Abstract
Over the past decade, thousands of long noncoding RNAs (lncRNAs) have been identified, many of which play crucial roles in normal physiology and human disease. LncRNAs can interact with chromatin and then recruit protein complexes to remodel chromatin states, thus regulating gene expression. However, how lncRNA-chromatin interactions contribute to their biological functions is largely unknown. Here, we collected and constructed an atlas of 188,647 lncRNA-chromatin interactions in human and mouse. All lncRNAs showed diverse epigenetic modification patterns at their binding sites, especially the marks of enhancer activity. Functional analysis of lncRNA target genes further revealed that lncRNAs could exert their functions by binding to both promoter and distal regulatory elements, especially the distal regulatory elements. Intriguingly, many important pathways were observed to be widely regulated by lncRNAs through distal binding. For example, NEAT1, a cancer lncRNA, controls 13.3% of genes in the PI3K-AKT signaling pathway by interacting with distal regulatory elements. In addition, “two-gene” signatures composed of a lncRNA and its distal target genes, such as HOTAIR-CRIM1, provided significant clinical benefits relative to the lncRNA alone. In summary, our findings underscored that lncRNA-distal interactions were essential for lncRNA functions, which would provide new clues to understand the molecular mechanisms of lncRNAs in complex disease.
Keywords: long noncoding RNA (long ncRNA, lncRNA); chromatin modification; epigenetics; gene regulation; transcription enhancer; survival
Introduction
Emerging evidence shows that more than 85% of the human genome is transcribed into RNA, whereas only about 2% is translated into proteins (1, 2) Among these noncoding transcripts, long noncoding RNAs (lncRNAs)5 have been implicated in an expanding number of biological and disease processes (3, 4). Deregulation of lncRNAs, such as HOTAIR and PVT1, has been observed in various cancers, representing a potential target for therapy in cancer and serving as an independent diagnosis and prognostic biomarkers (5–8). Despite intensive efforts to decipher lncRNA functions (9–12), understanding the detailed molecular mechanisms how lncRNAs elicit their functions remains, however, a major scientific challenge.
In general, lncRNAs can carry out their functions by diverse mechanisms, including signals, decoys, guides, and scaffolds (13). The ability to function as epigenetic modulators, which creates local chromatin states that facilitate or block the binding of other regulators through the recruitment of chromatin modifiers, has been extensively reported (13, 14). Recently, several high-throughput techniques were developed to generate the genome-wide map of lncRNA-chromatin interactions, including chromatin isolation by RNA purification (ChIRP-seq) (15), capture hybridization analysis of RNA targets (CHART-seq) (16), RNA antisense purification (RAP-seq) (17), and chromatin oligo affinity precipitation (ChOP-seq) (18). These deep sequencing-based experimental results globally deciphered the binding sites of a specific lncRNA in a high resolution, allowing to characterize its downstream molecular mechanisms. For example, using CHART-seq, Simon et al. (19) detected the high-resolution maps of Xist bindings on the X chromosome and revealed a hit-and-run mechanism of Xist during X-chromosome inactivation in mouse cell lines. Thus, comprehensive dissection of genome-wide lncRNA-chromatin interactions not only can help elucidate their functional roles but also provide important clues to understanding how lncRNAs contribute to the pathogenesis of complex disease through epigenetic modification.
Here, we prospectively collected publicly available ChIRP-seq (15), CHART-seq (16), RAP-seq (17), and ChOP-seq (18) data and identified 188,647 lncRNA-chromatin interactions in human and mouse. By combining a large scale of epigenetic profiling data, we comprehensively dissected the epigenetic patterns of lncRNA-binding sites. Our findings provided a new insight into the molecular mechanisms of lncRNAs, which could promote understanding of functions of lncRNAs and benefit clinical application in the future.
Results
Systematic identification of lncRNA-chromatin–binding sites
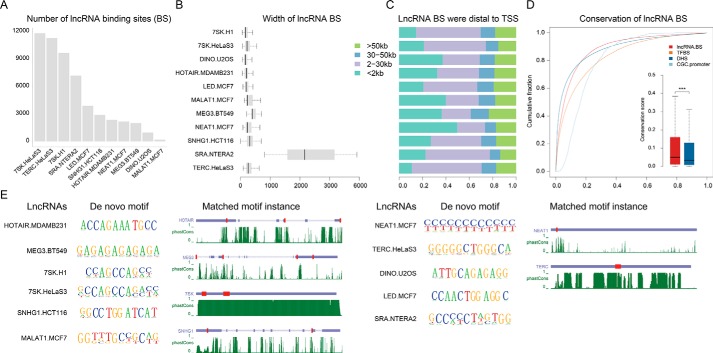
Through carefully literature searching, we collected 23 lncRNA-chromatin sequencing data ((ChIRP-seq (15), CHART-seq (16), and ChOP-seq (18)), involving 12 and 10 lncRNAs in human and mouse cell lines, respectively (Table S1). The majority of data contain two independent sequencing runs with “even” and “odd” probes. However, we observed striking differences in signaling intensity between even and odd signals (Fig. S1). To identify high-confidence lncRNA-binding sites, we adapted a method described by West et al. (20) by considering more stringent criteria (see “Experimental procedures” for details, Fig. S2). During this process, three lncRNAs (i.e. DACOR1, LincHSC2, and long noncoding RNAs transcribed by ERV-9 LTR retrotransposon) were removed due to lack of even and odd samples, low fold-enrichment, or lack of annotation in GENCODE version 27. As a result, a total of 77,031 and 111,616 lncRNA-chromatin interactions were identified in human and mouse, respectively (Fig. 1A and Fig. S3). On average, each lncRNA binds to about 6,420 and 12,401 sites in human and mouse, respectively (Fig. 1A and Fig. S3, “supporting Notes). For the human lncRNA 7SK with binding sites identified in H1 and HeLa-S3 cell lines, a high correlation was observed, indicating the high reproducibility of binding sites identified (Fig. S4). The average size of binding sites was smaller than 1 kb expected for SRA whose average size was around 2 kb (Fig. 1B). Most binding sites were distal to the transcription start sites (TSSs) of the closest genes (including protein-coding and noncoding genes) (Fig. 1C, Fig. S5). For these human lncRNAs, their binding sites showed significantly higher conservation than DNase I hypersensitive sites (DHSs) based on phatCons score (21) (p value <0.05, Wilcoxon rank-sum test, Fig. 1D) despite the overall low primary sequence conservation of lncRNAs (22). Motif discovery of lncRNA-binding sites revealed significant de novo DNA-binding motifs for each lncRNA using HOMER (23) (Fig. 1E), consistent with the findings in the previous studies (15, 18, 20). For example, both HOTAIR- and MEG3-binding sites were enriched in a GA-rich polypurine motif, which was identified as a shared feature of the polycomb response element (24). This supports the credibility of lncRNA-binding sites identified. In addition, these de novo motifs could match the primary sequences of lncRNAs and several motif instances occupied in the lncRNA sequence were identified (Fig. 1E, Table S2), which was probably due to the formation of DNA:RNA triplex (20, 24, 25).
Figure 1.
Systematic identification of lncRNA-chromatin–binding sites. A, the statistic of lncRNA- binding sites identified in human. B, lncRNA-binding sites in human were focal. C, most of the lncRNA-binding sites were distal to its closest genes. The binding sites occurred more than 2 kb from TSS were considered as distal. D, cumulative distributions of sequence conservation scores of lncRNA-binding sites, DHS, transcription factor-binding sites (TFBS), and promoters of CGC genes. The significance test was performed using a Wilcoxon rank-sum test (***, p value < 0.001; n.s., not significant, p value > 0.05). E, motif discovery and scanning of lncRNA-binding sites in human using HOMER and FIMO, respectively. For motif discovery, only the motif with the highest significant level was shown. For motif scanning, only the top 3 motif instance ranked by conservation were shown in the transcripts. The red boxes represented the matched motif instances in lncRNAs. The green lines represent conservation calculated by phastCons.
Diverse epigenetic modification patterns at lncRNA-binding sites
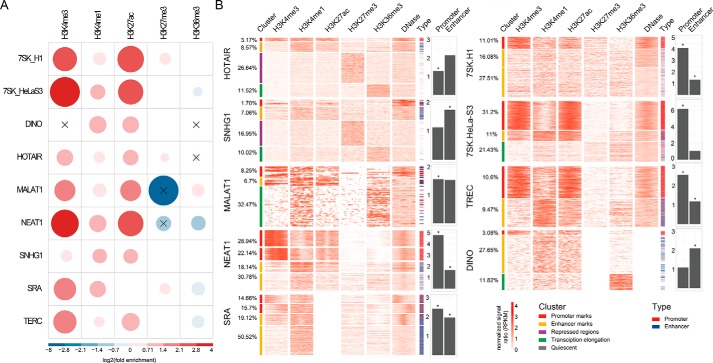
LncRNAs have emerged as key regulators of epigenetic modifications by recruiting chromatin-remodeling complexes to specific genomic loci (25). To explore the associations between lncRNA-binding sites and epigenetic modifications, we selected five core histone modifications (including H3K4me3, H3K4me1, H3K27ac, H3K27me3, and H3K36me3) with available data in the matched tissue/cell types (Table S3), and evaluated whether these histone markers were significantly enriched for lncRNA-binding sites using genome association tool (GAT) (26) (“Experimental procedures”). Only lncRNAs (9 human and 8 mouse lncRNAs) with at least two distinct epigenetic modification data available in the matched cell lines were used to analyze the associations between epigenetic marks and their DNA-binding sites. All of the lncRNAs showed diverse associations with at least two epigenetic marks (Fig. 2A and Fig. S6A). Nearly all human lncRNAs were significantly enriched in H3K4me3, associated primarily with active promoters (GAT, q-value <0.05), and all showed significant enrichments for H3K4me1 and H3K27ac, two markers for active enhancers (GAT, q-value <0.05).
Figure 2.
Complex regulatory role of lncRNAs at distinct genomic elements in human. A, fold-enrichment between lncRNA-binding sites and five core histone modifications were calculated by GAT and visualized using corrplot. The areas of circles show the value of log2-transformed fold-enrichment. The color intensity and the size of the circle are proportional to the value. The legend on the bottom side of the correlogram shows the log2-transformed fold-enrichment with their corresponding colors. The black cross in the circle means the association was not significant (GAT, q-value > 0.05). B, the cluster represents the groups of lncRNA-binding sites established by k-means classification using histone modification and open chromatin data. The heatmaps represent the normalized signal ratio of histone modification marks and open chromatin around lncRNA-binding sites at 10 bp resolution. The bars beside the heatmap represents the overlaps between lncRNA-binding sites and regulatory elements in the matched cell lines (blue for enhancers, red for promoters). The barplots represent the fold-enrichment between lncRNA-binding sites and regulatory elements determined by GAT in the matched cell lines (*, p value <0.05).
To further investigate the epigenetic patterns of lncRNA-binding sites, we calculated the levels of five core histone modifications and chromatin accessibility (DNase-seq) around lncRNA-binding sites (2 kb centered on the midpoint of binding peaks) at a 10-bp resolution in the matched cell lines (Table S3, “Experimental procedures”). By means of unsupervised k-means clustering analysis, we found that lncRNAs showed diverse epigenetic modification patterns at their binding sites, including promoter- (high H3K4me3), enhancer- (high H3K4me1 and low H3K4me3), repressed- (high H3K27me3), transcription elongation-associated (high H3K36me3), and quiescent patterns (low signal) (Fig. 2B and Fig. S6B). Focusing on a well-known lncRNA, HOTAIR, four distinct epigenetic modification patterns were observed. The majority of HOTAIR-binding sites (50.1%) were located within quiescent regions. A quarter of binding sites (26.64%) occurred at repressed regions (high H3K27me3) (Fig. 2B), consistent with previous reports that HOTAIR could mediate H3K27me3 by recruiting PRC2 complex (8). Notably, these binding sites also showed a moderate level of H3K4me1, indicating that many may represent poised enhancers. Interestingly, we also found 11.68% of HOTAIR-bindings sites exhibiting an active pattern (3.17% with high levels of H3K4me3 and H3K27ac; 8.57% with high levels of H3K4me1 and H3K27ac). As for NEAT1, the majority of binding sites (51.08%) showed high levels of H3K4me3, consistent with previous findings that NEAT1 substantially bound at promoters (24). Furthermore, we evaluated the associations between lncRNA-binding sites and regulatory elements (including promoters and enhancers) using GAT (26) (“Experimental procedures”). As a result, HOTAIR, MALAT1, NEAT1, SRA, 7SK, and TERC (both H1 and HeLa-S3 cell line) showed significant enrichments for promoters, whereas SNHG1, NEAT1, SRA, 7SK (H1 cell line), TERC, and DINO showed significant enrichments for enhancers in human (Fig. 2B). Taken together, our results showed diverse epigenetic modification patterns at lncRNA-binding sites, suggesting that lncRNAs may carry out more complex functions through regulating distinct types of regulatory elements.
Linking lncRNA-binding genomic regions with their potential target genes
Next, we explored the association between lncRNA-binding regions and potential target genes. For proximal lncRNA-binding regions, we directly assigned genes whose promoters (TSS ± 1 kb) overlap with lncRNA-binding regions to the corresponding lncRNAs as the promoter target genes (Fig. S7A, left). When lncRNAs interact with distal regulatory elements (>2 kb to nearest TSSs), we identified distal target genes by combining long-range chromosomal interaction data. Because there were not enough cell line-matched chromosomal interaction data available (only MCF7 and HeLa-S3), we aggregated long-range chromosomal interaction data from 24 cell types and tissues from the 4Dgenome database (27). After that, a total of 351,083 promoter-centered chromatin interactions (one end of an interaction should at least overlap with a promoter) were extracted. Finally, target genes of distal lncRNA-binding regions were determined by requiring distal lncRNA-binding regions interacting with gene promoters by a DNA loop (Fig. S7A, right).
As a result, we found that these lncRNAs had a large difference in the counts of target genes. An average of 3,100 protein-coding genes (PCGs) were identified for each lncRNA (Fig. S7B). LncRNA 7SK has the maximum number of target genes (8,051 PCGs), whereas MALAT1 has the minimum number of target genes (120 PCGs). Comparing promoter targets to distal targets, we found that, as expected, the numbers of distal targets were far more than promoter targets in most of the lncRNAs (the proportions of distal targets ranged from 53.5 to 94.4%). Interestingly, we observed significant overlaps between promoter and distal targets for NEAT1, MALAT1, TERC, SRA, 7SK (p value <10e-7, hypergeometric test, Fig. S8), suggesting that lncRNAs can target the same genes by binding to the promoter and distal regulatory elements. Furthermore, through analyzing publically available transcriptome dataset after knockdown of lncRNAs (MEG3, LED, DINO, NEAT1, and MALAT1, Table S4), a significant overlap between the deregulated genes by lncRNA manipulation and their target genes were found (p value <0.05, hypergeometric test, Fig. S9), suggesting that these lncRNAs can be involved in controlling or regulating the expression of their bound targets.
lncRNA-binding sites could explain lncRNA functions by binding to both promoters and distal regulatory elements
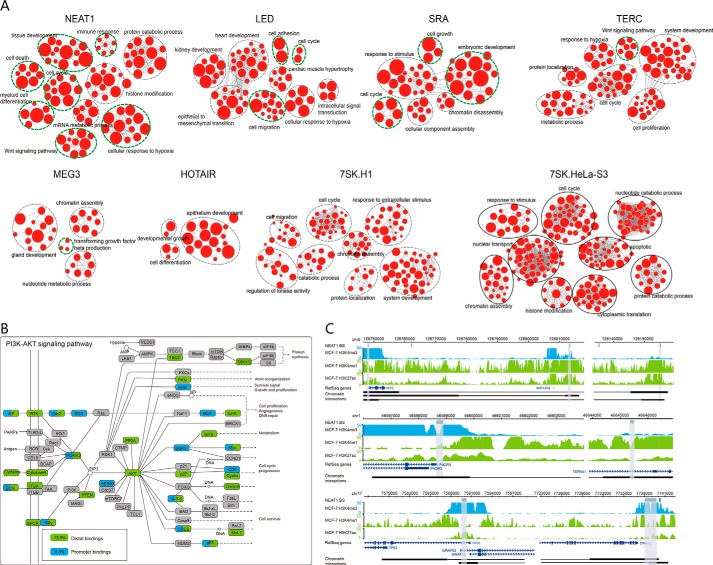
Subsequently, we performed GO and KEGG functional enrichment analysis for the target genes of each lncRNA. As a result, numerous biological processes were significantly enriched for some lncRNAs. Among these lncRNAs, NEAT1, LED, SRA, TERC, and MEG3 showed significant enrichments with their known functions (FDR < 0.05, hypergeometric test, Fig. 3A). Take the cancer-related lncRNA NEAT1 as an example (28), we found a mass of significantly enriched GO terms, involving cell differentiation, cell cycle, and cell death, further supporting previous observations (29–31). Similarly, KEGG pathway enrichment analysis also showed that the target genes could be enriched in various known pathways of lncRNAs (FDR < 0.05, hypergeometric test, Table S5), such as the PI3K-AKT signaling pathway (32) and MAPK signaling pathway (33) for NEAT1, transforming growth factor-β pathway for MEG3 (18), and WNT pathway for HOTAIR (34, 35) (Fig. S10, A and B).
Figure 3.
LncRNA bindings crucially contribute to lncRNA functions. A, functional enrichment results were visualized using Enrichmentmap plugin in Cytoscape. Node size was proportional to the size of the functional gene set. Clusters were manually circled and labeled. The green circles represented the known functions of lncRNAs. B, lncRNA NEAT1 may regulate the enriched KEGG pathway (PI3K-AKT signaling pathway) by both distal (13.3%, green) and promoter target genes (6.3%, blue). C, representative loci showed that target genes of NEAT1 could be targeted by both enhancers and promoter bindings, such as PIK3R3, MYC, and TP53.
To further understand how lncRNAs are involved in the regulation of pathways, we mapped target genes of lncRNAs to the enriched pathways. The PI3K-AKT signaling pathway plays a crucial role in cell cycle and cell survival and has been confirmed to be activated by NEAT1 (32). In the PI3K-AKT signaling pathway, about 17.0% (59/347) of the genes were associated with NEAT1-binding sites. Among these genes, 78.0% (46/59) are distal targets and 37.3% (22/59) are promoter targets (Fig. 3B). Notably, most of the upstream regulators (e.g. PI3KR3 and PTEN) and key downstream genes (e.g. GSK3B, MYC, CDK4, and TP53) in the PI3K-AKT signaling pathway were targeted by NEAT1, further supporting the crucial roles of NEAT1 in this pathway. Moreover, several target genes, such as MYC, PIK3R3, and TP53, were regulated by NEAT1 through binding to both enhancers and promoters (Fig. 3, B and C). Taken together, lncRNAs may exert their functions by binding both promoter and distal regulatory elements.
Prognostic implication of lncRNA-target genes in human cancer
LncRNAs could play crucial roles in cancer progression (36, 37), and some have been demonstrated to serve as prognostic factors, including HOTAIR, MALAT1, NEAT1, MEG3, and SNHG1 (38–42). Therefore, we re-evaluated the prognostic significance of seven lncRNAs (HOTAIR, LED, MALAT1, NEAT1, MEG3, TERC, and SNHG1) in cancer types matching with their detected cancer cells. We obtained gene (protein-coding genes and lncRNAs) expression data of 1,330 cancer patients derived from 3 tumor types (breast cancer, colorectal cancer, and cervical cancer) from the cancer genome atlas (TCGA). According to the median of lncRNA expression, the patients were divided into two groups (high-risk and low-risk groups). By log-rank test, we found that these lncRNAs did not significantly predict overall survival (OS) of cancer patients (Fig. 4A for HOTAIR, Fig. S11 for the other six lncRNAs). We thus wondered whether the prognostic benefits of lncRNAs may depend on their interacting targets. To address this issue, we evaluated the prognostic efficacy of pairs of lncRNAs and their target genes in cancers. At first, we divided cancer patients into four groups according to the median expression of lncRNAs and target genes: high expression of both lncRNAs and target genes; low expression of both of lncRNAs and target genes; high expression of lncRNAs and low expression of target genes; low expression of lncRNAs and high expression of target genes. Then, we evaluated the prognostic powers of pairs of lncRNAs and target genes (referring to distal and promoter target genes) by log-rank test (Fig. S12). We found that 193 pairs of HOTAIR-distal target genes, 674 pairs of LED-distal target genes, 22 pairs of MALAT1-distal target genes, 162 pairs of NEAT1-distal target genes, 98 pairs of MEG3-distal target genes, 988 pairs of TERC-distal target genes, and 315 pairs of SNHG1-distal target genes significantly predicting OS of patients (p value < 0.05, Table S6). We also found that 13, 51, 6, 135, 13, 361, and 23 lncRNA-promoter targets pairs in HOTAIR, LED, MALAT1, NEAT1, MEG3, TERC, and SNHG1, respectively, were significantly prognostic (p value < 0.05, Table S7).
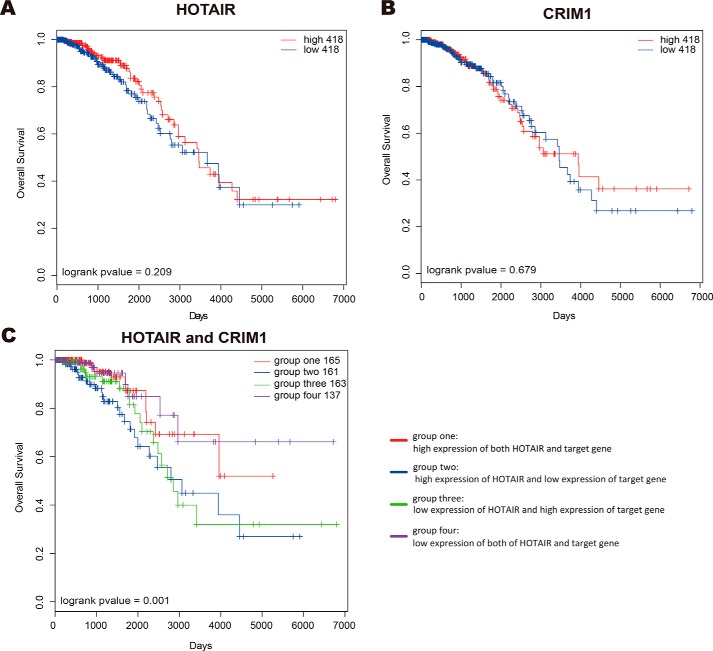
Figure 4.
Clinical benefits of HOTAIR and distal target CRIM1. A and B, Kaplan-Meier plot of overall survival of BRCA patients grouped by the median expression of HOTAIR and CRIM1, respectively. C, the BRCA patients were divided into four groups according to the median expression of HOTAIR and CRIM1: group one with high expression of HOTAIR and CRIM1 (red); group two with high expression of HOTAIR and low expression of CRIM1 (blue); group three with low expression of HOTAIR and high expression of CRIM1 (green); group four with low expression of both of HOTAIR and CRIM1 (purple).
To assess whether the survival prediction ability of these pairs of lncRNA-target genes was independent of other clinicopathologic factors in cancers, univariate and multivariable Cox regression analysis was performed. The covariables included age, gender, AJCC stage, and prognostic pairs of lncRNA-target genes. For HOTAIR, LED, MALAT1, NEAT1, MEG3, TERC, and SNHG1, there were 98, 205, 8, 54, 28, 289, and 80 lncRNA-distal target pairs, and 8, 20, 0, 37, 5, 97, and 3 lncRNA-promoter pairs independently predicting OS of patients (Tables S8 and S9). We also found that, among these significantly prognostic pairs, most distal/promoter target genes did not significantly predict OS of patients alone. For example, the combination of HOTAIR and its distal target gene CRIM1 was associated with prognosis in breast cancer (p value = 0.001, log-rank test, Fig. 4C). The breast cancer patients with high expression of HOTAIR and low expression CRIM1 had the worst survival, which remained statistically significant in multivariable Cox regression analysis with adjustment for age and AJCC tumor stage (HR: 2.078; 95% CI: 1.002–4.312, p value = 0.044, Table S10). In contrast, individual expression of either HOTAIR or CRIM1 was not sufficient to yield a statistically significant survival benefit (p value = 0.209 for HOTAIR, p value = 0.679 for CRIM1, log-rank test, Fig. 4B). Taken together, these observations suggested that a combination of lncRNAs with their targets could significantly improve the prognostic power of a single gene and could be potential powerful predictors of patient outcomes.
Discussion
To date, the understanding of the lncRNA mechanism that is essential for deciphering lncRNA functions is very limited. By integrating numerous lncRNA-chromatin interactions with a large number of epigenetic data, we found substantial binding of lncRNAs to the distal regulatory elements. Importantly, comprehensive analysis of biological functions and pathways showed that these distal binding events were highly responsible for lncRNA functions through chromatin loops. Many key components in important cancer pathways were targeted by lncRNAs via distal binding. To our knowledge, our results are the first to systematically underscore the importance of distal binding for lncRNA functions.
Epigenetic profiling of lncRNA-binding sites showed diverse epigenetic modification patterns, implying that lncRNAs had diverse chromatin preferences and were able to bind to different functional elements, including promoter-, enhancer-, repressed-, transcriptional elongation-associated and quiescent regions. As for HOTAIR, previous studies reported that HOTAIR could recruit PRC2 and LSD1 to the target gene promoters, inducing histone H3K27-trimethylation and H3K4-demethylation, and ultimately resulting in gene silencing (8, 25, 43). Consistently, our results showed that HOTAIR was enriched in promoters and H3K27me3 regions. In addition, about 11.74% of HOTAIR-binding sites showed significantly higher levels of H3K4me3, H3K4me1, and H3K27ac relative to the normal cell line. Because HOTAIR overexpression could also lead to loss of H3K27me3 at 34 gene promoters (8), HOTAIR might also have an active role in gene regulation, which should be tested in further studies. Recently, such versatile lncRNAs have been revealed by Wongtrakoongate et al. (44). They found that lncRNA SRA could interact with either TrxG or PRC2 to deliver either activating or silencing signals, or even both simultaneously, to establish bivalent domains. To explore the more comprehensive epigenetic modification patterns, it is very necessary to consider more epigenetic marks in the future (such as H3K27me1, H3K36me1, and H3K4me2).
Indeed, previous studies of Xist (X inactive specific transcript), one of the best-studied lncRNAs, reported that proper localization of Xist on chromatin is essential for its function (45). Perturbation of its RNA-binding domain or its interacting nuclear matrix protein SAFA can globally destroy the localization on the X chromosome, thus leading to loss of transcriptional silencing on the X chromosome (46). Similar phenomena were also observed in other lncRNAs. Our results further indicated that lncRNAs may substantially localize to DNA enhancers, and their functions may be highly associated with these lncRNA-enhancer interactions. By binding enhancers, lncRNAs could affect gene expression via spatial proximity, which is mediated by the 3D architecture of the genome (47).
LncRNA distal binding could greatly promote the understanding of lncRNA functions in cancers and provide clinical benefits. Our results showed that the known cancer lncRNA HOTAIR bound to enhancers of nearly all key genes, such as IGF1R, TRAF2, FGF family, WNT5A, DVL1, and CCND3, in cancer-associated pathways (including MAPK and WNT signaling pathway). Furthermore, some cancer-associated pathways were only regulated by lncRNA distal target genes, such as focal adhesion for DINO and Hippo signaling pathway for LED. Such substantial enhancer bindings offered new insights into how abnormal lncRNAs contribute to the pathogenesis of cancer. Moreover, the “two-gene” predictor, consisting of a specific lncRNA and its enhancer targets, such as HOTAIR-CRIM1 was strongly associated with the clinical outcome in breast cancer patients, which could be potential prognostic biomarkers. In contrast with previous studies (8), HOTAIR alone was insufficient to offer survival benefits. This improvement of prognosis may reflect the limitation of single lncRNA as prognostic biomarkers, further supporting the importance of lncRNA-enhancer interactions.
Notably, different numbers of lncRNAs-binding sites were identified for distinct lncRNAs. Different hybridization capture approaches and sequencing depth may affect lncRNA-binding sites. For different approaches, two previous studies compared the coverage of sequencing reads of roX2 CHART that uses a handful of short capture oligonucleotides and roX2 ChIRP signal that use a pool of oligonucleotides that tile to full-length of the RNA from S2 cells (48, 49). They found that different approaches for identification of roX2 chromatin interaction showed a high degree of similarity in overall signal profile. By down-sampling the sequence reads, we found that the number of lncRNA-binding sites increases rapidly as the depth of sequencing increases (Fig. S13A), consistent with previous reports (50). Higher depth of sequencing coverage is thus expected to identify more credible binding sites in the future. However, there was no significant correlation between the number of binding sites and the sequencing depth (Fig. S13, B and C). Especially, the lncRNAs with sufficient sequencing depth, such as NEAT1, MALAT1, and LED, only had the smallest number of peaks. The results might suggest that the differences in binding may be mainly dependent on the biological properties of specific lncRNAs.
Taken together, our results first revealed a comprehensive characterization of the diverse epigenetic patterns of lncRNA-binding sites and demonstrated the importance of lncRNA distal bindings for lncRNAs' functions. LncRNA-enhancer interactions should be integrative into the lncRNA-based functional studies, which could be of great value in revealing new mechanisms underlying the development of complex disease and identifying novel prognostic biomarkers.
Experimental procedures
Deep sequencing, peak calling, motif analysis, and visualization
In humans, 27 lncRNA-chromatin interaction data for 12 lncRNAs (ChIRP-, CHOP-, and CHART-seq), 52 ChIP-seq data for five histone modifications (H3K4me1, H3K4me3, H3K27ac, H3K27me3, and H3K36me3), and 8 DNase-seq data in 8 related tissue/cell types, were collected (Table S1). In mouse, 25 lncRNA-chromatin interaction data for 10 lncRNA, and 42 ChIP-seq data for five histone modifications, were collected (Table S3). For these sequencing data, raw reads were uniquely mapped to the reference genome (hg19 assembly) using Bowtie2 (version 2.0.6) (51). Peak detection was run using MACS2 (version 2.0.10.20130501, p value <1E-5) (52).
Identifying epigenetic modification sites
For H3K4me1, H3K4me3, and H3K27ac, gapped peaks were called, whereas for H3K27me3 and H3K36me3, broad peaks were called. The intersections between replicates for an epigenetic mark were extracted. Enhancers in a certain tissue/cell type were defined as the H3K4me1-gapped peaks more than 2 kb away from the TSSs annotated by GENCODE version 27 (53). Poised enhancers were defined as the enhancers overlapping with H3K27me3 peaks. Promoters were defined as 2-kb regions centered on the TSSs promoters determined by extending 1 kb in both directions from the TSS as defined.
Identifying lncRNA-binding sites
For ChIRP-, CHOP-, and CHART-seq, narrow peaks were called. Because a large difference between the two probe sets (even and odd probe sets) was observed, we adapted a method described by West et al. (20) to identify high-confidence lncRNA-binding sites. Additional criterion (Fig. S2) were further considered: 1) OverlapScore: the proportion of overlapping parts relative to the total peak length in even and odd samples. Thus, higher OverlapScores represent more reliable binding sites. OverlapScore should be larger than 0.5.
2) Considering the proximity of peaks between the even and odd samples, at least one peak summit from either the even samples or the odd samples should be located in their overlapping regions.
3) Similar signal intensities (the correlation coefficient >0.3) in the extended overlapping regions between the even and odd samples were required according to previous studies (15, 18, 54).
4) Due to anomalous, unstructured, or high signal in next-generation sequencing experiments independent of cell line or experiment, the binding regions should not overlap with the blacklist regions derived from ENCODE (55) and Memiya et al. (66) (https://sites.google.com/site/anshulkundaje/projects/blacklists).6
Specifically, for TERC, we considered the intersection between two conditions as the true peaks. For MEG3, as the raw data were not available (only merged bam could be downloaded from EBI database), we directly used the processed data offered by the author (Pearson correlation >0.3 and fold-enrichment against input >2). For lncRNA with more than two replicates (SHNG1 and SRA), we directly extracted the overlapping regions across all replicate samples using bedtools2 (version 2.0.6) (56). DACOR1 and LncRNA transcribed by ERV-9 LTR retrotransposon were excluded due to only one sample available and lack of annotation in GENCODE version 27 (53), respectively.
Sequence motif analysis
Sequences of the top 30% lncRNA-binding sites (ranked by fold-enrichment) within ±200 bp around peak summits were extracted and motif discovery analysis against these regions was performed using HOMER (23). Only motifs of the highest significance were reported. Motif locations were scanned in the lncRNAs exons using FIMO (57).
Evolutionary conservation analysis
The evolutionary conservation analysis was performed using UCSC 46-way phastCons vertebrate conserved elements (21).
Signal quantification and visualization
DeepTools2 (58) was used to create bigwigs by normalizing the ChIP-seq sample with the input samples of each cell line and the ratio of the normalized signal (RPKM) was calculated using the bamCompare function at 1 bp resolution. The signal heatmaps were made using the regions of ± 1kb around the midpoint of lncRNA-binding sites. For visualization purpose and the high correlation between even and odd samples (average PCC = 0.751), only one sample was shown in the heatmap.
Genomic intervals enrichment analysis
GAT (26) was used to assess the significances between lncRNA-binding sites and other genomic regions, including DNase I-hypersensitive regions, histone markers, enhancers, and promoters. 1000 simulations were performed with considering the different regional GC content.
Identified target genes of lncRNA-binding sites
The genes whose promoters were bound by lncRNAs were considered as promoter target genes. For distal lncRNA-bound regions (>2 kb to nearest TSS), we identified distal target genes by combining long-range chromosomal interaction data. First, we aggregated the chromatin interaction data across 24 cell types and tissues from the 4Dgenome database, involving 3,095,879 interactions among 17,239 genes (27). Then, 351,083 promoter-centered chromatin interactions (one end of an interaction should at least overlap with a promoter) were extracted. Interactions that connected two gene promoters were not used. Finally, target genes of distal lncRNA-binding regions were determined by requiring that distal lncRNA-binding regions interacted with gene promoters by the DNA loop.
GO and KEGG pathway enrichment analysis
Functional enrichment in Gene Ontology (GO) biological processes and KEGG pathways of lncRNA target genes were performed using the clusterProfiler R package (59), setting an FDR threshold of 0.05 for statistical significance. Visualization of GO enrichment was performed using Enrichment Map plugin version 2.0.1 (60) in Cytoscape version 3.7.0 (61). Similar GO terms were clustered together based on the similarity between each other using the overlap coefficient. Clusters were manually circled and labeled to highlight the prevalent biological functions among related GO terms. KEGG pathway visualization for relevant KEGG pathways was produced using the Bioconductor package Pathview (62).
Gene expression profiles data
We obtained protein-coding gene expression data (level 3) in 3 cancer types (breast cancer, colorectal cancer, and cervical cancer) as well-as the clinical data from the cancer genome atlas (TCGA) (63). The expression data of lncRNAs were collected from TANRIC (64). Genes and lncRNAs that had RPKM values above 0.1 in less than 90 and 30% of the samples were filtered out and removed from consideration, respectively (65).
Survival analysis
For survival analysis, overall survival was used as the end points. The Kaplan-Meier method was performed for visualization purposes and the differences between survival curves were calculated by a log-rank test. For lncRNA or target genes, patients were stratified into groups with high (median) and low (median) expression values. To explore the prognostic power of lncRNAs with their target genes, the patients were divided into four groups according to the median expression of lncRNAs and target genes: group one with high expression of lncRNAs and target genes; group two with high expression of lncRNAs and low expression of target genes; group three with low expression of lncRNAs and high expression of target genes; group four with low expression of both of lncRNAs and target genes. Univariate and multivariate Cox proportional hazards regression models were applied to estimate the prognostic benefits of pairs of lncRNAs-target genes. The p values smaller than 0.05 were considered to be statistically significant. All of the statistical analyses were performed using R software version 3.4.4.
Author contributions
G. Z., Y. L., and A. X. formal analysis; G. Z. visualization; G. Z. and L. X. writing-original draft; Y. L., A. X., J. S., H. Z., L. X., S. Z., and T. L. data curation; T. Z., Y. X., and X. L. supervision; T. Z., Y. X., and X. L. project administration; Y. X. and X. L. funding acquisition; Y. X. investigation.
Supplementary Material
This work was supported in part by the National Key R&D Program of China Grant 2018YFC2000100, National Natural Science Foundation of China Grants 31871336, 61873075, and 61573122, special funds for the construction of higher education in Heilongjiang Province Grant UNPYSCT-2016049, and Graduate Innovation Funds of Harbin Medical University Grant YJSCX2016-14HYD. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S13, Tables S1–S10, and notes.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- lncRNA
- long noncoding RNAs
- ChIRP-seq
- chromatin isolation by RNA purification
- CHART-seq
- capture hybridization analysis of RNA targets
- RAP-seq
- RNA antisense purification
- ChOP-seq
- chromatin oligo affinity precipitation
- TSS
- transcription start site
- DHS
- DNase I hypersensitive site
- GAT
- genome association tool
- PCG
- protein-coding gene
- OS
- overall survival
- GO
- Gene Ontology
- Xist
- X inactive specific transcript
- MAPK
- mitogen-activated kinase
- FDR
- false discovery rate.
References
- 1. Hangauer M. J., Vaughn I. W., and McManus M. T. (2013) Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9, e1003569 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G. K., Khatun J., et al. (2012) Landscape of transcription in human cells. Nature 489, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fatica A., and Bozzoni I. (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 4. Prensner J. R., and Chinnaiyan A. M. (2011) The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esteller M. (2011) Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 6. Prensner J. R., Iyer M. K., Balbin O. A., Chanasekaran S. M., Cao Q., Brenner J. C., Loxman B., Asangani I. A., Grasso C. S., Kominsky H. D., et al. (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29, 742–749 10.1038/nbt.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hessels D., Klein Gunnewiek J. M., van Oort I., Karthaus H. F., van Leenders G. J., van Balken B., Kiemeney L. A., Witjes J. A., and Schalken J. A. (2003) DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44, 8–15 10.1016/S0302-2838(03)00201-X [DOI] [PubMed] [Google Scholar]
- 8. Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., Wond D. J., Tsai M. C., Hung T., Argani P., Rinn J. L., Wang Y., et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu S. J., Horlbeck M. A., Cho S. W., Birk H. S., Malatesta M., He D., Attenello F. J., Villalta J. E., Cho M. Y., et al. (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu S., Li W., Liu J., Chen C. H., Liao Q., Xu P., Xu H., Xiao T., Cao Z., Peng J., et al. (2016) Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 34, 1279–1286 10.1038/nbt.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao T., Xu J., Liu L., Bai J., Xu C., Xiao Y., Li X., and Zhang L. (2015) Identification of cancer-related lncRNAs through integrating genome, regulome and transcriptome features. Mol. Biosyst. 11, 126–136 [DOI] [PubMed] [Google Scholar]
- 12. Wang L., Li J., Zhao H., Hu J., Ping Y., Li F., Lan Y., Xu C., Xiao Y., and Li X. (2016) Identifying the crosstalk of dysfunctional pathways mediated by lncRNAs in breast cancer subtypes. Mol. Biosyst. 12, 711–720 [DOI] [PubMed] [Google Scholar]
- 13. Mercer T. R., and Mattick J. S. (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- 14. Holoch D., and Moazed D. (2015) RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu C., Qu K., Zhong F. L., Arandi S. E., and Chang H. Y. (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 44, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simon M. D., Wang C. I., Kharchenko P. V., West J. A., Chapman B. A., Alekseyenko A. A., Borowsky M. L., Kuroda M. I., and Kingsont R. E. (2011) The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20497–20502 10.1073/pnas.1113536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engreitz J. M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E. S., Plath K., and Guttman M. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A. R., Hoberg E., Moustakas A., et al. (2015) MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6, 7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon M. D., Pinter S. F., Sarma K., Rutenberg-Schoenberg M., Bowman S. K., Kesner B. A., Maier V. K., Kingston R. E., and Lee J. T. (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West J. A., Davis C. P., Sunwoo H., Simon M. D., Sadreyev R. I., Wang P. I., Tolstorukov M. Y., and Kingston R. E. (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 55, 791–802 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hiller L. W., Richards S., et al. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnsson P., Lipovich L., Grandér D., and Morris K. V. (2014) Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071 10.1016/j.bbagen.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horard B., Tatout C., Poux S., and Pirrotta V. (2000) Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20, 3187–3197 10.1128/MCB.20.9.3187-3197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., Shi Y., Segal E., and Chang H. W. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heger A., Webber C., Goodson M., Ponting C. P., and Lunter G. (2013) GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29, 2046–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teng L., He B., Wang J., and Tan K. (2015) 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics 31, 2560–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng X., Zhang Y., Liu Y., Fang L., Li L., Sun J., Pan Z., Xin W., and Huang P. (2018) HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J. Cell. Biochem. 119, 3247–3256 10.1002/jcb.26481 [DOI] [PubMed] [Google Scholar]
- 29. Zeng C., Xu Y., Xu L., Yu X., Chang J., Yang L., Chen S., and Li Y. (2014) Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 14, 693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang P., Wu T., Zhou H., Jin Q., He G., Yu H., Xuan L., Wang X., Tian L., Sun Y., Liu M., and Qu L. (2016) Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J. Exp. Clin. Cancer Res. 35, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A. H., and Pierron G. (2014) NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183 10.1091/mbc.e13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu H., Li J., and Zhou Z. G. (2018) NEAT1 promotes cell proliferation in multiple myeloma by activating PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 6403–6411 [DOI] [PubMed] [Google Scholar]
- 33. Zhang F., Wu L., Quan J., Qu B., Xia S., La T., Wu Y., Ma J., Zeng J., Guo Q., Cui Y., et al. (2016) Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 75, 96–104 10.1016/j.jaut.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 34. Li J., Yang S., Su N., Wang Y., Yu J., Qiu H., and He X. (2016) Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/β-catenin pathway in human ovarian cancer. Tumour Biol. 37, 2057–2065 [DOI] [PubMed] [Google Scholar]
- 35. Guo X., Xiao H., Guo S., Li J., Wang Y., Chen J., and Lou G. (2019) Long noncoding RNA HOTAIR knockdown inhibits autophagy and epithelial-mesenchymal transition through the Wnt signaling pathway in radioresistant human cervical cancer HeLa cells. J. Cell. Physiol. 234, 3478–3489 [DOI] [PubMed] [Google Scholar]
- 36. White N. M., Cabanski C. R., Silva-Fisher J. M., Dang H. X., Govindan R., and Maher C. A. (2014) Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 15, 429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du Z., Fei T., Verhaak R. G., Su Z., Zhang Y., Brown M., Chen Y., and Liu X. S. (2013) Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kogo R., Shimamura T., Mimori K. Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., Miyano S., and Mori M. (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 [DOI] [PubMed] [Google Scholar]
- 39. Hirata H., Hinoda Y., Shahryari V., Deng G., Nakajima K., Tabatabai Z. L., Ishii N., and Dahiya R. (2015) Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 75, 1322–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen T., Wang H., Yang P., and He Z. Y. (2017) Prognostic role of long noncoding RNA NEAT1 in various carcinomas: a meta-analysis. Onco Targets Ther. 10, 993–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui X., Yi Q., Jing X., Huang Y., Tian J., Long C., Xiang Z., Liu J., Zhang C., Tan B., Li Y., and Zhu J. (2018) Mining prognostic significance of MEG3 in human breast cancer using bioinformatics analysis. Cell. Physiol. Biochem. 50, 41–51 [DOI] [PubMed] [Google Scholar]
- 42. Zhang M., Wang W., Li T., Yu X., Zhu Y., Ding F., Li D., and Yang T. (2016) Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed. Pharmacother. 80, 73–79 [DOI] [PubMed] [Google Scholar]
- 43. Ge X. S., Ma H. J., Zheng X. H., Ruan H. L., Liao X. Y., Xue W. Q., Chen Y. B., Zhang Y., and Jia W. H. (2013) HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 104, 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wongtrakoongate P., Riddick G. Fucharoen S., and Felsenfeld G. (2015) Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet. 11, e1005615 10.1371/journal.pgen.1005615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chu C., Zhang Q. C., da Rocha S. T., Flynn R. A., Bharadwaj J., Calabrese J. M., Magnuson T., Heard E., and Chang H. Y. (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hasegawa Y., Brockdorff N., Kawano S., Tsutui K., Tsutui K., and Nakagawa S. (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19, 469–476 [DOI] [PubMed] [Google Scholar]
- 47. Engreitz J. M., Ollikainen N., and Guttman M. (2016) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 48. Machyna M., and Simon M. D. (2018) Catching RNAs on chromatin using hybridization capture methods. Brief Funct. Genomics 17, 96–103 10.1093/bfgp/elx038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rutenberg-Schoenberg M., Sexton A. N., and Simon M. D. (2016) The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genomics Hum. Genet. 17, 69–94 [DOI] [PubMed] [Google Scholar]
- 50. ENCODE Project Consordium (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLos Biol. 9, e1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., Aken B. L., Barrell D., Zadissa A., Searle S., et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Léveillé N., Melo C. A., Rooijers K., Díaz-Lagares A., Melo S. A., Korkmaz G., Lopes R., Moqadam F. A., Maia A. R., Wijchers P. J., et al. (2015) Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 6, 6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quinlan A. R., and Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., and Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dundar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu G., Wang L. G., Han Y., and He Q. Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Merico D., Isserlin R., Stueker O., Emili A., and Bader G. D. (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Luo W., and Brouwer C. (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics, 29, 1830–1831 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cancer Genome Atlas Research, N., et al. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J. N., and Liang H. (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 75, 3728–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sanchez-Garcia F., Villagrasa P., Matsui J., Kotliar D., Castro V., Akavia U. D., Chen B. J., Saucedo-Cuevas L., Rodriguez Barrueco R., Llobet-Navas D., Silva J. M., and Pe'er D. (2014) Integration of genomic data enables selective discovery of breast cancer drivers. Cell 159, 1461–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Memiya H. M., Kundaje A., and Boyle A. P. (2019) The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.