ABSTRACT
Cancer-associated fibroblasts (CAFs) are activated fibroblasts and can interact with cancer cells to promote tumor progression. The process of how tumor cells reprogram normal fibroblasts (NFs) to tumor-promoting CAFs regulated by long non-coding RNA (lncRNA) remains incompletely understood. The tumor cells-released exosomes can induce reprogramming of NFs into CAFs. This study aimed to explore the role of melanoma-derived exosomes in regulating NF-CAF transition and to clarify whether lncRNA Gm26809 was involved in this process. The results showed that the exosomes secreted by melanoma cell B16F0 induced reprogramming of fibroblast NIH/3T3 cells into CAFs, as evidenced by increased expression of CAFs markers (α-SMA and FAP) and facilitated cell migration. Mechanistically, B16F0-secreted exosome delivered Gm26809 into NIH/3T3 cells where Gm26809 mediated reprogramming of fibroblast NIH/3T3 cells into CAFs. Furthermore, the conditioned medium from the co-culture of NIH/3T3 cells and B16F0-exosomes facilitated cell proliferation and migration in a melanoma cell line (Cloundman S91), and the effect was abrogated by Gm26809 knockdown in B16F0 cells. In summary, melanoma-derived exosomes facilitate melanoma cell proliferation and migration through reprogramming fibroblasts into tumor-promoting CAFs via transferring Gm26809.
KEYWORDS: Melanoma, cancer-associated fibroblasts, exosome, Gm26809
Introduction
Malignant melanoma is the most aggressive and the deadliest form of skin cancer [1]. According to the World Health Organization, 132,000 new cases of melanoma are diagnosed worldwide each year [2]. The major cause of melanoma mortality is metastasis to distant organs [3]. Despite the extensive improvement in the treatment, malignant melanoma still presents a major clinical challenge. Thus, a better understanding of mechanisms underlying melanoma metastasis and progression is urgently needed.
Fibroblasts are common cells of the connective tissue and can be activated in cancer, which are commonly known as cancer-associated fibroblasts (CAFs). CAFs are an important player in the tumor microenvironment that can interact with cancer cells to promote tumor metastasis and progression [4]. CAFs can communicate with cancer cells through various mechanisms [5]. Recently, increasing studies have underscored the important role of exosomes in the crosstalk between CAFs and cancer cells [5]. Exosomes are small lipid-bilayer-enclosed vesicles delivered by many cells and have recently been recognized as critical mediators in intercellular communication by transferring donor cell-specific lipids, proteins, and RNAs to recipient cells [6]. CAFs-secreted exosomes can be absorbed by cancer cells and modulate carcinogenesis by transferring specific cargos [7]. Correspondingly, the cancer cells-released exosomes can also induce reprogramming of normal fibroblasts (NFs) into CAFs [5,8].
Long non-coding RNAs (lncRNAs), a group of non-protein-coding RNAs with more than 200 nucleotides in length, are now recognized as potent regulators of various cellular processes [9–12]. Tumor-secreted exosomal lncRNAs play an important role in reprogramming the tumor microenvironment [13]. Exosome-transmitted lncRNAs can be taken up by recipient cells and modulate phenotypic and functional changes in the recipient cells [14]. Recent data suggest that lncRNAs in tumor-secreted exosomes play a role in inducing the NF-CAF transition [15].
Recent data showed that Gm26809 (ENSMU-SG00000097815), a newly discovered lncRNA, was significantly increased in cytotoxic T-lymphocytes CTLL2 upon stimulation with exosomes secreted from B16F0 melanoma cells (B16F0-exosomes) [16]. Given the concordance in mRNAs levels between in exosomes and in exosome-treated CTLL2 cells, it can be assumed that the increased lncRNA-Gm26809 observed in CTLL2 cells treated with B16F0-exosomes might be originated from the B16F0-exosomes, which requires further investigation. Thus, in this study, we sought to elucidate whether Gm26809 was enriched in B16F0-exosomes and whether Gm26809 was involved in melanoma exosomes-induced reprogramming of NFs into CAFs.
Materials and methods
Cell culture
Mouse primary epidermal melanocytes were purchased from Procell (#CP-M099; Wuhan, China) and cultured in complete medium (#CM-M099; Procell). Mouse embryonic fibroblast NIH/3T3 (#BNCC339681) and mouse melanoma cell lines B16F0 (#BNCC100927) and Cloundman S91 (#BNCC340255) were purchased from Bena Culture Collection (Beijing, China). B16F0 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% exosome-depleted fetal bovine serum (FBS; Gibco) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) in humidified air at 37°C with 5% CO2.
Isolation and identification of melanocyte- and melanoma-derived exosomes
Exosomes were isolated and purified from the cell supernatant from melanocytes and B16F0 melanoma cells by ultrahigh velocity centrifugation. Briefly, culture media samples were centrifuged at 500 × g for 10 min to remove dead cells and then at 2,000 × g for 20 min to remove residual cells and debris. Following additional centrifugation at 10,000 × g for 30 min, the supernatant was filtered with 0.22 μm filters (Corning, Corning, NY, USA) and then centrifuged again at 120,000 × g for 70 min. The precipitated exosome pellets were re-suspended in PBS for identification and either used immediately or stored at −80°C until required.
For identification, the morphologic characteristics of exosomes were observed by transmission electron microscopy (TEM; HT7700; HITACHI, Japan). The particle size distribution of exosomes was determined by Nano-ZS ZEN 3600 (Malvern Instruments, UK). Total protein was extracted from exosomes using Total exosome RNA and protein isolation kit (Invitrogen, Thermo Fisher Scientific, Inc) and the protein levels of exosomal surface markers CD9, CD63, and TSG101 were determined by western blot.
Cell infection
The lentivirus overexpressing Gm26809 (LV-Gm26809), the shRNA-expressing lentivirus targeting Gm26809 (LV-shGm26809) or their corresponding controls LV-negative control (NC) and LV-Scramble were constructed and infected into B16F0 cells or NIH/3T3 cells. RNA was extracted from B16F0 cells or NIH/3T3 cells and subjected to qRT-PCR to examine the overexpression and silencing efficiency of Gm26809.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells and exosomes using the TRIzol reagent (Takara, Dalian, China). Total RNA was reverse transcribed into cDNA by using PrimeScript RT Reagent Kit (Takara), followed by qRT-PCR using SYBR® Premix Ex Taq™ II (Takara). The relative expression level was calculated by the 2−ΔΔCt method and normalized to the internal control GAPDH.
Western blot
The cell lysates were extracted using the radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) and quantified using the bicinchoninic acid (BCA) Protein Assay Kit (Beyotime). Aliquots of 20 μg of protein were loaded per lane and separated by 10% SDS-PAGE gels and electroblotted onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Subsequently, the membranes were blocked with 5% fat-free milk and then incubated overnight at 4°C with primary antibodies against α-smooth muscle actin (α-SMA) and fibroblast activation protein (FAP) (1:1000, Abcam, Cambridge, MA, USA). After washing with Tris-Buffered Saline Tween-20 (TBST) three times, the membranes were then incubated with the horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2000; Santa Cruz Biotechnology, Dallas, TX, USA). Blots were examined by an Enhanced Chemiluminescence (ECL) Detection kit (Pierce Biotechnology, Rockford, IL, USA). The band intensity was analyzed by Image-Pro Plus 6.0 software. GAPDH served as the loading control.
Transwell migration assay
(1) To assess the effect of melanocyte- or B16F0-derived exosomes on the migrative capacity of NIH/3T3 cells, NIH/3T3 cells in 100 μl of serum-free medium were seeded in the upper chamber of cell culture inserts (24-well; Corning, USA). NC-Exo, B16F0-Exo, LV-NC-Exo, or LV-Gm26809-Exo in 600 μl of 10% FBS-DMEM were seeded in the lower chambers.
(2) To evaluate the effect of Gm26809 expression on NIH/3T3 cellular migration, NIH/3T3 cells which have been infected with LV-NC or LV-Gm26809 were seeded in the upper chamber, 600 μl of 10% FBS-DMEM was then added into the lower chambers.
(3) To assess the effect of culture media from co-culture of NIH/3T3 cells and B16F0-secreted exosomes on the migrative capacity of Cloundman S91 cells, Cloundman S91 cells were plated in 96-well culture plates and a total of 100 µl cell suspension in serum-free medium (5 × 104 cells/ml) was seeded into the upper chamber. NIH/3T3 cells treated with or without B16F0-Exo, LV-Scramble-Exo, and LV-shGm26809-Exo were seeded in 600 μl of 10% FBS-DMEM in the lower chambers.
After 24 h of incubation at 37°C, cells that had traversed to the reverse face were stained with 0.1% crystal violet for 20 min at room temperature. The number of migrated cells was imaged, counted and averaged from five randomly selected fields under a microscope (Nikon E100; Nikon Corp, Japan).
Cell proliferation assay
Cloundman S91 cells were plated in 96-well culture plates and cultured with the culture media from NIH/3T3 cells, which were stimulated with or without B16F0-Exo, LV-Scramble-Exo, and LV-shGm26809-Exo. Following incubation for 24, 48, and 72 h, cell proliferation was measured by 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer’s instruction. Cellular viability was examined by measuring the optical density (OD) at 490 nm by a microplate reader (Multiskan Mk3, Thermo Labsystems, Finland).
Exosome tracing
To monitor the interaction between exosomes and fibroblasts, the exosomes were labeled with a PKH26 red fluorescent labeling kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s instructions and co-cultured with NIH/3T3 cells for 4 h. The uptake of exosomes by NIH/3T3 cells was analyzed under a confocal microscope (Olympus FV1200, Japan).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7.0. Data were expressed as mean ± standard deviation (SD) from triplicates of independent experiments. Two-sided Student’s t test and one-way ANOVA were used to analyze the difference among groups. P values <0.05 were considered significant.
Results
Characterization of melanocyte- and melanoma-derived exosomes
The exosomes isolated from mouse melanocytes and B16F0 melanoma cells (NC-Exo and B16F0-Exo, respectively) were characterized by TEM, nanoparticle tracking analysis, and western blot. The yielded vesicles were consistent with exosomes in morphology (oval-shaped) under TEM (Figure 1a). Nanoparticle tracking analysis confirmed that most vesicles were consistent with exosomes in size (approximately 132 nm and 127 nm for NC-Exo and B16F0-Exo, respectively) (Figure 1b). Moreover, western blot analysis demonstrated that these vesicles were positive for exosomal surface markers CD9, CD63, and TSG101 (Figure 1c). These results verified that the vesicles were exosomes.
Figure 1.
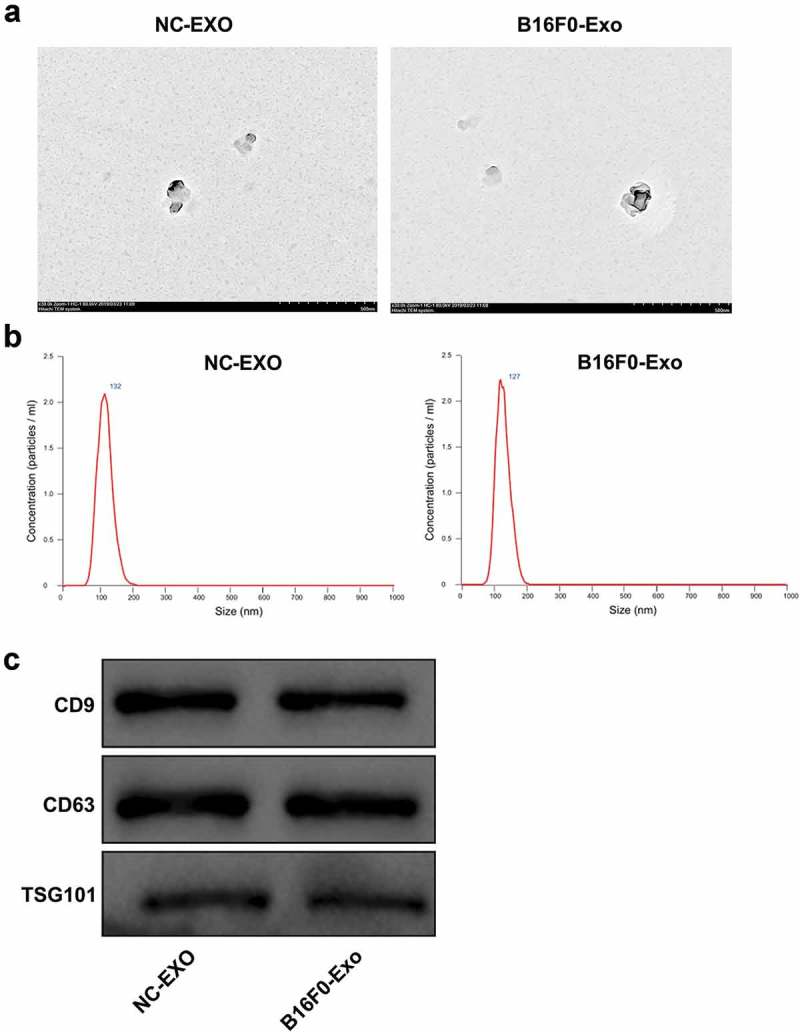
Characterization of melanocyte- and melanoma-derived exosomes.
Exosomes from mouse primary epidermal melanocytes and B16F0 melanoma cells (NC-Exo and B16F0-Exo, respectively) were isolated. (a) The morphological characteristics of exosomes visualized by transmission electron microscopy. Scale bar, 500 nm. (b) Nanoparticle tracking analysis of the size of exosomes. (c) The protein levels of exosomal surface markers CD9, CD63, and TSG101 determined by western blot.
Melanoma-derived exosomes transform fibroblasts into CAFs
To explore the role of melanoma-derived exosomes in regulating the transformation of fibroblasts into CAFs, PKH26-labeled fibroblasts (NIH/3T3 cells) were co-cultured with NC-Exo and B16F0-Exo for 24 h. The fluorescently stained cytoplasm of NIH/3T3 cells was observed under a confocal microscope, confirming the uptake of exosomes by NIH/3T3 cells (Figure 2d). Furthermore, qRT-PCR and western blot showed that B16F0-Exo treatment remarkedly upregulated mRNA and protein levels of CAF-specific markers α-SMA and FAP when compared with the NC-Exo treatment group (Figure 2a,b). Moreover, the migration capacity of NIH/3T3 cells was significantly enhanced by stimulation with B16F0-Exo (Figure 2c).
Figure 2.
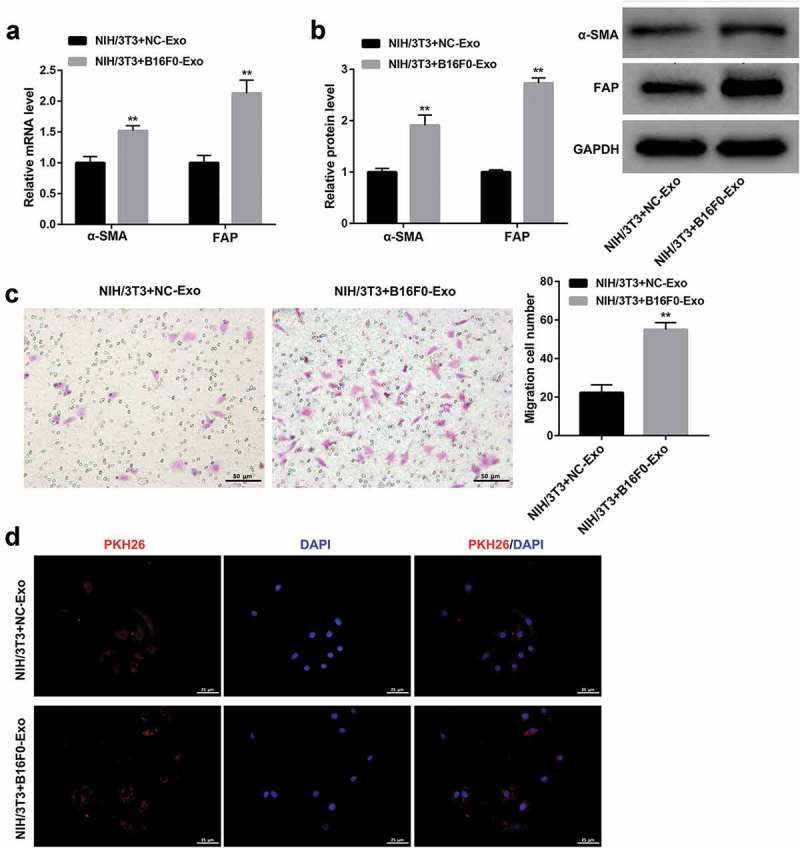
Melanoma-derived exosomes transform fibroblasts into CAFs.
NIH/3T3 cells were co-cultured with exosomes (20 μg/ml) extracted from mouse melanocytes (NC-Exo) and B16F0 cells (B16F0-Exo) for 24 h. (a, b) qRT-PCR and western blot were performed to examine the mRNA and protein levels of α-SMA and FAP, respectively. (c) Transwell migration assay was conducted to evaluate the migration ability of NIH/3T3 cells. Scale bar, 50 μm. (d) Confocal microscope images showed the uptake of PKH26-labeled exosomes in NIH/3T3 cells. PKH26: PKH26-labeled exosomes (red), DAPI: cell nuclei (blue). Scale bar, 25 μm. Values are represented as means ± SD (n = 3). **P< 0.01, vs. NIH/3T3+ NC-Exo.
Exosome-encapsulated high gm26809 expression enhances the melanoma exosomes-mediated transformation of fibroblasts into CAFs
Interestingly, Gm26809 expression was more abundant in B16F0-Exo than that in NC-Exo (Figure 3a). Accordingly, we further investigated whether Gm26809 was involved in the B16F0-Exo-mediated transformation of fibroblasts into CAFs. To this end, fibroblast NIH/3T3 cells were co-cultured with B16F0-Exo, LV-NC-Exo (exosomes from LV-NC-infected B16F0 cells), and LV-Gm26809-Exo (exosomes from LV-Gm26809-infected B16F0 cells). qRT-PCR analysis confirmed that Gm26809 expression was notably higher in LV-Gm26809-Exo than that in LV-NC-Exo (Figure 3b). In addition, the uptake of these exosomes by NIH/3T3 cells was confirmed under a confocal microscope (Figure 3f). We found that LV-Gm26809-Exo treatment significantly increased Gm26809 expression in NIH/3T3 cells (Figure 3c). Meanwhile, LV-Gm26809-Exo treatment significantly increased α-SMA and FAP protein levels (Figure 3d) and facilitated cell migration (Figure 3e) in NIH/3T3 cells. Together, these data indicated that Gm26809 overexpression in B16F0 cells increases exosomal Gm26809 from B16F0 cells, and the exosome-encapsulated high Gm26809 expression enhances the B16F0-Exo-mediated transformation of fibroblasts into CAFs.
Figure 3.
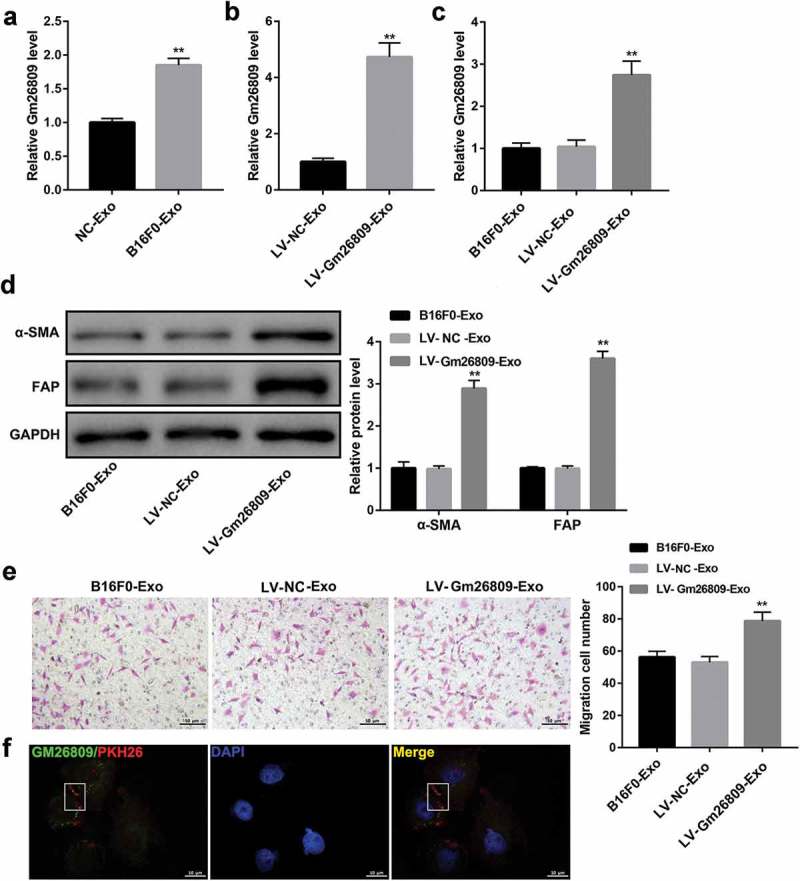
Gm26809 overexpression enhanced the B16F0-Exo-mediated transformation of fibroblasts into CAFs.
(a) qRT-PCR analysis of Gm26809 in NC-Exo and B16F0-Exo. (b) qRT-PCR analysis of Gm26809 in LV-NC-Exo and LV-Gm26809-Exo. qRT-PCR analysis of Gm26809 expression (c), western blot analysis of α-SMA and FAP protein levels (d), and cell migration determined by Transwell migration assay (E) in NIH/3T3 cells which were stimulated with B16F0-Exo, LV-NC-Exo and LV-Gm26809-Exo (20 μg/ml) for 24 h. (F) Confocal microscope images confirmed the uptake of exosomes in NIH/3T3 cells. FITC: FITC-tagged Gm26809 (green), PKH26: PKH26-labeled exosomes (red), DAPI: cell nuclei (blue). Scale bar, 10 μm. Values are represented as means ± SD (n = 3). **P< 0.01, vs. LV-NC-Exo.
Gm26809 overexpression in fibroblasts transforms fibroblasts into CAFs
To further elucidate the functional role of Gm26809 in regulating the transformation of fibroblasts into CAFs, we infected NIH/3T3 cells with LV-NC and LV-Gm26809. Data revealed that Gm26809 overexpression led to a significant increase in protein levels of CAF-specific markers α-SMA and FAP (Figure 4a). Furthermore, the migration capacity of NIH/3T3 cells was greatly promoted by Gm26809 overexpression (Figure 4B). Collectively, these data suggested that Gm26809 overexpression transforms fibroblasts into CAFs.
Figure 4.
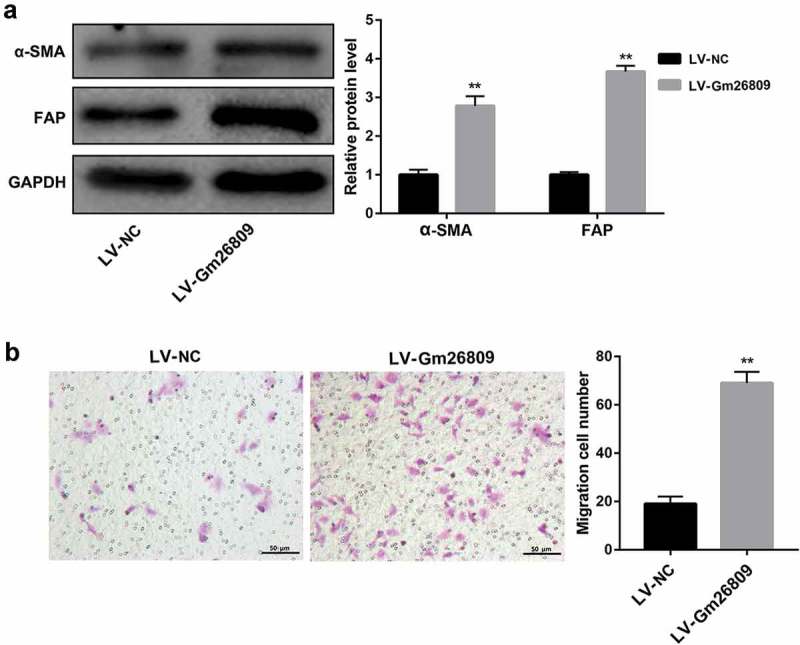
Gm26809 transforms fibroblasts into CAFs.
Western blot analysis of α-SMA and FAP protein levels (a), and cell migration determined by Transwell migration assay (b) in NIH/3T3 cells which were infected with LV-NC and LV-Gm26809. Values are represented as means ± SD (n = 3). **P< 0.01, vs. LV-NC.
Fibroblasts treated with melanoma-derived exosomes facilitate melanomagenesis, which can be rescued by Gm26809 knockdown
Finally, we determined whether fibroblasts-treated with melanoma-derived exosomes regulate melanoma cell proliferation and migration. To address this, the mouse melanoma cell line Cloundman S91 was cultured by the culture media from NIH/3T3 cells which were stimulated with B16F0-Exo, LV-Scramble-Exo, and LV-shGm26809-Exo. MTT assay and Transwell migration assay showed that treatment with the culture media from NIH/3T3 cells treated with B16F0-Exo greatly promoted Cloundman S91 cell proliferation (Figure 5a) and migration (Figure 5b) when compared with the untreated group, indicating that fibroblasts treated with melanoma-derived exosomes might promote tumorigenesis in vitro. More importantly, these changes were effectively abrogated by the culture media from NIH/3T3 cells treated with LV-shGm26809-Exo (Figure 5a,b). These data collectively indicated that melanoma-derived exosomes facilitate melanoma cell proliferation and migration through reprogramming fibroblasts into pro-tumor CAFs via transferring Gm26809.
Figure 5.
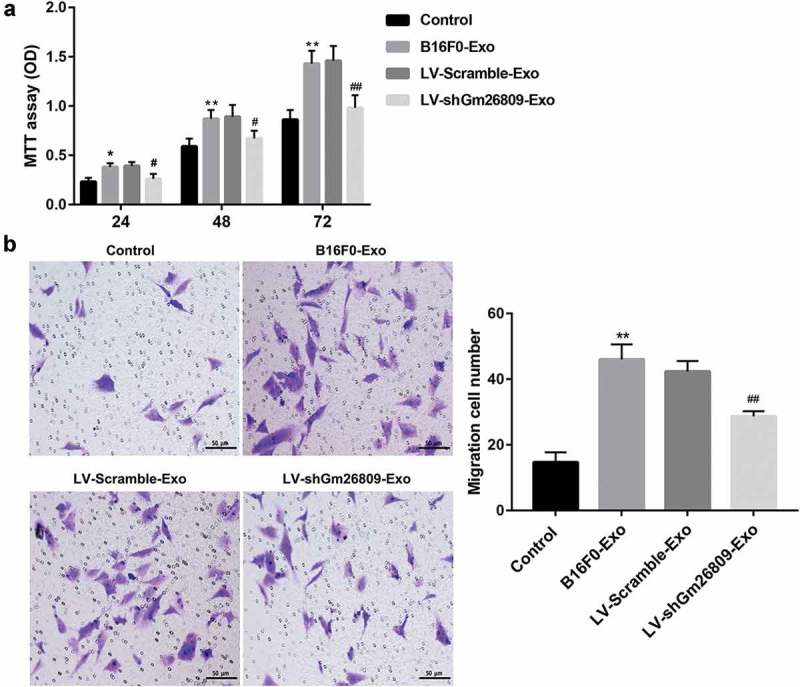
Fibroblasts treated with melanoma-derived exosomes facilitate melanomagenesis, which can be rescued by Gm26809 knockdown.
Mouse melanoma cells Cloundman S91 were cultured by the culture media of NIH/3T3 cells, which were stimulated with or without B16F0-Exo, LV-Scramble-Exo, and LV-shGm26809-Exo (20 μg/ml) for 24 h. (a) MTT assay was performed to evaluate Cloundman S91 cell proliferation. (b) Transwell migration assay was conducted to evaluate the migration ability of Cloundman S91 cells. Scale bar, 50 μm. Values are represented as means ± SD (n = 3). *P< 0.05, **P< 0.01, vs. Control. #P< 0.05, ##P< 0.01, vs. LV-Scramble-Exo.
Discussion
CAFs are derived from neighboring NFs or other cells that undergo a differentiation process induced by cancer cells and participate in tumor microenvironment construction to promote tumor proliferation and metastasis [5]. The cancer cells-released exosomes can induce reprogramming of NFs into CAFs. Exosomes are nanometer-sized vesicles that shuttle cargos such as proteins and RNAs between cells to mediate intercellular communication [17]. Our results showed that the B16F0-secreted exosomes increased expression of CAFs markers (α-SMA and FAP) [5] and facilitated NIH/3T3 cell migration. Our findings suggested that melanoma-secreted exosomes induced reprogramming of NFs into CAFs, which has been also reported by the few previous studies [8].
Exosomes derived from tumor cells play a crucial role in inducing NF-CAF transition via transferring RNA, proteins, and other bioactive materials [17,18]. For example, melanoma cell-secreted exosomal miR-155 can trigger NFs reprogramming into proangiogenic CAFs [8]. Apart from miRNAs, lncRNAs have also been shown to help cancer cells induce NFs to transform into CAFs [15]. For example, exosomal lncRNA CAF secreted from oral squamous cell carcinoma (OSCC) cells increased its expression levels in stromal fibroblasts where it induced the reprogramming of NFs into CAFs [15]. However, few studies examined the role of lncRNAs in the regulation of NF-CAF transition. Here, our qRT-PCR results confirmed our speculation that lncRNA Gm26809 was enriched in B16F0 cells-secreted exosomes. In addition, B16F0 exosomes-encapsulated lncRNA Gm26809 could be delivered into fibroblast NIH/3T3 cells and enhance the B16F0 exosomes-mediated transformation of fibroblasts into CAFs. Our results also provided direct evidence that lncRNA Gm26809 overexpression in NIH/3T3 cells induced reprogramming of fibroblast NIH/3T3 cells into CAFs. Together, these data provided the first evidence that melanoma (B16F0)-secreted exosomes induced reprogramming of NFs (NIH/3T3) into CAFs, at least partially, via lncRNA Gm26809 delivery.
Recent evidence indicates that CAFs are an important player in the tumor microenvironment that can communicate with cancer cells to promote tumor metastasis and progression [4]. On one hand, the cancer cells-released exosomes can trigger transform of NFs into CAFs [5,8], which was confirmed by our above-mentioned studies. On the other hand, CAFs promote tumor growth through various mechanisms, notably angiogenesis, metastasis and immune evasion [19–21]. In this study, we observed that the culture media from fibroblasts (NIH/3T3 cells) which were treated with B16F0-derived exosomes facilitates melanoma Cloundman S91 cell proliferation and migration, indicating that the culture media of CAFs induced by melanoma-secreted exosomes has a tumorigenic role in melanoma. Importantly, Gm26809 knockdown in B16F0 cells impaired the promoting effect of CAFs on melanoma cell proliferation and migration. Given that Gm26809 overexpression induced NFs reprogramming into CAFs, we concluded that Gm26809 knockdown in B16F0 cells led to reduction of Gm26809 in B16F0-secreted exosomes and in NIH/3T3 cells, impaired the B16F0 exosomes-induced NF-CAF transition and thus blocked the B16F0 exosomes-mediated promotion of melanoma cell proliferation and migration, which is the novelty of the present study. Our findings suggest that exosomal Gm26809 may be a potential target for controlling melanoma progression. However, more studies in other cells need to be carried out and gradually extended to animals so as to validate the functional role of Gm26809 in melanoma progression. The molecular mechanisms underlying the Gm26809 regulation of NF-CAF transition also require further investigation. In addition, further research is needed to investigate whether melanoma-derived exosomes transferred Gm26809 to other stromal cells and regulated cell behavior of other stromal cells.
In conclusion, our study demonstrates, for the first time to our knowledge, that melanoma-derived exosomes facilitate melanoma cell proliferation and migration through reprogramming fibroblasts into tumor-promoting CAFs via transferring Gm26809.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Ward EM, Thun MJ, Hannan LM, et al. Interpreting cancer trends. Ann N Y Acad Sci. 2006;1076:29–53. [DOI] [PubMed] [Google Scholar]
- [2].American Cancer Society Cancer Facts and Figures 2018 Atlanta American Cancer Society;. 2019.
- [3].Zbytek B, Carlson JA, Granese J, et al. Current concepts of metastasis in melanoma. Expert Rev Dermatol. 2008;3:569–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schoepp M, Strose AJ, Haier J.. Dysregulation of miRNA expression in cancer associated fibroblasts (CAFs) and its consequences on the tumor microenvironment. Cancers (Basel). 2017;9(6):pii: E54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang X, Li Y, Zou L, et al. Role of exosomes in crosstalk between cancer-associated fibroblasts and cancer cells. Front Oncol. 2019;9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang P, Zhou H, Lu K, et al. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Au Yeung CL, Co NN, Tsuruga T, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou X, Yan T, Huang C, et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2018;37:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fan Q, Yang L, Zhang X, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. [DOI] [PubMed] [Google Scholar]
- [10].Sigdel KR, Cheng A, Wang Y, et al. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J Immunol Res. 2015;2015:848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hombach S, Kretz M.. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. [DOI] [PubMed] [Google Scholar]
- [12].Kaushik SB, Kaushik N. Non-coding RNAs in skin cancers: an update. Noncoding RNA Res. 2016;1:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Leveille N, Baglio SR. Exosome-transferred lncRNAs at the core of cancer bone lesions. Crit Rev Oncol Hematol. 2019;139:125–127. [DOI] [PubMed] [Google Scholar]
- [14].Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. [DOI] [PubMed] [Google Scholar]
- [15].Ding L, Ren J, Zhang D, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39:397–406. [DOI] [PubMed] [Google Scholar]
- [16].Bland CL, Byrne-Hoffman CN, Fernandez A, et al. Exosomes derived from B16F0 melanoma cells alter the transcriptome of cytotoxic T cells that impacts mitochondrial respiration. Febs J. 2018;285:1033–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ringuette Goulet C, Bernard G, Tremblay S, et al. Exosomes induce fibroblast differentiation into cancer-associated fibroblasts through TGFbeta signaling. Mol Cancer Res. 2018;16:1196–1204. [DOI] [PubMed] [Google Scholar]
- [18].Dror S, Sander L, Schwartz H, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol. 2016;18:1006–1017. [DOI] [PubMed] [Google Scholar]
- [19].Weber CE, Kuo PC. The tumor microenvironment. Surg Oncol. 2012;21:172–177. [DOI] [PubMed] [Google Scholar]
- [20].Shiga K, Hara M, Nagasaki T, et al. Cancer-associated fibroblasts: their characteristics and their roles in tumor growth. Cancers (Basel). 2015;7:2443–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]


