Abstract
Genetic intratumoral heterogeneity is a natural consequence of imperfect DNA replication. Any two randomly selected cells, whether normal or cancerous, are therefore genetically different. We re-analyzed the extent of genetic heterogeneity within untreated cancers with particular regard to its clinical relevance. We found that homogeneity of predicted functional mutations in driver genes was the rule rather than the exception. In primary tumors with multiple samples, 97% of driver gene mutations in 38 patients were homogeneous. Moreover, among metastases from the same primary tumor, 100% of driver mutations in 17 patients were homogeneous. With a single biopsy of a primary tumor in 14 patients, the likelihood of missing a functional driver gene mutation that was present in all metastases was 2.6%. Furthermore, all functional driver gene mutations detected in the primary tumor were present among all metastases. Last, we found that individual metastatic lesions responded concordantly to targeted therapies in 91% of 44 patients. These data indicate that the cells within the primary tumors that gave rise to metastases are genetically homogeneous with respect to functional driver gene mutations and suggest that future efforts to develop combination therapies have the potential to be curative.
Introduction
Cancer is an evolutionary process spanning multiple decades. During the expansion of cell populations, intratumoral heterogeneity (ITH) arises as a natural consequence of imperfect DNA replication1–4. Whenever a cell divides, a few mutations across the whole genome are acquired. In a tumor comprised of billions of cells, every conceivable point mutation is expected to be present in at least a few cells. At the genetic level, not only is every cancer type different, but also every tumor of the same type and every cell of the same tumor are different. This extensive heterogeneity has been considered a major barrier to drug development and long-term disease control5–10. However, the success (even if short-lived) of several forms of targeted therapies suggests that intratumoral heterogeneity does not preclude initial therapeutic response. For example, in patients with metastases – who represent the majority of patients treated with therapeutic agents – it would be difficult to observe an objective response if some metastatic lesions did not harbor the targeted driver gene mutation in the vast majority of their cells. How can the successful responses to targeted therapies be reconciled with the intratumoral heterogeneity that has been observed in next generation sequencing studies?
Here we re-evaluate sequencing data in the literature with particular regard to the clinical significance of intratumoral heterogeneity. As a result of the different forms of tumor heterogeneity and the recent focus on subclonal heterogeneity, some discrepancies have arisen between the interpretations of observed heterogeneity and its clinical implications2,11. Other discrepancies arise from loose distinctions between functional driver gene mutations and passenger mutations because not every mutation within a bona fide driver gene actually drives tumorigenesis2,12–14. When these factors are taken into account, the sequencing data are in harmony with clinical experience. Homogeneity of true driver gene mutations emerges as the rule rather than the exception in treatment-naïve cancers.
Driver gene mutations and their role in tumor evolution
Before we begin with a quantitative description of tumor heterogeneity at the genetic level, we review some of the basic principles underlying the genetic determinants of cancer. Solid tumors typically require alterations of three driver genes to convert a normal cell to a cancer cell2,15–20. This number can vary among cancer types and individual patients. Each of these alterations promotes tumorigenesis by providing a selective growth advantage to the cells within their microenvironment. In other words, driver gene mutations result in an increase in cell division or a decrease in cell death, resulting in a net cell gain overall. Relatively small changes in the cell birth rate, b, or in the death rate, d, can dramatically alter the net growth rate, given by r = b − d21–26. For example, assume a tumor grows exponentially with a volume doubling time of 150 days27. The growth rate is then r = ln(2)/150 ≈ 0.5% per day. If the cells within the tumor divide every 4 days28,29, then b = 1/4 = 0.25 per day and the death rate is d = b − r = 0.245 per day according to the above given formula. Suppose a driver gene mutation then occurs in one cell of this tumor. A driver gene mutation causes an increase in the birth rate of on average 0.4%21, though some driver gene mutations can confer much stronger or weaker selective advantages30,31. A typical new birth rate is then b′ = b(1 + 0.004) = 0.251 per day. If the death rate is unchanged, then the new growth rate of this cell is r′ = b′ − d = 0.6% per day. The new mutation therefore increases the net growth rate by 20% per day (= 0.6%/0.5%). The number of these mutated cells will then double every ≈ 120 days (= ln(2)/0.006) as opposed to 150 days of the cells without this additional driver gene mutation. Over many months to years, this difference is sufficient for cells with driver gene mutations to progressively outgrow the cells without this new mutation in the tumor21,32–35.
Driver genes can be classified into well-defined signaling pathways and their effects depend on the tissue origin of the cells. A few dozens of driver genes are recurrently mutated across many cancer types. Most driver genes are recurrently mutated only in a few tissues and cancer types. Functional consequences of mutated driver genes complement each other, resulting in patterns of co-occurrence and mutual exclusivity among driver gene mutations36,37. In the case of oncogenes, a single missense mutation generally represents the genetic alteration responsible for activating it. In the case of tumor suppressor genes, inactivation typically requires two separate mutations. One of these mutations is usually intragenic (producing a stop codon, for example), while the other is often a large deletion that inactivates the other allele38,39. It is important to note that driver genes (e.g., NOTCH1 or CDH1) can act as oncogenes in one cancer type but as tumor suppressor genes in other cancer types2,40, reflecting the different signaling circuitries that define organogenesis.
The first driver gene mutation allows the formation of a small clonal expansion, creating a benign lesion17,41,42. These lesions typically grow to a size of a few million cells and are usually undetectable clinically. The second driver gene mutation results in a second wave of clonal local expansion, often leading to a clinically detectable, though still benign, tumor19,43–45. The third mutation endows the tumor cell not only with a further selective growth advantage but also with the ability to expand its environment by invading through the basement membrane19,45, thereby defining malignancy (a.k.a. cancer). Advanced tumors typically contain frequent gains and losses of focal genomic regions, chromosome-arms, and whole chromosomes39,46–48. Depending on the cancer type, whole-genome duplication (WGD) occurs in 10–80% of cancers which could lead to subsequent chromosomal alterations38,49. To date, it has been impossible to determine whether the rate of chromosome gains and losses (chromosome instability) increases during tumor progression. However, a new approach employing organoids should make this possible in the future50. Despite intense efforts, no genetic alterations have been identified that unambiguously endow the cell with the ability to metastasize51. The process of metastasis seems stochastic; once a cancer has developed (i.e., acquired invasive growth capability), it may only be a matter of time before a cell invades a vessel and seeds a distant metastasis13.
Driver gene mutations are “clonal” if they are present in virtually all cells of the cancer. Clonal mutations are also called “truncal” because they are in the trunk of the tumor evolutionary tree. Subclonal mutations represent those that are present in only a subset of the cancer’s cells. Subclonal mutations are sometimes described as “branched” because they occur on a branch of the tree when the evolutionary trajectory of the tumor can be assessed. Though three driver gene mutations appear to be sufficient for the development of a malignant solid tumor, more than three driver genes can be observed in cancers, because the evolutionary process of tumors never stops. These additional mutations can be clonal, but are more likely to be subclonal compared to the first three mutations driving the disease.
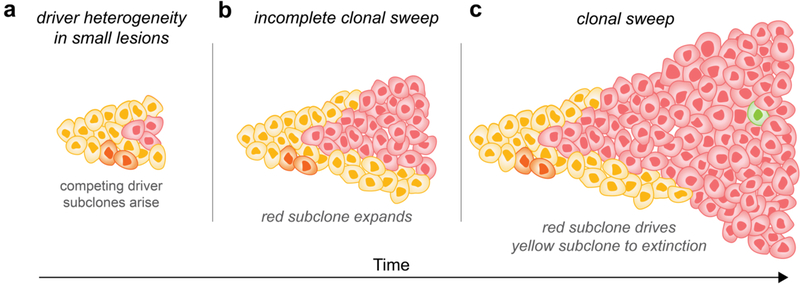
Heterogeneity among driver gene mutations in benign tumors and expanded clones occurs frequently18–20,45,52–54. However, this heterogeneity can essentially be erased in the primary tumor by a mutation that endows a strong growth advantage (e.g., the third mutation in the chain resulting in the advent of malignancy). In the parlance of population genetics, such a mutation results in a clonal sweep: the vast majority of the cells within the cancer descend from this mutant cell, which outcompeted other clones in the developing tumor (Fig. 1).
Fig. 1: Clonal sweeps give rise to driver gene mutation homogeneity.
Subclonal cells containing different driver genes emerge over time. Subclones of cells with different driver gene mutations are colored yellow, orange, red, or green. a | Driver gene mutation heterogeneity in a small lesion. b | Lesion grows with the expansion of both the yellow and the red subclones. Some subclones may progress, others remain stable or regress. c | The red subclone sweeps through the lesion and eradicates the preexisting driver gene heterogeneity harbored by the yellow subclone. New driver gene mutations in another subclone (green) may be acquired during the growth of the lesion.
Relatively few mutations result in a gain-of-function or loss-of-function of a driver gene to confer a selective growth advantage. These driver gene mutations are functional because they increase the rate of cell division or decrease the rate of cell death. Just because a mutation occurs in a driver gene does not mean that it drives tumorigenesis. Mutations in driver genes that are not functional should be considered passenger mutations because of their effectively neutral consequences on selection13,23,40,55. With sufficiently deep sequencing, essentially every possible point mutation in every gene will be observed. Many candidate driver gene lists already contain more than 1000 genes, and driver gene mutations are becoming increasingly likely to be false-positives. Each mutation in a driver gene needs to be carefully assessed before functional consequences should be indicated. In this review, we have attempted to rigorously distinguish mutations that are likely to be functional from those that are not.
A final and often unappreciated point about genetic heterogeneity is that it is not confined to tumors. Any two randomly selected normal cells from a healthy adult contain hundreds to thousands of genetic alterations that distinguish the two cells3,48,56–60. One can precisely quantify heterogeneity and evolutionary relationships through various metrics such as the Simpson index, the Shannon index, or the Jaccard similarity index26,61–66. Applied to normal cells and cancer cells, these metrics reveal that cancer cells are more similar to each other than normal cells are to each other. For example, the fraction of distinct genetic alterations between any two random cancer cells from a single cancer is much smaller than between any two random normal cells from a normal organ56,64. The reduced heterogeneity in cancer cells is a consequence of the clonal sweeps described above, wiping out all prior heterogeneity among the other clones (Fig. 1). Viewed in such a quantitative context and in comparison to normal cells, the extent of intratumoral genetic heterogeneity does not emerge as a distinctive feature of cancer. Nevertheless and independent of the degree of heterogeneity, the key questions addressed in this review remain the same: in which situations is genetic heterogeneity clinically important and why?
Forms of tumor heterogeneity
Genetic differences between cancers of two patients, each with a different tumor type (intertype heterogeneity) are well-known (e.g., mutations present in prostate cancer vs. in pancreatic cancer). Even cancers of the same type in two different individuals are genetically very different and may share very few or no somatic mutations (intratype heterogeneity). These differences are the basis for precision medicine: patients are treated with drugs that target the genetic alterations that are present in their particular tumor. This contrasts with conventional chemotherapeutics, in which all patients with a given tumor type are treated identically. Perhaps the epitome of personalized medicine is illustrated by the “tumor type-agnostic” approval of immune checkpoint inhibitors for cancers67. The drug pembrolizumab is now recommended for treatment of patients whose tumors are mismatch repair deficient, regardless of the tumor type. A similar tumor type-agnostic indication for patients with tumors bearing TRK mutations was recently approved68.
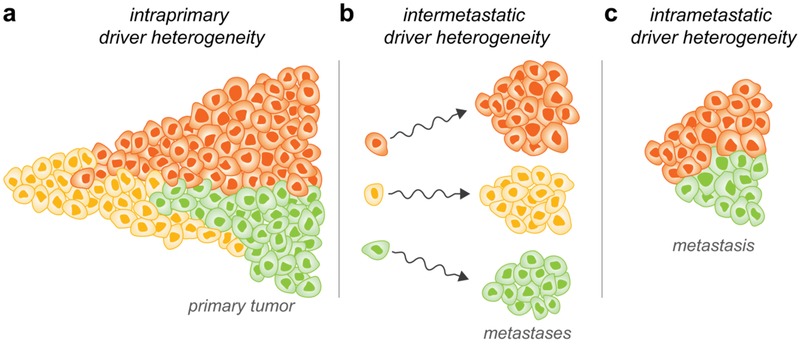
This review focuses on three forms of heterogeneity that affect the same cancer in a single induvidual2,69. Intraprimary heterogeneity refers to the genetic heterogeneity between two cells of the same primary tumor; intermetastatic heterogeneity refers to the genetic heterogeneity between the cells that seed distant metastasis, and intrametastatic heterogeneity refers to the genetic heterogeneity between two cells of the same metastasis (Fig. 2).
Fig. 2: Three forms of heterogeneity within a single patient.
Subclonal cells containing different driver genes emerge over time. Subclones of cells with different driver gene mutations are colored yellow, orange, or green. a | Intraprimary heterogeneity: Subclones containing different driver gene mutations expand in parallel. b | Intermetastatic heterogeneity: Cells with different driver gene mutations disseminate and colonize distant sites, leading to driver gene heterogeneity among the founding cells of different metastases. c | Intrametastatic heterogeneity: Mutations in the founding cells of a metastasis clonally expand so that they are present in all cells of the metastasis. However, additional driver gene mutations can be acquired during the growth process of the metastatic lesion. Whether intrametastatic heterogeneity can arise from the dissemination of new clones from one metastatic lesion to another is the subject of ongoing research80,140,141.
Intraprimary heterogeneity
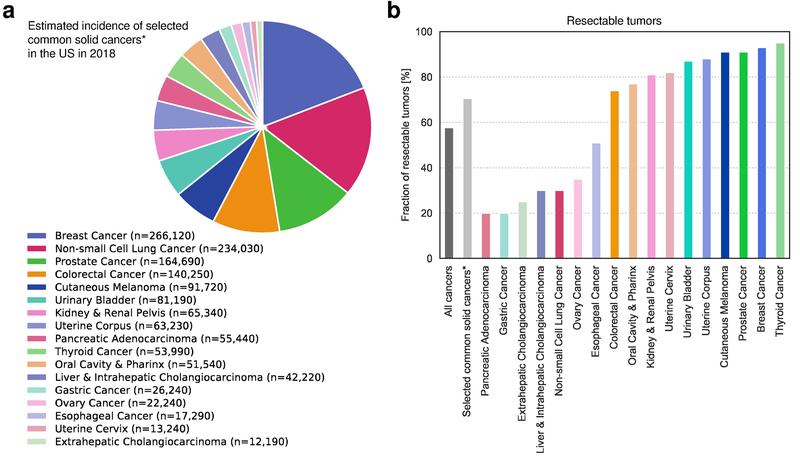
Intraprimary heterogeneity can directly affect patient outcomes only when the primary tumor cannot be excised. As depicted in Fig. 3, in ~57% of all newly diagnosed cancers in the US, the primary tumor is surgically resectable. For selected common solid cancers (representing 81% of all new cancer cases in the US), ~70% of primary tumors are surgically resectable and intraprimary heterogeneity is clinically irrelevant70. Nonetheless, intraprimary heterogeneity can sometimes provide important prognostic information61,66,71,72. Moreover, when the primary tumor cannot be completely resected, such as is nearly always the case in glioblastomas, intraprimary heterogeneity among driver genes can limit the response to therapies that target such driver genes. Such driver heterogeneity explains, for example, why agents that target the EGFR variant translocation have not achieved notable clinical success; not all cells within most glioblastomas contain this translocation73.
Fig. 3: Majority of primary tumors are surgically resectable at the time of diagnosis.
a | Estimated incidence of selected solid cancers in the United States in 201870. *Solid cancer types with more than 10,000 estimated new cases per year were selected. Selected cancer types represent approximately 81% of all new cancer cases in the US. Hematological cancers, cancer types with less than 10,000 estimated new cases per year, and cancers for which surgery is not routinely recommended (i.e. small-cell lung cancers), or for which the primary tumor often cannot be completely resected (i.e. glioblastoma) were excluded. b | Fraction of resectable primary tumors across cancer types in the US. Approximately 70% (984,506/1,400,960) of newly diagnosed cases of these solid cancer types (panel a) and approximately 57% (984,506/1,735,350) of all newly diagnosed cancer cases are resectable.
Intraprimary heterogeneity can be assessed through multi-region or single cell sequencing of primary tumors. Several studies of this sort have revealed driver gene mutations present only in a subset of the evaluated regions of some tumors74–81. Some of this heterogeneity can be explained by sequencing noise, low neoplastic cell content, or low sequencing depth in individual samples81–84. For example, if the depth of sequencing for a specific genomic position is only n = 10 reads, and the neoplastic content of the tumor DNA used for sequencing is 50%, then the probability of completely missing that mutation (k = 0) when it occurs in 100% of the cancer cells is (assuming that one of two alleles in the tumor cells is mutated, f = 0.5/2). The probability of missing at least one of three such mutations is if it is assumed that the mutation needs to be observed twice (k < 2) to ensure that it is real rather than an artifact of sequencing. Multi-sample analysis accentuates this problem because the probability that sequencing depth or neoplastic cell content is low in at least one sample strongly increases with the number of analyzed samples23. Most sequencing data analysis methods have been optimized for single-sample analysis, and few methods have been described to minimize artifacts in the context of multi-sample analysis82,85. Some of the newer methods to control for multi-sample analysis artifacts have been applied to the sequencing data described in this study (Supplementary Methods S1).
Minimal functional consequences of subclonal driver gene mutations
Intraprimary heterogeneity is conferred by subclonal mutations. But are the subclonal mutations that occur in driver genes functional? As noted above, just because a mutation occurs in a driver gene does not mean that the particular mutation actually drives tumorigenesis. To address this question, we reanalyzed data from 38 untreated primary epithelial tumors derived from six cancer types in which multi-region sequencing had been performed (13 ovarian86, 10 colorectal23,79,87,88, 9 breast78, 4 pancreatic64, 1 gastric89, and 1 endometrial cancer13). In each region, we classified mutations as present if their “present probability” was at least 80% according to the Bayesian inference model of Treeomics82 (Supplementary Methods S1). Mutations were classified as absent if their “absent probability” was at least 80%. Mutations that were present in at least one sample but not in all samples were classified as subclonal. Ambiguous mutations that did not reach these presence or absence probability thresholds in all samples of a patient were excluded from the analysis to minimize effects of low sequencing depth or low neoplastic cell content. We thereby identified 19 subclonal, non-synonymous mutations within the 299 driver genes listed in the TCGA consensus list40 (Supplementary Table S1; Fig. 4a). The number of subclonal mutations was considerably less than the number of clonal mutations (19 subclonal vs. 143 clonal in the same 38 cancers).
Fig. 4: Intratumoral heterogeneity in untreated primary tumors and among metastases.
Intraprimary heterogeneity analysis based on 96 samples from 38 subjects (13 ovarian86, 10 colorectal23,79,87,88, 9 breast78, 4 pancreatic64, 1 gastric89, and 1 endometrial cancers13; Supplementary Methods S1). Intermetastatic heterogeneity analysis based on 67 metastases samples of 17 subjects (6 pancreatic64, 4 endometrial13,104, 3 colorectal79, 2 breast103, 1 gastric89, 1 prostate105 cancers). a | Driver gene mutations present in all samples from a single primary tumor were more frequently predicted to be functional than those present in only a subset of the samples from a primary tumor (54% vs. 11%, P < 0.001). The fraction of subclonal functional driver gene mutations (11%) was not significantly different from the fraction of clonal or subclonal functional passenger gene mutations in the same tumor (3.3% and 2.3%). b | Mutations in driver genes that were present among all metastases samples of a subject were more frequently predicted to be functional than those present only in a subset of metastases samples (65% vs. 0%, P < 0.001). The fraction of subclonal functional driver gene mutations (0%) was not significantly different from the fraction of clonal and subclonal functional passenger gene mutations in the same samples (4.1% and 6.6%). c | On average 69% and 66% of the mutations per patient were clonal (homogeneous) among primary tumor samples and among metastases, respectively. Mutations in putative driver genes were significantly more homogeneous among primary tumor samples (90%, P < 0.001) and among metastases (84%, P < 0.0048) than mutations in all genes (sum of passenger genes and driver genes). Likely functional driver gene mutations were even more homogeneous among primary tumor samples (98%, P < 0.0042) and among metastases (100%, P < 0.0018) than other categories of mutations. Two-sided Fisher’s exact tests were used in panels a and b. Two-sided Wilcoxon rank-sum tests were used in panel c. Thick black bars denote 90% confidence interval. Numbers in brackets denote number of variants in each group. ** P < 0.01; *** P < 0.001.
To determine whether these 19 subclonal mutations were likely to be functional, we pooled information from various databases and used bioinformatic methods to create a two-phase algorithm, called LiFD (Likely Functional Driver; Supplementary Methods S1). In the first phase of LiFD, mutations that are annotated in OncoKB90, the catalog of validated oncogenic mutations (CGI, Cancer Genome Interpreter12), known cancer hotspots91, or present at least 4 times in COSMIC92 (Catalogue of Somatic Mutations in Cancer) were predicted to be functional. If a mutation was not annotated as functional in the first phase, we used CHASMplus14,93, FATHMM94, CanDrA95, CGI12, and VEP96 to predict the functional consequences of this mutation in the second phase of LiFD. If the majority of the methods that produced a result predicted functionality, we classified the mutation as likely functional (Supplementary Methods S1; Supplementary Fig. S1). This method was lenient in that it allowed mutations scored as significant as judged by only a subset of tools to be considered functional in LiFD.
Through LiFD, we found that clonal mutations in putative driver genes were significantly more likely to have functional consequences than subclonal driver gene mutations in the same tumors (Fig. 4a; Supplementary Table S1). Only two (11%) of the 19 subclonal mutations compared to 77 (54%) of the 143 clonal mutations were predicted to be functional (P < 0.001, two-sided Fisher’s exact test; Fig. 4a). The two likely functional subclonal driver gene mutations occurred in PTEN in the same patient, one in each of two regions of the cancer. When evaluated at the individual tumor level, 97% (37/38) of the tumors evaluated had no functional subclonal driver gene mutations (Fig. 4). On average, we found 2.1 likely functional driver gene mutations per primary tumor.
Survival analysis of patients with subclonal driver gene mutations
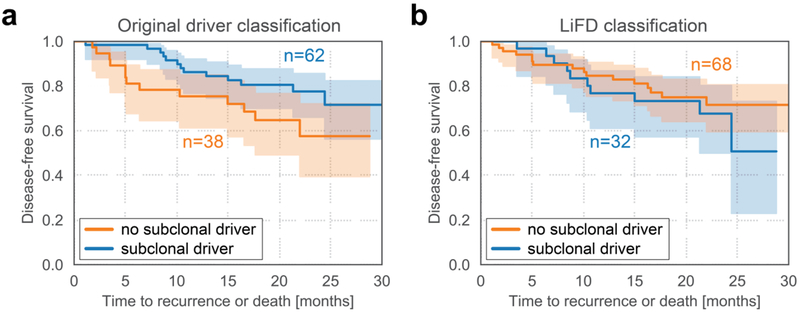
Another clinically important question is whether patients whose tumors have subclonal mutations in driver genes and are thus heterogeneous with respect to driver gene mutations have a worse prognosis than patients without such heterogeneity61,71,76,97–99. To address this question, we reanalyzed data from 100 early-stage non-small-cell lung cancers where 62 subjects were reported to have at least one subclonal driver gene mutation72 (either point mutation or short insertion or deletion). We did not find a statistically significant difference in disease-free survival between patients that exhibited subclonal driver gene mutations (n = 62) and those that did not (n = 38) based on the originally reported heterogeneity and driver classification (Fig. 5a). When the heterogeneity classification and the driver gene mutation classification described above were used (Supplementary Methods S1), the number of cancers harboring subclonal driver gene mutations decreased from 62 to 32. Nevertheless, no significant difference in patient outcomes was observed (Fig. 5b). Though it would be reasonable to expect that tumors that had acquired additional driver gene mutations would be more aggressive, allowing escape from host control and conferring worse survival, this was not the case. We again found that clonal mutations in driver genes were significantly more likely to be functional than subclonal ones although the high mutation rate in lung cancers complicates the driver functionality prediction (33% vs 20%, P < 0.001, two-sided Fisher’s exact test).
Fig. 5: Subclonal driver gene mutations did not lead to worse patient outcomes in patients with non-small-cell lung carcinomas.
Analysis based on data of Jamal et al.72. a | No statistically significant difference in disease-free survival between patients that harbored subclonal driver mutations (n = 62) and those that did not harbor any subclonal driver gene mutations (n = 38), according to the originally provided driver and heterogeneity classification. Shaded areas denote 90% confidence interval. The hazard ratio of subjects with subclonal driver gene mutations was 0.51 (95% CI: 0.24 − 1.1; P = 0.088, likelihood ratio test). b | When the LiFD algorithm for identifying functional driver gene mutations was applied, the number of patients that harbored subclonal driver gene mutations was 32 and the number of patients that did not harbor any functional driver gene mutations was 68. No statistically significant difference in disease-free survival between patients that harbored subclonal functional driver gene mutations and those that did not harbor subclonal functional driver gene mutations was observed. The hazard ratio of subjects with subclonal functional driver gene mutations was 1.4 (95% CI: 0.61 − 3.0; P = 0.46, likelihood ratio test). In panel b, a different heterogeneity classification was performed than in panel a (Supplementary Methods S1).
Single biopsies generally provide adequate information for precision medicine
Intraprimary tumor heterogeneity also informs the number of biopsies required for choosing the optimal targeted therapies for metastatic lesions. For example, if only a single region of a primary tumor is biopsied, what would be the probability of selecting a functional (and perhaps targetable) driver gene mutation that was not present in all metastases? Conversely, what would be the probability of missing a functional driver gene mutation that was present in all metastases if only a single biopsy were used for sequencing analysis? To address these questions, we reanalyzed data from 14 treatment-naïve subjects in whom at least one sample of the primary tumor and at least two distinct metastases were sequenced13. First, any detected functional driver gene mutations present in a primary tumor biopsy were also present among all metastases of that patient. Second, we found that the proportion of functional driver gene mutations present in all metastases but missing from a primary tumor biopsy was on average 2.6% (Supplementary Methods S1; Supplementary Table S2). These data support the conclusion that in most patients, a single biopsy of a primary tumor captures the information necessary for therapeutic choices about the treatment of extant or presumptive metastases. Because untreated samples of the primary tumor and of multiple metastases are rarely available, these analysis results are based on a relatively small cohort, representing only five cancer types. Further research will be required to determine the clinical scenarios in which multiple rather than single biopsies are advantageous. For example, when lesions are multi-focal, such as in the esophagus, evaluation of biopsies from several sites clearly provides useful information66,71,72,100.
Intermetastatic heterogeneity
Intermetastatic heterogeneity is the most important form of heterogeneity for patients with primary tumors that can be completely excised2,13,101. Intermetastatic heterogeneity of driver gene mutations determines whether all lesions have the capacity to respond to a given targeted therapeutic agent. If even a single lesion lacks the driver gene mutation being targeted, and therefore continues to grow following the initiation of therapy, it is much less likely that an objective response will be observed than if all lesions harbor the mutation102.
Most studies of intratumoral heterogeneity have focused on primary tumors although metastases are responsible for most cancer-related deaths. Moreover, intermetastatic heterogeneity provides a uniquely informative view of intraprimary heterogeneity. If intraprimary heterogeneity were important for treatment response of metastatic disease, some metastases would be derived from the subclones in the primary tumor that define this heterogeneity. In prior studies addressing this issue, patients have often been treated with toxic or mutagenic agents which complicate the interpretation of mutations observed in metastases. We therefore surveyed the literature for patients in which at least two distant treatment-naïve metastases underwent genome/exome-wide sequencing13. Across all cancer types, and among tens of thousands of patients in whom genome-wide sequencing was performed, only 17 subjects were found to fulfill these requirements13,64,79,89,103–107 (6 pancreatic, 4 endometrial, 3 colorectal, 2 breast, 1 gastric, 1 prostate cancer; Supplementary Methods S1). Using the LiFD classification framework, we found that 65% (44/68) of all clonal non-synonymous mutations in driver genes were predicted to be functional while no (0/14) subclonal mutations were predicted to be functional (Fig. 4b; Supplementary Table S3). Hence, all of the predicted functional driver gene mutations were present in all metastatic lesions of individual patients (Fig. 4c). The fraction of subclonal driver gene mutations predicted to be functional (0%) was not significantly different from the fraction of clonal or subclonal passenger gene mutations predicted to be functional (4.1% and 6.6%, respectively). We repeated this functional analysis with a more expansive driver gene list and obtained similar results (Supplementary Fig. S2, Supplementary Methods S1).
For previously treated metastases, varying degrees of intermetastatic heterogeneity of driver gene mutations have been reported within and across cancer types64,74,101,108–112. This is not unexpected because therapies create selective bottlenecks that unmask additional mutations. Which of these additional mutations in driver or passenger genes actively contribute to progression and resistance is often unclear, particularly when no functional analysis was performed. Moreover, the selective bottleneck enforced by a therapeutic agent can be very different from the selective bottlenecks operating during cancer initiation and progression: a potent driver mutation of cancer initiation may not contribute to resistance and a potent resistance mutation may not drive carcinogenesis. Some of the additionally observed driver gene mutations can be explained by their role in conferring resistance. For example, KRAS mutations following treatment with EGFR inhibitors, loss of PTEN following treatment with PIK3CA inhibitors, or FGFR2 mutations following treatment of cholangiocarcinoma patients with FGFR inhibitors6,9,27,113–117. The mutations that confer resistance to targeted agents such as these are lesion-specific, and can differ considerably among the metastases of a single patient. This type of intrametastatic heterogeneity is very important for selecting second line therapies. However, it is typically not relevant for selecting the initial therapies for newly diagnosed patients because it is usually only present in a tiny fraction of metastatic cells prior to therapy8,23.
Clinical correlates of intermetastatic heterogeneity
The success of targeted therapies for most cancers is dependent on the homogeneity of the targeted driver gene mutations among metastases. At present, all targeted therapies are based on oncogene alterations. Tumor suppressor gene alterations cannot yet be targeted by drugs because there is currently no way to restore the function of an inactivated protein. The conclusions described above, based on genome-wide sequencing of metastatic lesions in untreated patients, are strongly supported by sequencing studies of individual oncogenes in metastases from the same patients. For example, it has been shown that mutations in KRAS, NRAS, BRAF, and PIK3CA are nearly always concordant across all metastatic lesions in colorectal cancer patients118,119. The same EGFR mutations are similarly almost always found in all metastatic lesions in lung adenocarcinoma patients120, and the same BRAF and NRAS mutations are found in metastatic lesions of patients with melanomas121,122.
Perhaps most importantly, are the actual responses observed in patients treated with targeted therapies consistent with the predicted homogeneity among driver genes emphasized in this review? Very few published studies provide detailed data on the response of individual lesions to targeted therapies. In general, most studies report only the data required to meet RECIST criteria for response, i.e., whether the sum of the diameters of all measured lesions decreases or increases102. An objective response is reported if the sum of those diameters decreases by more than 30%, and tumor progression is reported if the sum increases by more than 20%. However, we gathered data from two clinical trials that more directly addressed the question considered here as a proof of concept, i.e., if one metastatic lesion responds to a targeted therapy, do all index metastatic lesions respond too, or do some lesions continue to grow?
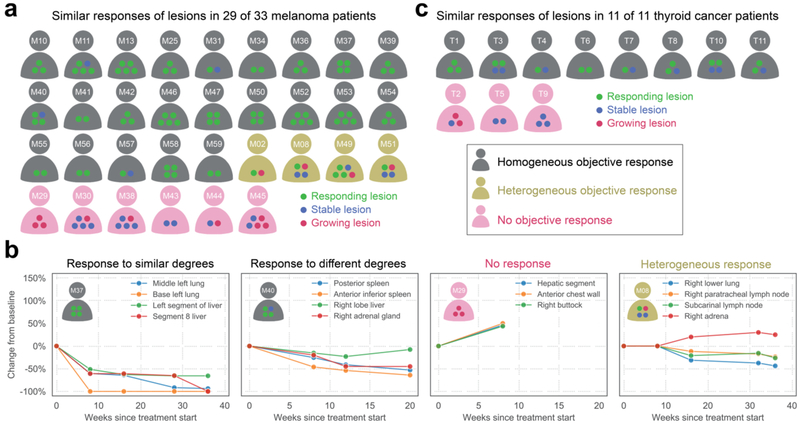
In the first of the two trials, 33 patients with melanoma and at least 2 index lesions were treated with targeted therapy123,124 (dabrafenib, trametinib, GSK2141795). All patients had a V600E mutation in BRAF in their primary tumors. Of these 33 patients, a decrease of ≥30% in diameter of at least one index lesion was observed in 27 patients (Fig. 6a; Supplementary Table 4). We then determined how often one of the other index lesions in these 27 patients grew during the initial treatment period, generally 8 to 16 weeks and prior to the emergence of resistance. We found that in 23 of the 27 patients, none of the other index lesions grew by ≥10% (representative examples in Fig. 6b).
Fig. 6: Lesions of individual patients respond similarly to targeted therapy.
Patients are represented by humanoid cartoons. Circles within the humanoids represent responding, stable, or growing lesions (green, blue, and red, respectively). A lesion was considered to respond if it shrank by at least 30% in diameter; a lesion was considered to grow if its diameter increased by at least 10%; and a lesion was considered to be stable if it did not grow by at least 10% or shrink by at least 30%. a | At least one lesion responded in 27 of 33 melanoma patients123,124. In 23 patients (gray humanoids), no lesion grew. In four patients (yellow humanoids), one of the lesions grew while the others responded, i.e., a heterogeneous response was observed. In six patients (red humanoids), no lesion responded. b | Examples of different types of responses to targeted therapy. All lesions responded in patient M37. One lesion responded less well than three other lesions in patient M40. None of the lesions responded in patient M29. One lesion responded, two lesions remained stable, and a fourth lesion grew in patient M08. c | At least on lesion responded in 8 of 11 thyroid cancer patients125. In eight patients (gray humanoids), no lesions grew. In three patients (red humanoids), no lesion responded. Additional information about these patients’ responses are provided in (Supplementary Tables S4–S5). In 91% (40/44) of the patients analyzed (with melanomas or thyroid cancers), all lesions responded similarly to targeted therapy.
In the second trial, 11 patients with metastatic thyroid cancers were treated with pazopanib (VEGFR/PDGFR/RAF inhibitor) and trametinib (MEK inhibitor)125. Of these 11 patients, a decrease of ≥30% in diameter of at least one index lesion was observed in 8 patients (Fig. 6c; Supplementary Table 5). In all 8 patients, none of the other index lesions grew by ≥10% during the first 6 months after the start of therapy or until end of treatment (whichever occurred first).
The conclusions from both trials are thereby similar. When an objective tumor response from a targeted therapy is observed in one metastatic lesion, it is common (89% of 35 patients) for all lesions in that patient to respond to the therapy. If we include cases where no lesion achieved an objective response, similar responses (regression, growth, or no stable diameters) of all index lesions were observed in 91% (40/44) of the patients. Note that this does not mean that all index lesions respond in an identical fashion. In some cases, all lesions regressed to the same extent (Fig. 6b). In other cases, the timing and degree of response varied. The timing and degree of the response is dependent on a host of factors other than the presence of the targeted driver gene mutation in the metastatic lesion. In particular, the timing and extent of a response depends on the vascularity in each tumor because this determines the dose actually delivered to the lesion. The microenvironment can also impact drug delivery and local immunity might play a role126. Moreover, the degree of a response depends on the number of cells in the lesion that contain a mutation that can confer resistance (intrametastatic heterogeneity). These additional factors are currently beyond the control of the oncologist. But unless all metastatic lesions contain the targeted mutation, a targeted therapy will usually not be very useful. Fortunately, these clinical results confirm the above noted sequencing studies which demonstrated in all 17 patients evaluated (and in all 67 distinct metastases from those patients), if one metastasis contained a predicted functional driver gene mutation, all the other metastases of the same patient contained the identical mutation.
Intrametastatic heterogeneity
Intrametastatic heterogeneity does not impact the initial response to therapy but is responsible for disease recurrence after a response6,8,9,115. Such recurrences result from mutations present in a small fraction of the cells within each metastasis prior to treatment; the larger the lesion, the more likely that such resistant cells exist8. Thus, treatment of relatively early metastatic states – with conventional chemotherapeutic agents, with targeted agents, or with immunotherapeutic drugs – are much more likely to be successful than treatment of bulky metastatic disease. Although we did not formally analyze intrametastatic heterogeneity here, a recent study of 2,520 metastases in which deep whole genome sequencing was performed showed that 96% of all driver gene point mutations were clonal38. Similarly, 95% of driver gene mutations were shared among the metastatic lesions of individual patients in a study of 100 clear-cell renal cell carcinoma patients127.
Conclusions
The results described above lead to several important conclusions. First, tumors are heterogeneous, but the term “heterogeneity” needs to be used with caution and nuance, as normal cells are heterogeneous too. We show that the extent of intraprimary heterogeneity of functional driver gene mutations is relatively small (mean of 2% per patient; Fig. 4c). We point out that unless the primary tumor cannot be excised, the extent of this heterogeneity is of little clinical consequence. More importantly from an oncologist’s viewpoint, the extent of heterogeneity of functional driver gene mutations among metastases of the same patient is minimal (Fig. 4c). Moreover, the sequencing data amassed in the literature is highly concordant with clinical experience (Fig. 6).
Multi-region sequencing enables a more refined inference of the genealogy of tumors, offering key insights into the nature of the tumorigenic process128,129. For example, such sequencing has allowed investigators to determine the time course of tumorigenesis, which is now generally acknowledged to take decades19,29,39,130 (a result also consistent with clinical experience). A particularly informative example of this principle was recently published: the first genetic alteration in kidney cancers occurs during early adulthood, decades prior to the onset of malignancy131. The growth of a primary tumor and its subclonal diversification occur much after the first genetic alterations, typically a few years to a decade before diagnosis39,131. Multi-region sequencing is also critical to evaluate the potential of targeted therapies to be effective in cancers that cannot be surgically excised in their entirety, such as brain tumors, or to forecast the future evolutionary trajectory of a tumor23,24,61,132 (Fig. 3). But for tumors that can be completely excised, sequencing of a single region from the primary tumor is generally adequate to find the clonal mutations susceptible to targeted therapies (Fig. 4; Supplementary Table S2).
Our study has several limitations. One of them is that our analyses of heterogeneity was limited to intragenic mutations (single base substitutions and small insertions and deletions). Other types of genetic alterations, as well as epigenetic alterations, undoubtedly play a role in tumorigenesis. For example, copy number alterations occur nearly ubiquitously in cancers and can confer selective advantages133. Unfortunately, the target genes selected for by such copy number alterations are notoriously difficult to identify71,133–137. With dramatic changes in copy numbers (such as occurs with true amplifications in ERBB2 or EGFR), the target gene can be identified. In the much more usual case of small changes in copy number (2 to 3-fold imbalances), it is unknown whether such copy number changes reflect a single underlying culprit gene, the combined effect of many genes131,133,135, or simply represent passenger alterations arising as a result of chromosomal instability39,49,138. Mutations in non-coding regions of the genome also play a role in certain cancers. However, except for mutations in the TERT promoter, individual non-coding mutations that drive tumorigenesis are rarely recurrent139. Moreover, similar to copy number changes, it is currently very challenging to determine whether a given non-coding mutation is functional; tools like those used here are not yet available for predicting the effects on tumorigenesis of non-coding mutations. The same challenges apply to the thousands of epigenetic changes that occur in every cancer. Unless these changes occur in the ~300 well-documented driver genes2,40, it is currently unfeasible to reliably discern which of them are likely to drive tumorigenesis, i.e., to cause a selective growth advantage in the actual human tumor microenvironment in which they occurred. Another limitation involves the difficulty of identifying functional genetic mutations. The LiFD classification framework combines the evidence of various databases and algorithms to minimize false-positives and false-negatives but is still only predictive rather than definitive. Finally, these studies of untreated cancers with numerously sampled primary tumors and metastases by necessity involved only a small number of cases. As sequencing becomes routine in clinical and research studies, we expect it will be possible to extend our type of evaluation to many other cancer cases.
The results reviewed here provide optimism for future targeted combination therapies. If intermetastatic heterogeneity in driver genes was routinely found, there would be little hope of achieving meaningful responses in most patients. We find that such heterogeneity is rare (Fig. 4), and this is compatible with the clinical success of targeted therapies in patients with metastatic disease (Fig. 6). Though these targeted therapies are not curative because of evolving resistance within metastases (intrametastatic heterogeneity), there is no theoretical reason why combinations of targeted therapies could not be curative9. Indeed, it has been shown that treatment of metastases with just two drugs for which no single alteration can confer cross-resistance should, in theory, cure most cancers8. These results apply not only to those driver gene mutations that are currently targeted but also to future targeted therapies – so such cures appear to be possible and eminently worthy of further pursuit.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all authors of the original publications for enabling this reanalysis by sharing their sequencing data, in particular Adam Bass, David Brown, Sotiriou Christos, Christina Curtis, William Gibson, Erling Hoivik, Marek Cmero, Chris Hovens, Tae-Min Kim, Sug-Hyung Lee, Marc Ryser, Sohrab Shah, Darryl Shibata, Matthew Stachler, Ruping Sun, and Allen Zhang. This study was supported by the National Institutes of Health grants K99CA229991 (J.G.R.), CA179991 (C.A.I.-D.), F31CA180682 (A.P.M.-M.), T32 CA160001–06 (A.P.M.-M.), CA43460 (B.V.), as well as by the Lustgarten Foundation for Pancreatic Cancer Research, The Sol Goldman Center for Pancreatic Cancer Research, The Virginia and D. K. Ludwig Fund for Cancer Research, an Erwin Schrödinger fellowship (J.G.R.; Austrian Science Fund FWF J-3996), a Landry Cancer Biology fellowship (J.M.G.), and the Office of Naval Research grant N00014-16-1-2914.
Footnotes
Competing interests statement
K.W.K. and B.V. are founders of Personal Genome Diagnostics and Thrive and advisors of Sysmex, Eisai, CAGE, Neophore. B.V. is also an advisor to Nexus. These companies and others have licensed technologies related to the work described in this paper from Johns Hopkins University. Some of these licenses are associated with equity or royalty payments to K.W.K. and B.V. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. Other authors declare no competing interests.
REFERENCES
- 1.Heppner GH Tumor Heterogeneity. Cancer Res 44, 2259–2265 (1984). [PubMed] [Google Scholar]
- 2.Vogelstein B et al. Cancer Genome Landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martincorena I & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal R, McGranahan N, Herrero J & Swanton C Deciphering Genetic Intratumor Heterogeneity and Its Impact on Cancer Evolution. Annu. Rev. Cancer Biol 1, 223–240 (2017). [Google Scholar]
- 5.Engelman JA et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Diaz LA Jr et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty KT et al. Combined BRAF and MEK Inhibition in Melanoma with BRAF V600 Mutations. N. Engl. J. Med 120929000030002 (2012). doi: 10.1056/NEJMoa1210093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozic I et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife 2, e00747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozic I & Nowak MA Resisting resistance. 1, 203–221 (2017). [Google Scholar]
- 10.McGranahan N et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med 7, 283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sottoriva A, Barnes CP & Graham TA Catch my drift? Making sense of genomic intra-tumour heterogeneity. Biochim. Biophys. Acta (BBA)-Reviews Cancer 1867, 95–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamborero D et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 10, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiter JG et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokheim C & Karchin R Enhanced context reveals the scope of somatic missense mutations driving human cancers. Cell Syst 9, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B & Kinzler KW The Path to Cancer--Three Strikes and You’re Out. N. Engl. J. Med 373, 1895–1898 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Tomasetti C, Marchionni L, Nowak MA, Parmigiani G & Vogelstein B Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci USA 112, 118–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hruban RH, Goggins M, Parsons J & Kern SE Progression model for pancreatic cancer. Clin. Cancer Res 6, 2969–2972 (2000). [PubMed] [Google Scholar]
- 18.Cross W et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol 2, 1661–1672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makohon-Moore AP et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature 561, 201–205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun 9, 2884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozic I et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA 107, 18545–18550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bozic I, Gerold JM & Nowak MA Quantifying Clonal and Subclonal Passenger Mutations in Cancer Evolution. PLoS Comput. Biol 12, e1004731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun R et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat Genet 49, 1015–1024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams MJ et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nat. Genet 50, 895–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wodarz D & Komarova NL Dynamics of Cancer: Mathematical Foundations of Oncology. (World Scientific Publishing, Singapore, 2014). [Google Scholar]
- 26.Altrock PM, Liu LL & Michor F The mathematics of cancer: integrating quantitative models. Nat. Rev. Cancer 15, 730–745 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Amikura K, Kobari M & Matsuno S The time of occurrence of liver metastasis in carcinoma of the pancreas. Int. J. Pancreatol 17, 139–146 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Jones S et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA 105, 4283–4288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yachida S et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vermeulen L et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995–998 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Cannataro VL, Gaffney SG & Townsend JP Effect sizes of somatic mutations in cancer. JNCI J. Natl. Cancer Inst 110, 1171–1177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowell PC The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Greaves M & Maley CC Clonal evolution in cancer. Nature 481, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiter JG, Bozic I, Allen B, Chatterjee K & Nowak MA The effect of one additional driver mutation on tumor progression. Evol. Appl 6, 34–45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiter JG, Bozic I, Chatterjee K & Nowak MA TTP: Tool for Tumor Progression in Computer Aided Verification, Lecture Notes in Computer Science 8044, 101–106 (Springer Berlin; Heidelberg, 2013). [Google Scholar]
- 36.Landau DA et al. Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Vega F et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priestley P et al. Pan-cancer whole genome analyses of metastatic solid tumors. bioRxiv 415133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstung M et al. The evolutionary history of 2,658 cancers. bioRxiv 161562 (2017). [Google Scholar]
- 40.Bailey MH et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fearon ER & Vogelstein B A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Kurman RJ & Shih I-M The Origin and pathogenesis of epithelial ovarian cancer-a proposed unifying theory. Am. J. Surg. Pathol 34, 433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy SJ et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 145, 1098–1109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiter JG & Iacobuzio-Donahue CA Pancreatic carcinogenesis - several small steps or one giant leap? Nat Rev Gastroenterol Hepatol 14, 7–8 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Teixeira VH et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med 25, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lengauer C, Kinzler KW & Vogelstein B Genetic instabilities in human cancers. Nature 396, 643 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Beroukhim R et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama A et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Bielski CM et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet 50, 1189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolhaqueiro ACF et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet 51, 824 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Massagué J & Obenauf AC Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung S-H et al. Whole-exome sequencing identifies recurrent AKT1 mutations in sclerosing hemangioma of lung. Proc Natl Acad Sci USA 113, 10672–10677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtius K, Wright NA & Graham TA Evolution of Premalignant Disease. Cold Spring Harb. Perspect. Med a026542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuboki Y et al. Single-cell sequencing defines genetic heterogeneity in pancreatic cancer precursor lesions. J. Pathol (2018). doi: 10.1002/path.5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winters IP, Murray CW & Winslow MM Towards quantitative and multiplexed in vivo functional cancer genomics. Nat. Rev. Genet 19, 741–755 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Blokzijl F et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bae T et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359, 550–555 (201AD). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lodato MA et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee-Six H et al. The landscape of somatic mutation in normal colorectal epithelial cells. bioRxiv (2018). doi: 10.1101/416800 [DOI] [PubMed] [Google Scholar]
- 60.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maley CC et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer 17, 605–619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durrett R, Foo J, Leder K, Mayberry J & Michor F Intratumor heterogeneity in evolutionary models of tumor progression. Genetics 188, 461–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almendro V et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 6, 514–527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Makohon-Moore AP et al. Limited heterogeneity of known driver gene mutations among the metastases of individual pancreatic cancer patients. Nat. Genet 49, 358–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenbloom DIS, Camara PG, Chu T & Rabadan R Evolutionary scalpels for dissecting tumor ecosystems. Biochim Biophys Acta 1867, 69–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maley CC et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet 38, 468–473 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Lemery S, Keegan P & Pazdur R First FDA approval agnostic of cancer site--when a biomarker defines the indication. N Engl J Med 377, 1409–1412 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Drilon A et al. Efficacy of larotrectinib in TRK fusion--positive cancers in adults and children. N. Engl. J. Med 378, 731–739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makohon-Moore A & Iacobuzio-Donahue CA Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegel RL, Miller KD & Jemal A Cancer statistics, 2018. CA. Cancer J. Clin 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Andor N et al. Pan-cancer analysis of the extent and consequences of intra-tumor heterogeneity. Nat. Med 22, 105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jamal-Hanjani M et al. Tracking the evolution of non--small-cell lung cancer. N. Engl. J. Med 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Francis JM et al. EGFR Variant Heterogeneity in Glioblastoma Resolved through Single-Nucleus Sequencing. Cancer Discov 4, 956–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bashashati A et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J. Pathol 231, 21–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gerlinger M et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet 46, 225–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Bruin EC et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yates LR et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21, 751–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim T-M et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res 21, 4461–4472 (2015). [DOI] [PubMed] [Google Scholar]
- 80.McPherson A et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet 48, 758–767 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Zhang J et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346, 256–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reiter JG et al. Reconstructing metastatic seeding patterns of human cancers. Nat. Commun 8, 14114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi W et al. Reliability of whole-exome sequencing for assessing intratumor genetic heterogeneity. Cell Rep 25, 1446–1457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zare F, Dow M, Monteleone N, Hosny A & Nabavi S An evaluation of copy number variation detection tools for cancer using whole exome sequencing data. BMC Bioinformatics 18, 286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salari R et al. Inference of tumor phylogenies with improved somatic mutation discovery. J. Comput. Biol 20, 933–944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang AW et al. Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell 173, 1755–1769 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Sottoriva A et al. A Big Bang model of human colorectal tumor growth. Nat. Genet 47, 209–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ryser MD, Min B-H, Siegmund KD & Shibata D Spatial mutation patterns as markers of early colorectal tumor cell mobility. Proc Natl Acad Sci USA 115, 5774–5779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pectasides E et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 8, 37–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chakravarty D et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol 1, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang MT et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forbes S et al. COSMIC: High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer. Curr. Protoc. Hum. Genet 1–37 (2016). doi: 10.1002/cphg.21 [DOI] [PubMed] [Google Scholar]
- 93.Masica DL et al. CRAVAT 4: cancer-related analysis of variants toolkit. Cancer Res 77, e35–e38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shihab HA, Gough J, Cooper DN, Day INM & Gaunt TR Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics 29, 1504–1510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mao Y et al. CanDrA: cancer-specific driver missense mutation annotation with optimized features. PLoS One 8, e77945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McLaren W et al. The ensembl variant effect predictor. Genome Biol 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Løes IM et al. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int. J. Cancer 139, 647–656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith JC & Sheltzer JM Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. Elife 7, e39217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwarz RF et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med 12, e1001789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gokulan RC, Garcia-Buitrago MT & Zaika AI From genetics to signaling pathways: molecular pathogenesis of esophageal adenocarcinoma. BBA-Rev Cancer (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naxerova K & Jain RK Using tumour phylogenetics to identify the roots of metastasis in humans. Nat. Rev. Clin. Oncol 12, 258–272 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Eisenhauer EA et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Brown D et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat. Commun 8, 14944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gibson WJ et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet 48, 848–855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong MKH et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 6, 6605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanborn JZ et al. Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc Natl Acad Sci USA 112, 10995–11000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee J-Y et al. Tumor evolution and intratumor heterogeneity of an epithelial ovarian cancer investigated using next-generation sequencing. BMC Cancer 15, 85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turajlic S & Swanton C Metastasis as an evolutionary process. Science 352, 169–175 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Kumar A et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao Z-M et al. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci USA 113, 2140–2145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hunter KW, Amin R, Deasy S, Ha N-H & Wakefield L Genetic insights into the morass of metastatic heterogeneity. Nat. Rev. Cancer 18, 211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sharma SV, Bell DW, Settleman J & Haber DA Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Juric D et al. Convergent loss of PTEN leads to clinical resistance to a PI (3) K$α$ inhibitor. Nature 518, 240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khan KH et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov 8, 1270–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kobayashi S et al. EGFR mutation and resistance of non--small-cell lung cancer to gefitinib. N. Engl. J. Med 352, 786–792 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Goyal L et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion--positive cholangiocarcinoma. Cancer Discov 7, 252–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vakiani E et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J. Clin. Oncol 30, 2956–2962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brannon AR et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 15, 454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yatabe Y, Matsuo K & Mitsudomi T Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J. Clin. Oncol 29, 2972–2977 (2011). [DOI] [PubMed] [Google Scholar]
- 121.Colombino M et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol 30, 2522–2529 (2012). [DOI] [PubMed] [Google Scholar]
- 122.Boursault L et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One 8, e70826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ribas A et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAFV600-mutated melanoma: a phase 1b study. Lancet Oncol 15, 954–965 (2014). [DOI] [PubMed] [Google Scholar]
- 124.Algazi AP et al. SWOG S1221: A phase 1 dose escalation study co-targeting MAPK-dependent and MAPK-independent BRAF inhibitor resistance in BRAF mutant advanced solid tumors with dabrafenib, trametinib, and GSK2141795 (ClinicalTrials.gov ). J. Clin. Oncol 35, 2578 (2017). [Google Scholar]
- 125.Kurzrock R et al. The VEGF Receptor Tyrosine Kinase Inhibitor Pazopanib in Combination with the MEK Inhibitor Trametinib in Advanced Solid Tumors and Differentiated Thyroid Cancers. Clin. Cancer Res (in press) (2019). doi: 10.1158/1078-0432.CCR-18-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pommier A et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 360, eaao4908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Turajlic S et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Naxerova K et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schwartz R & Schäffer AA The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet 18, 213–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jones S et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci 105, 4283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mitchell TJ et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 173, 611–623.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beerenwinkel N, Schwarz RF, Gerstung M & Markowetz F Cancer evolution: mathematical models and computational inference. Syst. Biol 64, e1–e25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davoli T et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Santarius T, Shipley J, Brewer D, Stratton MR & Cooper CS A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10, 59 (2010). [DOI] [PubMed] [Google Scholar]
- 135.Xue W et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA 109, 8212–8217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Solimini NL et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science 337, 104–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Knouse KA, Davoli T, Elledge SJ & Amon A Aneuploidy in cancer: Seq-ing answers to old questions. Annu. Rev. Cancer Biol 1, 335–354 (2017). [Google Scholar]
- 138.Lengauer C, Kinzler KW & Vogelstein B Genetic instabilities in human cancers. Nature 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- 139.Rheinbay E et al. Recurrent and functional regulatory mutations in breast cancer. Nature 547, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.El-Kebir M, Satas G & Raphael BJ Inferring parsimonious migration histories for metastatic cancers. Nat. Genet 50, 718–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Heyde A, Reiter JG, Naxerova K & Nowak MA Consecutive seeding and transfer of genetic diversity in metastasis. Proc Natl Acad Sci USA (2019). doi: 10.1073/pnas.1819408116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rubinsteyn A et al. hammerlab/pyensembl: Version 1.0.1. (2016). doi: 10.5281/zenodo.154747 [DOI] [Google Scholar]
- 143.Rubinsteyn A et al. varcode v0.4.15. (2016). doi: 10.5281/zenodo.58031 [DOI] [Google Scholar]
- 144.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tokheim CJ, Papadopoulos N, Kinzler KW, Vogelstein B & Karchin R Evaluating the evaluation of cancer driver genes. Proc Natl Acad Sci USA 113, 14330–14335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.