ABSTRACT
High specificity accompanied with the ability to recruit immune cells has made recombinant therapeutic antibodies an integral part of drug development. Here we present a generic approach to generate two novel IgG-derived antibody formats that are based on a modification of the CrossMab technology. MoAbs harbor two heavy chains (HCs) resulting in one binding entity and one fragment crystallizable region (Fc), whereas DuoMabs are composed of four HCs harboring two binding entities and two Fc regions linked at a disulfide-bridged hinge. The latter bivalent format is characterized by avidity-enhanced target cell binding while simultaneously increasing the ‘Fc-load’ on the surface. DuoMabs were shown to be producible in high yield and purity and bind to surface cells with affinities comparable to IgGs. The increased Fc load directed at the surface of target cells by DuoMabs modulates their antibody-dependent cell-mediated cytotoxicity competency toward target cells, making them attractive for applications that require or are modulated by FcR interactions.
KEYWORDS: CrossMab, domain exchange, cancer therapy, IGF-1R, ADCC, antibody
Introduction
One mode-of-action of therapeutic antibodies is the induction of antibody-dependent cell-mediated cytotoxicity (ADCC) by cells displaying Fcγ receptors (FcγRs), such as natural killer (NK) cells and macrophages.1–3 The event that initiates ADCC is FcγR binding to the Fc region of antibodies. The presence and accessibility of Fc on the surface of target cells, as well as its FcR-binding competence, are essential for ADCC and determines its efficacy.4,5 Hence, ADCC induction can be modulated by engineering the Fc of antibodies in regions that are important for FcR-interactions.5,6 Modulation of those interactions is not limited to substitutions in the amino acid sequence of the Fc (CH2 and CH3 domains), but can also be achieved by altering the glycosylation on the Fc because FcγRIIIa binding involves glycosylated CH2.4,7–9 Alterations of the glycosylation pattern, via production in cell lines with modified glycosylation pathways, in turn directly influence (and hence enhance) FcγR interactions, as in the case of obinutuzumab, a glycoengineered ADCC-enhanced Type II CD20-binding antibody.6,10,11 This molecule has increased potency compared to rituximab in vitro and in vivo,6,12,13 received the US Food and Drug Administration’s ‘breakthrough therapy’ designation, and was approved for therapy in 2013.14
ADCC reactions are not only influenced by epitope-dependent presentation of Fc (i.e., Fc-FcγR-interactions), but also depend on the efficacy and/or ratio between single domain engagement, coupled domain engagements and avidity.15 As a consequence, it is reasonable to assume that increased amounts of antibodies bound to the surface of target cells elicit more potent responses. Thus, the number of Fc regions exposed on cell surface matters.16 Elevated levels of Fc accessible on cell surfaces provide more targets for FcγR binding.17 In addition, spatial proximity such as crosslinking of FcγR leads to enhanced downstream signaling, leading to the release of cytotoxic mediators and cytokines. Because of that, increasing the number of Fc on target cells may serve as an alternative approach to increase ADCC. This, however, can be limited by the number of antigens on the surface of target cells. Those eventually become saturated by the targeting antibody because a regular IgG can maximally deliver one Fc per two antigen-binding fragment (Fab) binding sites upon bivalent binding or one Fc per binding site upon monovalent antigen binding. This limitation of the Fc load by saturation can only be overcome by increasing the number of Fc entities per binding event to cell surfaces.
Here, we describe a novel CrossMab-derived antibody format (termed DuoMab) composed of four engineered heavy chains (HCs). DuoMabs possess two antigen-binding CrossFab arms and two Fc regions attached to the Fab arms at the hinge region in a similar geometry (C-terminal after the hinge) as standard IgGs. DuoMabs demonstrate biophysical behavior similar to standard IgGs and similar pharmacokinetics (PK) in vivo. ADCC analyses reveal that increased numbers of Fc deposited by DuoMabs lead to enhanced ADCC potency toward target cells.
Results
MoAbs and DuoMabs are CrossMab-containing IgG derivatives
The key modules for generating the CrossMabCH1-CL are Fab arm derivatives with swapped CH1 and CL domains, either serving as light chain (LC) or, when fused to hinge-Fc regions, as knob or hole HCs of bispecific antibodies.18-22 Based on the same principle, MoAbs and DuoMabs can be generated by fusing ‘CrossFab’ arms comprising VL-CH1 and VH-CL domains via hinge to wildtype Fc regions and co-expression of those entities without complementary LCs. The scheme depicted in Figure 1 shows that such entities can assemble upon co-expression in a dimeric manner to form MoAbs or in a tetrameric manner to form DuoMabs. MoAbs carry one antigen-binding entity and one Fc region. DuoMabs are composed of two antigen-binding CrossFab arms and two Fc regions that are connected to each other at their hinge regions.
Figure 1.
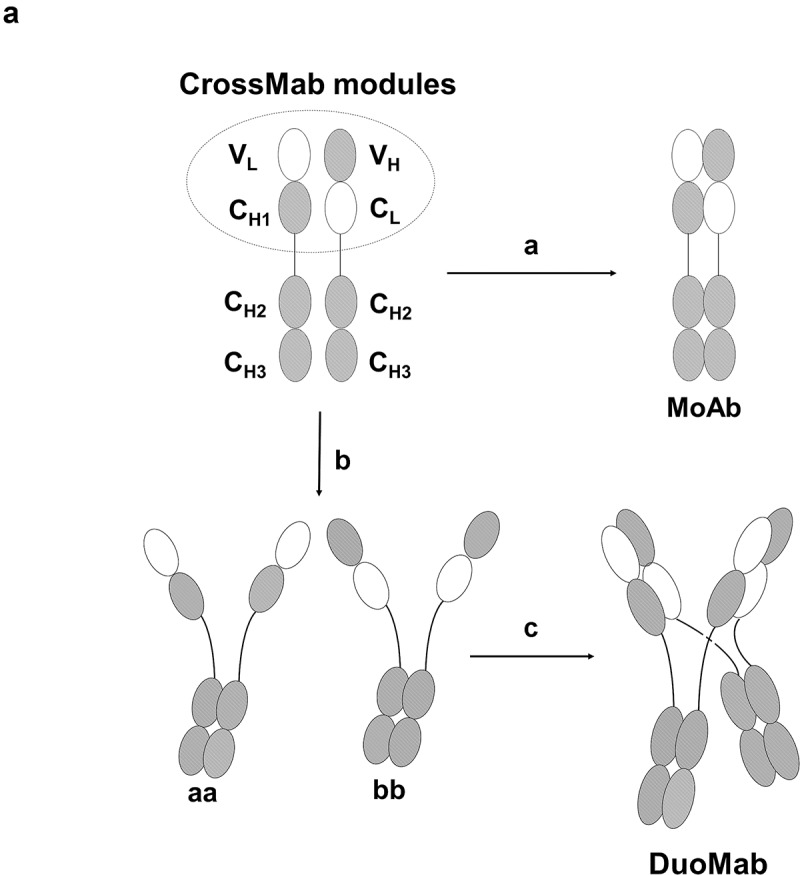
(a) Assembly of MoAbs and DuoMabs. Heterodimerization of two different crossed heavy chains (a with b) generates the monovalent MoAb (route A). Homodimerization of two identical chains (a with a and b with b) generates putative intermediates aa and bb (route b) that associate to form DuoMabs (route c).
As anticipated, transient co-expression of complementary-crossed HCs (without “knob-into-hole” mutations) leads to the secretion of two products into cell culture supernatants, irrespective of which antibody specificity was applied to generate the molecules (cMet, IGF-1R and ErbB3). The domain borders of the crossed HCs are depicted in Figure 2a. Protein-A affinity chromatography and subsequent separation of both by size-exclusion chromatography (SEC) (Figure 2b), analysis by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 2c) and further characterization by SEC with multi-angle laser light scattering (SEC-MALLS) (Figure 2d,e) reveal that those represent either MoAbs or DuoMabs as entities with defined size. The SDS-PAGE hints that the disulfide bridges (e.g., in the IGF1R in Figure 2c) are not connected in a subset of the molecules even though their H-chains are assembled to proper dimers (a phenomenon that can also be observed on SDS-PAGE to some degree for regular IgGs). This leads to disassembly of DuoMabs to Moab-sized entities in the presence of SDS.
Figure 2.
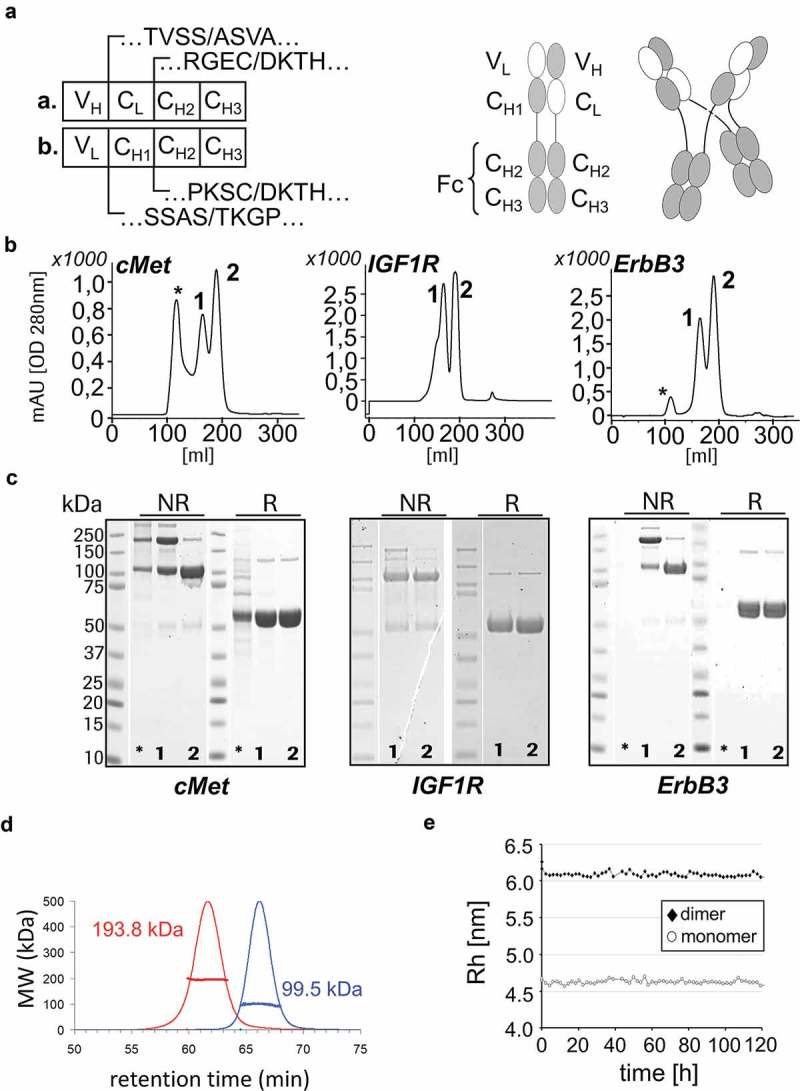
Co-expression and secretion of MoAbs and DuoMabs. Cells that express crossed HC entities assemble them into MoAbs and DuoMabs. Domain borders of two HCs (a./b.) are depicted in (a). Both entities become simultaneously secreted into culture supernatants from which they are purified by Protein-A affinity and SEC. Simultaneous presence of both entities is shown by analytical SEC (1 = DuoMab, 2 = MoAb, * = high molecular weight species, aggregates) (b) and SDS-PAGE (c) for three different antibodies. NR = non-reduced; R = reduced. (d) SEC-MALLS confirms MoAbs and DuoMabs as entities of defined composition and size. (e) SEC-MALLS analysis over extended period shows hydrodynamic radii as Rh values. The measurement clearly confirms two distinct species representing MoAb (marked as “monomer”) and DuoMab (marked as “dimer”).
Directed MoAb and DuoMab generation enforced by CH3 knob-into-hole and IgG-IgA exchange mutations
Co-expression of complementary-crossed HCs without additional knob or hole mutations produced both MoAbs as well as DuoMabs in the same cell. Those proteins become simultaneously secreted into culture supernatants from which they can be purified as separate entities by simple SEC (Figure 2). To simplify production, we nevertheless desired directed expression of either MoAb or DuoMab without significant presence of the other entity. This was achieved by introducing asymmetric mutations into the CH3 domains. Adding regular “knobs-into-holes” mutations into complementary crossed HCs favors predominantly heterodimerization leading to MoAb assembly,23 which is also indicated by an almost baseline-separated SEC profile (Figure 3a). In comparison to the MoAb/DuoMab ratio in Figure 2, the SEC profiles in Figure 3 reveal a clear shift toward MoAb formation. The presence of the correct chain size is confirmed by clear bands in the SDS-PAGE analysis (Figure 3b).
Figure 3.
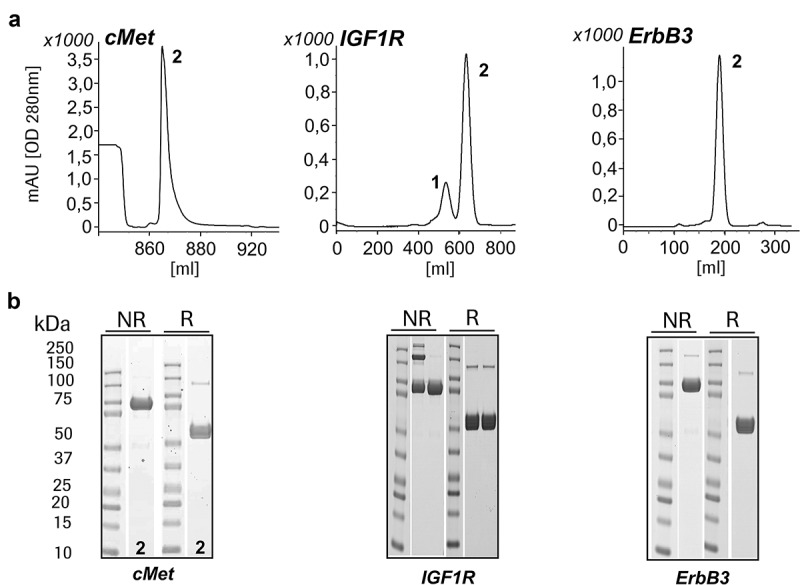
Introduction of the ‘knobs-into-holes’ mutations in the CH3 domains affects the assembly of MoAbs and DuoMabs. Modifications that favor heterodimerization (e.g., “knobs-into-holes” mutations) will increase generation of MoAbs. (a) SEC and (b) SDS-PAGE reveals preferential MoAb assembly (1 = DuoMab, 2 = MoAb). The shown SEC profiles reflect the chromatogram of a preparative purification (in order to show all species without pooling just the desired and show them). Due to the fact that some molecules express better than others (different binders), we obtained different yields after affinity chromatography. The height and retention times differ due to the use of different SEC columns according to the amount of protein.
In contrast, preferential expression of DuoMab tetramers requires two sets of Fcs that are not able to form heterodimers. This was achieved via fusion of CrossFab entities to IgG1-IgA1 hybrid-Fc regions (IgG-Fc does not bind to IgA-Fc). Those hybrid Fc (hyb.) in combination with wildtype Fc (Fc) regions assemble preferentially in the form of tetrameric HCs, as shown schematically in Figure 4a. Also depicted is the underlying modular setup of the IgG1 and IgA1 fusion. The product molecule is confirmed by both the SEC profile and the SDS-PAGE analysis as a clear peak in Figure 4b and bands of expected molecular weight in Figure 4c. An alternative approach to enforce the homodimerization of the Fc parts (a:a and b:b) is the introduction of a defined set of mutations (so-called “Yin-Yang”(Supplementary Figure S1)). All antibodies described in the following sections were generated with the hybrid Fc approach.
Figure 4.
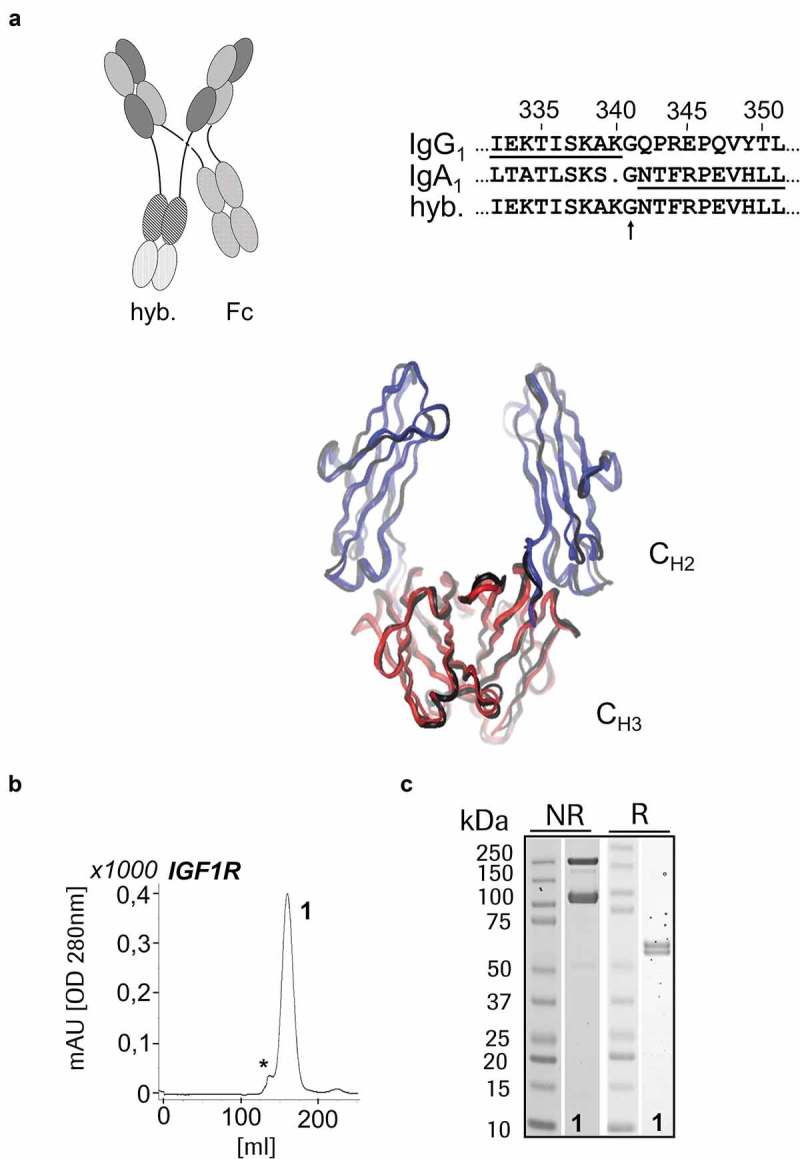
IgG-IgA hybrid-based DuoMab: (a) Modifications that maintain attractive interactions between identical chains but lead to repulsion of different chains (e.g., Fc-domains taken from different isotypes make hybrid Fcs that preferentially assemble, whereas wt-Fcs assemble as shown before) increase the level of DuoMabs. IgG1/IgA1 hybrid sequences (crossover point indicated) for a preferential generation of heavy chain dimers that subsequently tetramerize to form DuoMabs. Expression and preferential assembly of IGF-1R-binding DuoMabs demonstrated by SEC (b) and SDS-PAGE (c) (1 = DuoMab).
The tetrameric conformation was confirmed by negative stain transmission electron microscopy (NS-TEM) (Figure 5a) and small-angle X-ray scattering (SAXS). We further confirmed the correct intact and reduced masses (data not shown) and the inter-chain disulfide bonds of DuoMabs (supplementary Figure S2) by electrospray ionization mass spectrometry. Additionally, SAXS data indicate that DuoMabs adopt a near-planar X-shaped tetrameric structure in solution (Figure 5b), in agreement with the NS-TEM data.
Figure 5.
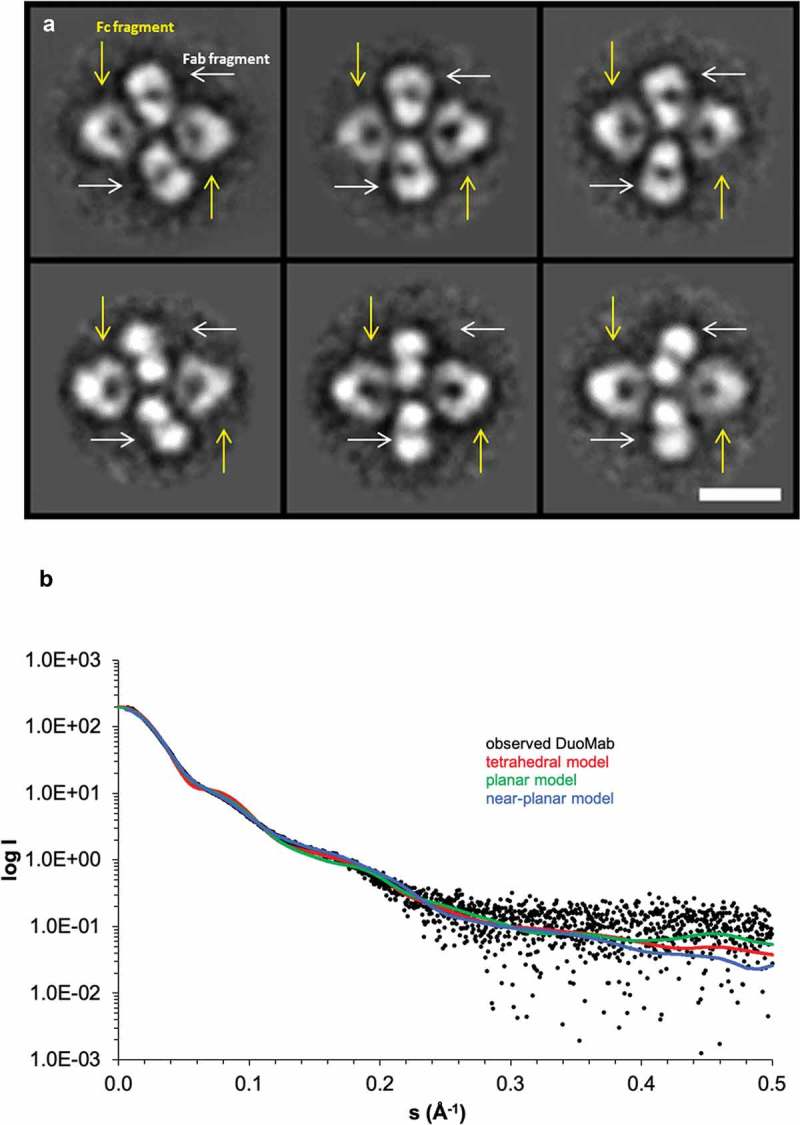
Tetrameric scaffolds of DuoMabs are confirmed by class averages derived from micrographs recorded with negative stain transmission electron microscopy on antigen cMET binding DuoMabs (a, top row), as well as such binding antigen ErbB3 (a, bottom row). The Fc parts can be discriminated from Fabs because of their triangular shape and hole (yellow arrows in a). The Fabs are found in side as well as top views, with or without their central hole (white arrows in a). Scale bar represents 10 nm; all boxes are 31 by 31 nm. Small-angle X-ray scattering (SAXS) based on the ErbB3-specific DuoMab indicates that DuoMabs have a near-planar structure (b). Fit of calculated scattering profiles of different DuoMab models to the observed scattering profile of DuoMab: tetrahedral homology model (red, χ2 = 4.8), planar homology model (green, χ2 = 2.0), near-planar model obtained by a combination of rigid body and flexible modeling (blue, χ2 = 1.5).
DuoMabs retain cell surface target engagement activities of parent antibodies
IGF-1R-binding DuoMabs based on wildtype IgG1 Fc were subjected to antigen-binding analyses and receptor signaling analyses to assess if their target binding properties are retained compared to their parental IgG molecules. For these comparisons, we focused on DuoMabs which bind in a bivalent manner, similar to their parent IgG counterparts (MoAbs bind monovalent and hence cannot be compared to bivalent IgGs. The analyses we performed on monovalent MoAb format are nevertheless provided in Figure S3). Surface plasmon resonance (SPR) assays confirmed that antigen-binding activity was retained as in the parent IgG (KD = 2–4 nM). Fluorescence-activated cell sorting-based cell-binding analyses on IGF-1R expressing A549 cells demonstrated equal binding efficacy of DuoMabs when compared to parental IgGs (Figure 6a). Internalization assays indicated similar internalization properties of parental IgG and DuoMab derivatives (Figure 6b). To assess the effects of cell surface binding on signaling of the cognate receptor, we also analyzed the effects on IGF-1R phosphorylation in NIH3T3 cells that overexpressed human IGF-1R. Figure 6c reveals inhibition efficacies of IGF-1R phosphorylation in the same range when comparing parent IgG with DuoMab. The shift seen could be explained by a different geometry (e.g., hinge flexibility) between the molecules. Thus, the IGF1-R binding DuoMab retains target binding as well as inhibition of receptor signaling activities compared to its parental IgG counterpart.
Figure 6.
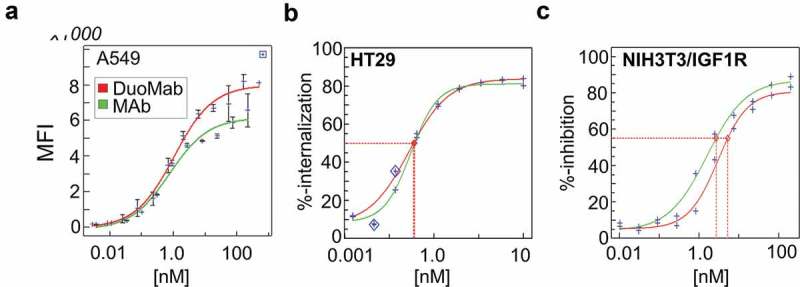
DuoMabs retain cell surface target engagement activities of parent antibodies. (a) Binding of <IGF-1R> DuoMab (EC50 = 1.02nM) and IgG (Mab) (EC50 = 1.00 nM) to A549 cells. (b) Internalization of DuoMab (EC50 = 0.028 nM) and IgG (EC50 = 0.30 nM) into HT29 cells (c) Inhibition of IGF-1R-signaling (phosphorylation) by IgG (IC50 = 2.7nM) and DuoMab (IC50 = 5.3nM). Nonlinear-dose-response curve fits are shown. The small change in ‘c’ (red vs. green curve) might be caused by altered geometry/flexibility in the hinge-region of DuoMab versus IgG. However, overall activities in all these assays do not differ to a significant degree (we consider the phosphorylation activities to be undistinguishable within the assay limitations).
The DuoMab elicits enhanced binding to FcγRIIIa and enhanced ADCC potencies compared to parent IgGs
ADCC is triggered when cell-bound IgGs bind to Fc receptors (in particular FcγRIIIa) on immune cells. The presence of two Fcs in one DuoMab molecule might therefore increase FcγRIIIa binding compared to an IgG with one Fc region, and thereby enhance ADCC. To test this theory, binding of the DuoMab to FcγRIIIa was assessed by an SPR-based binding assay, using FcγRIIIa (V158) as ‘ligand’ for the immobilized IgG or DuoMab. Figure 7a shows that the parental IgG and DuoMab display different binding properties to FcγRIIIa. Increased signals and retention of the DuoMab compared to the IgG can be explained by an avidity-enhanced binding mode of the DuoMab, or by the mutations introduced for generating the hybrid Fc generation or a combination of both.24
Figure 7.
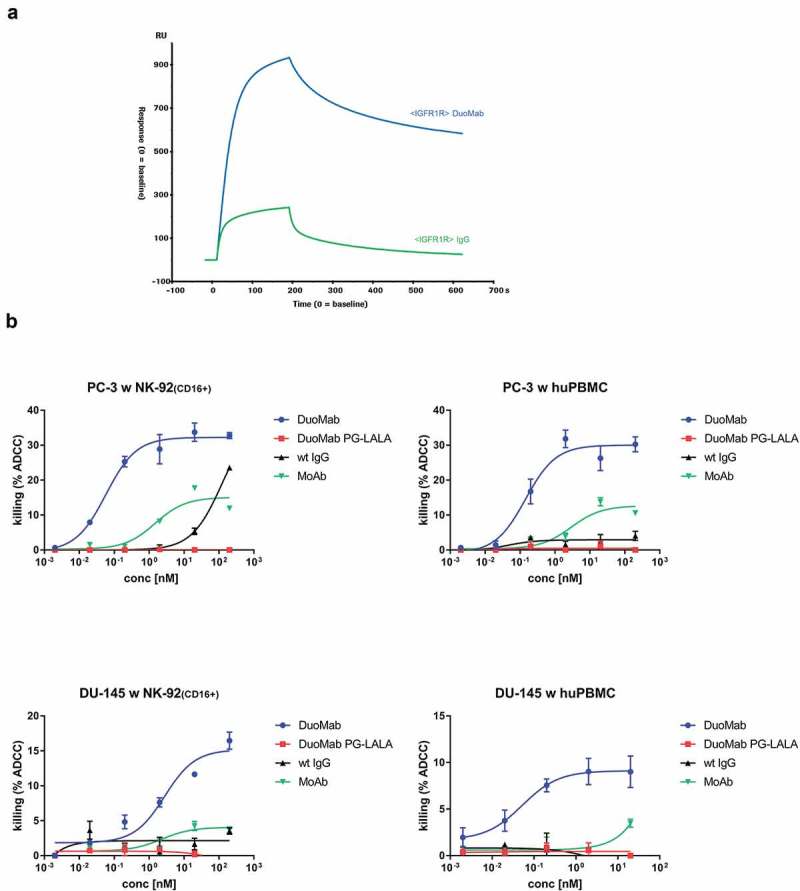
(a) DuoMabs display higher FcγRIIIa binding compared to wt IgGs. (b) Increasing the Fc load on cell surfaces by application of DuoMabs results in enhanced ADCC compared to IgGs. IGF-1R expressing DU-145 and PC-3 cells were co-cultivated with CD16-positive NK-92 cells (1:5 ratio) or PBMCs (1:25 ratio) and exposed to either IGF-1R-binding DuoMabs, DuoMabs (PG-LALA), IgGs or MoAbs at different concentrations. All experiments were repeated three times. huPBMC were purified from the blood of two healthy donors. Shown are mean values from triplicates and standard deviations of representative experiments. Nonlinear dose-response curve fits are shown.
To evaluate if modulation of FcγRIIIa binding through addition of a second Fc translates to increased ADCC, co-incubation assays with peripheral blood mononuclear cells (PBMCs) or CD16-overexpressing NK-92 cells were performed.25 IGF-1R-expressing PC-3 and DU-145 cells were used as target cells. Figure 7b shows ADCC induction after exposure to either the parent IGF-1R-binding IgG or its corresponding MoAb and DuoMab derivatives. In all setups DuoMabs mediated the strongest ADCC, followed by MoAbs. This finding was expected as both entities can double the Fc load on target cells compared to the parental wild-type IgG. Thus, increasing the Fc load on cell surfaces by application of DuoMabs results in enhanced ADCC. DuoMabs that carry the PG-LALA (L234A, L235A, P329G) mutation, and hence cannot bind FcγRIIIa, did not induce any ADCC, which confirms the proposed mode of action.
DuoMab behaves similar to IgG in preclinical rodent PK and cytokine release assays
Among other aspects, serum half-life of antibodies and antibody derivatives is determined by their ability to bind neonatal Fc receptor (FcRn), which – in turn – depends on accessibility and FcRn-binding competency of their Fc regions.26,27 To determine whether an additional Fc per molecule results in altered PK properties, we assessed the PK of DuoMabs in NMRI mice in direct comparison to that of parent IgGs. The results of these analyses show that DuoMabs have PK parameters in the same range as regular IgGs (serum half-life) (Table 1). Thus, duplication of the Fc region and/or changes in the geometry or proximity to their Fc have no major impact on the PK properties of DuoMabs.
Table 1.
Comparison of the PK properties of the DuoMab and the parent IgG.
| IGF1R Mab | IGF1R DuoMab | ||
|---|---|---|---|
| C0 | µg/mL | 81.9 | 201.56 |
| Cmax | µg/mL | 80.7 | 195.9 |
| Tmax | h | 0.5 | 0.5 |
| AUC0-inf | h*µg/mL | 9349 | 12096 |
| Term t1/2 | H | 106.2 | 83.5 |
| CI | mL/min/kg | 0.018 | 0.013 |
| Vss | L/kg | 0.16 | 0.1 |
Sustmann et al., Table 1
Eradication of tumor cells by ADCC is a desired effect of antibody-based tumor therapies, but it is accompanied by the release of cytokines such as interleukins, tumor necrosis factor (TNF) and interferon. Therefore, whole blood (human) cytokine assays were performed to compare the degree of cytokine release triggered by DuoMabs in comparison to parent IgGs. Figure 8 shows the levels of released IL-6, IL-8, TNF and IFNγ as a consequence of 24hr co-cultures of heparinized whole blood (six healthy human donors, data presented relative to control IgG1 antibody cetuximab). The results of these analyses reveal cytokine secretion in a dose-dependent manner for the parental IgG as well as for the DuoMab derivative. However, despite the increased Fc load in DuoMabs, which increases ADCC competency and cytokine release, none of the measured cytokines were found at elevated levels compared to the wildtype IgG.
Figure 8.
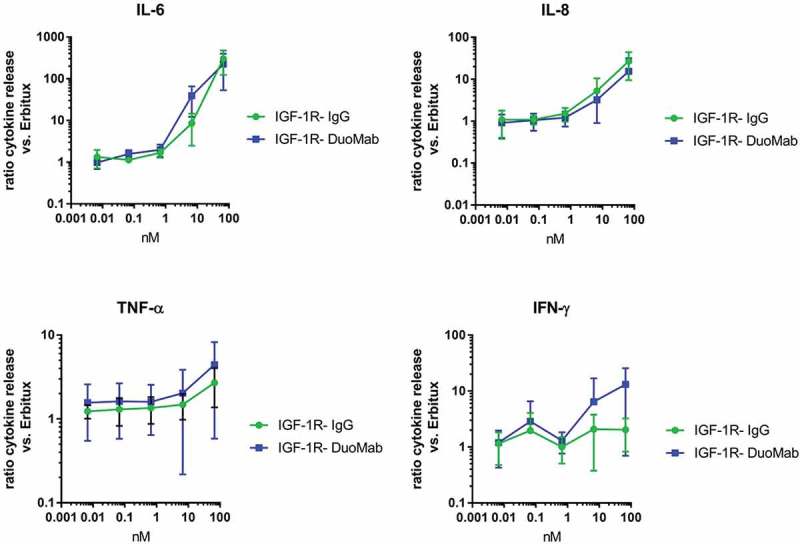
Release of IL-6, IL-8, TNF and IFN-γ as a consequence of exposure to IGF-1R binding IgG or corresponding DuoMab is presented as mean value (+SEM) of determinations from six donors (<IGF-1R> vs cetuximab as reference).
Discussion
Here, we describe two novel CrossMab-based antibody formats, a monovalent MoAb format and a bivalent DuoMab format with a unique and novel structure. Both formats are generally applicable for binders with various specificities. Antibody derivatives that bind IGF-1R were used here to describe in more detail their generation, characterization and functionalities.
The DuoMab format applies Fab region domain crossover modules that were originally designed for the generation of CrossMabs with orthogonal Fc domains. The equilibrium in the production of these two molecules from a production cell line can be varied by different Fc heterodimerization technologies, either the use of the “knobs-into-holes” mutations to generate preferentially MoAbs or alternatively two different Fc isotypes or orthogonal pairs of Fc mutations to preferentially generate the DuoMabs. The resulting DuoMabs harbor two Fab-like (crossed) binding arms and two Fc parts that are linked to each other via hinge regions. One particular feature that differentiates DuoMab from an IgG and most other bsAbs is that it is only composed of HCs and does not contain any light chains. The Fv functionalities (VH/VL) are integrated into the HCs. Another variation of the DuoMabs from most other antibodies derivatives is a quadruple connection in the hinge region between binding arms and Fc portions. Two hinges, each consisting of two disulfide-linked entities, are crosswise connected to Fc regions. The high density and close proximity of disulfides might make them susceptible to aggregation and less flexible in regard to antigen binding. Potential high-order entities, such as daisy-chained molecules as described for IgG2 antibodies by Wypych et al.,28 could be separated by SEC. In general, no major aggregation problems were observed as indicated by a small “aggregate peak” in our preparative SEC. Moreover, no reduced binding capacity was observed for the DuoMabs, and the lower flexibility of the hinge region may also be advantageous depending of the choice of the targeted antigen. Since the chain pairing in the molecule assembly is exclusively based on interactions in the Fc, the approach is not restricted to a certain specificity of the variable domains, but can rather be adapted for any desired target antigen. A further advantage of this format is the full compatibility with standard upstream- and downstream processes for production of therapeutic antibodies. Finally, it could potentially also be used to generate bispecific DuoMab that recognize two different antigens and harbor two Fc domains.
In addition to the ‘amount’ of Fcs directed at target cells, composition and/or clustering of Fc regions are relevant for their functionality. In this regard, the choice of Fc sequences (e.g., IgG1, IgG2, IgA, IgM, other designed oligomer-formats) can directly influence therapeutic applicability of recombinant antibody derivatives. Activity is further modulated by the binding capability of those modules to Fc receptors, including FcRn (to modulate PK properties), and FcγRs to trigger immune-mediated effector functionalities such as ADCC, complement-dependent cytotoxicity, or trogocytosis.29–31 The DuoMab format that combines two Fc on one antibody opens novel possibilities with the option to combine and co-deliver in close proximity hybrid or mutated Fcs with different features in one molecule.
ADCC can be observed for DuoMabs in direct comparison to their regular IgG counterparts. ADCC is a natural immune defense mechanism against foreign or aberrant protein structures such as those that are displayed on pathogen-infected cells. This mechanism has been successfully exploited by therapeutic antibodies, which mimic the opsonization of cell surfaces and recruit ADCC-executing immune cells to the desired target cell, e.g., tumor cells.32 Exemplarily, the IgG-derived drug rituximab, an established medication against lymphomas and leukemia, triggers ADCC through the activation of NK cells via its Fc. The interaction of the Fc with the FcγRIIIa receptor on immune cells is crucial for the initiation of ADCC.5 The currently established approaches to enhance ADCC that are not based on the principle of enhancing Fc load include Fc alterations that enhance the affinity between NK cell and antibody. One such approach involves modifying the protein glycosylation of Fc to confer enhanced FcR binding, and thereby enhanced NK cell activation. The antibody obinutuzumab, which, like rituximab, binds the CD20 antigen on cancer cells and which demonstrates enhanced ADCC due to glycoengineering, serves as example for that principle.11,33 Other approaches make use of mutations to strengthen the Fc-FcγRIIIa interaction,6 or increase/multiply the numbers of Fc regions.34,35
In the underlying work, we developed a generic approach to obtain antibodies with IgG-like binding characteristics but enhanced ADCC properties by simply doubling the Fc region. Regarding the limited number of antigens on a target cell, the Fc load can be theoretically doubled, which increases ADCC capacity.16 We showed that indeed a significantly higher ADCC reaction occurred with DuoMabs, as compared with the parental IgG behavior. It may be possible to further enhance the ADCC potency of such constructs by additional glycoengineering of DuoMab entities. Such approaches may encompass selection of a modified expression host or introduction of amino acid point mutations in the Fc.
The use of MoAbs is also a way to increase the Fc load if antigen density is an issue, in the same stoichiometry as DuoMab – one binding site, one Fc. However, monovalent binding requires antibodies with high monovalent affinity. DuoMabs have the same stoichiometry, but do not require very high affinity and rely on avidity effects.
In conclusion, we show that DuoMabs can be produced as defined molecules by expression and purification technologies that are established for standard antibodies. DuoMabs possess PK behavior similar to IgGs and can be applied to enhance ADCC-mediated killing of tumor cells by increasing the Fc loads on surfaces of target cells.
Materials & methods
Transient expression of immunoglobulin variants in HEK293 cells
Antibodies were expressed by transient transfection of human embryonic kidney 293-F cells using the FreeStyle™ 293 Expression System according to the manufacturer’s instruction (Invitrogen, USA). For transfection, all plasmids were applied in equal molar amounts. Antibody containing cell culture supernatants were harvested 7 days after transfection by centrifugation at 14000 g for 30 min and filtered through a sterile filter (0.22 µm).
Purification of antibodies
Cell culture supernatants were applied to a HiTrap ProteinA HP (5 ml) column (GE Healthcare, Sweden) equilibrated with phosphate-buffered saline (PBS) buffer (10 mM Na2HPO4, 1 mM KH2PO4, 137 mM NaCl and 2.7 mM KCl, pH 7.4). Products were eluted with 0.1 M citrate buffer, pH 2.8 and the protein-containing fractions were neutralized with 0.1 ml 1 M Tris, pH 8.5. Then, the eluted protein fractions were loaded on a Superdex200 HiLoad 120 ml 16/60 or 26/60 gel filtration column (GE Healthcare, Sweden) equilibrated with 20 mM histidin, 140 mM NaCl, pH 6.0. The displayed SEC profiles reflect the chromatogram of a preparative purification. Due to the fact that some molecules express better than others (different binders), we obtained different yields after affinity chromatography. The height and retention times differ due to usage of different SEC columns according to the amount of protein.
Analysis of purified proteins
Antibodies were analyzed by SDS-PAGE in the presence and absence of a reducing agent (5 mM 1,4-dithiotreitol). The NuPAGE® Pre-Cast gel system (Invitrogen, USA) was used according to the manufacturer’s instruction (4-12% Tris-Glycine gels) and subsequently stained with Coomassie brilliant blue. The aggregate content of antibody samples was analyzed by high-performance SEC using a Superdex 200 analytical size-exclusion column (GE Healthcare, Sweden) in 200 mM KH2PO4, 250 mM KCl, pH 7.0 running buffer at 25°C.
Mass spectrometry
Intact-deglycosylated masses were determined by electrospray ionization mass spectrometry. 100 µg protein was deglycosylated with 50 mU N-Glycosidase F (PNGaseF, Roche Applied Science, Germany) in 100 mM KH2PO4/K2HPO4, pH 7 at 37°C for 12–24 h at a protein concentration of up to 2 mg/ml and subsequently desalted via HPLC on a Sephadex G25 column (GE Healthcare). The masses of the respective antibody chains were determined after deglycosylation and reduction. 50 µg antibody in 115 µl was incubated with 60 µl 1M tris(2-carboxyethyl)phosphine (TCEP) and 50 µl 8 M guanidine-hydrochloride and subsequently desalted. Mass spectrometry analysis was performed on a Q-Star Elite MS system equipped with a TriVersa NanoMate chip-based electrospray system (Advion, UK).
For the determination of the inter-chain disulfide bridging of the DuoMab, 250 µg protein was denatured in 250 mM Tris-HCl, 5 M Gua-HCl, pH 8, re-buffered in 50 mM Tris-HCl, pH 7.5 using NAP-5 columns (GE Healthcare Life Sciences, Germany) and digested with 5 µg endoproteinase Lys-C (Roche Applied Science, Germany) at 37°C for 19 h. Finally, the reaction was stopped by adding trifluoroacetic acid to 0.4%. The samples were analyzed by electrospray ionization mass spectrometry using a maXis 4G UHR-QToF instrument (Bruker Daltonics, USA) equipped with a TriVersa NanoMate. Dipeptides were reduced with TCEP.
SEC-MALLS
SEC-MALLS was used to determine the approximate molecular weight of proteins in solution. The following instrumentation was used: Dionex Ultimate 3000 HPLC; column: Superose6 10/300 (GE Healthcare); eluent: 1 x PBS; flow rate: 0.25 mL/min; detectors: OptiLab REX (Wyatt Inc., Dernbach), MiniDawn Treos (Wyatt Inc., Dernbach). Molecular weights were calculated with the Astra software, version 5.3.2.13. Since the measured molecular weight (MoAb = 99.5 kDa; DuoMab = 193.8 kDa) fits to the expected theoretical mass (MoAb = 98.2 kDa, DuoMab = 196.2 kDa), we did not use different columns. Due to the assumptions that the proteins have the same refractive index in solution, we have not done any adjustment of dn/dc values.
Surface plasmon resonance
The binding properties of the antibodies were analyzed by SPR technology using a Biacore instrument (Biacore, GE-Healthcare, Uppsala). An anti-human IgG antibody (goat anti-human IgG Fcg-fragment, Jackson ImmunoResearch, Cat#109-006-098) was immobilized on the surface of a CM5 biosensorchip using amine-coupling chemistry. The flow cells were activated with a 1:1 mixture of 0.1 M N-hydroxysuccinimide and 0.1 M 3-(N,N- dimethylamino)propyl-N-ethylcarbodiimide at a flow rate of 5 μl/min. The anti-human IgG antibody was injected in sodium acetate, pH 5.0 at 10 μg/ml. A reference control flow cell was treated in the same way, but with vehicle buffers only instead of the capturing antibody. Surfaces were blocked with an injection of 1 M ethanolamine/HCl pH 8.5. The IGF-1R antibodies were diluted in HBS-P and injected. All interactions were performed at 25°C. The regeneration solution of 3 M Magnesium chloride was injected for 60 s at 5 μl/min flow to remove any non-covalently bound protein after each binding cycle. Signals were detected at a rate of one signal per second. For affinity measurements, human FcγIIIa was immobilized to a CM-5 sensor chip by capturing the His-tagged receptor to an anti-His antibody (Penta-His, Qiagen), which was coupled to the surface by standard amine-coupling and blocking chemistry on a SPR instrument (Biacore T100). After FcγRIIIa capturing, 50 nM IGF-1R antibody was injected at 25°C at a flow rate of 5 µL/min. The chip was afterward regenerated with a 60 s pulse of 10 mM glycine-HCl, pH 2.0 solution.
ADCC assay
IGF-IR-expressing DU-145 and PC-3 cells (1.5 x 105 cells/ml) were seeded out in RPMI1640 media supplemented with 2 mM glutamine and 10% fetal calf serum (FCS) in 96-well plates. Cells were incubated overnight at cell culture conditions. The following day, NK-92-CD16+ (E:T ratio 5:1) or PBMC (E:T ratio 25:1) and the respective antibody solution were added to a final volume of 200 µl.25 PBMC were purified from fresh whole blood of healthy human donors (collected after informed consent at the Blood Donation Center, SRK, Basel or Roche Infirmary Services, Switzerland or Roche Gesundheitsdienst Penzberg, Germany) according to the manufacturer’s recommendations using Ficoll® Paque Plus (GE Healthcare, Chicago, IL, USA) and Leucosep™ centrifuge tubes (Greiner Bio-one, Kremsmünster, Austria). RPMI1640 media supplemented with 2 mM glutamine and 5% FCS was used as assay media. Plates were incubated for 14 h at cell culture conditions. Next, 100 µl of each well was transferred into a white, clear bottom 96-well plate using a liquidator. 100 µl of the detection solution from the Cytotoxicity Detection Kit (LDH; Roche, 11 644 793 001) was added. Plates were incubated in the dark at ambient temperature for 15 min. Total lysis (100% killing) was achieved by adding a final concentration of 2% Triton-X-100 to the control wells. Both, the preparation of the reagents and the calculation of the specific lysis was performed according to the manufacturer’s instructions.
IGF-1R internalization assay
HT29 cells at 1.5 × 104 cells/well were incubated in a 96-well microtiter plate in RPMI with 10% FCS overnight at 37°C and 5% CO2 in order to allow attachment of the cells. The medium was aspirated and 100 μl anti-IGF-1R antibody diluted in RPMI + 10% FCS was added in a concentration range from 10 nM to 2 pM in 1:3 dilution steps. The cells were incubated with antibody for 18 h at 37°C. Afterward, the medium was again removed and 120 µl MES lysis buffer (25 mM MES pH 6.5 + Complete) were added. For enzyme-linked immunosorbent assay (ELISA), 96-well streptavidin-coated polystyrene plates (Nunc) were loaded with 100 μl biotinylated monoclonal anti-human IGF-1R (in-house production) diluted 1:200 in 3% bovine serum albumin (BSA)/PBST (final concentration 2.4 µg/ml) and incubated under constant agitation for 1 h at room temperature. Afterward, cells were washed three times with 200 µl PBST. 100 µl of the cell lysate solution were added per well, again incubated for 1 h at room temperature on a plate shaker, and washed three times with 200 µl PBST. After removal of the supernatant, 100 µl/well of polyclonal anti-human IGF-1Rα Ra-C20-IgG (Santa Cruz #sc-713) diluted 1:750 in 3% BSA/PBST was added followed by the same incubation and washing intervals as described above. In order to detect the specific antibody bound to IGF-1R, 100 µl/well of a polyclonal horseradish-peroxidase (HRP)-coupled rabbit antibody (Cell Signaling #7074) diluted 1:4000 in 3% BSA/PBST were added. After another hour, unbound antibody was again removed by washing thoroughly 6 times as described above. For quantification of bound antibody, 100 µl/well 3,3ʹ-5,5ʹ-tetramethylbenzidin (Roche, BM-Blue ID.-Nr.11484281) was added and incubated for 30 min at room temperature. The colorogenic reaction is finally stopped by adding 25 µl/well 1M H2SO4 and the light absorption is measured at 450 nm. Cells not treated with antibody are used as a control for 0% downregulation, lysis buffer as background control.
IGF-1R autophosphorylation assay (IGF-1 stimulation)
Targeting IGF-1R by IGF-1R antibodies results in inhibition of IGF-1-induced autophosphorylation. Autophosphorylation as a consequence of exposure to the wildtype IGF-1R DuoMab compared to the parental IGF-1R IgG1 antibody was investigated. IGF1R phosphorylation was assessed essentially as previously described.36 Briefly, 3T3-IGF-1R cells were treated with different concentrations of the respective antibody for 30 min. Then, the mixture was incubated for 10 min with 10 nM recombinant human IGF-1 (PreProtech) before lysis (Cell Lysis Kit, Bio-Rad). Levels of phosphorylated IGF-1R protein were determined by a phospho-IGF-1R specific ELISA, combining human IGF-1R capture with phospho-tyrosine specific detection antibodies (Tyr1135/1136)/insulin receptor beta (Tyr1150/1151)(19H7) antibody, Cell Signaling followed by anti-rabbit IgG-POD (Cell Signaling) and development with 3,3′-5,5′-tetramethylbenzidin (Roche) as previously described.36
Negative stain transmission electron microscopy
Protein solutions at concentration c = 1 mg/ml were diluted to about c = 10 µg/ml in D-PBS for analysis with NS-TEM. 5 µL of diluted sample solutions were adsorbed for 1 min to glow discharged, parlodion and carbon-film coated 200 mesh copper TEM grids. Excess sample was removed with blotting paper. The grids were washed 5 times on milliQ water drops, negatively stained with 2% uranyl acetate, blotted again and finally air-dried. Micrographs were recorded with a Philips CM10 TEM (Philips, Eindhoven, the Netherlands) operated at 80 kV, at a nominal magnification of 130,000 x, using a 2000 by 2000 pixel side-mounted CCD Camera (Veleta; Olympus soft imaging solutions GmbH, Münster, Germany), corresponding to a pixel size of 3.85 Å at the specimen level. Image analysis was performed on manually selected particles from recorded micrographs using the EMAN2 image-processing package. A total of 1027 and 832 particles were extracted from 36 to 46 micrographs, respectively, from sample cMET and ErbB3. Extracted particles were aligned, and classified by multivariate statistical analysis yielding 8 class averages. Classes resulting from an average of 112 to 160 single particles with the best signal-to-noise ratio were selected and presented.37
Cytokine release assays
After informed consent was provided, venous blood from healthy blood donors was collected in vacutainer tubes with heparin as anticoagulant (Blood Donation Center, SRK, Basel and Roche Infirmary Services, Switzerland) and kept at room temperature until initiation of the assay.195μl of blood was added in triplicates to U-bottom wells of 96-well plates containing 5μl of the respective antibody in a concentration range from 0.1 to 100 µg/ml. After incubation for 24 h at 37°C cells and plasma were separated by centrifugation at 1800g for 5 min. Plasma samples were stored at −70°C until analysis of TNF, IL-6 and IL-8 with a 9-plex MSD® 96-well Multi-ArrayTM (MesoScaleDiscovery®). Cytokine concentrations were determined as mean values with standard deviations of triplicates, unless otherwise noted. Analysis of cytokines was performed using MSD Discovery Workbench® (v 3.0) software (Mesoscale®). Data were plotted as a mean ratio compared to the cytokine levels upon cetuximab treatment (as reference control).
PK analyses
Female NMRI mice (23–32 g body weight) were used. For a single intravenous dose of 10 mg/kg, the mice were allocated to three groups with 2–3 animals each. Blood samples are taken from group 1 at 0.5, 168 and 672 h, from group 2 at 24 and 336 h and from group 3 at 48 and 504 h after dosing. Blood samples of about 100 µL were obtained by retrobulbar puncture. Serum samples of at least 40 µl were obtained from blood after 1 h at room temperature by centrifugation (9300 x g) at room temperature for 2.5 min. Serum samples were frozen directly after centrifugation and stored frozen at – 20°C until analysis. The human antibody concentrations were determined with an ELISA using 1% mouse serum. Biotinylated monoclonal antibody against human Fcγ (mAb<hFcγPAN>IgG-Bi) was bound to streptavidin-coated microtiter plates, serum samples (in various dilutions) and reference standards, respectively, were added and bound to the immobilized mAb<hFcγPAN>IgG-Bi. Then, digoxigeninylated monoclonal antibody against human Fcγ (mAb<hFcγPAN>IgG-Dig) was added. The human antibodies were detected via anti-Dig-HRP antibodies. ABTS-solution was used as the substrate for HRP. The specificity of the used capture and detection antibody, which does not cross-react with mouse IgG, enables quantitative determination of human antibodies in mouse serum samples, applying the corresponding sample for reference. The presented PK parameters were calculated by non-compartmental analysis, using the PK evaluation program WinNonlinTM, version 5.2.1.
Small-angle X-ray scattering
To prepare samples suitable for small-angle X-ray scattering (SAXS) measurements, purified DuoMab was additionally purified via SEC on a Superdex 200 10/300 GL column (GE Healthcare) in 20 mM histidine-HCl pH 6.0, 140 mM NaCl, and concentrated to yield a sample in concentration range from 1 to 7 mg/ml. The flowthrough of the concentration step was used as buffer reference for SAXS measurements. SAXS data were collected at beamline X33 at EMBL/DESY, Hamburg. Scattering profiles of BSA and lysozyme were measured as reference for molecular mass determination. The ATSAS package22 was used to process and analyze data. Theoretical scattering profiles from tetrahedral and planar homology models were calculated and fitted to measured profiles with CRYSOL.38,39 The best fit to the scattering data was obtained using BUNCH by modeling DuoMab as a two-fold symmetric dimer of Fab and Fc fragments connected by a flexible linker consisting of the hinge region.40
Abbreviations
| ADCC | antibody-dependent cellular toxicity |
| ELISA | enzyme linked immonosorbent assay |
| Fab | antibody fragment composed of Fd and LC |
| HC | antibody heavy chain |
| IFG-1R | insulin growth factor 1 receptor |
| LC | antibody light chain |
| MALLS | multi angle laser light scattering |
| NK | natural killer |
| NS-TEM | negative stain transmission electron microscopy |
| PK | pharmacokinetic |
| SAXS | small-angle X-ray scattering |
| SEC | size exclusion chromatography |
| SPR | surface plasmon resonance |
Acknowledgments
SD is a student member, CS and CK are mentors of the international graduate program ‘i-Target’: Immunotargeting of Cancer.
Disclosure of Potential Conflicts of Interest
The specified authors are employed by Roche, CS, SD, JTR, HK, MM, MEL, BB, KPK, UB, WS and CK hold patents relating to antibody engineering technology. Roche has an interest in and develops targeted therapies.
Supplementary Material:
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Fanger MW, Shen L, Graziano RF, Guyre PM.. Cytotoxicity mediated by human Fc receptors for IgG. Immunol Today. 1989;10:92–99. doi: 10.1016/0167-5699(89)90234-X. [DOI] [PubMed] [Google Scholar]
- 2.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, Chan C, Chung HS, Eivazi A, Yoder SC, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci USA. 2006;103:4005–10. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol . 2010;10:301. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells. 2011;16:1071–80. doi: 10.1111/j.1365-2443.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellner C, Otte A, Cappuzzello E, Klausz K, Peipp M. Modulating Cytotoxic Effector Functions by Fc Engineering to Improve Cancer Therapy. Transfus Med Hemother. 2017;44:327–36. doi: 10.1159/000479980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mössner E, Brünker P, Moser S, Püntener U, Schmidt C, Herter S, Grau R, Gerdes C, Nopora A, van Puijenbroek E, et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct- and immune effector cell-mediated B-cell cytotoxicity. Blood. 2010;4393–402. doi: 10.1182/blood-2009-06-225979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefferis R, Lund J, Pound JD. IgG-Fc-mediated effector functions: molecular definition of interaction sites for effector ligands and the role of glycosylation. Immunol Rev. 1998;163:59–76. [DOI] [PubMed] [Google Scholar]
- 8.Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25:1325–34. doi: 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umaña P, Jean–Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat. Biotechnol. 1999;17:176. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- 10.Ferrara C, Stuart F, Sondermann P, Brünker P, Umaña P. The Carbohydrate at FcγRIIIa Asn-162: AN ELEMENT REQUIRED FOR HIGH AFFINITY BINDING TO NON-FUCOSYLATED IgG GLYCOFORMS. J Biol Chem. 2006;281:5032–36. doi: 10.1074/jbc.M510171200. [DOI] [PubMed] [Google Scholar]
- 11.Umana P, Ekkehard M, Peter B, Gabriele K, Ursula P, Suter T, Grau R. GA101, a novel humanized type II CD20 antibody with glycoengineered Fc and enhanced cell death induction, exhibits superior anti-tumor efficacy and superior tissue B cell depletion in vivo. Blood. 2007;110:2348. [Google Scholar]
- 12.Golay J, Da Roit F, Bologna L, Ferrara C, Leusen JH, Rambaldi A, Klein C, Introna M. Glycoengineered CD20 antibody obinutuzumab activates neutrophils and mediates phagocytosis through CD16B more efficiently than rituximab. Blood. 2013;122:3482–91. doi: 10.1182/blood-2013-05-504043. [DOI] [PubMed] [Google Scholar]
- 13.Alduaij W, Ivanov A, Honeychurch J, Cheadle EJ, Potluri S, Lim SH, Shimada K, Chan CHT, Tutt A, Beers SA, et al. Novel type II anti-CD20 monoclonal antibody (GA101) evokes homotypic adhesion and actin-dependent, lysosome-mediated cell death in B-cell malignancies. Blood. 2011;117:4519–29. doi: 10.1182/blood-2010-07-296913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans JB, Syed BA. Next-generation antibodies. Nat Rev Drug Discovery. 2014;13:413. doi: 10.1038/nrd4255. [DOI] [PubMed] [Google Scholar]
- 15.Teeling JL, Mackus WJ, Wiegman LJ, van Den Brakel JH, Beers SA, French RR, van Meerten T, Ebeling S, Vink T, Slootstra JW, et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J Immunol. 2006;177:362–71. doi: 10.4049/jimmunol.177.1.362. [DOI] [PubMed] [Google Scholar]
- 16.Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: clinical response, cellular immunity, and immunoescape. J Clin Oncol: Off J Am Soc Clin Oncol. 2010;28:4390–99. doi: 10.1200/JCO.2009.27.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preithner S, Elm S, Lippold S, Locher M, Wolf A, Silva A, Baeuerle PA, Prang NS. High concentrations of therapeutic IgG1 antibodies are needed to compensate for inhibition of antibody-dependent cellular cytotoxicity by excess endogenous immunoglobulin G. Mol Immunol. 2006;43:1183–93. doi: 10.1016/j.molimm.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci. 2011;108:11187–92. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenn S, Schiller CB, Griese JJ, Duerr H, Imhof-Jung S, Gassner C, Moelleken J, Regula JT, Schaefer W, Thomas M, et al. Crystal structure of an anti-Ang2 CrossFab demonstrates complete structural and functional integrity of the variable domain. PLoS One. 2013;8:e61953. doi: 10.1371/journal.pone.0061953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein C, Lammens A, Schafer W, Georges G, Schwaiger M, Mossner E, Hopfner K-P, Umaña P, Niederfellner G. Epitope interactions of monoclonal antibodies targeting CD20 and their relationship to functional properties. MAbs. 2013;5:22–33. doi: 10.4161/mabs.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regula JT, Imhof-Jung S, Mølhøj M, Benz J, Ehler A, Bujotzek A, Schaefer W, Klein C, Bradbury A. Variable heavy–variable light domain and Fab-arm CrossMabs with charged residue exchanges to enforce correct light chain assembly. Protein Eng Des Sel. 2018;31:289–99. doi: 10.1093/protein/gzy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein C, Schaefer W, Regula JT, Dumontet C, Brinkmann U, Bacac M, Umaña P. Engineering therapeutic bispecific antibodies using CrossMab technology. Methods. 2018;154:21–31. [DOI] [PubMed] [Google Scholar]
- 23.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–21. [DOI] [PubMed] [Google Scholar]
- 24.Monnet C, Jorieux S, Urbain R, Fournier N, Bouayadi K, De Romeuf C, Behrens CK, Fontayne A, Mondon P. Selection of IgG variants with increased fcrn binding using random and directed mutagenesis: impact on effector functions. Front Immunol. 2015;6:39. doi: 10.3389/fimmu.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes CA, Nicolini VG, Herter S, van Puijenbroek E, Lang S, Roemmele M, Moessner E, Freytag O, Friess T, Ries CH, et al. GA201 (RG7160): a novel, humanized, glycoengineered anti-EGFR antibody with enhanced ADCC and superior in vivo efficacy compared with cetuximab. Clin Cancer Res. 2013;19:1126–38. doi: 10.1158/1078-0432.CCR-12-0989. [DOI] [PubMed] [Google Scholar]
- 26.Hinton PR, Xiong JM, Johlfs MG, Tang MT, Keller S, Tsurushita N. An engineered human IgG1 antibody with longer serum half-life. J Immunol. 2006;176:346–56. doi: 10.4049/jimmunol.176.1.346. [DOI] [PubMed] [Google Scholar]
- 27.Keizer RJ, Huitema ADR, Schellens JHM, Beijnen JH. Clinical Pharmacokinetics of Therapeutic Monoclonal Antibodies. Clin Pharmacokinet. 2010;49:493–507. doi: 10.2165/11531280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 28.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Mathieu M, Brezski RJ. IgG Fc engineering to modulate antibody effector functions. Protein Cell. 2018;9:63–73. doi: 10.1007/s13238-017-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HI, Yoon HW, Jung ST. The Highly Evolvable Antibody Fc Domain. Trends Biotechnol. 2016;34:895–908. doi: 10.1016/j.tibtech.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Cook EM, Lindorfer MA, van der Horst H, Oostindie S, Beurskens FJ, Schuurman J, Zent CS, Burack R, Parren PWHI, Taylor RP. Antibodies that efficiently form hexamers upon antigen binding can induce complement-dependent cytotoxicity under complement-limiting conditions. J Immunol. 2016;197:1762–75. doi: 10.4049/jimmunol.1600648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iannello A, Ahmad A. Role of antibody-dependent cell-mediated cytotoxicity in the efficacy of therapeutic anti-cancer monoclonal antibodies. Cancer Metastasis Rev. 2005;24:487–99. doi: 10.1007/s10555-005-6192-2. [DOI] [PubMed] [Google Scholar]
- 33.Goede V, Klein C, Stilgenbauer S. Obinutuzumab (GA101) for the treatment of chronic lymphocytic leukemia and other B-cell non-hodgkin’s lymphomas: a glycoengineered type II CD20 antibody. Oncology Research and Treatment. 2015;38:185–92. doi: 10.1159/000381524. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Chen Y, Pelletier M, Cvitkovic R, Bonnell J, Chang CY, Koksal AC, O’Connor E, Gao X, Yu X-Q, et al. Enhancement of antibody functions through Fc multiplications. Mabs. 2017;9:393–403. doi: 10.1080/19420862.2017.1281505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms BR MA, US), Kohli N (Brighton, MA, US), Lugovskoy A (Woburn, MA, US), Geddie M (Somerville, MA, US), Krauland EM (Lebanon, NH, US), Roach WG (Lebanon, NH, US), Su S (Boston, MA, US), Abu-yousif A (Boston, MA, US). ANTI-C-MET tandem Fc bispecific antibodies. Cambridge (MA): Merrimack Pharmaceuticals, Inc; 2016. [Google Scholar]
- 36.Schanzer JM, Wartha K, Croasdale R, Moser S, Kunkele KP, Ries C, Scheuer W, Duerr H, Pompiati S, Pollman J, et al. A novel glycoengineered bispecific antibody format for targeted inhibition of epidermal growth factor receptor (EGFR) and insulin-like growth factor receptor type I (IGF-1R) demonstrating unique molecular properties. J Biol Chem. 2014;289:18693–706. doi: 10.1074/jbc.M113.528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Konarev PV, Volkov VV, Petoukhov MV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr. 2006;39:277–86. doi: 10.1107/S0021889806004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svergun D, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–73. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- 40.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–50. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


