Abstract
Observational and interventional studies for HIV cure research often use single-copy assays to quantify rare entities in blood or tissue samples. Statistical analysis of such measurements presents challenges due to tissue sampling variability and frequent findings of 0 copies in the sample analysed. We examined four approaches to analysing such studies, reflecting different ways of handling observations of 0 copies: (A) replace observations of 0 copies with 1 copy; (B) add 1 to all observed numbers of copies; (C) treat observations of 0 copies as left-censored at 1 copy; and (D) leave the data unaltered and apply a method for count data, negative binomial regression. Because research seeks to estimate general patterns rather than individuals’ values, we argue that unaltered use of 0 copies is suitable for research purposes and that altering those observations can introduce bias. When applied to a simulated study comparing preintervention to postintervention measurements within 12 participants, methods A–C showed more attenuation than method D in the estimated intervention effect, less chance of finding P < 0.05 for the intervention effect and a lower chance of including the true intervention effect within the 95% confidence interval. Application of the methods to actual data from a study comparing multiply-spliced HIV RNA among men and women estimated smaller differences by methods A–C than by method D. We recommend that negative binomial regression, which is readily available in many statistical software packages, be considered for analysis of studies of rare entities that are measured by single-copy assays.
Keywords: HIV, latent reservoir, rare entities, statistical bias
Introduction
Measurements of rare entities in blood or tissue samples are important tools in HIV-related cure research, particularly for assessing the latent reservoir, latency reversal and residual viraemia in effectively treated study participants [1–10]. Such assays include measurements of cell-associated unspliced [11] and multiply-spliced HIV RNA [7], integrated DNA [12,13], total DNA [12,14] and low levels of HIV RNA in plasma [15,16]. These differ from most measurements used in clinical research because they measure very rare entities, often with a substantial fraction of samples having no copies at all. Here, we use the term single-copy assay to mean a measurement method that quantifies how many copies are present in a tissue sample with reasonable precision even when there is zero or only one. Statistical analysis of data from such assays presents some specific issues summarised in Box 1.
Box 1. Statistical issues for single-copy assays.
-
1.
Tissue or blood sampling. The tissue sample or blood volume assayed is only a small fraction, often <0.1%, of what is present in the participant's body, which means that it will not perfectly represent what is true for the participant as a whole. For blood sampling, we might assume perfect mixing, so that the sample is a random fraction of the person's entire bloodstream, but this still leaves the number of copies present in the sample subject to random variation that follows a Poisson distribution. This variation is inevitable, even for an assay with no measurement error, and it can be quite substantial for the low copy numbers often sought by single-copy assays.
-
2.
Assay input varies. In many cases, the number of cells or volume of plasma assayed will differ for different samples. For example, cell yields may vary, depending on blood volume and the CD4+ T cell count, or a damaged tube or short blood draw may limit the volume of plasma available to be assayed for a particular participant. Statistical analyses will be more accurate if they account for this variation, or at least prevent it from introducing distortions.
-
3.
Precision varies. The inevitable sampling variation noted in point 1 cannot be assumed to be identical for all study samples. Instead, some will be more precise than others, depending on the number of copies found and the input to the assay. Accounting for such differences in precision can improve statistical analyses of research studies.
-
4.
Zero copies. Because of sampling variation and the rarity of the target entities, single-copy assays may indicate that no copies were present in some of the tissue samples assayed. This seems unacceptable to many investigators, because they know that some entities must have been present in the participant, even if absent from a particular sample. Zeroes are also problematic because interest often focuses on relative changes or differences. This leads to analysis on the logarithmic scale, where 0 copies would become infinite and so would preclude many simple statistical approaches. For example, fold change cannot be calculated for a participant whose baseline sample had zero copies.
Many investigators may ignore issues 1–3 and make adjustments for issue 4 so that familiar statistical methods can be used. We evaluate here three such strategies, along with an alternative, negative binomial regression, which readily handles all of these statistical issues.
Focus on research
To properly evaluate possible statistical analysis approaches, we must keep in mind two crucial aspects of the goals of research.
First, we wish to learn about generalisable patterns rather than individual participants. For example, we may want to estimate the mean or median of an HIV reservoir measure in a population, the difference between two groups or the effect of an experimental treatment. For research purposes, random variations that are sometimes upward and sometimes downward can average out to produce acceptable overall results. Conversely, improving the accuracy of only selected observations can distort the overall results. To illustrate this, we consider a very simple example where we wish to estimate the mean copies per million cells (CPMC) in a population. Suppose every person in the population actually has a true CPMC of exactly 1. We recruit some participants and assay exactly 1 million cells from each of them with a single-copy assay. On average, 37% of the participants will have 0 copies in their million cells assayed, due to Poisson sampling variation. If we decide to count those participants as having 1 CPMC, because each person's true CPMC must be higher than zero, then we will have improved the measurement for each person in that 37% from a value that is too low to one that is exactly correct. Nevertheless, when we proceed to estimate the population mean using the ‘improved’ data, we will on average estimate the mean to be 1.37 CPMC instead of the correct value of 1 CPMC. In contrast, if we leave the zero results as zero, even though they must be lower than the entire person's true value, we will on average estimate the correct mean. In general, selective adjustment of only some of the data is likely to disrupt analyses of generalisable patterns, particularly if all the adjustments are in the same direction.
Second, the importance of an effect or difference depends heavily on how large it is, not just whether it exists. Despite the focus often given to whether P<0.05, that is only a small part of the information that a study provides [17–19]. Notably, the effect of a treatment may be too small to be important even when a study finds a small P-value for it, and conversely, an effect may be large enough to be important even if it has P>0.05. Good estimation of effect size is important for maximising the information obtained from expensive assays, highly selected individuals and trials that ask participants to accept the risks of experimental interventions. The uncertainty around the estimate is also important, as often shown by the standard error or a confidence interval (CI). Notably, synthesis of evidence via meta-analysis usually ignores P-values from individual studies, instead focusing exclusively on the estimated effect sizes and their uncertainty. For evaluating different analysis approaches, we therefore care about not only the statistical power but also how far off they typically are (e.g. median absolute error), how much they systematically underestimate or overestimate effects (bias), and how good the accompanying uncertainty measures are (e.g. CI coverage).
Statistical analysis methods
For simplicity in describing and discussing analysis methods, we consider measurement of copies per some generic ‘input’. When a specific form of input is desirable for illustration, we will focus on a measurement of CPMC. We also focus on the common situation where interest focuses on relative differences or changes, so that analysis on a logarithmic scale is appropriate. Box 2 describes four approaches.
Box 2. Methods for statistical analysis of studies using a single-copy assay.
-
A.
Zero copies treated as 1. Replace all observations of 0 copies with 1 copy. Calculate log(copies/input) for all observations and analyse this by standard methods for normally distributed data, such as t-tests and linear regression. For example, see Reference 20.
-
B.
Add 1 to copies. Calculate log((1+copies)/input) and analyse this by standard methods for normally distributed data.
-
C.
Zero copies treated as left-censored. Consider observations of 0 copies as indicating that log(copies/input) might be any value less than log(1/input). Analyse the resulting data by methods for left-censored, normally distributed data [21–23]. For example, see Reference 10.
-
D.
Negative binomial regression. Analyse the observed copies (including 0) by negative binomial regression. Include input as an ‘exposure’ variable, or equivalently loge(input) as an ‘offset’ (depending on the statistical software used), in order to effectively model loge(copies/input). For example, see Reference 7.
We discuss for each method both theoretical considerations and the results of applying them to simulated data sets that have a known true effect of interest. We randomly generated 1000 replicates of a study evaluating within-person changes in CPMC from before to after a treatment in N=12 participants. We chose N=12 because it is a common choice for early studies, where small sample sizes are often appropriate [24]. Baseline loge(CPMC) was normally distributed with mean loge(3) and SD 0.75. The 12,000 simulated baseline true CPMC values had a 2.5 percentile of 0.7, a median of 3.0 and a 97.5 percentile of 12.8. We randomly generated observed numbers of copies from a Poisson distribution with a mean equal to the input number of cells times the true CPMC. The array of true CPMC values produced 0 copies in 12% of observations with cell input of 1 million. Within-person changes in loge(CPMC) were normally distributed with mean loge(0.25) and SD 0.75, independent of baseline CPMC. The simulated true changes in CPMC had a 2.5 percentile of a 17.6-fold reduction, a median of a 4.0-fold reduction and a 97.5 percentile of a 1.1-fold increase. The resulting array of true CPMC values after intervention produced 0 copies in 45% of post-treatment observations with cell input of 1 million. This is a substantial decrease, while not so overwhelming as to be obvious regardless of analysis method.
We then generated four versions of observed data for each simulated participant based on four different measurement scenarios. We made two different assumptions about input to the assays: either that input was always equal to 1 million cells or that it varied at random with a uniform distribution between 500,000 and 1,500,000 cells. We also assumed either that copies were counted exactly without error or that there was a measurement error for all non-zero numbers of copies that was normally distributed on the loge scale with a mean of 0 and an SD of 0.3. This corresponds to many assays where there may be a clear negative when no copies are present, but positive signals are translated to copies using an estimated standard curve, which typically produces non-integer values [12,16].
We tallied the performance of the four analysis methods over 1000 simulated data sets. Bias was shown by comparing the median estimated fold reduction to the true value (a fourfold reduction). Another measure of the quality of the estimated effects is the median of the absolute difference between the estimates and the true value. We evaluated the accompanying uncertainty of each estimate of the treatment effect by tallying how often the 95% CI contained the true value (known as the ‘coverage’ probability). For a 95% CI, this should ideally be 95%. We also tabulated the power, which is the proportion of the analyses that had P-values of <0.05. Table 1 summarises the results obtained by each method. We have not considered non-parametric analyses because they do not provide quantitative effect estimates.
Table 1.
Summary of analysis results for 1000 simulated studies of before–after differences in copies per million cells, using four different analysis methods (see Box 2 and text)
| Method | Median estimated fold reduction | Median absolute error (log10 scale) | 95% CI coverage (%)* | Power (%) |
|---|---|---|---|---|
| Input fixed at 1,000,000 cells. Copies counted exactly. | ||||
| A. 0 copies reset to 1, paired t-test | 2.03 | 0.29 | 27.1 | 81.9 |
| B. Add 1 to copies, paired t-test | 2.08 | 0.28 | 22.3 | 86.4 |
| C. 0 copies treated as left-censored | 2.66 | 0.18 | 77.1 | 87.5 |
| D. Negative binomial regression | 3.20 | 0.15 | 88.1 | 89.5 |
| Input fixed at 1,000,000 cells. Copies measured with error. | ||||
| A. 0 copies reset to 1, paired t-test | 2.03 | 0.29 | 29.2 | 77.0 |
| B. Add 1 to copies, paired t-test | 2.11 | 0.28 | 25.6 | 83.4 |
| C. 0 copies treated as left-censored | 2.78 | 0.17 | 80.6 | 83.6 |
| D. Negative binomial regression | 3.30 | 0.15 | 87.4 | 85.8 |
| Input varies from 500,000 to 1,500,000 cells. Copies counted exactly. | ||||
| A. 0 copies reset to 1, paired t-test | 2.01 | 0.30 | 24.5 | 75.7 |
| B. Add 1 to copies, paired t-test | 2.08 | 0.28 | 23.2 | 82.9 |
| C. 0 copies treated as left-censored | 2.71 | 0.17 | 76.6 | 85.7 |
| D. Negative binomial regression | 3.23 | 0.14 | 87.5 | 87.3 |
| Input varies from 500,000 to 1,500,000 cells. Copies measured with error. | ||||
| A. 0 copies reset to 1, paired t-test | 2.00 | 0.30 | 26.0 | 72.3 |
| B. Add 1 to copies, paired t-test | 2.09 | 0.28 | 28.2 | 79.5 |
| C. 0 copies treated as left-censored | 2.77 | 0.16 | 81.6 | 81.7 |
| D. Negative binomial regression | 3.23 | 0.15 | 87.4 | 85.0 |
CI: confidence interval.
Each study includes N = 12 persons, and the true mean reduction in log(copies per million cells) corresponds to a fourfold reduction in copies per million cells.
*For CIs, the ideal value for coverage is 95%.
Method A can arise from consideration of the lower limit of detection (LLOD) of the assay. For a single-copy assay as defined here, detection is limited only by the absence of any copies in the sample assayed, so for any given input, the LLOD is 1/input. Method A therefore is equivalent to the common practice of treating ‘undetectable’ results as being equal to the limit of detection [21]. As described in the previous section, this will cause bias, and the general strategy of treating undetectable results as if they were observed values of the LLOD has been cogently criticised [21,25]. Because there is no accounting for assay input, 0 copies with low input will end up being counted as a higher value than 1 copy obtained from a higher input. In situations with a highly variable input, an ad hoc way to mitigate this problem is to exclude measurements with very low input, but this requires an arbitrary threshold for what input is ‘too low’. An intuitively appealing variation, excluding samples with low input only if they turn out to have 0 copies, introduces additional bias by selectively excluding lower (zero) values.
The theoretical disadvantages of method A are reflected in the results in Table 1, where we have applied the paired t-test command in Stata version 13.1 (StataCorp, College Station, TX, USA) to logarithmically transformed values, obtaining the estimated mean change and its 95% CI in addition to the P-value. Method A tended to estimate only half as much within-person change as was typically present, and its CIs excluded the true value about 80% of the time. A variation on method A [26] that replaces 0 copies with 0.5 copies performed better, closer to method C. It was least biased for the case with fixed input and with measurement error, where it had a median estimated effect of 2.61, 95% CI coverage of 69.0% and power of 82.3%.
Method B also converts observations of 0 copies to 1 copy, but it preserves the distinction between 0 and 1 observed copies by also altering all the other observations. While this applies a consistent transformation to all data, it does not perfectly preserve the interpretation of results in terms of fold effects, which is often an important reason for using logarithmic transformation. In order to obtain an interpretable quantitative estimate of the effect of treatment, we can nevertheless treat the analysis results as if they were from an unmodified logarithmic transformation. This produces the results shown in Table 1, where method B is only slightly better than method A: it is typically off by about twofold and its CI only rarely includes the true value. In addition, method B can still count 0 copies with low input as if it were a higher observed rate than 1 copy with a higher input. If input varies systematically, such as might occur when comparing different tissues or cell types, this could spuriously make the lower input case appear to have higher rates.
Method C treats observations of 0 copies as being left-censored observations of 1 copy, meaning that log(copies/input) could be any number less than log(1/input). This follows an approach that was advocated by Marschner et al.[21] for HIV viral load assays that had fairly high LLODs. That approach was an important improvement over treating undetectable results as equal to the LLOD, and it has been generalised for application in mixed-effects models [22,23,27]. For single-copy assays, however, this approach does not match the information actually provided by the data. A result of 0 copies indicates that no copies were present in the particular sample assayed, not that some fractional number less than 1 was present. In addition, observing 0 copies does not provide strong evidence that the person's true CPMC is below 1/input: the upper 95% CI bound would be 3/input [28], and there is no sharp demarcation between possible and impossible values. Thus, method C does not match the information about either the particular sample assayed or the CPMC in the person. An additional drawback of method C is that, as with methods A and B, it does not account for varying input. The analyses summarised in Table 1 used Proc NLmixed in SAS version 9.4 (SAS Institute, Cary, NC, USA), with a random person effect and a fixed effect quantifying within-person change from pretreatment to post treatment [27]. The results are better than those for methods A and B, but still with considerable downward bias in the intervention effect estimate and poorer performance overall than method D.
Method D uses a count model, which is a natural choice for the number of copies present in the samples. As noted in Box 1, Poisson variation in the number of copies present in a tissue sample is inevitable, and it will be of non-negligible magnitude for rare entities. Although Poisson regression is a simple count model, it may often be too optimistic about random variation because of additional sources of variability, such as person-to-person differences and assay measurement error. We therefore focus here on negative binomial regression, which generalises the Poisson distribution to also allow for such additional sources of variability [29]. It can be used to model rates such as CPMC by employing a standard modification to account for the denominator (e.g. the ‘per million cells’ in ‘CPMC’). Appendix A provides details of how this is done and shows how to implement it in the popular statistical packages Stata and SAS. The variability of the observed copies around their modelled expectation is assumed to follow a negative binomial distribution. This model matches biological intuition in that all study participants are assumed to have a non-0 (but possibly small) true CPMC, and observations of 0 copies are assumed to have arisen via sampling variability. Observations of 0 copies can therefore be included without any ad hoc modifications. Observations that are likely to be less precise (due to lower observed number of copies and/or lower input) are automatically given less influence on the model results. Notably, observations with 0 copies and low input are appropriately downweighted without any need for a cutoff defining when input is too low and observations should be excluded. We fit the models with the menbreg command in Stata version 13.1 (StataCorp). This command allowed us to include a normally distributed random intercept to reflect the repeated observations (preintervention and post intervention) on the same participants. The models estimate multiplicative effects on the expected number of copies in each sample, along with an overdispersion parameter reflecting variation in excess of that expected from Poisson sampling variability alone.
The theoretical advantages of method D are reflected in the results in Table 1, where it had the best performance on all the metrics. It still had some bias, typically estimating an attenuated ~3.2-fold decrease instead of the true 4.0-fold decrease, but the bias is smaller with this method than with any of the others. Similarly, the CI coverage is less than the ideal 95%, but it is better than for any other method. Although the negative binomial distribution was originally defined for integers, the method readily generalises to non-integer numbers of observed ‘copies’ by calculation of the likelihood with the mathematical gamma function in place of factorial terms (see Reference 30, page 203). The workability of this generalisation is reflected in the Table 1 cases with measurement error, where none of the non-0 observed values are integers. Thus, negative binomial regression can be applied directly to observed data without rounding whenever assays produce non-integer numbers of copies.
The bias that was seen even with method D results from the combination of two factors: (1) imprecision in the measurements caused by Poisson sampling variability and (2) the person-to- person variation in the effect of the intervention (see Appendix B). If the simulated studies examine 100 million input cells instead of 1 million, sampling variation is mostly eliminated. This eliminates the bias while also making observation of 0 copies very rare and consequently making all four methods roughly equivalent. In contrast, the bias remains largely unchanged if cell input remains at 1 million and the number of participants is increased 10-fold to N=120. (Thus, for HIV clinical trials, increasing the number of participants may not mitigate the statistical challenges shown here. Increasing tissue or blood sampling would reduce the bias of these methods but will likely encounter limitations of cost, acceptability and feasibility.) When the simulations are done with every person having exactly a fourfold reduction, the bias for method D is also eliminated. Bias for the other methods is reduced slightly, but with methods A and B still having median estimates of <2.3-fold and method C having a median estimate of <3.1-fold. We have not presented this more favourable case as the primary set of results because we believe that person-to-person variation in intervention effects will usually occur.
Null simulations
We also ran simulations identical to those for Table 1, except with the intervention having no effect. In this case, the null hypothesis is true, and P-values should therefore be <0.05 about 5% of the time. Table 2 shows that all the methods were close to this theoretical expectation, except that method D found P < 0.05 too often when measurement error was present. The Stata command for these method D analyses uses a normal approximation to compute P-values. If we instead use a t distribution with 11 degrees of freedom, then the per cent with P < 0.05 would be about right: 5.3% for fixed input and 5.4% for varying input. This, however, comes at the cost of excessive conservatism when measurement error is not present, with P < 0.05 only 1.8% of the time with fixed input and 2.5% of the time with varying input. Thus, in this challenging situation with a very small sample size, there was no ideal solution for method D.
Table 2.
Summary of analysis results for 1000 simulated studies similar to those summarised in Table 1, but with intervention having no effect
| Method | Per cent with P < 0.05* |
|---|---|
| Input fixed at 1,000,000 cells. Copies counted exactly. | |
| A. 0 copies reset to 1, paired t-test | 5.2 |
| B. Add 1 to copies, paired t-test | 5.5 |
| C. 0 copies treated as left-censored | 5.9 |
| D. Negative binomial regression | 3.9 |
| Input fixed at 1,000,000 cells. Copies measured with error. | |
| A. 0 copies reset to 1, paired t-test | 5.4 |
| B. Add 1 to copies, paired t-test | 4.9 |
| C. 0 copies treated as left-censored | 5.8 |
| D. Negative binomial regression | 8.0 |
| Input varies from 500,000 to 1,500,000 cells. Copies counted exactly. | |
| A. 0 copies reset to 1, paired t-test | 5.5 |
| B. Add 1 to copies, paired t-test | 5.0 |
| C. 0 copies treated as left-censored | 6.4 |
| D. Negative binomial regression | 4.8 |
| Input varies from 500,000 to 1,500,000 cells. Copies measured with error. | |
| A. 0 copies reset to 1, paired t-test | 5.0 |
| B. Add 1 to copies, paired t-test | 5.9 |
| C. 0 copies treated as left-censored | 6.4 |
| D. Negative binomial regression | 7.3 |
*The ideal value for this is 5%.
Example
A recent study [31] examined sex differences in multiply-spliced HIV RNA among effectively treated persons with HIV. This measurement provides a good illustration of the issues discussed here, because samples from 24 of the 52 participants (46%) had 0 copies, and the input to the assay ranged widely from 62,000 to 3,288,000 resting CD4 cells, with a median of 1,485,000.
Table 3 shows the results of applying the four Box 2 methods; for the reasons noted earlier, the study itself used negative binomial regression (method D). Although this is a between-person comparison, the results are qualitatively similar to the within-person scenario shown in Table 1. Methods A–C all produce smaller estimated differences and larger P-values than method D.
Table 3.
Estimated differences in multiply-spliced HIV RNA per million resting CD4 cells, from a study comparing 26 women with 26 men, all with effectively treated HIV [31]
| Method | Estimated male : female ratio | 95% Confidence interval | P-value |
|---|---|---|---|
| A. 0 copies reset to 1, unpaired t-test | 2.38 | 1.07–5.27 | 0.034 |
| B. Add 1 to copies, unpaired t-test | 2.11 | 0.98–4.53 | 0.055 |
| C. 0 copies treated as left-censored | 2.73 | 0.87–8.56 | 0.084 |
| D. Negative binomial regression | 6.17 | 1.95–19.6 | 0.002 |
Discussion
For studies measuring rare entities via single-copy assays, results can vary substantially, depending on the methods used for statistical analysis and how observations of 0 copies are handled. Negative binomial regression handles the specific challenges noted in Box 1, and its theoretical advantages manifested as expected in our simulations and when applied to a data set from an actual study. Null hypothesis testing, however, was either too liberal in some cases or too conservative in other cases, depending on whether a normal or t-distribution was used for the P-value calculations. Although the negative binomial distribution is classically defined for integer counts, negative binomial regression can handle continuous values (as reflected in our simulations with measurement error), and our simulations verified that measurement error producing non-integer numbers of copies had little impact on the advantages of negative binomial regression over the other methods evaluated. Thus, this approach can be implemented using standard statistical software, such as Stata [30], SAS [32] or R [33,34], regardless of whether the assay produces exact integer numbers of copies.
When a single-copy assay indicates that no copies of the target entity were present in the sample analysed, labs often report the value that would have been produced if 1 copy had been present, usually preceded by a ‘<’ symbol. A statistician taking this at face value would naturally be led to method A (if no < was included) or method C (if < was included). Statisticians must therefore ensure that they understand the actual information that assays provide; if it is a single-copy assay in the sense addressed here, then analysis of the actual copies measured, including zeroes, can be accomplished by negative binomial regression. As we have discussed, including an observed zero does not assume that the participant had no entities in his or her entire body or that an additional sample would necessarily also have had 0 copies. It simply makes use of the actual result for the sample that was actually assayed, treating observed zeroes as resulting from Poisson sampling variability.
Regardless of the method used, assessment of potential overly influential observations or outliers will often be relevant for the study of rare entities. When most samples contain copy numbers in the single digits, an observation with hundreds of copies could disrupt any quantitative statistical analysis. In many cases, such observations will warrant special investigation or handling, such as exclusion or Winsorizing [35]. We also caution against ‘P- hacking’ [36]. Although we evaluated four different analysis methods here, analysing an actual study by applying all four and then presenting only the one with the smallest P-value would be a poor approach.
We have focused here on just a few relatively simple situations, with the goal of pointing out potential difficulties with analysis of single-copy assays and the potential advantages of using negative binomial regression. Additional research in this area could investigate a wider variety of situations. In more extreme situations than those considered here, such as even smaller sample sizes or even higher proportions of zero observations, study-specific simulations may be useful for choosing an analysis method. Some attenuation of the estimated intervention effect was present in Table 1 even for the negative binomial regression analyses, so development of improved methods for analysing such studies would be worthwhile.
We have considered here measurement methods that seek to determine how many copies of a rare entity are present in a tissue or blood sample. Statistical methods for count data are therefore a natural choice, and negative binomial regression is a flexible type of count model that is implemented in many statistical software packages and can be applied when the data include zero counts and non-integer numbers of copies. It readily deals with the Box 1 issues, while the other methods all have both theoretical drawbacks and poorer performance in the situations examined here. We therefore recommend that researchers using single-copy assays to measure rare entities consider negative binomial regression for statistical analysis.
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers U19 AI096109, UM1AI126611, K08AI116344, and UM1 AI068634), amfAR, the Foundation for AIDS Research (grant numbers 108842-55-RGRL and 109301), and the Bill and Melinda Gates Foundation (grant number OPP1115400).
Conflicts of interest
The authors do not have any conflicts of interest to declare.
Appendix A:
Details for method D and syntax for implementing it in Stata and SAS
Negative binomial regression can model rates such as CPMC, defined as copies/input, by including the amount of input as an ‘exposure’ or ‘offset’ variable, meaning that loge(input) is included in the predictive model as a fixed term (its regression coefficient is fixed at 1 instead of being estimated). For the simulated study data used here, the model for each given participant i (i=1 to 12) at time j (before or after the intervention, j=0 or 1) is
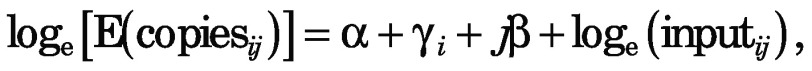 |
where E is the expectation (mean), α is the fixed overall intercept term, γi is the person-specific random intercept and β is the regression coefficient for the effect of the treatment. This model is mathematically equivalent to
 |
which shows how it models loge(copies/input) as desired.
We now describe examples of how to implement method D on data similar to the simulations described in the main text. The methods require that the data set be in ‘long’ format with two observations per participant, one for before intervention and one for after, and we use the following variables:
ID = identification number for each participant
After = 1 if the observation is for the postintervention measurement
=0 if the observation is for the preintervention (baseline) measurement
Copies = measured # of copies of target entity present in the sample assayed
InputCells = # of cells in the sample assayed
logInputCells = loge(InputCells)
Stata:
menbreg Copies After, exposure(InputCells) || ID:
SAS:
proc glimmix;
class ID;
model Copies = After / dist=negbin offset=logInputCells solution cl; random int / subject=ID;
run;
Appendix B
Discussion of bias for method D
The factors leading to bias even for method D may seem counterintuitive, along with the fact that increasing N does not eliminate the bias. While a detailed mathematical analysis of this is beyond the scope of this paper, we offer here a possible conceptual explanation.
Because the relation between loge(CPMC) and CPMC is nonlinear, the symmetric normal distributions of true loge(CPMC) values correspond to asymmetric log-normal distributions of true CPMC values. The mean CPMC is therefore larger than the exponentiated value of the mean loge(CPMC), and the difference between the two is more pronounced for the postintervention values of loge(CPMC) because they have a larger variance. Consequently, the ratio of the mean true CPMC at baseline to the mean true CPMC after intervention is only about 3, instead of the fourfold difference that is typical within subjects. Thus, if this were a cross-sectional study comparing two groups, the correct value for negative binomial regression would be 3 because it estimates multiplicative effects on the mean number of copies. (This would not occur if the change in true loge(CPMC) from baseline to postintervention were the same for everyone.)
Inclusion of random intercepts in the model attempts to focus on the within-person changes by accounting for within-person correlation. Ideally, this would change the correct result for negative binomial regression from threefold to the fourfold typical change as specified for the simulations. We believe, however, that the correct result moves only part way toward fourfold because Poisson sampling variation attenuates the within-person correlation between baseline and postintervention copies to a value that is less than the within-person correlation in true CPMC values. This could cause the ‘typical’ within-person change, as defined for negative binomial regression, to be less than the fourfold change that we defined as being the most meaningful typical within-person change in the simulation specifications. Increasing the sample size would not change this. On the other hand, increasing the input to 100 million cells would substantially reduce the relative variability in the observed counts, making them correspond closely to the true CPMC values. This would mitigate the attenuation of the correlation, making the estimate that accounts for within-person correlation in the number of copies correspond better to the desired fourfold reduction.
References
- 1. Palmer S, Malclarelli F, Wiegand A et al. . Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA 2008; 105: 3879– 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gay CL, Bosch RJ, Ritz J et al. . Clinical trial of the anti-PD-L1 antibody BMS-936559 in HIV-1 infected participants on suppressive antiretroviral therapy. J Infect Dis 2017; 215: 1725– 1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Archin NM, Liberty AL, Kashuba AD et al. . Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487: U1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rasmussen TA, Tolstrup M, Brinkmann CR et al. . Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1: E21. [DOI] [PubMed] [Google Scholar]
- 5. Vibholm L, Schleimann MH, Hojen JF et al. . Short-course toll-like receptor 9 agonist treatment impacts innate immunity and plasma viremia in individuals with human immunodeficiency virus infection. Clin Infect Dis 2017; 64: 1686– 1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sogaard OS, Graversen ME, Leth S et al. . The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 2015; 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elliott JH, McMahon JH, Chang CC et al. . Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2015; 2: E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elliott JH, Wightman F, Solomon A et al. . Activation of HIV transcription with short- course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10: e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gandhi RT, McMahon DK, Bosch RJ et al. . Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13: e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riddler SA, Aga E, Bosch RJ et al. . Continued slow decay of the residual plasma viremia level in HIV-1-infected adults receiving long-term antiretroviral therapy. J Infect Dis 2016; 213: 556– 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pasternak AO, Jurriaans S, Bakker M et al. . Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS ONE 2009; 4: e8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vandergeeten C, Fromentin R, Merlini E et al. . Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol 2014; 88: 12385– 12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chomont N, El-Far M, Ancuta P et al. . HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15: U92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewin SR, Murray JM, Solomon A et al. . Virologic determinants of success after structured treatment interruptions of antiretrovirals in acute HIV-1 infection. J Acquir Immune Defic Syndr 2008; 47: 140– 147. [PubMed] [Google Scholar]
- 15. Somsouk M, Dunham RM, Cohen M et al. . The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+T cell recovery: a randomized crossover trial. PLoS One 2014; 9: e116306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palmer S, Wiegand AP, Maldarelli F et al. . New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 2003; 41: 4531– 4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amrhein V, Greenland S, McShane B. Retire statistical significance. Nature 2019; 567: 305– 307. [DOI] [PubMed] [Google Scholar]
- 18. Wasserstein RL, Schirm AL, Lazar NA. Moving to a world beyond "p<0.05". Am Stat 2019; 73: 1– 19. [Google Scholar]
- 19. Gardner MJ, Altman DG. Confidence-intervals rather than P-values – estimation rather than hypothesis-testing. Br Med J (Clin Res Ed) 1986; 292: 746– 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer M, Joos B, Niederoest B et al. . Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy. Retrovirology 2008; 5: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marschner IC, Betensky RA, DeGruttola V et al. . Clinical trials using HIV-1 RNA-based primary endpoints: statistical analysis and potential biases. J Acquir Immune Defic Syndr Hum Retrovirol 1999; 20: 220– 227. [DOI] [PubMed] [Google Scholar]
- 22. Hughes JP. Mixed effects models with censored data with application to HIV RNA levels. Biometrics 1999; 55: 625– 629. [DOI] [PubMed] [Google Scholar]
- 23. Vaida F, Liu L. Fast implementation for normal mixed effects models with censored response. J Comput Graph Stat 2009; 18: 797– 817. Implemented in the R package lmec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bacchetti P, Deeks SG, McCune JM. Breaking free of sample size dogma to perform innovative translational research. Sci Transl Med 2011; 3: 87ps24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helsel DR. Fabricating data: how substituting values for nondetects can ruin results, and what can be done about it. Chemosphere 2006; 65: 2434– 2439. [DOI] [PubMed] [Google Scholar]
- 26. Hong F, Aga E, Cillo AR et al. . Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 2016; 54: 902– 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thiebaut R, Jacqmin-Gadda H. Mixed models for longitudinal left-censored repeated measures. Comput Methods Programs Biomed 2004; 74: 255– 260. [DOI] [PubMed] [Google Scholar]
- 28. Eypasch E, Lefering R, Kum CK et al. . Probability of adverse events that have not yet occurred – a statistical reminder. BMJ 1995; 311: 619– 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedan A. Analysis of count data using the SAS system ( 2001). Available at: www2.sas.com/proceedings/sugi26/p247-26.pdf ( accessed July 2019).
- 30. StataCorp Stata multilevel mixed-effects reference manual release 15 ( 2017). Available at: www.stata.com/manuals/me.pdf ( accessed July 2019).
- 31. Scully EP, Gandhi M, Johnston R et al. . Sex-based differences in HIV-1 reservoir activity and residual immune activation. J Infect Dis 2019; 219: 1084– 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. SAS Institute The GLIMMIX procedure ( 2017). Available at: support.sas.com/documentation/onlinedoc/stat/143/glimmix.pdf ( accessed July 2019).
- 33. Bolker B. glmer.nb ( 2017). Available at: www.rdocumentation.org/packages/lme4/versions/1.1-15/topics/glmer.nb ( accessed July 2019).
- 34. Fournier DA, Skaug HJ, Ancheta J et al. . AD Model Builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Methods Softw 2012; 27: 233– 249. [Google Scholar]
- 35. Rivest LP. Statistical properties of Winsorized means for skewed distributions. Biometrika 1994; 81: 373– 383. [Google Scholar]
- 36. Head ML, Holman L, Lanfear R et al. . The extent and consequences of P-hacking in science. PLoS Biol 2015; 13: e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]


