Abstract
In this issue, Sepulveda et al. (2018) discovered an interesting role of Hsp47 in regulating the unfolded protein response (UPR) wherein Hsp47 binds to IRE1α and displaces BiP, thereby activating the IRE1α arm of the UPR pathway by a previously undetermined mechanism.
The process of protein folding in the endoplasmic reticulum (ER) is complex and involves numerous players whose goal is to permit the secretion of properly folded proteins while retaining misfolded proteins and targeting them for degradation. This delicate balance between protein folding and misfolded protein degradation can be disrupted due to an influx of protein synthesis or an accumulation of misfolded proteins resulting in ER stress, which activates the unfolded protein response (UPR) in an attempt to restore ER homeostasis (Walter and Ron 2011). In mammalian cells, UPR activation can be achieved via three well-characterized signal transducers, one of which is Inositol-Requiring Enzyme 1 alpha (IRE1α). IRE1α is a central regulator of ER proteostasis capable of interacting with components of the ER stress recovery pathway, as well as those of the apoptotic pathway; however, the nature of the IRE1α interactome is not well characterized. Sepulveda et al. identified the ER chaperone Hsp47 as a novel IRE1α interacting partner and demonstrated a role for it in activating IRE1α.
IRE1α is a type I ER transmembrane protein that, unlike the other two UPR signal transducers (ATF6 and PERK), is present in protozoan and metazoan cells alike and thus is both highly conserved and characterized. The cytosolic tail of IRE1α, which contains a serine/threonine kinase and a RNase domain, dimerizes upon misfolded protein accumulation. This dimerization event results in IRE1α autophosphorylation and activates the RNase domain. The activation of the RNase domain leads to the unconventional splicing of XBP1 mRNA, which modulates the expression of ER components that respond to ER stress (Hetz Papa 2017). One of the consequences of activating the UPR in response to stress is the global attenuation of protein synthesis. This is achieved in part by the Regulated IRE1-Dependent Decay (RIDD) pathway, which results in the degradation of various mRNAs by the endonuclease activity of IRE1α in an attempt to alleviate ER stress. However, under prolonged ER stress, IRE1α oligomerizes and upregulates pro-inflammatory and pro-apoptotic proteins. As a result, IRE1α is able to modulate cell fate as a function of stress sensing, although the mechanism by which IRE1α is regulated to function in these distinct pathways is poorly understood, as is the mechanisms by which the ER senses misfolded proteins.
Evidence using in vitro approaches suggests that the binding of the chaperone BiP/Grp78 to the IRE1α luminal domain results in allosteric inhibition of the dimerization of the cytoplasmic tail, thus keeping IRE1α inactive under ER stress-free conditions (Carrara et al. 2015). Specifically, the Ron laboratory recently determined that the BiP ATPase domain is recruited to the luminal domain of IRE1α by the ERdj4 J-domain, resulting in the displacement of ERdj4 and returning IRE1α to its monomeric form thus inactivating the stress sensor (Amin-Wetzel et al. 2017). Additionally, an ER stress sensing mechanism has been proposed in yeast by Walter and colleagues wherein the IRE1p luminal domain senses ER stress by directly associating with misfolded proteins through a proposed luminal ligand-binding groove resulting in allosteric activation of IRE1α (Karagöz et al. 2017).
While IRE1α has been known to regulate ER stress as a result of the UPR, we are only beginning to understand the mechanistic details of its regulation. The study by Sepulveda et al. in this issue of Molecular Cell uncovers a novel role for the ER chaperone Hsp47 in IRE1α regulation, specifically one in which it physically associates with IRE1α and displaces BiP, as well as identifying additional IRE1α -associating proteins (Sepulveda et al. 2017). The role of Hsp47 in proteostasis is further conveyed as its overexpression enhanced the splicing of XBP1 mRNA with measurable consequences in the induction of UPR-controlled genes such as EDEM1. In addition to contributing to the protein-folding/rescue branch of the UPR, Hsp47 also modulates global protein synthesis as its overexpression enhances mRNA decay through the RIDD pathway. Interestingly, Hsp47, whose role has previously been attributed to collagen trafficking, appears to enhance the phosphorylation and oligomerization of IRE1α, and thus its activation, possibly as a result of its direct association with the IRE1α luminal domain. To further demonstrate the conserved role of Hsp47 in regulating IRE1α, studies were carried out in D. melanogaster larvae and mouse models of ER stress both of which were consistent with the proposed function of Hsp47 in enhancing mRNA splicing and modulating apoptosis through IRE1α.
While the study by Sepulveda et al. provides new insight into the regulation of IRE1α by the chaperone Hsp47, it also raises interesting questions for future consideration. The collagen-specific Hsp47 chaperone belongs to the serine-protease inhibitor (Serpin) family, although it lacks inhibitory function (Ito and Nagata 2017). Unlike other members of the Serpin family, the active form of Hsp47 is the latent fold. Essentially, Hsp47 takes on a loop-inserted form, which in other Serpins is the inactive low energy form. Hsp47 binds collagen in the ER and releases it in the Golgi as a result of a pH-induced conformational change, thus raising the question of which Hsp47 conformational fold binds to IRE1α and is that different from the collagen-bound form (Saga et al. 1987)? Additionally, upon displacement from IRE1α is the released Hsp47 active in terms of binding to and chaperoning collagen? Furthermore, ablating the level of Hsp47 results in Golgi fragmentation and inhibition of O-glycosylation induces Hsp47 expression, both of which suggest a possible function of Hsp47 in Golgi stress (Miyata et al. 2013). Since this link remains unclear, it would be of interest to investigate whether they are connected to the role of Hsp47 as an IRE1α regulator. For instance, does Hsp47 sense Golgi stress and remediate it through its activity with IRE1α? Lastly, Nagata and colleagues have recently identified a Hsp47 small molecule that inhibits the interaction between Hsp47 and collagen. It would be of interest to determine if this probe would inhibit IRE1α (Ito et al. 2017). In conclusion, these findings by Sepulveda et al. provide us with further insight into the intricate pathways in which the cell has evolved to cope with stress.
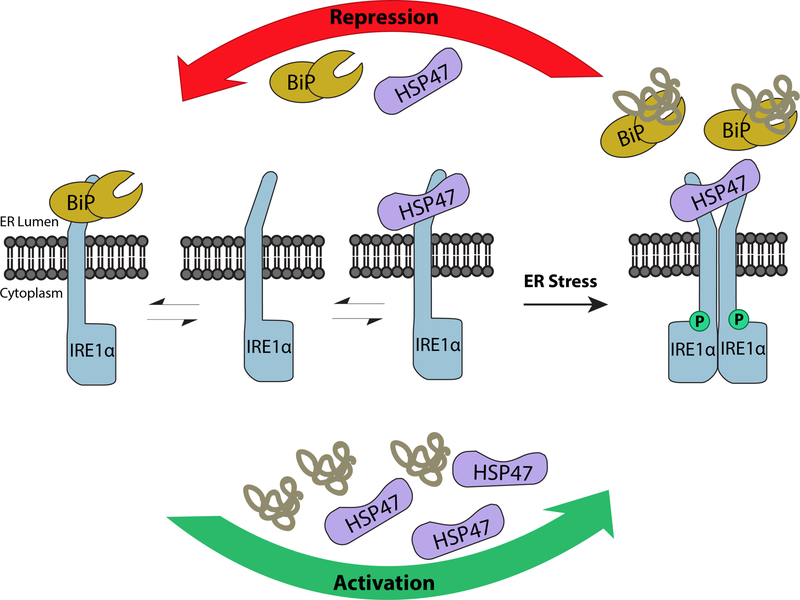
Repression and Activation of IRE1α.
Under normal ER conditions, monomeric IRE1α is maintained in an inactive form through BiP binding (left panel). Basal levels of Hsp47 compete with BiP for IRE1α binding to keep the protein at basal homeostasis (central panel). Upon ER stress, Hsp47 accumulates and induces IRE1α activation in part by displacing BiP (bottom and right panel). The accumulated misfolded proteins facilitate BiP dissociation from IRE1α. As a result, IRE1α dimerizes resulting in its autophosphorylation, both of which are necessary for its downstream activity. Once the stress is alleviated, or under prolonged stress conditions, Hsp47 dissociates from the IRE1α complex, returning the protein to its inactive form (top panel).
Authors Funding:
Chemistry-Biology Interface program training grant (T32GM008515 to L.L.) and the National Institutes of Health under award GM086874 (to D.N.H.)
References
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP, and Ron D (2017). Cell. 7, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, and Ali MM (2015). Elife. 4 doi: 10.7554/eLife.03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, and Papa FR (2017). Mol. Cell 69, 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Ito S, and Nagata K (2017). Semin. Cell Dev. Biol 62, 142–151. [DOI] [PubMed] [Google Scholar]
- Ito S, Ogawa K, Takeuchi K, Takagi M, Yoshida M, Hirokawa T, Hirayama S, Shin-Ya K, Shimada I, Doi T, et al. (2017). J. Biol. Chem 8, 20076–20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Acosta-Alvear D, Nguyen HT, Lee CP, Chu F, and Walter P (2017). Elife. 6 pii: e30700. doi: 10.7554/eLife.30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Mizuno T, Koyama Y, Katayama T, and Tohyama M (2013). PLoS One. 8, (7):e69732. doi: 10.1371/journal.pone.0069732. Print 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga S, Nagata K, Chen WT, and Yamada KM (1987). J. Cell. Biol 105, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda D, Rojas-Rivera D, Rodríguez DA, Groenendyk J, Köhler A, Lebeaupin C, Ito S, Urra H, Carreras-Sureda A, Hazari Y et al. (2017). Mol. Cell [DOI] [PubMed] [Google Scholar]
- Walter P, and Ron D (2011). Science. 334, 1081–1086. [DOI] [PubMed] [Google Scholar]