Abstract
Introduction
Cell-free deoxyribonucleic acid DNA (cf-DNA) in urine is promising due to the advantage of urine as an easily obtained and non-invasive sample source over tissue and blood. In clinical practice, it is important to identify non-invasive biomarkers of chronic kidney disease (CKD) in monitoring and surveillance of disease progression. Information is limited, however, regarding the relationship between urine and plasma cf-DNA and the renal outcome in CKD patients.
Methods
One hundred and thirty-one CKD patients were enrolled between January 2016 and September 2018. Baseline urine and plasma cell-free mitochondrial DNA (cf-mtDNA) and cell-free nuclear DNA (cf-nDNA) were isolated using quantitative real-time PCR. Estimated glomerular filtration rate (eGFR) measurement was performed at baseline and 6-month follow-up. Favorable renal outcome was defined as eGFR at 6 months minus baseline eGFR> = 0. Receiver operator characteristics (ROC) curve analysis was performed to assess different samples of cf-DNA to predict favorable renal outcomes at 6 months. A multivariate linear regression model was used to evaluate independent associations between possible predictors and different samples of cf-DNA.
Results
Patients with an advanced stage of CKD has significantly low plasma cf-nDNA and high plasma neutrophil gelatinase-associated lipocalin (NGAL) levels. Low urine cf-mtDNA, cf-nDNA levels and low plasma NGAL were significantly correlated with favorable renal outcomes at 6 months. The urine albumin-creatinine ratio (ACR) or urine protein-creatinine ratio (PCR) level is a robust predictor of cf-mtDNA and cf-nDNA in CKD patients. Baseline urine levels of cf-mtDNA and cf-nDNA could predict renal outcomes at 6 months.
Conclusions
Urinary cf-mtDNA and cf-nDNA may provide novel prognostic biomarkers for renal outcome in CKD patients. The levels of plasma cf-nDNA and plasma NGAL are significantly correlated with the severity of CKD.
Electronic supplementary material
The online version of this article (10.1186/s12882-019-1549-x) contains supplementary material, which is available to authorized users.
Keywords: Cell-free mitochondrial deoxyribonucleic acid, Cf-mtDNA, Cell-free nuclear deoxyribonucleic acid, Cf-nDNA, Neutrophil gelatinase-associated lipocalin, NGAL, Chronic kidney disease
Background
Chronic kidney disease (CKD) is a global public health problem affecting up to 10% of the population worldwide [1], although guidelines for the clinical staging of CKD have been established [2]. The natural history of the earlier phases of CKD are highly unpredictable and the early identification of CKD and timely detection of progression are truly global challenges [3, 4]. It is important to recognize the risk factors of CKD, and better approaches for the prevention, early detection, and treatment of CKD [5].
The kidney is a highly energetic organ and rich in mitochondria and numerous studies have shown that mitochondrial dysfunction contributes to different types of kidney diseases, including diabetic and nondiabetic nephropathy [6]. Proteinuria, caused by a primary insult to the kidney, induces oxidative stress in renal tubular cells and causes mitochondrial dysfunction [7, 8], which leads to cellular damage by reactive oxygen species generation as well as epithelial–mesenchymal transition (EMT) [9, 10]. To date, CKD is characterized by mitochondrial dysfunction, oxidative stress, and aberrant autophagy [11, 12] in addition to significant changes in the activation of transforming growth factor-β, p53, hypoxia-inducible factor, chronic inflammation, and traditional vascular dysfunction [11]. By the experimental models of CKD, preventing mitochondrial dysfunction inhibits renal tubular cell EMT and renal fibrosis [13]. However, only a relatively small number of translational studies have shown the clinical relevance of these mechanisms between renal diseases and mitochondrial dysfunction in humans [6, 14, 15].
Free circulating nucleic acids, cell-free deoxyribonucleic acid (cf-DNA), were discovered in human plasma in 1948 by Mandel and Metais [16]. Some studies showed the damaged mitochondria release their DNA content into the systemic circulation [17, 18]. Thus, cell-free mitochondrial deoxyribonucleic acid (cf-mtDNA) is easily detected in plasma, and has been explored as a biomarker of various diseases [19–21]. Recently, increased plasma levels of cf-mtDNA have been reported to correlate with the severity of injury in patients sustaining polytrauma [22] and the severity of stroke [23], be a prognostic marker of acute myocardial infarction [24] and intensive care unit patients [25, 26]. Alterations of cf-mtDNA in the blood also might be associated with several systemic diseases, including primary mitochondrial disorders, carcinogenesis, and hematologic diseases [27, 28]. In a community-based population cohort, higher plasma cf-mtDNA was associated with a lower incidence of CKD independent of traditional risk factors [20] and was associated with a lower prevalence of microalbuminuria [29]. Apart from blood, cf-DNA could also be detected in urine. Urine cf-DNA originates either from cells shedding into urine from genitourinary tract, or from cell free DNA in circulation passing through glomerular filtration [30]. The presence of genetic information in urine has been observed in some clinical studies. For examples, the urinary cf-mtDNA level had statistically significant correlations with the peak serum creatinine level and the duration of hospitalization in a study of acute kidney injury (AKI) [30]. Patients who required temporary dialysis also tended to have higher urinary cf-mtDNA levels than those without dialysis [31], but no relationship between the urinary cf-mtDNA level and renal outcomes has been reported. A recent study showed that urinary cf-mtDNA level was increased in mice after 10–15 min of ischemia, and that the level correlated with the duration of ischemia [32]. Otherwise, platelet and leukocyte counts in samples are important sources of variation when cf-DNA is measured in DNA extracted from whole blood [27, 28]. Emerging data showed that measuring cf-DNA extracted from whole blood (nuclear deoxyribonucleic acid, cf-nDNA) could yield different results from peripheral blood mononuclear cells or buffy coat (cf-mtDNA), because of the presence of mitochondrial DNA in platelets [27, 28].
Since the majority of urine cf-DNA originates from apoptosis or necrosis of cells exfoliated from urogenital system [30]. And the minority are originating from blood circulation, which contains important genomic information from various positions all over the body [33, 34]. We postulate that urine cf-DNA might be regarded as a marker which combined genetic information from urogenital system and systemic. We hypothesis that urinary cf-DNA might be a better novel biomarker than circulating cf-DNA in predicting renal outcome in CKD patients. To clarify the clinical application of cf-mtDNA and cf-nDNA in CKD, we studied the correlation between cf-mtDNA and cf-nDNA, both urine and plasma, and the stage of CKD or prognosis of renal outcomes.
Methods
The Institutional Review Board of Changhua Christian Hospital approved the experimental protocols (approval number 140306) and all the participants provided written informed consent to participate the study. All patients of the study joined our nationwide preventive multidisciplinary program, also regulated by the Clinical Care Program Certification and Joint Commission International, for early CKD or pre-ESRD (end stage renal disease). We investigated patients who were enrolled in our CKD care program between January 2016 and September 2018.The goals for patients’ blood pressure, glucose and lipid control were based on the KDOGI guidelines.
Overall, of 131 patients with CKD, 7 patients dropped-out for the duration of follow-up less than 6 months, and 52 volunteers were recruited from the Nephrology Clinic at Changhua Christian Hospital, a tertiary referral hospital in Taiwan. The duration of follow-up for CKD was more than 6 months in all patients. The patients with following criteria were excluded: infection, acute fever, hepatic or cardiac disease, endocrinopathy, glomerulonephritis proved by biopsy or treatment with steroids or immunosuppressants, surgery, trauma, missing data at baseline, prior kidney transplant, acute kidney injury and a history of RRT or hospitalization for any cause in the past 3 months. The amount of proteinuria was calculated by urinary protein-creatinine ratio (PCR, mg/g) or albumin-creatinine ratio (ACR, mg/g). Microalbuminuria was established when two out of three ACR determinations were found to be within the range of 30–300 mg/g in a 6-month period. We calculated the glomerular filtration rate (eGFR) of the patients according to the CKD Epidemiology Collaboration equation (eGFRCKD-EPI) [35]. The stages of CKD were defined as follows: stage 1, eGFR > 90 ml/min/1.73 m2; stage 2, eGFR 60–89 ml/min/1.73 m2; stage 3a, eGFR 45–59 ml/min/1.73 m2; stage 3b, eGFR 30–44 ml/min/1.73 m2; stage 4, eGFR 15–29 ml/min/1.73 m2 and stage 5, eGFR < 15 ml/min/1.73 m2 or maintenance on RRT. We divided the population to early CKD (stages 1–3a) and advanced CKD (stages 3b–5) in the study, based on our national CKD care program.
Studies in CKD always address primary outcomes of death, ESRD, or doubling of baseline serum creatinine. Coresh et al. extended use of percentage reduction in eGFR as a surrogate for hard outcomes and they reported that longer–term follow-up, more than 1 year, was strongly predictive of ESRD and death [36]. We, therefore, examined the change of eGFR as a surrogate for hard outcomes within 6 months. For study simplicity, favorable renal outcome was defined as eGFR at 6 months minus baseline eGFR> = 0; unfavorable renal outcome indicated eGFR at 6 months minus baseline eGFR <0.
Blood, plasma and urine sampling
Participants’ venous blood samples, following 8-h overnight fasting, were obtained and first morning urine samples were collected. Aliquots of urine were immediately frozen at − 80 °C until further analysis, but specimen reserved for no longer than 1 month. All assays were undertaken in duplicate with intra-assay variation coefficient less than 5%. Urinary albumin concentration was measured by an immunoturbidimetric method (Roche Diagnostics GmbH, Mannheim, Germany).
DNA isolation and qPCR
The baseline data of urine and plasma cell-free mitochondrial DNA (cf-mtDNA) and cell-free nuclear DNA (cf-nDNA) were isolated from 131 patients with CKD in our study. Blood collection were drawn from each subject in the morning after overnight fasting 8 h. For each subject, 5 ml of whole blood was withdrawn from an antecubital vein and quickly delivered into an EDTA-K3-containing plastic tube. Plasma was collected by centrifugation of blood at 2500 rpm for 10 min, divided into several aliquots, and stored in − 80 °C until analysis. Urine samples were placed on 4 °C, centrifuged at 12000 rpm for 15 min within 8 h of collection, and the urine supernatants were separated and stored at − 80 °C until extraction DNA. Plasma and Urine cfDNA was extracted urine the Viral DNA mini Kit (AccuBioMed, Co, Ltd., Taiwan), following the protocol of manufacturer.
Leukocyte mitochondrial DNA copy number
We used a LightCycler® 480 Instrument (Roche, Mannheim, Germany) to measure mitochondrial copy number (MCN) in leukocytes. Briefly, we extracted total DNA using the Gentra Puregene DNA kit (Qiagen, Hilden, Germany). Real-time polymerase chain reaction (PCR) was used to amplify the ND1 gene of mtDNA and β-globin of nuclear DNA, respectively. The relative MCN of mtDNA was normalized to the β-globin gene.
Quantification of urine and plasma cell-free DNA by real-time PCR
Baseline urine and plasma cf-mtDNA and cf-nDNA were quantified by real-time PCR using the LightCycler® 480 Instrument (Roche, Mannheim, Germany) using specific primers to amplify the β-globin (forward: 5′-GTG CAC CTG ACT CCT GAG GAG A-3′, reverse: 5′-CCT TGA TAC CAA CCT GCC CAG-3′) and MT-ND1 (forward: AACATACCCATGGCCAACCT, reverse: AGCGAAGGGTTGTAGTAGCCC) genes from urine and plasma DNA, and standard regression analyses were used to derive the amount of urine and plasma nuclear DNA and mitochondrial DNA. The DNA concentrations were expressed as genome equivalents (GE)/mL of urine and plasma, where 1 GE was equivalent to 6.6 ng DNA [37]. Serially diluted human genomic DNA solution were used for preparing a six-point calibration curve [38].
Plasma neutrophil gelatinase-associated lipocalin (NGAL)
Plasma NGAL concentrations were measured using a commercially available assay kit (Immunology Consultants Laboratory, Inc., Oregon, USA).
Statistical analysis
Results are presented as the median (interquartile range) or number (proportion, %). All data were analyzed by the ANOVA test, if p with significant will receive further post hoc test analysis. A multivariate linear regression model was used to evaluate independent associations between possible predictors and cf-mtDNA or cf-nDNA. Receiver operator characteristics (ROC) curve analysis was performed to assess different samples of cf-DNA and NGAL to predict renal disease related outcomes. Youden’s index is the sum of sensitivity and specificity minus one, which is the most commonly used criterion for cut-off point selection in the context of ROC curve analysis. The maximum value of the index may be used as a criterion for selecting the optimum cut-off point. We used logistic regression to analyze the significant predictors of dissimilar renal outcome at 6 months. Statistical methods were employed to determine the appropriate number of patient and control samples required to ensure meaningful and statistically significant data (with a power of 80%, a sample size of 38 was sufficient to detect an observed difference). All statistical analyses were performed using IBM SPSS 20 (SPSS, Inc., Chicago, IL, USA) and a qualified statistician was employed to determine which tests should be used and whether they performed the analysis. In all analyses, P-values of < 0.05 were considered statistically significant.
Results
The demographic and clinical characteristics of the varying stages of chronic kidney diseases
Table 1 shows significant differences in age, renal functions, phosphate and low-density lipoprotein levels among varying stages of chronic kidney diseases and the further post hoc test analysis showed CKD stage related. High incidence, 37 to 59%, of hypertension encounters in varying stage CKD patients. Concerning DM, the incidence is increasing comparable to CKD stage worsen, 12% DM in early stage CKD and 54% in stage 5 CKD.
Table 1.
Demographic characteristics of varying stages of chronic kidney disease
| Variables | Stage 1,2a | Stage 3Ab | Stage 3Bc | Stage 4,5d | p value | Post hoc |
|---|---|---|---|---|---|---|
| n = 23 | n = 28 | n = 31 | n = 42 | |||
| Age (year) | 47.1 ± 12.7 | 53.6 ± 9.4 | 54.9 ± 10.5 | 58.7 ± 6.1 | 0.002* | a < c,d |
| Gender (M/F) | 3/20 | 8/20 | 22/9 | 29/13 | 0.212 | |
| Hypertension (%) | 42% | 37% | 59% | 54% | 0.191 | |
| Diabetes mellitus (%) | 12% | 11% | 23% | 54% | 0.160 | |
| Systolic blood pressure (mm Hg) | 138 ± 18 | 144 ± 28 | 138 ± 24 | 142 ± 15 | 0.943 | |
| Diastolic blood pressure (mm Hg) | 90 ± 15 | 91 ± 4 | 84 ± 18 | 84 ± 12 | 0.784 | |
| BMI (kg/m2) | 27.8 ± 5.4 | 28.2 ± 6.1 | 26.2 ± 6.1 | 24.8 ± 4.3 | 0.172 | |
| BUN (mg/dL) | 14.3 ± 4.4 | 17.7 ± 3.8 | 31.4 ± 13.0 | 64.5 ± 27.1 | <0.001** | a,b < c < d |
| eGFR (ml/min/1.73 m^2) | 84.4 ± 20.2 | 52.1 ± 4.4 | 31 ± 9.6 | 11.5 ± 7.3 | <0.001** | a > b > c > d |
| Phosphate (mg/dL) | 3.5 ± 0.4 | 3.5 ± 0.6 | 4.0 ± 0.7 | 5.0 ± 1.1 | <0.001** | a,b,c < d |
| Cholesterol (mg/dL) | 198.3 ± 50.4 | 171.5 ± 31.8 | 196.6 ± 47.3 | 153.3 ± 44.5 | 0.001** | a,c > d |
| HDL-C (mg/dL) | 47.8 ± 14.3 | 43.3 ± 9.8 | 46.1 ± 12.6 | 46.7 ± 21.2 | 0.677 | |
| LDL-C (mg/dL) | 118.2 ± 45.3 | 104.6 ± 31.4 | 114.9 ± 41.3 | 80.8 ± 26.7 | 0.040* | c > d |
| Fasting Glucose (mg/dL) | 118.3 ± 46.1 | 106 ± 21.8 | 112.6 ± 43.5 | 94.9 ± 24.6 | 0.263 | |
| HbA1C (%) | 6.3 ± 1.4 | 6.0 ± 0.6 | 6.2 ± 1.1 | 6.1 ± 0.9 | 0.778 | |
| Urine A/C ratio (mg/g) | 438.0 ± 826.4 | 535.7 ± 1018.4 | 2121.3 ± 2540.7 | 1548.0 ± 2377.2 | 0.058 | |
| WBC (×103/μL) | 7.1 ± 2.4 | 6.2 ± 1.9 | 6.4 ± 2.1 | 6.8 ± 2.0 | 0.323 | |
| Uric Acid (mg/dL) | 7.1 ± 1.2 | 7.0 ± 1.4 | 7.5 ± 2.6 | 7.0 ± 1.2 | 0.629 | |
| Platelet(×103/μL) | 230.2 ± 72.6 | 209.2 ± 59.5 | 201.9 ± 51.1 | 195.1 ± 58.2 | 0.159 |
P-value by One-Way ANOVA follow with Bonferroni multiple comparisons at type I error of 0.05 level
*P <0.05, **P <0.01. Gender (M/F), female/male; BMI, body mass index; BUN, blood urea nitrogen; HDL, high-density lipoprotein; LDL, low-density lipoprotein; HbA1C, glycated hemoglobin; WBC, white blood count; eGFR, estimated glomerular filtration rate; urine A/C ratio, urine albumin-creatinine ratio
Post hoc: aindicates group stage 1,2; bindicates group stage 3a; cindicates group stage 3b; dindicates group stage 4,5
The levels of cell-free mitochondrial (cf-mtDNA) and nuclear deoxyribonucleic acid (cf-nDNA), both urine and plasma, in varying stages of CKD patients
The plasma levels of neutrophil gelatinase-associated lipocalin (NGAL) significantly increase among the advanced stages of CKD (Table 2, Additional file 1: Fig. S1A). The plasma levels of cf-nDNA decrease as the renal function deterioration (Additional file 1: Figure S1B), in contrast to cf-mtDNA and mitochondrial copy number (MCN), which is not significantly changed among varying stages of CKD patients. There was no significant changes in urinary cf-mtDNA and cf-nDNA levels for varying stages of CKD. Urinary 8-hydroxy- 2-deoxyguanosine (8-OHdG), a symbolic marker of oxidative stress, is also not significantly changed in our study. In post hoc analysis, the data reveals that base plasma cf-nDNA is significant higher in early CKD (stage 1 and 2) than other stages and base plasma NGAL level is higher in advanced CKD (stage 3b, 4 and 5) than early CKD.
Table 2.
The levels of different parameters in varying stages of CKD
| Stage 1,2 a | Stage 3 b | Stage 3 c | Stage 4,5 d | P value | Post hoc tests | |
|---|---|---|---|---|---|---|
| Plasma cf-mtDNA (GE/mL) | 523 ± 710 | 373 ± 360 | 420 ± 664 | 112 ± 134 | 0.209 | |
| Plasma cf-nDNA (GE/mL) | 1269 ± 1195 | 698 ± 527 | 439 ± 569 | 104 ± 70.01 | <0.001** | a > b,c,d |
| Urine cf-mtDNA (GE/mL) | 0.43 ± 0.69 | 0.98 ± 3.10 | 2.78 ± 10.12 | 6.49 ± 17.85 | 0.220 | |
| Urine cf-nDNA (GE/mL) | 2.83 ± 4.60 | 4.49 ± 12.09 | 15.39 ± 44.01 | 8.01 ± 12.08 | 0.351 | |
| Plasma NGAL (ng/mL) | 394 ± 175 | 489.6 ± 174.9 | 744.2 ± 272.1 | 1096 ± 488.7 | <0.001** | a,b,<c,d |
| MCN (per cell) | 90.7 ± 66.9 | 94.9 ± 77.01 | 92.9 ± 89.9 | 110.5 ± 112.7 | 0.901 | |
| Urine 8-OH dG/Cr | 3.73 ± 1.81 | 3.78 ± 2.93 | 4.15 ± 2.65 | 2.99 ± 1.53 | 0.416 |
P-value by One-Way ANOVA follow with Bonferroni multiple comparisons at type I error of 0.05 level
*P <0.05, **P <0.01
Post hoc a indicates group stage 1,2; b indicates group stage 3a; c indicates group stage 3b; d indicates group stage 4,5
MCN mitochondrial copy number, NGAL neutrophil gelatinase-associated lipocalin; 8-OH dG 8-hydroxy-2-deoxyguanosine, cf-mtDNA cell-free mitochondrial DNA, cf-nDNA cell-free nuclear DNA, GE/mL genome equivalents/mL
The difference of baseline parameters between different (favorable vs. unfavorable) renal outcome at 6 months (Table 3)
Table 3.
The difference of baseline parameters between favorable and unfavorable renal outcome at 6 months
| Favorable renal outcome (n = 53) |
Unfavorable renal outcome (n = 70) |
P value | |
|---|---|---|---|
| Plasma cf-mtDNA (GE/mL) | 441.1 ± 525.6 | 370.6 ± 641.9 | 0.538 |
| Plasma cf-nDNA (GE/mL) | 760.9 ± 732.9 | 554.1 ± 850.9 | 0.184 |
| Urine cf-mtDNA (GE/mL) | 0.489 ± 0.751 | 3.745 ± 12.197 | 0.027* |
| Urine cf-nDNA (GE/mL) | 1.764 ± 2.388 | 15.119 ± 40.791 | 0.009* |
| MCN (per cell) | 87.21 ± 69.40 | 101.55 ± 95.75 | 0.356 |
| Urine 8-OH dG/Cr | 3.62 ± 2.44 | 4.03 ± 2.44 | 0.346 |
| NGAL (ng/mL) | 585.24 ± 251.62 | 780.85 ± 391.39 | 0.032* |
*P < 0.05, Student’s test
MCN, mitochondrial copy number; NGAL, neutrophil gelatinase-associated lipocalin; 8-OH dG, 8-hydroxy-2-deoxyguanosine; cf-mtDNA, cell-free mitochondrial DNA; cf-nDNA, cell-free nuclear DNA; GE/mL, genome equivalents /mL
Favorable renal outcome indicated eGFR at 6 months minus baseline eGFR> = 0; unfavorable renal outcome indicated eGFR at 6 months minus baseline eGFR <0
There are significantly lesser levels of urinary cf-mtDNA, cf-nDNA and plasma NGAL in the favorable renal outcome group. The levels of urinary 8-hydroxy-2-deoxyguanosine (8-OHdG), plasma cf-mtDNA and plasma cf-nDNA are not significantly different between favorable and unfavorable renal outcome groups.
The correlation between urinary cf-nDNA, cf-mtDNA and variables
As univariate analysis for the correlation between urinary cf-nDNA, cf-mtDNA and variables (Tables 4 and 5, Additional file 2: Figure S2 and Additional file 3: Figure S3), the urinary cf-nDNA is significant correlation with urine PCR, urine ACR, urine protein, cf-mtDNA and plasma NGAL. Concurrently, the urinary cf-mtDNA is significant correlation with urine PCR, urine ACR, urine protein and cf-mtDNA. In the multivariate analysis of predictors, we demonstrate that both urine P/C ratio and A/C ratio are the significant predictors for urinary cf-mtDNA and cf- nDNA levels (Tables 6 and 7).
Table 4.
Univariate correlation between urinary cf-nDNA and variables
| Variable | Rho correlation coefficient | P value |
|---|---|---|
| Urine A/C ratio (mg/g) | 0.408 | 0.005* |
| Urine P/C ratio (mg/g) | 0.314 | 0.007* |
| Urine cf-mtDNA (GE/mL) | 0.448 | <0.001** |
| Urine protein (mg/dL) | 0.246 | 0.029* |
| NGAL (ng/mL) | 0.322 | 0.001** |
*P < 0.05, **P <0.01; Spearman's rho correlation
NGAL, neutrophil gelatinase-associated lipocalin (ng/mL); cf-mtDNA, cell-free mitochondrial DNA (GE/mL); GE/mL, genome equivalents/mL; urine A/C ratio, urine albumin-creatinine ratio (mg/g); urine P/C ratio, urine protein-creatinine ratio (mg/g)
Table 5.
Univariate correlation between urinary cf-mtDNA and variables
| Variable | Rho correlation coefficient | P value |
|---|---|---|
| Urine A/C ratio (mg/g) | 0.375 | 0.008* |
| Urine P/C ratio (mg/g) | 0.349 | 0.002** |
| Urine cf-nDNA (GE/mL) | 0.448 | <0.001** |
| Urine protein (mg/dL) | 0.167 | 0.125 |
| NGAL (ng/mL) | 0.168 | 0.066 |
*P < 0.05, **P < 0.01; Spearman’s rho correlation
NGAL, neutrophil gelatinase-associated lipocalin (ng/mL); cf-nDNA, cell-free nuclear DNA (GE/mL); GE/mL, genome equivalents/mL; urine A/C ratio, urine albumin-creatinine ratio (mg/g); urine P/C ratio, urine protein-creatinine ratio (mg/g)
Table 6.
Multivariate Predictors of urinary cf- nDNA
| Predictors | Unstandardized Coefficients | Standardized Coefficients | p value | |
|---|---|---|---|---|
| B | Std. Error | Beta | ||
| (Constant) | −14.839 | 24.831 | 0.576 | |
| NGAL | −0.017 | 0.024 | −0.071 | 0.502 |
| Urine protein | −0.144 | 0.150 | −0.463 | 0.380 |
| Urine A/C ratio | −0.074 | 0.019 | −2.214 | 0.013* |
| Urine P/C ratio | 0.085 | 0.014 | 3.526 | 0.002** |
R2 = 0.960; *P <0.05, **P <0.01
NGAL, neutrophil gelatinase-associated lipocalin (ng/mL); cf-nDNA, cell-free nuclear DNA (GE/mL); GE/mL, genome equivalents/mL; urine A/C ratio, urine albumin-creatinine ratio (mg/g); urine P/C ratio, urine protein-creatinine ratio (mg/g)
Table 7.
Multivariate Predictors of urinary cf- mtDNA
| Predictors | Unstandardized Coefficients | Standardized Coefficients | p value | |
|---|---|---|---|---|
| B | Std. Error | Beta | ||
| (Constant) | −1.790 | 2.295 | 0.471 | |
| NGAL | −0.001 | 0.002 | −0.049 | 0.597 |
| Urine protein | −0.018 | 0.014 | −0.537 | 0.261 |
| Urine A/C ratio | −0.007 | 0.002 | −1.867 | 0.015* |
| Urine P/C ratio | 0.008 | 0.001 | 3.277 | 0.002** |
R2 = 0.969; *P < 0.05, **P < 0.01
NGAL, neutrophil gelatinase-associated lipocalin (ng/mL); cf-mtDNA, cell-free mitochondrial DNA (GE/mL); GE/mL, genome equivalents/mL; urine A/C ratio, urine albumin-creatinine ratio (mg/g); urine P/C ratio, urine protein-creatinine ratio (mg/g)
Evaluation of urine cf-mtDNA, and urine cf-nDNA levels as predictors of CKD patient outcomes after 6 months
Figure 1 shows the urinary cf-mtDNA and cf-nDNA levels as predictors of CKD patient outcomes after 6 months. The areas under the curves (AUC) were as follows: urine cf-mtDNA: 0.685 (0.586–0.784, P = 0.001*), and urine cf-nDNA: 0.730 (0.637–0.823, P < 0.001*). Both are better than plasma NGAL (data not shown). The optimal Youden’s index-based cut-off point was estimated. Urine cf-mtDNA cut-off value was 0.893 GE/mL, with sensitivity 0.860, 1 – specificity 0.545, and Youden’s index was 0.315. Meanwhile, urinary cf-nDNA cut-off value was 3.116 GE/mL, with sensitivity 0.907 and 1 – Specificity 0.530, and Youden’s index was 0.377. Via logistic regression analysis, we confirmed that both urine cf-mtDNA and urine cf-nDNA could be the significant predictors for dissimilar renal outcome (favorable vs. unfavorable) at 6 months (Table 8).
Fig. 1.
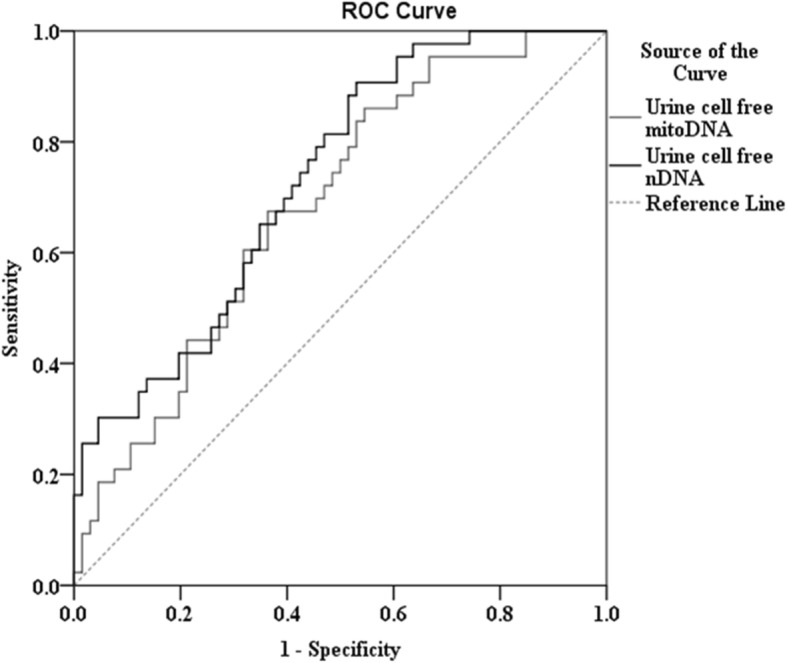
Evaluation of urinary cf-nDNA and urine cf-mtDNA as predictors of CKD patient outcomes after 6 months. The areas under the curves (AUC) were as follows: urine cf-mtDNA: 0.685 (0.586–0.784, P = 0.001*), and urine cf-nDNA: 0.730 (0.637–0.823, P < 0.001*)
Table 8.
Logistic regression analysis of predictors of favorable renal outcome at 6 months
| B | S.E. | P value | Odds ratio | 95% C.I. | ||
|---|---|---|---|---|---|---|
| Module 1 | ||||||
| Baseline eGFR | .010 | .011 | .377 | 1.010 | .988 | 1.031 |
| Gender (male) | .044 | .494 | .929 | 1.045 | .396 | 2.753 |
| Age | .023 | .020 | .249 | 1.023 | 0.984 | 1.063 |
| NGAL | .001 | .001 | .152 | 1.001 | 1.000 | 1.003 |
| Urine cf-mtDNA | .596 | .266 | .025* | 1.815 | 1.076 | 3.059 |
| Constant | −2.543 | 1.632 | .119 | .079 | ||
| Module 2 | ||||||
| Baseline eGFR | .027 | .014 | .061 | 1.027 | .999 | 1.056 |
| Gender (male) | .278 | .587 | .636 | 1.320 | .417 | 4.175 |
| Age | .054 | .024 | .027* | 1.056 | 1.006 | 1.107 |
| NGAL | .002 | .001 | .110 | 1.002 | 1.000 | 1.004 |
| Urine cf-nDNA | .255 | .102 | .012* | 1.290 | 1.057 | 1.575 |
| Constant | −5.688 | 2.182 | .009 | .003 | ||
*P < 0.05. Module 1: test urine cf-mtDNA as a covariate; Module 2: test urine cf-nDNA as a covariate
NGAL, neutrophil gelatinase-associated lipocalin (ng/mL); cf-mtDNA, cell-free mitochondrial DNA (GE/mL); cf-nDNA, cell-free nuclear DNA (GE/mL); GE/mL, genome equivalents/mL
Discussion
To the best of our knowledge, this study is the first to show the clinical significant correlation between urine cf-mtDNA, urine cf-nDNA and divergent renal outcome at 6 months. We also propose that both urine PCR and ACR could significantly predict urinary cf-nDNA and cf- mtDNA levels.
Our study showed greater amounts of cf-nDNA in earlier stage CKD, but no correlation between urinary nuclear and mitochondria cf-DNA and CKD staging. Otherwise, there were greater quantities of urinary cf-nDNA and cf-mtDNA in the unfavorable renal outcome group. Our findings were consistent with the results from a diabetes population study that showed mtDNA was readily detectable in urinary supernatant and kidney tissue, and that its levels correlated with renal function and scarring in DN [39]. However, their study did not measure plasma cf-DNA levels or a correlation between urinary nuclear cf-DNA levels and DN prognosis.
The death of cells in the tissues is an active process that supports the homeostasis of tissues [40] and increase of cf-DNA in plasma might occur due to the enhancement of programmed cell death [41]. In AKI models, the activation of autophagy provided a significant contribution to the elimination of damaged cells from tissues [41]. Our CKD cohort findings of more plasma cf-DNA in earlier stage CKD suggested that low plasma cf-DNA in later stages, even stages 4–5, indicated the deregulation or decompensation of programmed cell death by multifactorial mechanisms. As well-known, the process of autophagy is generally assumed to be a mechanism of survival or a cytoprotective mechanism that removes damaged organelles, proteins, and other macromolecules [42, 43]. An animal study showed that kidney epithelium and podocytes were sufficient to trigger a degenerative disease of the kidney with many of the manifestations of human focal segmental glomerulosclerosis, following the prevention of autophagic flux [11].
According to the results of a number of studies, cell death accompanied by the release of cf-DNA fragments into the blood and urine can occur through autophagy as well as mechanisms of apoptosis and necrosis [42, 43]. A variety of stress stimuli can induce autophagy process, such as infection, oxidative stress, starvation, hypoxia etc. The stimulation of autophagy by these stimuli produced cellular energy stress and activated 50-adenosine monophosphate activated protein kinase (AMPK) by sensing increases in AMP:ATP and ADP:ATP ratios [43, 44]. In a DN study, Dr. Wei found a statistically significant inverse correlation between urinary supernatant and intra-renal mtDNA levels [39]. These findings were inter-related to our results where high urinary cfDNA levels might be a marker to predict kidney tissue injury in CKD patients and a worse renal outcome. According multivariate analysis, we suggested that urine P/C ratio and A/C ratio are both the significant predictors for urinary cf-nDNA and cf-mtDNA levels. However, we did not find a statistically significant correlation between urinary cfDNA and CKD staging, possibly because of the relatively small population enrolled.
The detection of microalbuminuria is a standard method to diagnose the early stages of DKD; however, some patients with microalbuminuria have advanced renal disease [45]. Microalbuminuria is not as sensitive as invasive renal biopsy. There is an unmet need to identify non-invasive biomarkers of DKD in its early stages [45–49]. Our AUC analysis showed that urinary cf-nDNA and cf-mtDNA levels were reliable to predict renal function outcomes within 6 months.
Our findings were consistent with the positive correlation between NGAL levels and CKD staging. The NGAL gene product is a protein (23–26 kDa) induced by triggers of acute kidney injury [50, 51]. NGAL is rapidly released from renal tubular cells in response to various insults to the kidney. In contrast, NGAL was recently shown to be useful in the quantitation and prediction of CKD [52, 53]. Bolignano et al. reported that NGAL closely reflected the entity of renal impairment and represented an independent risk marker for the progression of CKD [52]. Liu et al., however, demonstrated that urine NGAL levels did not predict progressive CKD [54]. NGAL is a member of the lipocalin family of proteins that has been extensively studied in acute kidney injury (AKI). NGAL is a robustly expressed protein in the kidney following ischemic or nephrotoxic injury in animals [55] and humans [56]. Via AUC analysis, we demonstrated that cfDNA might be more sensitive than well-known CKD biomarkers such as NGAL (data not shown).
There are several potential limitations to this study. First, a relatively low number of patients were investigated. Secondary, we conducted a cohort study for our clinical analysis of less than 12 months duration. Third, there was subject-to-selection bias and information on exposure was subject to observation bias from the statistic model. Only large-scale collaborative multicenter or international studies will identify important risk factors. Finally, other hard outcomes in more than 6 months, beside eGFR surrogate, could be conducted in future study.
Conclusions
In conclusion, both urinary cf-mtDNA and cf-nDNA should be novel biomarkers to predict the prognosis of chronic renal diseases. The levels of plasma cf-nDNA and NGAL were significantly correlated with the severity of CKD.
Supplementary information
Additional file 1. Figure S1. Box-whisker plots for plasma NGAL (Figure S1A) and plasma cf-nDNA (Figure S1B) levels in varying stages of CKD.
Additional file 2. Figure S2. Scatter-plots for correlation analysis between urinary cf-nDNA and different variables. Figure S2A Correlation between urine cf-nDNA and urine protein/creatinine ratio. Figure S2B Correlation between urine cf-nDNA and urine albumin/creatinine ratio. Figure S2C Correlation between urine cf-nDNA and urine protein ratio. Figure S2D Correlation between urine cf-nDNA and plasma NGAL. Figure S2E Correlation between urine cf-nDNA and urine cf-mtDNA.
Additional file 3. Figure S3. Scatter-plots for correlation analysis between urinary cf-mtDNA and different variables. Figure S3A Correlation between urine cf-mtDNA and urine albumin/creatinine ratio. Figure S3B Correlation between urine cf-mtDNA and urine protein/creatinine ratio.
Acknowledgements
This project was supported by the Changhua Christian Hospital, Changhua, Taiwan. We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Abbreviations
- 8-OH dG
8-hydroxy-2-deoxyguanosine
- ACR
albumin-creatinine ratio
- AKI
acute kidney injury
- cf-DNA
cell-free DNA
- cf-mtDNA
cell-free mitochondrial DNA
- cf-nDNA
cell-free nuclear DNA
- CKD
chronic kidney disease
- DNA
deoxyribonucleic acid
- eGFR
estimated glomerular filtration rate
- eGFRCKD-EPI
CKD- Epidemiology Collaboration equation
- EMT
epithelial–mesenchymal transition
- ESRD
end stage renal disease
- GE
genome equivalents
- KDIGO
Kidney Disease Improving Global Outcomes
- NGAL
neutrophil gelatinase-associated lipocalin
- PCR
protein-creatinine ratio
- ROC
Receiver operator characteristics
Authors’ contributions
C-HH, C-CC, and C-SL designed research; C-CC, C-LK, and C-SH conducted research; P-FC and C-LW analyzed data; C-HH, C-CC wrote the paper; C-HH, C-CC, and C-SL had primary responsibility for final content. All authors read and approved the final manuscript.
Funding
This study was supported by 104-CCH-ICO-002 and 103-CCH-IRP-009 from Changhua Christian Hospital, Changhua, Taiwan. This funding body had no role in the design of this study and will not have any role during its collection, analyses, interpretation of the data, or decision to submit results.
Availability of data and materials
All data and materials are availability in the draft.
Ethics approval and consent to participate
All experimental protocols were approved by the Institutional Review Board of Changhua Christian Hospital (approval number 140306) and all the participants provided written informed consent to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chin-San Liu and Ching-Hui Huang contributed equally to this work.
Contributor Information
Chia-Chu Chang, Email: chiachuchang0312@gmail.com.
Ping-Fang Chiu, Email: 68505@cch.org.tw.
Chia-Lin Wu, Email: 143843@cch.org.tw.
Cheng-Ling Kuo, Email: 97026@cch.org.tw.
Ching-Shan Huang, Email: 149390@cch.org.tw.
Chin-San Liu, Email: liu48111@gmail.com.
Ching-Hui Huang, Email: 28071@cch.org.tw.
References
- 1.Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res. 2017;179:24–37. doi: 10.1016/j.trsl.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levey AS, Eckardt KU, Tsukamoto Y, Levin A, Coresh J, Rossert J, et al. Definition and classification of chronic kidney disease: a position statement from kidney disease: improving global outcomes (KDIGO) Kidney Int. 2005;67:2089–2100. doi: 10.1111/j.1523-1755.2005.00365.x. [DOI] [PubMed] [Google Scholar]
- 3.Meguid El Nahas A, Bello AK. Chronic kidney disease: the global challenge. Lancet. 2005;365:331–340. doi: 10.1016/S0140-6736(05)17789-7. [DOI] [PubMed] [Google Scholar]
- 4.Bello AK, Nwankwo E, El Nahas AM. Prevention of chronic kidney disease: a global challenge. Kidney Int Suppl. 2005;98:S11–S17. doi: 10.1111/j.1523-1755.2005.09802.x. [DOI] [PubMed] [Google Scholar]
- 5.Gansevoort RT, Correa-Rotter R, Hemmelgarn BR, Jafar TH, Heerspink HJ, Mann JF, et al. Chronic kidney disease and cardiovascular risk: epidemiology, mechanisms, and prevention. Lancet. 2013;382:339–352. doi: 10.1016/S0140-6736(13)60595-4. [DOI] [PubMed] [Google Scholar]
- 6.Che R, Yuan Y, Huang S, Zhang A. Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Phys Renal Phys. 2013;306:F367–F378. doi: 10.1152/ajprenal.00571.2013. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, Chen Y, Zhang P, Huang S, Zhu C, Ding G, et al. Mitochondrial dysfunction accounts for aldosterone-induced epithelial-to-mesenchymal transition of renal proximal tubular epithelial cells. Free Radic Biol Med. 2012;53:30–43. doi: 10.1016/j.freeradbiomed.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Zhang A, Jia Z, Guo X, Yang T. Aldosterone induces epithelial mesenchymal transition via ROS of mitochondrial origin. Am J Physiol Ren Physiol. 2007;293:F723–F731. doi: 10.1152/ajprenal.00480.2006. [DOI] [PubMed] [Google Scholar]
- 9.Emma F, Montini G, Parikh SM, Salviati L. Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nat Rev Nephrol. 2016;12:267–280. doi: 10.1038/nrneph.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C, Huang S, Yuan Y, Ding G, Chen R, Liu B, et al. Mitochondrial dysfunction mediates aldosterone-induced podocyte damage. A therapeutic target of PPAR gamma. Am J Pathol. 2011;178:2020–2031. doi: 10.1016/j.ajpath.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Wei Q, Liu J, Yi M, Liu Y, Liu H, et al. AKI on CKD: heightened injury, suppressed repair, and the underlying mechanisms. Kidney Int. 2017;92:1071–1083. doi: 10.1016/j.kint.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawakami T, Gomez IG, Ren S, Hudkins K, Roach A, Alpers CE, et al. Deficient autophagy results in mitochondrial dysfunction and FSGS. J Am Soc Nephrol. 2015;26:1040–1052. doi: 10.1681/ASN.2013111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong W, Mao S, Yu J, Song J, Jia Z, Huang S, et al. NLRP3 deletion protects against renal fibrosis and attenuates mitochondrial abnormality in mouse with 5/6 nephrectomy. Am J Phys Renal Phys. 2016;310:F1081–F1088. doi: 10.1152/ajprenal.00534.2015. [DOI] [PubMed] [Google Scholar]
- 14.Hallan S, Sharma K. The role of mitochondria in diabetic kidney disease. Curr Diab Rep. 2016;16:61. doi: 10.1007/s11892-016-0748-0. [DOI] [PubMed] [Google Scholar]
- 15.Higgins GC, Coughlan MT. Mitochondrial dysfunction and mitophagy: the beginning and end to diabetic nephropathy? Br J Pharmacol. 2014;171:1917–1942. doi: 10.1111/bph.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandel P, Metais P. Les acides du plasma sanguin chez l’homme. C R Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- 17.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–U142. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tin A, Grams ME, Ashar FN, Lane JA, Rosenberg AZ, Grove ML, et al. Association between mitochondrial DNA copy number in peripheral blood and incident CKD in the atherosclerosis risk in communities study. J Am Soc Nephrol. 2016;27:2467–2473. doi: 10.1681/ASN.2015060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H, Ye H, Sun Z, Shen X, Song Z, Wu X, et al. Circulatory mitochondrial DNA is a pro-inflammatory agent in maintenance hemodialysis patients. PLoS One. 2014;9:e113179. doi: 10.1371/journal.pone.0113179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo Y.M., Rainer T.H., Chan L.Y., Hjelm N.M., Cocks R.A.. (2000) Plasma DNA as a prognostic marker in trauma patients. Clin Chem 46, 319–323. PMID: 10702517. [PubMed]
- 23.Rainer TH, Lam NYL. Circulating nucleic acids and critical illness. Ann N Y Acad Sci. 2006;1075:271–277. doi: 10.1196/annals.1368.035. [DOI] [PubMed] [Google Scholar]
- 24.Antonatos D, Patsilinakos S, Spanodimos S, Korkonikitas P, Tsigas D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann N Y Acad Sci. 2006;1075:278–281. doi: 10.1196/annals.1368.037. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10:R60. doi: 10.1186/cc4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saukkonen K, Lakkisto P, Varpula M, Varpula T, Voipio-Pulkki LM, Pettilä V, et al. Association of cell-free plasma DNA with hospital mortality and organ dysfunction in intensive care unit patients. Intensive Care Med. 2007;33:1624–1627. doi: 10.1007/s00134-007-0686-z. [DOI] [PubMed] [Google Scholar]
- 27.Hurtado-Roca Y, Ledesma M, Gonzalez-Lazaro M, Moreno-Loshuertos R, Fernandez-Silva P, Enriquez JA, et al. Adjusting MtDNA quantification in whole blood for peripheral blood platelet and leukocyte counts. PLoS One. 2016;11:e0163770. doi: 10.1371/journal.pone.0163770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urata M, Koga-Wada Y, Kayamori Y, Kang D. Platelet contamination causes large variation as well as overestimation of mitochondrial DNA content of peripheral blood mononuclear cells. Ann Clin Biochem. 2008;45:513–514. doi: 10.1258/acb.2008.008008. [DOI] [PubMed] [Google Scholar]
- 29.Lee JE, Park H, Ju YS, Kwak M, Kim JI, Oh HY, et al. Higher mitochondrial DNA copy number is associated with lower prevalence of microalbuminuria. Exp Mol Med. 2009;41:253–258. doi: 10.3858/emm.2009.41.4.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu T, Li J. (2017) Clinical applications of urinary cell-free DNA in cancer: current insights and promising future. Am J Cancer Res 7, 2318–2332. PubMed PMID: 29218253; PubMed Central PMCID: PMC5714758. [PMC free article] [PubMed]
- 31.Ho PWL, Pang WF, Luk CCW, Ng JKC, Chow KM, Kwan BCH, et al. Urinary mitochondrial DNA level as a biomarker of acute kidney injury. Kidney Dis. 2017;3:78–83. doi: 10.1159/000475883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitaker RM, Stallons LJ, Kneff JE, Alge JL, Harmon JL, Rahn JJ, et al. Urinary mitochondrial DNA is a biomarker of mitochondrial disruption and renal dysfunction in acute kidney injury. Kidney Int. 2015;88:1336–1344. doi: 10.1038/ki.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botezatu I, Serdyuk O, Potapova G, Shelepov V, Alechina R, Molyaka Y, et al. (2000) Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem 46, 1078–1084. PMID: 10926886. [PubMed]
- 34.Lichtenstein AV, Melkonyan HS, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci. 2001;945:239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. [DOI] [PubMed] [Google Scholar]
- 35.Levey Andrew S., Coresh Josef, Balk Ethan, Kausz Annamaria T., Levin Adeera, Steffes Michael W., Hogg Ronald J., Perrone Ronald D., Lau Joseph, Eknoyan Garabed. National Kidney Foundation Practice Guidelines for Chronic Kidney Disease: Evaluation, Classification, and Stratification. Annals of Internal Medicine. 2003;139(2):137. doi: 10.7326/0003-4819-139-2-200307150-00013. [DOI] [PubMed] [Google Scholar]
- 36.Coresh J, Turin TC, Matsushita K, Sang Y, Ballew SH, Appel LJ, et al. Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA. 2014;311:2518–2531. doi: 10.1001/jama.2014.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo Y. M. Dennis, Tein Mark S.C., Lau Tze K., Haines Christopher J., Leung Tse N., Poon Priscilla M.K., Wainscoat James S., Johnson Philip J., Chang Allan M.Z., Hjelm N. Magnus. Quantitative Analysis of Fetal DNA in Maternal Plasma and Serum: Implications for Noninvasive Prenatal Diagnosis. The American Journal of Human Genetics. 1998;62(4):768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García Moreira V, Prieto García B, de la Cera Martínez T, Alvarez Menéndez FV. Elevated transrenal DNA (cell-free urine DNA) in patients with urinary tract infection compared to healthy controls. Clin Biochem. 2009;42:729–731. doi: 10.1016/j.clinbiochem.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Wei PZ, Kwan BCH, Chow KM, Cheng PMS, Luk CCW, Li PKT, et al. Urinary mitochondrial DNA level is an indicator of intra-renal mitochondrial depletion and renal scarring in diabetic nephropathy. Nephrol Dial Transplant. 2018;33:784–788. doi: 10.1093/ndt/gfx339. [DOI] [PubMed] [Google Scholar]
- 40.Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol. 2015;7:a006080. doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tower J. Programmed cell death in aging. Ageing Res Rev. 2015;23:90–100. doi: 10.1016/j.arr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madeo F, Zimmermann A, Kroemer G, Kroemer G. Essential role for autophagy in life span extension. J Clin Invest. 2015;125:85–93. doi: 10.1172/JCI73946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Ku YH, Kim Y, Kim Y, Kim K, Lee JY, et al. Differential proteome profiling using iTRAQ in microalbuminuric and normoalbuminuric type 2 diabetic patients. Exp Diabetes Res. 2012;2012:168602. doi: 10.1155/2012/168602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varghese SA, Powell TB, Budisavljevic MN, Oates JC, Raymond JR, Almeida JS, et al. Urine biomarkers predict the cause of glomerular disease. J Am Soc Nephrol. 2007;18:913–922. doi: 10.1681/ASN.2006070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Z, Liu X, Li M, Shao C, Tao J, Sun W, et al. Differential urinary glycoproteome analysis of type 2 diabetic nephropathy using 2D-LC-MS/MS and iTRAQ quantification. J Transl Med. 2015;13:371. doi: 10.1186/s12967-015-0712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chien HY, Chen CY, Chiu YH, Lin YC, Li WC. Differential microRNA profiles predict diabetic nephropathy progression in Taiwan. Int J Med Sci. 2016;13:457–465. doi: 10.7150/ijms.15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harder JL, Hodgin JB, Kretzler M. Integrative biology of diabetic kidney disease. Kidney Dis (Basel) 2015;1:194–203. doi: 10.1159/000439196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nickolas TL, O’Rourke MJ, Yang J, Sise ME, Canetta PA, Barasch N, et al. Sensitivity and specificity of a single emergency department measurement of urinary neutrophil gelatinase-associated lipocalin for diagnosing acute kidney injury. Ann Intern Med. 2008;148:810–819. doi: 10.7326/0003-4819-148-11-200806030-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paragas N, Qiu A, Zhang Q, Samstein B, Deng SX, Schmidt-Ott KM, et al. The Ngal reporter mouse detects the response of the kidney to injury in real time. Nat Med. 2011;17:216–222. doi: 10.1038/nm.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolignano D, Lacquaniti A, Coppolino G, Donato V, Campo S, Fazio MR, et al. Neutrophil gelatinase-associated lipocalin (NGAL) and progression of chronic kidney disease. Clin J Am Soc Nephrol. 2009;4:337–344. doi: 10.2215/CJN.03530708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malyszko J, Malyszko JS, Bachorzewska-Gajewska H, Poniatowski B, Dobrzycki S, Mysliwiec M. Neutrophil gelatinase associated lipocalin is a new and sensitive marker of kidney function in chronic kidney disease patients and renal allograft recipients. Transplant Proc. 2009;41:158–161. doi: 10.1016/j.transproceed.2008.10.088. [DOI] [PubMed] [Google Scholar]
- 54.Liu KD, Yang W, Anderson AH, Feldman HI, Demirjian S, Hamano T, et al. Chronic renal insufficiency cohort (CRIC) study investigators: urine neutrophil gelatinase–associated lipocalin levels do not improve risk prediction of progressive chronic kidney disease. Kidney Int. 2013;83:909–914. doi: 10.1038/ki.2012.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra, J., Qing, M., Prada, A., Zahedi, K., Yang, Y., Barasch, J., et al. (2003) Identification of NGAL as a novel early urinary marker for ischemic renal injury. J Am Soc Nephrol 14, 2534–2543. PMID: 14514731. [DOI] [PubMed]
- 56.Mori K, Lee HT, Rapoport D, Drexler I, Foster K, Yang J, et al. Endocytic delivery of lipocalin-siderophore-iron complex rescues the kidney from ischemia-reperfusion injury. J Clin Invest. 2005;115:610–621. doi: 10.1172/JCI23056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1. Box-whisker plots for plasma NGAL (Figure S1A) and plasma cf-nDNA (Figure S1B) levels in varying stages of CKD.
Additional file 2. Figure S2. Scatter-plots for correlation analysis between urinary cf-nDNA and different variables. Figure S2A Correlation between urine cf-nDNA and urine protein/creatinine ratio. Figure S2B Correlation between urine cf-nDNA and urine albumin/creatinine ratio. Figure S2C Correlation between urine cf-nDNA and urine protein ratio. Figure S2D Correlation between urine cf-nDNA and plasma NGAL. Figure S2E Correlation between urine cf-nDNA and urine cf-mtDNA.
Additional file 3. Figure S3. Scatter-plots for correlation analysis between urinary cf-mtDNA and different variables. Figure S3A Correlation between urine cf-mtDNA and urine albumin/creatinine ratio. Figure S3B Correlation between urine cf-mtDNA and urine protein/creatinine ratio.
Data Availability Statement
All data and materials are availability in the draft.


