tukey's post hoc test (CA1, 1.0; dentate gyrus, 0.2 ± 0.1; thalamus, 0.3 ± 0.1; neocortex, 0.5 ± 0.1; cerebellum, 0.05 ± 0.04) with significant differences between the CA1 and dentate gyrus (p < 0.001), CA1 and thalamus (p = 0.001), CA1 and neocortex (p = 0.02), CA1 and cerebellum (p < 0.001), and neocortex and cerebellum (p = 0.03). n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001. Scale bars = 100 μm. Molecular mass markers are shown in kilodaltons. All error bars represent mean ± S.E.
To confirm the brain region–specific nature of phosphorylated TRIP8b, we investigated TRIP8b and pSer237 levels in several brain regions using immunoblotting (Fig. 4, C and D). These data show that phosphorylated TRIP8b is enriched in the CA1 region (relative pSer237 level to total TRIP8b, 1.0) compared with the levels found in the dentate gyrus (0.2 ± 0.1), thalamus (0.3 ± 0.1), neocortex (0.5 ± 0.1), and cerebellum (0.05 ± 0.04) (p < 0.05, n = 3). Taken together, these results demonstrate that TRIP8b isoform 1a-4 is phosphorylated at Ser237 and highly enriched in the distal dendrites of CA1 pyramidal cells.
Phosphorylated TRIP8b at Ser237 enhances binding to HCN channels
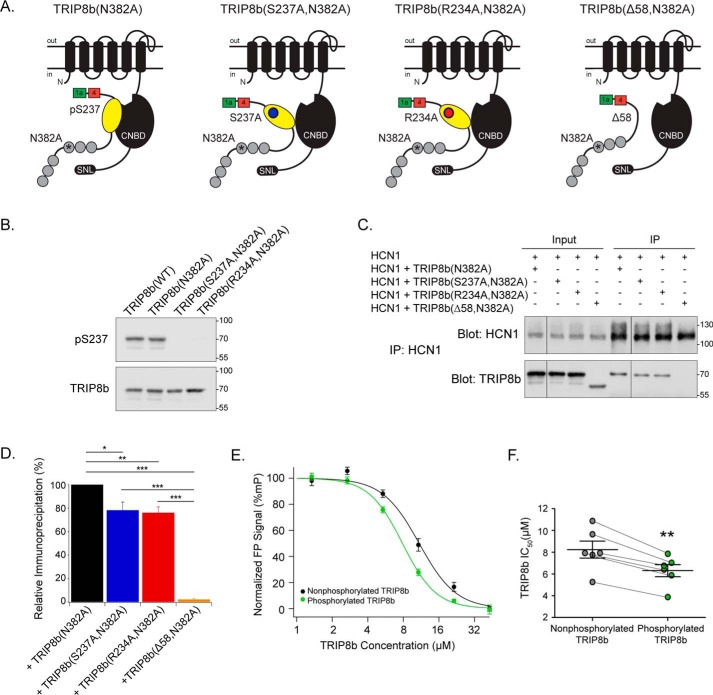
We reasoned that phosphorylation of Ser237 could regulate the strength of the TRIP8b–HCN interaction, given that the residue is located within the nano domain of TRIP8b responsible for CNBD binding. We investigated this possibility using coimmunoprecipitation. To isolate the effect of pSer237 specifically on CNBD binding, TRIP8b constructs with an ablated C-terminal tail TRIP8b–HCN interaction were used. These constructs contain a mutation of a key residue in the third TPR domain (N382A, previously referred to as TPR3-N13A) which disrupts the downstream interaction with the HCN channel C-terminal tail (21). TRIP8b constructs containing the N382A mutation alone, a phosphoablative mutation (S237A,N382A), a kinase consensus sequence mutation (R234A,N382A), or a negative control that disrupts both interaction sites (Δ58,N382A) were analyzed (21) (Fig. 5A). Because the arginine located at position 234 on TRIP8b is a critical residue in the consensus motifs for both CaMKIIα and PKA, the construct containing the R234A mutation is also predicted to prevent phosphorylation (phosphoablative) (56, 57). WT TRIP8b was basally phosphorylated in HEK293T cells, whereas both phosphoablative constructs show reduced signal from the α-pSer237 antibody (Fig. 5B). These constructs were transfected into HEK293T cells along with HCN1, HCN1 protein was immunoprecipitated from the lysates, and the level of TRIP8b bound to the channel was quantified (Fig. 5, C and D). Compared with TRIP8b(N382A), both phosphoablative constructs demonstrated reduced binding to HCN1 (relative enrichment of TRIP8b(N382A), 100; TRIP8b(S237A,N382A), 78.2 ± 7.0; TRIP8b(R234A,N382A), 76.1 ± 5.0; TRIP8b(Δ58,N382A), 2.1 ± 1.0; p < 0.05, n = 4).
Figure 5.
Phosphorylated TRIP8b at Ser237 enhances HCN1 CNBD binding. A, pictured is one subunit of an HCN channel with TRIP8b, indicating the TRIP8b mutant constructs used: TRIP8b(N382A), TRIP8b(S237A,N382A), TRIP8b(R234A,N382A), and TRIP8b(Δ58,N382A). The N382A mutation was described previously as TPR3-N13A (21). B, HEK293T cells were transiently transfected with TRIP8b(WT), TRIP8b(N382A), TRIP8b(S237A, N382A), or TRIP8b(R234A, N382A) constructs, and cell lysates were immunoblotted with antibodies against pSer237 and TRIP8b. The pSer237 antibody recognizes TRIP8b(WT) and TRIP8b(N382A), but both phosphoablative constructs demonstrate a reduced signal (n = 3). C, HEK293T cells were transiently transfected with the indicated constructs. HCN1 was immunoprecipitated (IP), and HCN1 and TRIP8b were immunoblotted. D, relative immunoprecipitation of HCN1 + TRIP8b(N382A): 100, +TRIP8b(S237A,N382A): 78.2 ± 7.0, +TRIP8b(R234A,N382A): 76.1 ± 5.0, +TRIP8b(Δ58,N382A): 2.1 ± 1.0. One-way analysis of variance with Tukey's post hoc test, n = 4. Significant differences identified between +TRIP8b(N382A) and +TRIP8b(S237A,N382A): p = 0.02, +TRIP8b(N382A) and +TRIP8b(R234A,N382A): p = 0.0098, +TRIP8b(N382A) and +TRIP8b(Δ58,N382A): p < 0.001, +TRIP8b(S237A, N382A) and +TRIP8b(Δ58,N382A), p < 0.001, +TRIP8b(R234A, N382A) and +TRIP8b(Δ58,N382A): p < 0.001. E, a fluorescence polarization assay was performed with fixed concentrations of CNBD387–591 (0.625 μm) and 8-f-cAMP (10 nm). Purified TRIP8b protein was incubated with CaMKIIα, with ATP (phosphorylated TRIP8b), or without ATP (nonphosphorylated TRIP8b) and titrated. F, quantification of IC50 with phosphorylated (6.3 ± 0.6 μm) and nonphosphorylated (8.2 ± 0.8 μm) TRIP8b, n = 6, p = 0.001; matched samples were analyzed with a paired Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. All error bars represent mean ± S.E. Molecular mass markers are shown in kilodaltons.
To confirm these results, we performed an additional in vitro fluorescence polarization binding assay to analyze the interaction between TRIP8b and the CNBD of HCN channels (58). This assay utilizes the change in polarization of a small, fluorescently tagged ligand (cAMP) whose polarization increases after binding a much larger protein (CNBD) (59). cAMP displacement serves as a readout of the affinity of TRIP8b binding because cAMP and TRIP8b compete for CNBD binding. We utilized fixed concentrations of FITC-tagged cAMP (8- f-cAMP, 10 nm) and the CNBD of HCN1 (CNBD387–591, 0.625 μm) and titrated increasing concentrations of a TRIP8b fragment conserved across all isoforms that includes the full nano domain through the C terminus of the protein (TRIP8b219–602). Prior to titration, TRIP8b219–602 was incubated with CaMKIIα, with or without ATP, at 30 °C for 30 min. Titrating increasing concentrations of phosphorylated TRIP8b increased the affinity of TRIP8b for the HCN1 CNBD (IC50, 6.3 ± 0.6 μm) compared with nonphosphorylated TRIP8b (8.2 ± 0.8 μm) (p = 0.001, n = 6; Fig. 5, E and F). There are no additional CaMKIIα motifs near the CNBD-binding region of TRIP8b. Thus, TRIP8b phosphorylated at Ser237 binds to the HCN1 CNBD to a greater degree than nonphosphorylated TRIP8b.
Phosphoablative TRIP8b constructs prevent hyperpolarization of HCN channel gating
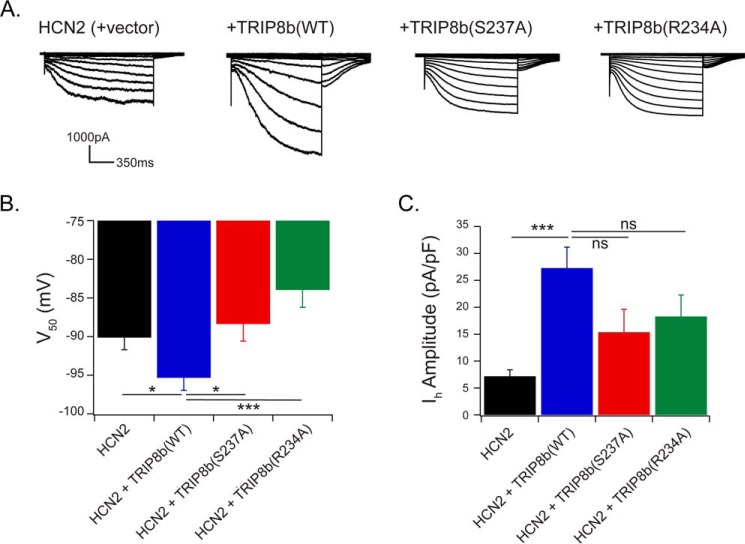
The change in affinity for HCN1 resulting from TRIP8b Ser237 phosphorylation could alter HCN channel gating or current density. To test this idea, we carried out patch clamp recordings from HCN channels. TRIP8b hyperpolarizes channel activation and enhances the current density of both HCN1 and HCN2 channels; hence, we used HEK293 cells that stably express HCN2 and exhibit robust currents. HEK293 cells (in which TRIP8b Ser237 is phosphorylated under basal conditions) stably expressing HCN2 were transfected with GFP and either TRIP8b(WT), TRIP8b(S237A), TRIP8b(R234A), or a control vector. As shown previously, expression of HCN2 with TRIP8b hyperpolarized the voltage of half activation (V50; HCN2 + vector: −90.1 ± 1.6 mV, HCN2 + TRIP8b(WT): −95.3 ± 1.6 mV; p < 0.05; n = 15, 13, respectively) (Fig. 6B). However, both phosphoablative constructs prevented TRIP8b-mediated hyperpolarization of channel gating (HCN2 + TRIP8b(S237A), −88.3 ± 2.2 mV; HCN2 + TRIP8b(R234A), −83.9 ± 2.3 mV; p < 0.05; n = 14, 15, respectively). Maximum tail current obtained following transfection with TRIP8b(WT) enhanced Ih amplitude compared with HCN2 alone (HCN2 + vector, 7.2 ± 1.2 pA/pF; HCN2 + TRIP8b(WT), 27.3 ± 3.9 pA/pF; p < 0.001; n = 15, 13, respectively). Both phosphoablative TRIP8b constructs generated a current density that was not significantly different from the TRIP8b(WT) condition but appears to be in an intermediate position between the HCN2 alone and +TRIP8b(WT) conditions (HCN2 + TRIP8b(S237A), 15.4 ± 4.3 pA/pF; HCN2 + TRIP8b(R234A), 18.3 ± 4.0 pA/pF; p > 0.05; n = 14, 15, respectively) (Fig. 6C). The data therefore support the interpretation that Ser237 phosphorylation is necessary for TRIP8b-mediated hyperpolarization of channel gating.
Figure 6.
TRIP8b with a phosphoablative mutation prevents hyperpolarization of HCN2 gating. A, representative current traces from HEK293 cells stably expressing HCN2 and transiently transfected with GFP and TRIP8b(WT), TRIP8b(S237A), TRIP8b(R234A), or control vector. Whole-cell recordings were performed with cells held at −40 mV in voltage clamp and stepped from −40 to −120 mV. B, quantification of V50; HCN2 + vector: −90.1 ± 1.6 mV (n = 15), HCN2 +TRIP8b(WT): −95.3 ± 1.6 mV (n = 13), HCN2 + TRIP8b(S237A): −88.3 ± 2.2 mV (n = 14), HCN2 + TRIP8b(R234A): −83.9 ± 2.3 mV (n = 15). Independent Student's t tests were performed to compare TRIP8b(WT) to each condition; +TRIP8b(WT) and + vector: p = 0.03, +TRIP8b(WT) and +TRIP8b(S237A): p = 0.02, +TRIP8b(WT) and +TRIP8b(R234A): p < 0.001. C, maximum tail current was divided by the cell capacitance to obtain Ih current amplitude for each condition. HCN2 + vector: 7.2 ± 1.2 pA/pF (n = 15), HCN2 + TRIP8b(WT): 27.3 ± 3.9 pA/pF (n = 13), HCN2 + TRIP8b(S237A): 15.4 ± 4.3 pA/pF (n = 14), HCN2 + TRIP8b(R234A): 18.3 ± 4.0 pA/pF (n = 15). Student's t tests were similarly performed to compare TRIP8b(WT) with each condition; +TRIP8b(WT) and +vector: p < 0.001, +TRIP8b(WT) and +TRIP8b(S237A): p = 0.054, +TRIP8b(WT) and +TRIP8b(R234A): p = 0.1. *, p < 0.05; **, p < 0.01; ***, p < 0.001. All error bars represent mean ± S.E.
TRIP8b Ser237 phosphorylation and CaMKIIα activity are reduced in epilepsy
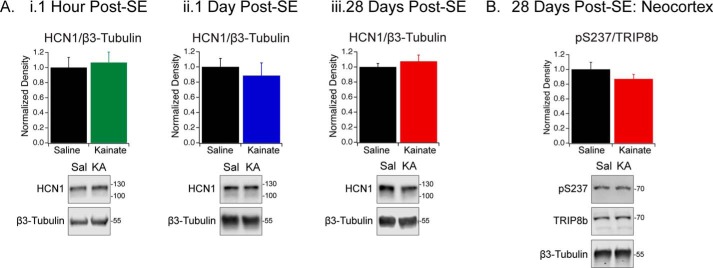
A prior report identified that total HCN1 protein is unchanged in chronic epilepsy; instead, the channels are mislocalized and the interaction with TRIP8b is disrupted (47). Using the KA model of TLE, we measured hippocampal HCN1 protein expression 1 h, 1 day, and 28 days post-status epilepticus (SE). Because seizures arise 2–4 weeks post-SE in this model, these time points correspond to the acute phase, the latent phase before spontaneous seizures arise, and the chronic phase exhibiting spontaneous seizures, respectively (60, 61). We observed no change in total HCN1 protein in the hippocampus during epileptogenesis (HCN1/β3-tubulin 1 h post-SE: Sal (1.0 ± 0.1, n = 5), KA (1.1 ± 0.1, n = 6), p > 0.05; 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.9 ± 0.2, n = 5), p > 0.05; 28 days post-SE: Sal (1.00 ± 0.04, n = 5), KA (1.1 ± 0.1, n = 5), p > 0.05) (Fig. 7A and Table S1).
Figure 7.
Hippocampal HCN1 protein expression and neocortical phosphorylated TRIP8b expression are unchanged during epileptogenesis. A, HCN1 protein was immunoblotted in hippocampal lysates harvested 1 h (i), 1 day (ii), and 28 days (iii) post-SE. HCN1 levels from Sal- and KA-treated rats were normalized to β3-tubulin. Immunoblot band density was quantified using an unpaired Student's t test; 1 Sal and 1 KA animal are shown. HCN1/β3-tubulin; 1 h post-SE: Sal (1.0 ± 0.1, n = 5), KA (1.1 ± 0.1, n = 6), p > 0.05; 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.9 ± 0.2, n = 5), p > 0.05; 28 days post-SE: Sal (1.00 ± 0.04, n = 5), KA (1.1 ± 0.1, n = 5), p > 0.05) (Table S1). Full blots are shown in Fig. S2. B, the neocortex was harvested from Sal- or KA-treated rats 28 days post-SE. Immunoblots were probed with antibodies against pSer237, TRIP8b, and β3-tubulin. Immunoblot band density was quantified using an unpaired Student's t test; 1 Sal and 1 KA animal are shown. pSer237/TRIP8b; 28 days post-SE: Sal (1.0 ± 0.1, n = 5), KA (0.9 ± 0.1, n = 5), p > 0.05 (Table S2). Full blots are shown in Fig. S2. All error bars represent mean ± S.E. Molecular mass markers are shown in kilodaltons.
Several kinases and phosphatases have altered activity during epileptogenesis, including increased calcineurin and reduced CaMKIIα activity (43, 51, 52). Thus, we analyzed the level of TRIP8b Ser237 phosphorylation to determine whether it is also altered during epileptogenesis. In the KA model of TLE, seizures are generated in the hippocampus and secondarily generalize to other brain regions, including the neocortex (62, 63). We quantified the level of Ser237 phosphorylation in the neocortex of rats in chronic epilepsy and observed no change in Ser237 phosphorylation 28 days post-SE (pSer237/TRIP8b 28 days post-SE: Sal (1.0 ± 0.1, n = 5), KA (0.9 ± 0.1, n = 5), p > 0.05) (Fig. 7B and Table S2).
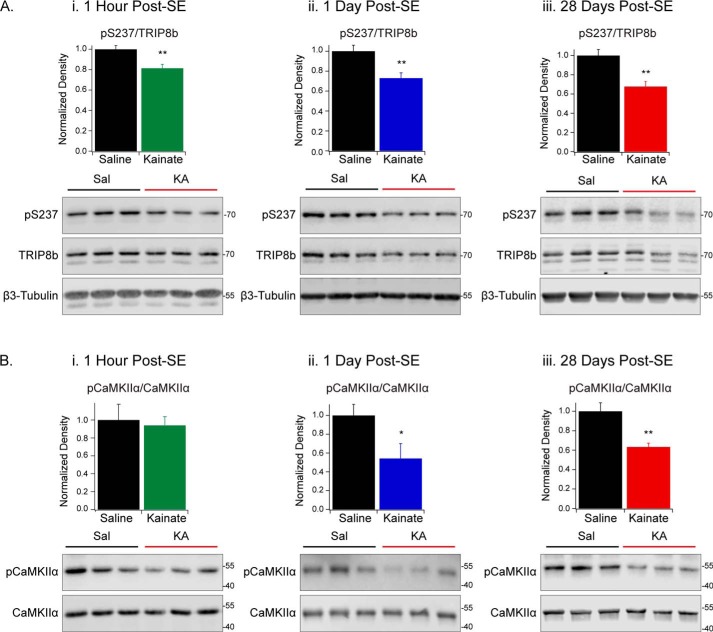
We then measured the level of TRIP8b phosphorylation at Ser237 in the hippocampus to determine whether changes take place within the epileptogenic zone. Immediately following SE, the level of phosphorylated TRIP8b at Ser237 was reduced in KA-treated animals compared with saline-treated animals (pSer237/TRIP8b, 1 h post-SE: Sal (1.00 ± 0.04, n = 5), KA (0.81 ± 0.04, n = 6), p < 0.01) (Fig. 8A, i, and Table S3). To estimate CaMKIIα activity, we used an antibody targeting CaMKIIα that is phosphorylated at Thr286, which corresponds to the activated kinase. Although the level of pThr286-CaMKIIα was unchanged, the loss of Ser237 phosphorylation could be a result of enhanced phosphatase activity that is reported following SE (pCaMKIIα/CaMKIIα 1 h post-SE: Sal (1.0 ± 0.2, n = 5), KA (0.9 ± 0.1, n = 6), p > 0.05) (Fig. 8B, i, and Table S3) (52). Both 1 day and 28 days post-SE, TRIP8b phosphorylation at Ser237 and pThr286-CaMKIIα were reduced in the hippocampus of KA-treated animals (pSer237/TRIP8b 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.7 ± 0.1, n = 5), p < 0.01; pSer237/TRIP8b 28 days post-SE: Sal (1.0 ± 0.1, n = 10), KA (0.7 ± 0.1, n = 11), p < 0.01; pCaMKIIα/CaMKIIα 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.5 ± 0.2, n = 5), p < 0.05); pCaMKIIα/CaMKIIα 28 days post-SE: Sal (1.0 ± 0.1, n = 5), KA (0.63 ± 0.04, n = 5), p < 0.01) (Fig. 8, A and B, and Table S3). These data establish that phosphorylated TRIP8b at Ser237 is reduced throughout the time course of epileptogenesis and that the change is localized to the epileptogenic zone.
Figure 8.
Reduction in TRIP8b phosphorylation and CaMKIIα activity in the KA model of TLE. A and B, hippocampi were harvested from Sal- or KA-treated rats 1 h (i), 1 day (ii), or 28 days (iii) post-SE. Immunoblots were probed with antibodies against pSer237, TRIP8b, and β3-tubulin (A) or pCaMKIIα and CaMKIIα (B). Immunoblot band density was quantified using unpaired Student's t test; 3 Sal and 3 KA animals are shown. A, pSer237/TRIP8b: 1 h post-SE: Sal (1.00 ± 0.04, n = 5), KA (0.81 ± 0.04, n = 6), p < 0.01; 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.7 ± 0.1, n = 5), p < 0.01; 28 days post-SE: Sal (1.0 ± 0.1, n = 10), KA (0.7 ± 0.1, n = 11), p < 0.01. B, pCaMKIIα/CaMKIIα, 1 h post-SE: Sal (1.0 ± 0.2, n = 5), KA (0.9 ± 0.1, n = 6), p > 0.05; 1 day post-SE: Sal (1.0 ± 0.1, n = 6), KA (0.5 ± 0.2, n = 5), p < 0.05); 28 days post-SE: Sal (1.0 ± 0.1, n = 5), KA (0.63 ± 0.04, n = 5), p < 0.01) (Table S3). *, p < 0.05; **, p < 0.01. Full blots are shown in Fig. S3. All error bars represent mean ± S.E. Molecular mass markers are shown in kilodaltons.
Discussion
In this study, we have shown that TRIP8b phosphorylation regulates HCN channel binding and function. TRIP8b binding results in a unique CNBD conformation, one that both hyperpolarizes gating and reduces cAMP affinity for the CNBD (20, 21, 26, 28). The identity of the specific amino acid residues responsible for the TRIP8b interaction with the CNBD has been the subject of several recent in vitro studies, one of which identified the nano domain as necessary and sufficient for TRIP8b binding to the HCN CNBD (54). Residue Ser237 is located within the nano domain, where it is predicted to influence the effect of TRIP8b on channel gating and trafficking (21, 22, 26, 29, 54).
We conclude that TRIP8b can be phosphorylated at residue Ser237 by CaMKIIα and PKA and that the presence of the phosphate group increases the affinity of TRIP8b for the HCN CNBD (Figs. 2D and 5). Our in vitro experiments using purified proteins produced results that were consistent with those seen in other expression systems, suggesting that additional proteins are unlikely to mediate the increased affinity. The negatively charged TRIP8b nano domain is thought to be drawn to the positively charged residues of the HCN CNBD. Immediately C-terminal to TRIP8b Ser237 is a series of negatively charged residues, EEEFE (239–243). Reversing the charge of this stretch has been shown previously to reduce TRIP8b binding to the HCN1 CNBD (64). Additionally, reversing the charge of two positively charged lysine residues (665 and 666) in the HCN2 CNBD reduces the binding affinity between the CNBD and TRIP8b (27). Thus, we reason that addition of a negatively charged phosphate group to Ser237 contributes to stabilization of TRIP8b binding to the CNBD. In addition to the effect of electrostatic interactions, dephosphorylation at Ser237 could interfere with the structural changes that naturally occur within the TRIP8b nano domain and the CNBD to reduce binding (54).
This work demonstrates that phosphorylation of Ser237 contributes to the high-affinity TRIP8b-HCN binding that is essential for dendritic trafficking and surface expression of HCN channels in the distal dendrites of CA1 pyramidal neurons. Distal dendritic enrichment of HCN1 in the hippocampus depends on an intact CNBD interaction (involving a stretch of TRIP8b that includes Ser237) as well as CaMKIIα activity (22, 48). The homeostatic increase in Ih observed following long-term potentiation is also dependent on CaMKIIα activity, which raises the possibility that Ser237 phosphorylation may be enhanced and contribute to Ih up-regulation in these instances (65). Because both TRIP8b-HCN binding sites are necessary for successful dendritic trafficking, there may be either a minimum strength of interaction required or some essential conformation of the proteins that is modulated by phosphorylation. The strength of the two TRIP8b-binding interactions involves allosteric modulation by TRIP8b, whereby binding to HCN at either site increases the affinity of the other (58). Thus, the loss of Ser237 phosphorylation we observed in epilepsy may not only affect the CNBD interaction but propagate downstream to weaken the C-terminal tail interaction as well. The role of phosphorylated Ser237 in dendritic channel enrichment is supported by data indicating that TRIP8b is not phosphorylated in regions containing somatic or presynaptic HCN channels but specifically phosphorylated in cells exhibiting dendritic channel enrichment. One possibility not addressed here is that Ser237 phosphorylation could be a consequence rather than a cause of dendritic channel targeting. Additional experiments will be necessary to distinguish whether Ser237 phosphorylation regulates both dendritic channel targeting and surface trafficking or only one aspect of TRIP8b function.
TRIP8b binding to the HCN channel CNBD generates hyperpolarization of channel activation, but the net result is enhanced channel trafficking and increased current density in CA1 (21, 22). Our electrophysiology data demonstrate that TRIP8b phosphorylation at Ser237 is necessary for hyperpolarization of channel gating. Phosphoablative TRIP8b constructs prevent this shift in activation voltage, which is consistent with our in vitro binding experiments that indicate that these constructs have a lowered affinity for the CNBD.
Implications for TLE
We observed that Ser237 phosphorylation was altered in a rat model of TLE, raising the possibility that these changes could be involved in producing HCN channel dysfunction. Both pilocarpine and KA models of TLE demonstrate reduced HCN channel function and dendritic expression in chronic epilepsy (42, 44, 46, 47). We noted that 1 h, 1 day, and 28 days post-SE, TRIP8b phosphorylation is significantly reduced. Given that Ser237 is dephosphorylated within an hour of SE, it is possible that this abnormality is essential to epileptogenesis. Although the loss of phosphorylation 1 h post-SE cannot be explained by a reduction in CaMKIIα activity, it may be due to increased calcineurin activity, which has been observed following SE (52). However, loss of TRIP8b phosphorylation during the latent and chronic phases correlates with reduced CaMKIIα activity and may be maintained by dysregulation of this kinase pathway. The reduction of TRIP8b phosphorylation at early time points may initiate dissociation of the TRIP8b–HCN complex that is observed in chronic epilepsy, leading to reduced dendritic channel trafficking and Ih (47). We identified reduced phosphorylation in the hippocampus, but interestingly, phosphorylation levels remained stable in the neocortex in chronic epilepsy. The data suggest that the reduction of Ser237 phosphorylation is specific to the epileptogenic zone and may play a role in development of epilepsy in the hippocampus.
In summary, this report identifies a posttranslational modification of TRIP8b at residue Ser237 as a regulator of HCN channel function under physiological conditions and TLE. Restoring TRIP8b phosphorylation at this site may strengthen HCN1 channel binding and dendritic trafficking to enhance Ih in TLE.
Experimental procedures
Immunoprecipitation for mass spectrometry
All animal use procedures were in strict accordance with the Guide for the Care and Use of Laboratory Animals described by the National Institutes of Health and approved by the University of California Davis Institutional Animal Care and Use Committee. Adult female Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA), were injected with either saline or kainic acid to induce acute seizures using a single i.v. dose of 10 mg/kg. For KA-treated rats, behaviors were scored according to the Racine scale, and rats were euthanized immediately after reaching level 5 seizures (66). Hippocampi from both treatment groups were harvested and homogenized on ice in 2 ml of ice-cold homogenization buffer (20 mm Tris-HCl (pH 8.0), 320 mm sucrose and protease inhibitors). The homogenate was then diluted to 0.5 mg/ml and solubilized in ice-cold buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 5 mm NaF, 1% Triton-X-100, 1 mm NaOV4, and protease inhibitors. Insoluble material was pelleted at 16,000 × g for 10 min at 4 °C and discarded. The resulting lysate was then subjected to immunoprecipitation overnight at 4 °C with a rabbit TRIP8b polyclonal antibody (generated against a C-terminal His-tagged fragment corresponding to aa 396–462 of TRIP8b). Immunopurified products were recovered by incubation with DynaBeads for 1 h at 4 °C, followed by three washes using ice-cold solubilization buffer and 1 wash with ice-cold detergent-free solubilization buffer. The beads were then removed to a fresh microfuge tube for nine more additional washes with ice-cold detergent free buffer. The final immunoprecipitation products were then eluted by boiling in 50 μl reducing SDS sample buffer.
Sample preparation for mass spectrometry
Immunoprecipitation products were resolved on a 7.5% SDS-PAGE gel and visualized by colloidal blue Coomassie stain. Bands corresponding to ≈66 kDa were excised for in-gel digestion for both saline- and KA-treated tissues as reported previously (66). Briefly, samples were destained with 50% acetonitrile and washed extensively with distilled water. Proteins were then reduced in 10 mm DTT at 56 °C for 1 h and alkylated in 5 mm iodoacetamide at room temperature for 30 min. Gel pieces were then washed and dehydrated prior to rehydration with trypsin solution (Promega, Madison, WI) at 12.5 ng/ml in 50 mm ammonium bicarbonate and incubated overnight at 37 °C. Digested peptide mixtures were extracted three times with 10% formic acid for 15 min on ice and 100% acetonitrile for 5 min at room temperature. Extracted peptides were desalted and stored on StageTip C18 at 4 °C (or room temperature) until LC-MS/MS.
LC-MS/MS analysis
LC-MS/MS analysis was performed at the UC Davis Proteomics Core Facility. LC separation was performed on a Nano Acquity UHPLC (Waters Corp.) with a Proxeon nanospray source. The digested peptides were reconstituted in 2% acetonitrile/0.1% TFA, and roughly 3 μg of each sample was loaded onto a 100 μm × 25 mm Magic C18 100 Å 5U reverse-phase trap column and desalted online before being separated on a 75 μm × 150 mm Magic C18 200 Å 3U reverse-phase column. Peptides were eluted using a gradient of 0.1% formic acid (A) and 100% acetonitrile (B) with a flow rate of 300 nL/min. A 120-min gradient was run with 5% to 35% B over 100 min, 35% to 80% B over 8 min, 80% B for 1 min, and 80% to 5% B over 1 min and finally held at 5% B for 10 min. Each of the gradients was followed by a 1-h column wash.
Mass spectra were collected on an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) in a data-dependent mode with one MS precursor scan followed by 15 MS/MS scans. A dynamic exclusion of 15 s was used. MS spectra were acquired with a resolution of 70,000 and a target of 1 × 106 ions or a maximum injection time of 30 ms. MS/MS spectra were acquired with a resolution of 17,500 and a target of 5 × 104 ions or a maximum injection time of 50 ms. Peptide fragmentation was performed using higher-energy collision dissociation with a normalized collision energy value of 27. Unassigned charge states as well as +1 and ions >+5 were excluded from MS/MS fragmentation.
Animals
All additional animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees of Northwestern University and Vanderbilt University. Adult male and female C57BL/6 mice (2–12 months) were obtained from The Jackson Laboratory (Ellsworth, ME). Adult male Sprague-Dawley rats (2–3 months) were obtained from Charles River Laboratories (Wilmington, MA).
Phospho-specific antibody pSer237
Phosphorylated TRIP8b antibody was generated by YenZym Antibodies, LLC (South San Francisco, CA). The immunogen used was a 15-amino acid peptide spanning residues 234–247 (RNHpSLEEEFERAKA) containing the phosphorylated serine residue. Rabbits were immunized with the synthesized peptide conjugated to keyhole limpet hemocyanin, and the elicited antiserum was subjected to peptide affinity absorption and purification to isolate the phosphospecific antibody components. The antibody is referred to as pSer237. All TRIP8b residues described in this manuscript use the Rattus norvegicus isoform 1a-4 (UniProt Q925N3) as a reference sequence.
Immunoblotting
Protein extracts were obtained from mouse or rat brain regions as indicated, homogenized, and sonicated briefly on ice in 20 mm HEPES (pH 7.4), 100 mm NaCl, 5 mm EDTA, and 5 mm EGTA with Halt protease and phosphatase inhibitor mixture (ThermoFisher) and 1% Triton X-100 and then centrifuged at 21,000 × g for 10 min at 4 °C (Figs. 2C; 3B; 4C; 7, A, iii, and B; and 8, A, iii, and B, iii) or 20 mm HEPES (pH 7.4), 320 mm sucrose, 5 mm EDTA, and 5 mm EGTA with Halt protease and phosphatase inhibitor mixture (ThermoFisher) and then centrifuged at 750 × g for 10 min at 4 °C (Figs. 7A, i and ii, and 8, A, i and ii, and B, i and ii). Protein concentration was determined using a Bradford protein assay (ThermoFisher), and samples were boiled in Laemmli sample buffer (Bio-Rad) containing β-mercaptoethanol (BME) at 95 °C for 5 min. Immunoblotting was performed as described previously (47). The proteins were resolved by SDS-PAGE and transferred to Immobilon-P PVDF membranes (Millipore, Billerica, MA) and incubated with blocking solution for 1 h at room temperature (5% milk or 5% BSA and 0.1% Tween 20 in TBS (TBS-T)). Primary antibodies were diluted in blocking solution: mouse α-TRIP8b (1:1500, NeuroMab clone N212/17; RRID: AB_10698035), custom rabbit α-TRIP8b C terminus (1:1000), mouse anti-α-tubulin (1:1000, Millipore, 05-829; RRID: AB_310035), custom rabbit anti-pSer237 (1:1000, YenZym), mouse anti-total CaMKIIα (1:1500, ThermoFisher, MA1–048, RRID: AB_325403), rabbit anti-pThr286 CaMKIIα/β (1:1500, ThermoFisher, PA1-4614, RRID: AB_2259386), custom rabbit anti-HCN1 (1:1000), and mouse anti-β3-tubulin (1:10,000, Sigma-Aldrich, T2200, RRID: AB_262133). Primary antibodies were incubated at room temperature for 1 h or overnight at 4 °C. Secondary antibodies were prepared in blocking solution and incubated at room temperature for 1 h and imaged either using the near-IR detection system (Li-Cor Biosciences, IRDye 800CW, 680RD) or Pierce ECL substrate (ThermoFisher). Band intensities were quantified using Li-Cor ImageStudio software.
Phosphatase treatment
Hippocampal lysates from WT mice were incubated with 1.5 units/50-μl reaction of CIP (New England Biolabs) for 2 h at 37 °C before Western blotting as above.
HEK293T cell transfection and preparation
HEK293T cells were maintained in DMEM with 10% FBS and 1% penicillin streptomycin. Cells were plated into 6-well tissue culture plates and transiently transfected 24 h later with an empty vector or TRIP8b construct using TransIT-LT1 (MirusBio) according to the manufacturer's instructions at a ratio of 1:3, HCN:TRIP8b. The TRIP8b(S237A) mutation was generated by two-stage PCR amplification using 5′-ggaaccatgccttggaagaggagtttg-3′ and 5′-cttccaaggcatggttcctggagag-3′ and reinserted into pxEGFP-C1-TRIP8b at EcoRI/BamHI sites. The TRIP8b(N382) mutation was generated as described previously (21), and the TRIP8b(R234A) mutation was generated using the QuikChange XL II kit with 5′-gaagtgccctgctctccgcgaaccattccttggaag-3′ and 5′-cttccaaggaatggttcgcggagagcagggcacttc-3′ (Agilent). 48 h post-transfection, cells were lysed with TEEN-TX (50 mm Tris, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, and 0.1% Triton X-100) containing Halt protease and phosphatase inhibitor mixture (ThermoFisher). Cell lysates were sonicated briefly on ice and centrifuged at 21,000 × g for 10 min. The supernatant was then used for Western blotting as above.
Recombinant protein purification
TRIP8b(219–602) and HCN1 CNBD(387–591) were cloned and generated as described previously (58). Protein constructs were purified as described elsewhere using nickel-nitrilotriacetic acid chromatography (Qiagen, Hilden, Germany) (67). Protein purity was analyzed by Coomassie staining, and protein concentration was detected by Bradford assay (ThermoFisher).
In vitro protein kinase assay
Purified TRIP8b(219–602) (residue numbering based on mouse TRIP8b isoform 1a-4) was incubated with CaMKIIα enzyme (New England Biolabs, P6060), CaCl2, calmodulin, and CaMKIIα protein kinase buffer (according to the manufacturer's instructions) with or without 200 μm ATP for 30 min at 30 °C. Purified TRIP8b(219–602) protein was also incubated with PKA (New England Biolabs, P6000) and protein kinase buffer with or without 200 μm ATP for 30 min at 30 °C. The reactions were terminated in Laemmli sample buffer (Bio-Rad) containing BME and boiled at 95 °C for 5 min. The protein was resolved by SDS-PAGE and immunoblotted using the indicated antibodies.
Immunohistochemistry
Immunohistochemistry was performed as described previously (68). Mice were deeply anesthetized with isoflurane and transcardially perfused with ice-cold PBS followed by 4% paraformaldehyde. Brains were removed and post-fixed for 48 h at 4 °C. Coronal sections (30 μm) were made using a vibratome (Leica), and antigen retrieval was performed with 10 mm sodium citrate (pH 9.0) for 10 min at 80 °C. Slices were allowed to cool, washed with PBS, and immersed in blocking solution (5% normal goat serum and 0.03% Triton X-100 in PBS) for 1 h at room temperature with gentle agitation. The following primary antibodies were diluted in blocking solution and applied overnight at 4 °C: custom guinea pig anti-HCN1 (1:3000), mouse anti-TRIP8b (1:1000, NeuroMab, clone N212/17, RRID: AB_10698035), rabbit anti-pSer237 (1:1000, YenZym), custom guinea pig anti-HCN2 (1:1000), mouse anti-Olig2 (1:1000, Millipore, MABN50, RRID: AB_10807410), and mouse anti-TRIP8b exon 4 (1:1000, NeuroMab, clone N212/3, RRID: AB_10671951). Sections were then washed three times for 5 min each and incubated with fluorescently labeled secondary antibodies (Invitrogen) in blocking solution for 1 h at room temperature. Sections were then washed an additional three times with 4′,6-diamidino-2-phenylindole included in the final wash. The tissue was mounted on glass slides using PermaFluor (ThermoFisher). Images were acquired on an Olympus Fv10i confocal microscope (FluoView Software) using a ×10 or ×60 oil immersion objective with or without ×2 digital zoom. Imaging work was also performed at the Northwestern University Center for Advanced Microscopy, generously supported by NCI, National Institutes of Health CCSG P30 CA060553, awarded to the Robert H. Lurie Comprehensive Cancer Center, on a Nikon A1R confocal microscope using a ×20 objective. Images were also acquired in part through use of the Vanderbilt Cell Imaging Shared Resource (supported by National Institutes of Health Grants CA68485, DK20593, DK58404, DK59637, and EY08126). WT and KO tissues used for comparison were prepared simultaneously, and images were acquired and presented with identical settings using Fiji software.
Coimmunoprecipitation
HEK293T cells were maintained and transfected as described above. 48 h post-transfection, cells were washed with PBS and lysed in 600 μl of TEEN-TX buffer (50 mm Tris, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, and 0.1% Triton X-100) supplemented with Halt protease and phosphatase inhibitor mixture (ThermoFisher). For coimmunoprecipitation from hippocampal lysate, the hippocampus was dissected in ice-cold PBS and lysed in 20 mm HEPES (pH 7.4), 100 mm NaCl, 5 mm EDTA, and 5 mm EGTA with Halt protease and phosphatase inhibitor mixture (ThermoFisher) and 1% Triton X-100. Lysates were sonicated briefly on ice and centrifuged at 21,000 × g for 10 min. A 50-μl aliquot of the supernatant (labeled “input”) was added to 12.5 μl of Laemmli sample buffer (Bio-Rad) containing BME and boiled at 95 °C for 5 min. Custom guinea pig anti-HCN1 antibody (4–5 μl for HEK293T samples) or mouse anti-TRIP8b exon 4 (5 μl for hippocampal samples, NeuroMab, clone N212/3, RRID: AB_10671951) was added to 450 μl of the remaining lysate and rotated at 4 °C overnight. The next day, 40 μl of protein A/G agarose bead slurry (ThermoFisher) was washed three times in TEEN-TX, added to the antibody-coupled sample, and nutated for 2 h at 4 °C. The beads were then washed in TEEN-TX buffer five times, 5 min each time, nutating at 4 °C, and then eluted by boiling in 40 μl of Laemmli sample buffer (Bio-Rad) containing BME at 95 °C for 5 min. Samples were resolved by SDS-PAGE (HEK293T cells: 8 μl of input and eluate; hippocampi: 10 μl of input, supernatant, and eluate). Densitometry of band intensity was performed using Li-Cor ImageStudio software, and the level of TRIP8b elution was divided by HCN1 elution for each condition.
Fluorescence polarization
CNBD and 8-f-cAMP oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). The HCN1 CNBD fragment used spans residues 387–591 of the mouse HCN1 isoform and contains a maltose-binding protein tag to enhance protein purification yield. Fluorescence polarization experiments were performed as described previously with modifications (58). Prior to these experiments, TRIP8b protein was incubated with CaMKII enzyme (New England Biolab), CaCl2, CaM, and protein kinase buffer according to manufacturer's instructions with or without 200 mm ATP. A series of 2-fold TRIP8b dilutions was then titrated into 0.625 μm HCN1 CNBD peptide and 10 nm 8-f-cAMP with sterile filtered PBS and 1 mm DTT. Samples were loaded in triplicate in black 384-well microtiter plates (Corning, Corning, NY), and polarization measurements were obtained using a Tecan microplate reader (Tecan, Zurich, Switzerland) at the Structural Biology facility at Northwestern University. We acknowledge staff and instrumentation support from the Structural Biology Facility at Northwestern University, the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, and NCI, National Institutes of Health CCSG P30 CA060553. Curve fitting and data analysis were performed using Igor Pro (WaveMetrics). Curves were fit by the Hill equation based on a previous report (58).
Electrophysiology
Electrophysiology was performed as described in Ref. 20 with modifications. HEK293 cells stably expressing HCN2 were generated by transfecting cells with mouse HCN2 in the pcDNA3 plasmid using TransIT-LT1 (MirusBio) transfection reagent according to the manufacturer's protocol. 24 h post-transfection, G418 (0.5 mg/ml final concentration, Sigma-Aldrich) was added to the culture medium. Cells were maintained in G418 solution for 6 weeks for selection of cells stably expressing the pcDNA-HCN2 construct, and then aliquots were frozen in culture medium with 5% DMSO (Sigma) and stored in liquid nitrogen.
Cells were plated on autoclaved 12 mm-diameter German-glass coverslips coated with poly-l-lysine (0.1 mg/ml, Sigma) and washed with Hank's balanced salt medium (without calcium or magnesium). Cells were transiently transfected 18–24 h later with EGFP plus a control vector, TRIP8b(WT), TRIP8b(S237A), or TRIP8b(R234A), using Lipofectamine 2000 (according to the manufacturer's instructions with modifications). Lipofectamine exposure was limited to 60 min, followed by a rinse and replacement using preconditioned culture medium. Whole-cell recordings were performed 24–48 h after transfection with pipettes made from borosilicate glass using a vertical puller (Narishige) with a final resistance of ∼2.5–3.5 megaohms. Cells were held at −40 mV in voltage clamp and progressively stepped (−10 mV) from −40 to −120 mV. Extracellular solution consisted of 145 mm NaCl, 10 mm KCl, 10 mm glucose, 10 mm HEPES, 2 mm CaCl2, and 1 mm MgCl2 buffered to pH 7.4 and an osmolarity of 312–315 mOsmol. The intracellular pipette solution contained 135 mm potassium gluconate, 10 mm MgCl2, 0.1 mm CaCl2, 1 mm EGTA, 10 mm HEPES, and 2 mm Mg-ATP buffered to pH 7.3 and an osmolarity of 295–305 mOsmol. Maximal tail current amplitude populations measured from each cell were fit to the Boltzmann equation as described in Ref. 20. Maximal Ih current density was calculated by dividing the maximal tail current by the cell capacitance, as described in Ref. 24. Currents were recorded via WinWCP software (University of Strathclyde), a MultiClamp 700A amplifier (Molecular Devices), and a National Instruments USB6221 interface card. The sampling rate was 10 kHz. Voltage clamping and series resistance were monitored throughout each experiment, and cells were discarded if the series resistance rose during the experiment. Two-tailed t tests were used to compare each group with the TRIP8b(WT)-transfected condition to test hypotheses generated by a pilot experiment. Expression of TRIP8b(WT) and its aforementioned mutants during the electrophysiology experiments described above were confirmed by subjecting cell samples of the exact same transfected conditions described above to Western blotting using an α-TRIP8b antibody (data not shown).
Kainic acid injections
Rats were allowed to acclimate in the on-site animal facility for at least 1 week prior to the experiment with food and water ad libitum. SE was induced by repeated low-dose intraperitoneal injections of KA dissolved in sterile saline (5 mg/kg; Tocris, Ellisville, MO) every 30–45 min. To reduce mortality, rats received full doses (5 mg/kg) or half-doses (2.5 mg/kg) of KA according to seizure progression. Control rats (no KA) were given equivalent injections of sterile saline. Seizures were scored using a modified Racine scale (69): stage 1, behavioral arrest with facial automatisms; stage 2, head nodding; stage 3, forelimb clonus; stage 4, forelimb clonus with rearing; stage 5, rearing and falling. Status epilepticus was terminated as described previously (47) using diazepam (10 mg/kg i.p. injection or up to 15 mg/kg as needed to terminate SE) (70). To prevent dehydration and minimize discomfort, animals were given subcutaneous injections of warmed lactated Ringer's solution as needed and monitored closely in the days following SE induction. For the 1 h and 1 day post-SE experiments, 11 rats were injected with saline, and 14 rats were injected with KA (3 of 14 died in SE). These rats were divided into five control rats and six KA rats for the 1 h post-SE experiment and six control rats and five KA rats for the 1 day post-SE experiment. For the 28 days post-SE experiment, five rats were injected with saline, and eight rats were injected with KA (one rat died in SE, and two rats died prior to the 28-day time point), leaving five KA rats for analysis.
Statistics
Data were analyzed using Student's t test for two groups or one-way analysis of variance with Tukey's post hoc test for pairwise comparisons among three or more groups. Electrophysiology data were analyzed using Student's t test for comparison between TRIP8b(WT) and each additional condition in a pairwise manner as indicated. Data are represented as mean ± S.E. (*, p < 0.05, **, p < 0.01, ***, p < 0.001).
Author contributions
K. M. F., K. A. L., Y. H., R. J. H., and D. M. C. conceptualization; K. M. F., K. A. L., I. E. M., R. J. H., and D. M. data curation; K. M. F., K. A. L., I. E. M., R. J. H., D. M., and J. S. T. formal analysis; K. M. F., D. M., and J. S. T. writing-original draft; K. M. F., K. A. L., Y. H., I. E. M., R. J. H., D. M., J. S. T., G. T. S., and D. M. C. writing-review and editing; Y. H., G. T. S., and D. M. C. resources; Y. H., J. S. T., and D. M. C. funding acquisition; G. T. S. supervision.
Supplementary Material
Acknowledgment
We thank Dr. Brett Phinney at the University of California Davis Proteomics Core Facility.
This work was supported by the National Institutes of Health Grants RO1-NS059934, RO1MH106511, and R21MH113262 (to D. M. C.) and Vanderbilt Institute for Clinical and Translational Research Award VR52450 (to Y. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S3.
- TLE
- temporal lobe epilepsy
- CNBD
- cyclic nucleotide binding domain
- HCN channel
- hyperpolarization-activated cyclic nucleotide-gated channel
- TPR
- tetratricopeptide repeat
- TKO
- TRIP8b knockout
- CaMKII
- calcium/calmodulin-dependent protein kinase II
- aa
- amino acids
- CIP
- calf intestinal phosphatase
- SE
- status epilepticus
- Sal
- saline
- BME
- β-mercaptoethanol
- KA
- kainic acid
- RRID
- research resource identifier.
References
- 1. Téllez-Zenteno J. F., and Hernandez-Ronquillo L. (2012) A review of the epidemiology of temporal lobe epilepsy. Epilepsy Res. Treat. 2012, 630853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Engel J., Jr. (2001) Mesial temporal lobe epilepsy: what have we learned? Neuroscientist 7, 340–352 10.1177/107385840100700410 [DOI] [PubMed] [Google Scholar]
- 3. Zack M. M., and Kobau R. (2017) National and State Estimates of the Numbers of Adults and Children with Active Epilepsy: United States, 2015. MMWR Morb. Mortal Wkly. Rep. 66, 821–825 10.15585/mmwr.mm6631a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. French J. A. (2007) Refractory epilepsy: clinical overview. Epilepsia 48, 3–7 10.1111/j.1528-1167.2007.00992.x [DOI] [PubMed] [Google Scholar]
- 5. Bernard C., Anderson A., Becker A., Poolos N. P., Beck H., and Johnston D. (2004) Acquired dendritic channelopathy in temporal lobe epilepsy. Science 305, 532–535 10.1126/science.1097065 [DOI] [PubMed] [Google Scholar]
- 6. Depienne C., Trouillard O., Saint-Martin C., Gourfinkel-An I., Bouteiller D., Carpentier W., Keren B., Abert B., Gautier A., Baulac S., Arzimanoglou A., Cazeneuve C., Nabbout R., and LeGuern E. (2009) Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J. Med. Genet. 46, 183–191 [DOI] [PubMed] [Google Scholar]
- 7. Butler K. M., Moody O. A., Schuler E., Coryell J., Alexander J. J., Jenkins A., and Escayg A. (2018) De novo variants in GABRA2 and GABRA5 alter receptor function and contribute to early-onset epilepsy. Brain 141, 2392–2405 10.1093/brain/awy171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wierschke S., Lehmann T. N., Dehnicke C., Horn P., Nitsch R., and Deisz R. A. (2010) Hyperpolarization-activated cation currents in human epileptogenic neocortex. Epilepsia 51, 404–414 10.1111/j.1528-1167.2009.02275.x [DOI] [PubMed] [Google Scholar]
- 9. Straessle A., Loup F., Arabadzisz D., Ohning G. V., and Fritschy J. M. (2003) Rapid and long-term alterations of hippocampal GABAB receptors in a mouse model of temporal lobe epilepsy. Eur. J. Neurosci. 18, 2213–2226 10.1046/j.1460-9568.2003.02964.x [DOI] [PubMed] [Google Scholar]
- 10. Poolos N. P., and Johnston D. (2012) Dendritic ion channelopathy in acquired epilepsy. Epilepsia 53, 32–40 10.1111/epi.12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Notomi T., and Shigemoto R. (2004) Immunohistochemical localization of Ih channel subunits, HCN1–4, in the rat brain. J. Comp. Neurol. 471, 241–276 10.1002/cne.11039 [DOI] [PubMed] [Google Scholar]
- 12. Magee J. C. (1998) Dendritic hyperpolarization-activated currents modify the integrative properties of hippocampal CA1 pyramidal neurons. J. Neurosci. 18, 7613–7624 10.1523/JNEUROSCI.18-19-07613.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magee J. C. (1999) Dendritic Ih normalizes temporal summation in hippocampal CA1 neurons. Nat. Neurosci. 2, 848 10.1038/12229 [DOI] [PubMed] [Google Scholar]
- 14. Poolos N. P., Migliore M., and Johnston D. (2002) Pharmacological upregulation of h-channels reduces the excitability of pyramidal neuron dendrites. Nat. Neurosci. 5, 767–774 10.1038/nn891 [DOI] [PubMed] [Google Scholar]
- 15. Kim C. S., Chang P. Y., and Johnston D. (2012) Enhancement of dorsal hippocampal activity by knockdown of HCN1 channels leads to anxiolytic- and antidepressant-like behaviors. Neuron 75, 503–516 10.1016/j.neuron.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen S., Wang J., and Siegelbaum S. A. (2001) Properties of hyperpolarization-activated pacemaker current defined by coassembly of HCN1 and HCN2 subunits and basal modulation by cyclic nucleotide. J. Gen. Physiol. 117, 491–504 10.1085/jgp.117.5.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wainger B. J., DeGennaro M., Santoro B., Siegelbaum S. A., and Tibbs G. R. (2001) Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 411, 805–810 10.1038/35081088 [DOI] [PubMed] [Google Scholar]
- 18. Santoro B., Liu D. T., Yao H., Bartsch D., Kandel E. R., Siegelbaum S. A., and Tibbs G. R. (1998) Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell 93, 717–729 10.1016/S0092-8674(00)81434-8 [DOI] [PubMed] [Google Scholar]
- 19. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., and Gouaux E. (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205 10.1038/nature01922 [DOI] [PubMed] [Google Scholar]
- 20. Lewis A. S., Schwartz E., Chan C. S., Noam Y., Shin M., Wadman W. J., Surmeier D. J., Baram T. Z., Macdonald R. L., and Chetkovich D. M. (2009) Alternatively spliced isoforms of TRIP8b differentially control H channel trafficking and function. J. Neurosci. 29, 6250–6265 10.1523/JNEUROSCI.0856-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han Y., Noam Y., Lewis A. S., Gallagher J. J., Wadman W. J., Baram T. Z., and Chetkovich D. M. (2011) Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J. Biol. Chem. 286, 20823–20834 10.1074/jbc.M111.236125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han Y., Heuermann R. J., Lyman K. A., Fisher D., Ismail Q. A., and Chetkovich D. M. (2017) HCN-channel dendritic targeting requires bipartite interaction with TRIP8b and regulates antidepressant-like behavioral effects. Mol. Psychiatry 22, 458–465 10.1038/mp.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Santoro B., Piskorowski R. A., Pian P., Hu L., Liu H., and Siegelbaum S. A. (2009) TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron 62, 802–813 10.1016/j.neuron.2009.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piskorowski R., Santoro B., and Siegelbaum S. A. (2011) TRIP8b splice forms act in concert to regulate the localization and expression of HCN1 channels in CA1 pyramidal neurons. Neuron 70, 495–509 10.1016/j.neuron.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fisher D. W., Han Y., Lyman K. A., Heuermann R. J., Bean L. A., Ybarra N., Foote K. M., Dong H., Nicholson D. A., and Chetkovich D. M. (2018) HCN channels in the hippocampus regulate active coping behavior. J. Neurochem. 146, 753–766 10.1111/jnc.14539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santoro B., Hu L., Liu H., Saponaro A., Pian P., Piskorowski R. A., Moroni A., and Siegelbaum S. A. (2011) TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J. Neurosci. 31, 4074–4086 10.1523/JNEUROSCI.5707-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saponaro A., Pauleta S. R., Cantini F., Matzapetakis M., Hammann C., Donadoni C., Hu L., Thiel G., Banci L., Santoro B., and Moroni A. (2014) Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. U.S.A. 111, 14577–14582 10.1073/pnas.1410389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bankston J. R., DeBerg H. A., Stoll S., and Zagotta W. N. (2017) Mechanism for the inhibition of the cAMP dependence of HCN ion channels by the auxiliary subunit TRIP8b. J. Biol. Chem. 292, 17794–17803 10.1074/jbc.M117.800722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeBerg H. A., Bankston J. R., Rosenbaum J. C., Brzovic P. S., Zagotta W. N., and Stoll S. (2015) Structural mechanism for the regulation of HCN ion channels by the accessory protein TRIP8b. Structure 23, 734–744 10.1016/j.str.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bankston J. R., Camp S. S., DiMaio F., Lewis A. S., Chetkovich D. M., and Zagotta W. N. (2012) Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. U.S.A. 109, 7899–7904 10.1073/pnas.1201997109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis A. S., Vaidya S. P., Blaiss C. A., Liu Z., Stoub T. R., Brager D. H., Chen X., Bender R. A., Estep C. M., Popov A. B., Kang C. E., Van Veldhoven P. P., Bayliss D. A., Nicholson D. A., Powell C. M., et al. (2011) Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J. Neurosci. 31, 7424–7440 10.1523/JNEUROSCI.0936-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang B., Sander T., Craven K. B., Hempelmann A., and Escayg A. (2008) Mutation analysis of the hyperpolarization-activated cyclic nucleotide-gated channels HCN1 and HCN2 in idiopathic generalized epilepsy. Neurobiol. Dis. 29, 59–70 10.1016/j.nbd.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DiFrancesco J. C., Barbuti A., Milanesi R., Coco S., Bucchi A., Bottelli G., Ferrarese C., Franceschetti S., Terragni B., Baruscotti M., and DiFrancesco D. (2011) Recessive loss-of-function mutation in the pacemaker HCN2 channel causing increased neuronal excitability in a patient with idiopathic generalized epilepsy. J. Neurosci. 31, 17327–17337 10.1523/JNEUROSCI.3727-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nava C., Dalle C., Rastetter A., Striano P., de Kovel C. G., Nabbout R., Cancès C., Ville D., Brilstra E. H., Gobbi G., Raffo E., Bouteiller D., Marie Y., Trouillard O., Robbiano A., et al. (2014) De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 46, 640–645 10.1038/ng.2952 [DOI] [PubMed] [Google Scholar]
- 35. DiFrancesco J. C., Castellotti B., Milanesi R., Ragona F., Freri E., Canafoglia L., Franceschetti S., Ferrarese C., Magri S., Taroni F., Costa C., Labate A., Gambardella A., Solazzi R., Binda A., et al. (2019) HCN ion channels and accessory proteins in epilepsy: genetic analysis of a large cohort of patients and review of the literature. Epilepsy Res. 153, 49–58 10.1016/j.eplepsyres.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 36. Dibbens L. M., Reid C. A., Hodgson B., Thomas E. A., Phillips A. M., Gazina E., Cromer B. A., Clarke A. L., Baram T. Z., Scheffer I. E., Berkovic S. F., and Petrou S. (2010) Augmented currents of an HCN2 variant in patients with febrile seizure syndromes. Ann. Neurol. 67, 542–546 10.1002/ana.21909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marini C., Porro A., Rastetter A., Dalle C., Rivolta I., Bauer D., Oegema R., Nava C., Parrini E., Mei D., Mercer C., Dhamija R., Chambers C., Coubes C., Thévenon J., et al. (2018) HCN1 mutation spectrum: from neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain 141, 3160–3178 10.1093/brain/awy263 [DOI] [PubMed] [Google Scholar]
- 38. Bonzanni M., DiFrancesco J. C., Milanesi R., Campostrini G., Castellotti B., Bucchi A., Baruscotti M., Ferrarese C., Franceschetti S., Canafoglia L., Ragona F., Freri E., Labate A., Gambardella A., Costa C., et al. (2018) A novel de novo HCN1 loss-of-function mutation in genetic generalized epilepsy causing increased neuronal excitability. Neurobiol. Dis. 118, 55–63 10.1016/j.nbd.2018.06.012 [DOI] [PubMed] [Google Scholar]
- 39. Nolan M. F., Malleret G., Dudman J. T., Buhl D. L., Santoro B., Gibbs E., Vronskaya S., Buzsáki G., Siegelbaum S. A., Kandel E. R., and Morozov A. (2004) A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell 119, 719–732 10.1016/j.cell.2004.11.020 [DOI] [PubMed] [Google Scholar]
- 40. Santoro B., Lee J. Y., Englot D. J., Gildersleeve S., Piskorowski R. A., Siegelbaum S. A., Winawer M. R., and Blumenfeld H. (2010) Increased seizure severity and seizure-related death in mice lacking HCN1 channels. Epilepsia 51, 1624–1627 10.1111/j.1528-1167.2010.02554.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang Z., Walker M. C., and Shah M. M. (2009) Loss of dendritic HCN1 subunits enhances cortical excitability and epileptogenesis. J. Neurosci. 29, 10979–10988 10.1523/JNEUROSCI.1531-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jung S., Jones T. D., Lugo J. N. Jr, Sheerin A. H., Miller J. W., D'Ambrosio R., Anderson A. E., and Poolos N. P. (2007) Progressive dendritic HCN channelopathy during epileptogenesis in the rat pilocarpine model of epilepsy. J. Neurosci. 27, 13012–13021 10.1523/JNEUROSCI.3605-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jung S., Bullis J. B., Lau I. H., Jones T. D., Warner L. N., and Poolos N. P. (2010) Downregulation of dendritic HCN channel gating in epilepsy is mediated by altered phosphorylation signaling. J. Neurosci. 30, 6678–6688 10.1523/JNEUROSCI.1290-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jung S., Warner L. N., Pitsch J., Becker A. J., and Poolos N. P. (2011) Rapid loss of dendritic HCN channel expression in hippocampal pyramidal neurons following status epilepticus. J. Neurosci. 31, 14291–14295 10.1523/JNEUROSCI.1148-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McClelland S., Flynn C., Dubé C., Richichi C., Zha Q., Ghestem A., Esclapez M., Bernard C., and Baram T. Z. (2011) Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann. Neurol. 70, 454–464 10.1002/ana.22479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arnold E. C., McMurray C., Gray R., and Johnston D. (2019) Epilepsy-induced reduction in HCN channel expression contributes to an increased excitability in dorsal, but not ventral, hippocampal CA1 neurons. eNeuro 6, ENEURO.0036-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin M., Brager D., Jaramillo T. C., Johnston D., and Chetkovich D. M. (2008) Mislocalization of h channel subunits underlies H channelopathy in temporal lobe epilepsy. Neurobiol. Dis. 32, 26–36 10.1016/j.nbd.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shin M., and Chetkovich D. M. (2007) Activity-dependent regulation of h channel distribution in hippocampal CA1 pyramidal neurons. J. Biol. Chem. 282, 33168–33180 10.1074/jbc.M703736200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williams A. D., Jung S., and Poolos N. P. (2015) Protein kinase C bidirectionally modulates Ih and hyperpolarization-activated cyclic nucleotide-gated (HCN) channel surface expression in hippocampal pyramidal neurons. J. Physiol. 593, 2779–2792 10.1113/JP270453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poolos N. P., Bullis J. B., and Roth M. K. (2006) Modulation of h-channels in hippocampal pyramidal neurons by p38 mitogen-activated protein kinase. J. Neurosci. 26, 7995–8003 10.1523/JNEUROSCI.2069-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Churn S. B., Kochan L. D., and DeLorenzo R. J. (2000) Chronic inhibition of Ca2+/calmodulin kinase II activity in the pilocarpine model of epilepsy. Brain Res. 875, 66–77 10.1016/S0006-8993(00)02623-8 [DOI] [PubMed] [Google Scholar]
- 52. Kurz J. E., Sheets D., Parsons J. T., Rana A., Delorenzo R. J., and Churn S. B. (2001) A significant increase in both basal and maximal calcineurin activity in the rat pilocarpine model of status epilepticus. J. Neurochem. 78, 304–315 10.1046/j.1471-4159.2001.00426.x [DOI] [PubMed] [Google Scholar]
- 53. Lundby A., Secher A., Lage K., Nordsborg N. B., Dmytriyev A., Lundby C., and Olsen J. V. (2012) Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 3, 876 10.1038/ncomms1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saponaro A., Cantini F., Porro A., Bucchi A., DiFrancesco D., Maione V., Donadoni C., Introini B., Mesirca P., Mangoni M. E., Thiel G., Banci L., Santoro B., and Moroni A. (2018) A synthetic peptide that prevents cAMP regulation in mammalian hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. Elife 7, e35753 10.7554/eLife.35753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Obenauer J. C., Cantley L. C., and Yaffe M. B. (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 10.1093/nar/gkg584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. White R. R., Kwon Y. G., Taing M., Lawrence D. S., and Edelman A. M. (1998) Definition of optimal substrate recognition motifs of Ca2+-calmodulin-dependent protein kinases IV and II reveals shared and distinctive features. J. Biol. Chem. 273, 3166–3172 10.1074/jbc.273.6.3166 [DOI] [PubMed] [Google Scholar]
- 57. Rust H. L., and Thompson P. R. (2011) Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem. Biol. 6, 881–892 10.1021/cb200171d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lyman K. A., Han Y., Heuermann R. J., Cheng X., Kurz J. E., Lyman R. E., Van Veldhoven P. P., and Chetkovich D. M. (2017) Allostery between two binding sites in the ion channel subunit TRIP8b confers binding specificity to HCN channels. J. Biol. Chem. 292, 17718–17730 10.1074/jbc.M117.802256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rossi A. M., and Taylor C. W. (2011) Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6, 365–387 10.1038/nprot.2011.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drexel M., Preidt A. P., and Sperk G. (2012) Sequel of spontaneous seizures after kainic acid-induced status epilepticus and associated neuropathological changes in the subiculum and entorhinal cortex. Neuropharmacol. 63, 806–817 10.1016/j.neuropharm.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chauvière L., Doublet T., Ghestem A., Siyoucef S. S., Wendling F., Huys R., Jirsa V., Bartolomei F., and Bernard C. (2012) Changes in interictal spike features precede the onset of temporal lobe epilepsy. Ann. Neurol. 71, 805–814 10.1002/ana.23549 [DOI] [PubMed] [Google Scholar]
- 62. Sheybani L., Birot G., Contestabile A., Seeck M., Kiss J. Z., Schaller K., Michel C. M., and Quairiaux C. (2018) Electrophysiological evidence for the development of a self-sustained large-scale epileptic network in the kainate mouse model of temporal lobe epilepsy. J. Neurosci. 38, 3776–3791 10.1523/JNEUROSCI.2193-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lothman E. W., and Collins R. C. (1981) Kainic acid induced limbic seizures: metabolic, behavioral, electroencephalographic and neuropathological correlates. Brain Res. 218, 299–318 10.1016/0006-8993(81)91308-1 [DOI] [PubMed] [Google Scholar]
- 64. Hu L., Santoro B., Saponaro A., Liu H., Moroni A., and Siegelbaum S. (2013) Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J. Gen. Physiol. 142, 599–612 10.1085/jgp.201311013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fan Y., Fricker D., Brager D. H., Chen X., Lu H. C., Chitwood R. A., and Johnston D. (2005) Activity-dependent decrease of excitability in rat hippocampal neurons through increases in I(h). Nat. Neurosci. 8, 1542–1551 10.1038/nn1568 [DOI] [PubMed] [Google Scholar]
- 66. Baek J. H., Rubinstein M., Scheuer T., and Trimmer J. S. (2014) Reciprocal changes in phosphorylation and methylation of mammalian brain sodium channels in response to seizures. J. Biol. Chem. 289, 15363–15373 10.1074/jbc.M114.562785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Han Y., Lyman K. A., Clutter M., Schiltz G. E., Ismail Q. A., Cheng X., Luan C. H., and Chetkovich D. M. (2016) Method for identifying small molecule inhibitors of the protein-protein interaction between HCN1 and TRIP8b. J. Vis. Exp. 10.3791/54540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Heuermann R. J., Jaramillo T. C., Ying S. W., Suter B. A., Lyman K. A., Han Y., Lewis A. S., Hampton T. G., Shepherd G. M. G., Goldstein P. A., and Chetkovich D. M. (2016) Reduction of thalamic and cortical Ih by deletion of TRIP8b produces a mouse model of human absence epilepsy. Neurobiol. Dis. 85, 81–92 10.1016/j.nbd.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Racine R. J. (1972) Modification of seizure activity by electrical stimulation: II: motor seizure. Electroencephalogr. Clin. Neurophysiol. 32, 281–294 10.1016/0013-4694(72)90177-0 [DOI] [PubMed] [Google Scholar]
- 70. Hall A. M., Brennan G. P., Nguyen T. M., Singh-Taylor A., Mun H. S., Sargious M. J., and Baram T. Z. (2017) The role of Sirt1 in epileptogenesis. eNeuro 4, ENEURO.0301-16.2017 10.1523/ENEURO.0301-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.