Abstract
The endoplasmic reticulum (ER) is a complex, multi-functional organelle, comprised of a continuous membrane and lumen that is organized into a number of functional regions. It plays various roles including protein translocation, folding, quality control, secretion, calcium signalling and lipid biogenesis. Cellular protein homeostasis is maintained by a complicated chaperone network, and the largest functional family within this network consists of proteins containing tetratricopeptide repeats (TPRs). TPRs are well-studied structural motifs that mediate intermolecular protein-protein interactions, supporting interactions with a wide range of ligands or substrates. Seven TPR-containing proteins have thus far been shown to localize to the ER and control protein organization and homeostasis within this multi-functional organelle. Here we discuss the roles of these proteins in controlling ER processes and organization. The crucial roles that TPR-containing proteins play in the ER are highlighted by diseases or defects associated with their mutation or disruption.
Keywords: endoplasmic reticulum, TPR domains, chaperones, protein translocation, glycosylation
A historical perspective of endoplasmic reticulum morphology, organization and function
In eukaryotic cells, cellular processes are frequently organized into specific subcompartments or organelles. The endoplasmic reticulum (ER) is comprised of a single lumen and contiguous membrane that spreads throughout the cell, commonly making it the largest organelle within a cell. The ER was first observed in 1902 by Emilio Veratti when he used Camillo Golgi’s staining method and found a cellular structure distinct from the Golgi apparatus. It was not until 1953 that the ER was named and acknowledged to be an organelle. With the development of electron microscopy, Keith Porter and George Palade validated the presence of a net-like (reticulum) structure within (endo) the cytoplasm (plasmic) via high-resolution electron microscopy (EM) imaging (Palade & Porter 1954; Veratti 1961). Palade also developed a method to isolate ER-derived “microsomes” and in combination with microscopy-based analyses, extensive biochemical assays were performed leading to the discovery that the ER participates in a number of essential processes including protein folding, quality control, trafficking, secretion and homeostasis, calcium signalling, and lipid biogenesis and regulation (Palade & Siekevitz 1956; Veratti 1961; Caro 1964; Stein & Stein 1967; Mazzarello et al. 2003; Lamriben et al. 2016). Studies in the last few decades have shown that different subdomains characterized by the enrichment of specific factors exist within this singular luminal space of the ER, creating functional sub-regions (Lynes & Simmen 2011).
Early microscopic and biochemical observations showed two distinct domains of the ER: the rough ER, characterized by the presence of ribosomes, and the smooth ER that is devoid of ribosomes. Extensive studies on the rough ER have provided numerous breakthroughs in understanding its function but progress on the smooth ER has been slower.
The rough ER membrane is easily distinguishable in EM images as it is studded with ribosomes that provides the rough appearance. Using combined EM and autoradiography, the flow of proteins from the cytoplasm, through the ER, and onwards to the Golgi was observed and provided primary evidence that the ER is the site of entry into the secretory pathway (Caro 1964). Ribosomes are nestled onto translocons (Sec61) embedded in the ER membrane that provide the conduit for translocation into the ER lumen (Park & Rapoport 2012). Proteins resident to the rough ER include translocon-associated factors such as proteins involved in signal sequence processing or glycosylation, as well as molecular chaperones (i.e. BiP, calnexin and calreticulin) and oxidoreductases (i.e. PDI and ERp57) that assist the folding and maturation of the newly synthesized polypeptides (Johnson & van Waes 1999; Alder et al. 2005; Shibatani et al. 2005; Harada et al. 2009). The signal sequence peptidase and glycosyltransferase complexes, as well as chaperones and oxidoreductases, are known to participate in co-translational events in the rough ER but they may also be observed in other regions of the ER (Chen et al. 1995; Molinari & Helenius 1999; Molinari & Helenius 2000; Daniels et al. 2003; Gilchrist et al. 2006).
Over the last two decades, work pertaining to the smooth ER has identified and characterized several subdomains including ER exit sites (ERES) and the ER quality control compartment (ERQC-C) (Budnik & Stephens 2009; Benyair, Ogen-Shtern, & Lederkremer 2015). ERES mediate the export of secretory cargo from the ER and generate the ER-Golgi intermediate (ERGIC) compartment (Jensen & Schekman 2011). This portion of the ER is part of the transitional or intermediary ER, characterized by its overlap with the rough ER and its transition into the Golgi vesicular network. An emerging subdomain of the ER is the ERQC-C. In order to ensure proper protein processing, nascent proteins of different folding states must be segregated by distinguishing properly folded proteins at ERES for export to the Golgi from those that are terminally misfolded for targeting for ER-associated degradation (ERAD) in the ERQCC (Benyair, Ogen-Shtern, & Lederkremer 2015). It has been suggested that a number of the ER quality control factors, as well as accumulated misfolded secretory cargo, are also collected into peri-centrosomal vesicles, or quality control vesicles (QVCs) (Benyair, Ogen-Shtern, Mazkereth, et al. 2015). However, these membrane enclosed QVCs and the localization of ERAD and ERQC factors are not well defined.
The first role shown to be associated with the ER was calcium storage as Ca2+ was observed to be sequestered in the sarcoplasmic reticulum (the term for the ER in muscle cells) (Mazzarello et al. 2003). The mechanisms by which the ER sequesters and releases Ca2+ gave essential insight into the studies of muscle contraction, signaling, fertilization and neurotransmission (Hales et al. 1974; McGraw et al. 1980). Whereas some calcium binding proteins, like calreticulin, or re-uptake pumps, like SERCA, appear to be widely distributed throughout the ER, IP3 receptors and ryanodine receptor calcium channels are often found clustered within the smooth ER (Satoh et al. 1990; Takei et al. 1992).
To maintain cellular homeostasis, organelles are connected by vesicle-mediated trafficking and membrane contact sites where two heterologous membranes are closely apposed (~within 30 nm) but do not fuse. The ER forms membrane contact sites with multiple membrane systems including the plasma membrane, mitochondria, Golgi, endosomes, and lipid droplets (Zhang & Hu 2016). Association of the ER with other organelles generally involves portions of the smooth ER and protein-protein interactions and/or protein-phospholipid interactions between the ER and the apposed organelle. The areas and shapes of such contacts are dynamic and may correlate with the functional demands of the contact, which include lipid and calcium exchange, fission and organelle movement.
The use of microscopic radioautography demonstrated the intracellular movement of labelled lipids, showing lipids were observed in the rough and smooth elements of the ER then in mitochondria and Golgi shortly thereafter. It was concluded that these lipid particles represented lipoproteins that formed in the ER, were processed in the Golgi and then transported into vacuoles to be dispersed into circulation (Stein & Stein 1967). Lipid and calcium exchange and regulation between the ER and rest of the cell was later determined to be attributed mainly to the ER forming physical membrane contacts with the plasma membrane and mitochondria (Pichler et al. 2001; Hayashi et al. 2009; Giorgi et al. 2009).
The domain of the smooth ER found in close contact with the plasma membrane is termed the plasma-associated membrane (PAM) (Pichler et al. 2001). A significant role of the PAM, also known as the cortical or peripheral ER, is calcium exchange and lipid transfer. The mitochondria-associated membranes (MAMs) are important for energy metabolism and regulated cell-death, therefore they are pivotal for cellular function and survival. MAMs also enable highly efficient transmission of calcium from the ER to mitochondria to stimulate oxidative metabolism (Rizzuto et al. 1998; Boehning et al. 2003; Simmen et al. 2010). Additionally, the smooth ER is now known to provide the point of origin for entire organelles such as the peroxisome, Russell bodies, and lipid droplets (Kopito & Sitia 2000; Kalantari et al. 2010; Mast et al. 2010).
A multitude of cellular functions rely on adaptor proteins and multi-protein complexes to mediate compartmentalization and organellar contact sites. A number of ER resident factors utilize protein-protein interaction motifs to nucleate multi-protein complexes directing, at least in part, the formation of ER subdomains (Zhang & Hu 2016). A better understanding of the employment of adaptor proteins and protein-protein interaction motifs could lead to a more complete understanding of ER functional organization. This review aims to describe how proteins containing adaptor motifs, specifically the TPR, contribute to the functional organization of the ER.
Structure and function of TPR domains
Adapter or scaffold proteins commonly utilize repeat motifs to nucleate protein complexes since repeats allow a variety of binding surfaces with minimal sequence variation. They are comprised of tandem repetitions of a short structural motif and have been used as molecular recognition tools in a wide variety of applications (Sawyer et al. 2013). Examples of repeat proteins are Leucine Rich Repeats (LRR), ankyrin repeats (Ank), the WD40 repeat, the armadillo repeat (ARM), and the TPR. Each of these motifs has its own signature, a conserved set of amino acids that specify the repeat structure (Good et al. 2011; Sawyer et al. 2013). The TPR is of particular interest because they are found in numerous proteins, serving as interaction modules and multi-protein complex mediators, and recently TPR-containing proteins were found to comprise approximately one third of the mammalian protein chaperone network in the cell (Zeytuni & Zarivach 2012; Brehme et al. 2014).
Since their identification, TPR-containing proteins have been found in all kingdoms of life and to regulate a number of diverse biological processes including protein folding and import, organelle targeting and vesicle fusion. A single TPR motif consists of 34 amino acids in a degenerate consensus sequence defined by a jig saw pattern of small and large hydrophobic amino acids. Though this general consensus sequence or pattern describes TPR motifs, no residue is invariant; however, residue type is highly conserved at three positions (8, 20 and 27) relative to the motif’s N-terminal residue (D’Andrea and Regan, 2003; Magliery and Regan, 2005).
The canonical unit of the TPR adopts a basic helix turn helix fold, and adjacent TPR units form a series of repeating antiparallel α-helices (Figure 1A). This yields an overall super-helical structure affected by the residue found between adjacent motifs and the number of repeats next to one another. A positive correlation was determined between protein thermostability and the number of TPR motifs using a non-natural recombinant TPR-containing protein with 20 sequential TPRs (Kajander et al. 2007). Additionally, increasing numbers of TPR motifs confer kinetic folding cooperativity and were shown to involve the thermodynamic interplay between the stability of individual repeats as well as the interaction between repeats (Javadi & Main 2009).
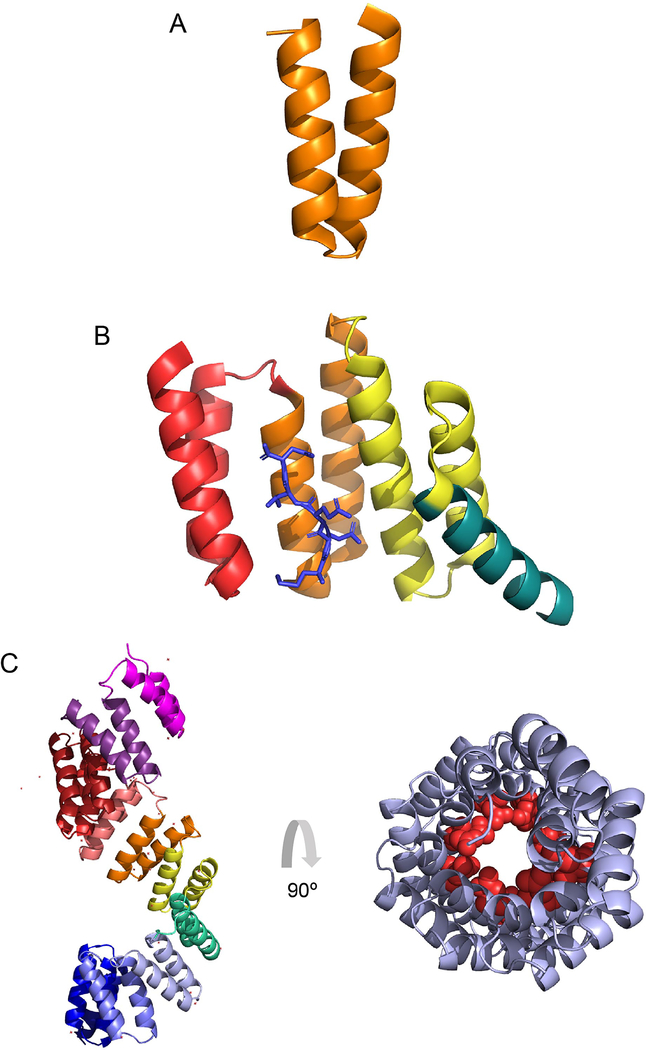
Figure 1. Tetratricopeptide repeat structures.
(A) A single tetratricopeptide repeat (TPR) motif (orange) consists of 34 amino acids that folds into two anti-parallel alpha helices. (B) TPR motifs are commonly found in clusters of three. The crystal structure (1ELR) represents the TPR domain of Hsp70/Hsp90 Organizing Protein (HOP), which binds to the chaperone Hsp90. The single TPR motifs are indicated in red, orange and yellow, with the C-terminal capping helix in turquoise. The C-terminal heptamer of Hsp90, which is the ligand for this cluster, is shown in blue. (C) The crystal structure (1W3B) represents the TPR region of OGT. It forms a superhelix with individual motifs indicated in pink, purple, brick red, red, orange, yellow, green, periwinkle, blue and violet from N-terminus to C-terminal, respectively (left panel). The structure in (C-right panel) is the 11.5 TPR motifs from (C- left panel) rotated 90 degrees, which demonstrates the superhelical nature of the tandem repeats. Conserved Asn (red spheres), thought to mediate target protein interaction, as seen in importin-α, line the inner surface of the superhelix.
TPR-containing proteins use different binding pockets to bind diverse ligands that usually do not share secondary structure or sequence similarities. Although a defined rule set is lacking, binding is usually highly specific. The crystal structures of many TPR-domain containing proteins show two anti-parallel α-helices packed in tandem arrays to form a structure with an amphipathic groove which can bind a target peptide (Grove et al., 2008; Zeytuni and Zarivach, 2012). This is not, however, the only mode of target recognition by TPR domains, with short amino acid insertions and alternative TPR motif conformations also shown to contribute to protein interactions, highlighting diversity in TPR domains and the versatility of this structure in mediating biological events (Allan & Ratajczak 2011). In nature, TPR motifs can be found in tandem arrays of 2–16 sequential motifs (D’Andrea & Regan 2003; Jiménez et al. 2012). Clusters of three TPR tandem motifs are the most common, and they form a distinct curve, binding the ligand on the concave side (Figure 1B) (Scheufler et al. 2000). The binding of ligands for clusters of three TPRs seems to be more specific, whereas when higher numbers of repeats are found adjacent to one another, the binding is more promiscuous (D’Andrea & Regan 2003; Jínek et al. 2004).
To obtain such diverse binding, the TPRs utilize their distinct fold to serve as an interaction platform. This platform can exhibit different surface residues in each binding surface, yielding specificity by a combination of factors (Zeytuni & Zarivach 2012). Residue type affects the electrostatic nature of the binding surface by contributing positive and negative charges. Additionally, residues with different hydrophobicity and size can support hydrophobic interactions between TPR domains and ligands. The secondary structure of TPR-bound ligands varies between extended coil to α-helix conformation, or a combination of the two. Elongated conformation maximizes the ligand surface presented to the TPR domain, as well as optimizes H-bond formation and thereby facilitates the specific recognition of the short amino acid stretches. Seven proteins containing TPRs have thus far been shown to localize to the ER and contribute to its diverse cellular functions (Figure 2) (Yan et al. 2002; Kaneko & Nomura 2003; Racapé et al. 2011; Sunryd et al. 2014; Preissler et al. 2015; Li et al. 2018). The roles and functions of these seven proteins will be discussed below.
Figure 2. ER TPR-containing proteins.
Seven TPR-containing proteins have been shown to localize to the ER. N-terminal signal sequences (black squares) target these proteins to the ER and the names, positions of tetratricopeptide repeats (TPRs) (orange squares) and hydrophobic domains (blue squares) are designated. SEL1L has a fibronectin-like type II domain at its N-terminus indicated in green. ERdj6’s J-domain (magenta rectangle) and FICD’s FIC domain (yellow rectangle) are located at the C-terminus of each protein.
Functional roles for ER TPR-containing proteins
Protein translocation across the ER membrane
Protein translocation is a process by which a polypeptide chain moves across a membrane (Blobel 1980; Schnell & Hebert 2003). The protein is frequently directed to its target membrane by an N-terminal signal sequence and protein translocation can occur co-translationally or post-translationally. TPR-containing proteins have been shown to be essential for a number of cellular translocation processes as cells or organisms lacking these proteins exhibit growth and survival defects (Harkness et al. 1994; Sun et al. 2014).
One of the well-studied TPR-containing protein import systems involves the translocation of proteins across the outer mitochondrial membrane. The majority of mitochondrial proteins are synthesized by cytosolic ribosomes and then imported into the organelle post-translationally. TOM (the translocase of the outer mitochondrial membrane) is a multi-protein complex that contains surface receptors for pre-protein recognition and a translocon that serves as the membrane conduit to cross the outer mitochondrial membrane. The translocase complex is comprised of at least seven proteins, including TOM70 and TOM20, both of which contain TPR motifs. Mitochondrial targeting requires the cytosolic molecular chaperones Hsc70/Hsp70 and Hsp90, and their chaperone activity and substrate binding is regulated by a number of cochaperones, of which some contain TPR motifs (discussed below in the chaperone networks section). TOM70, the mitochondrial import receptor, is also a member of the TPR co-chaperone family as it contains a conserved TPR-clamp domain comprised of three TPR motifs for interaction with Hsc70 and Hsp90 (Young et al. 2003). This interaction between TOM70 and chaperones has been shown to be important for preprotein import preparation and the initiation of the import process (Fan et al. 2006). TOM70 also contains an additional two clusters of TPRs (each containing three TPR motifs), which may be involved in preprotein recognition and translocation (Wu & Sha 2006). TOM20 contains a single TPR motif, which is unique in TPR functional analysis. The structure of TOM20 reveals that the single TPR motif along with a third helix form a hydrophobic groove, which is the binding site for N-terminal presequence peptides aiding in transport cargo recognition (Muto et al. 2001). These findings show the important roles that TPR-containing proteins play in mitochondrial protein import and how the different TPR functional domains exhibit various binding specificities.
In the secretory pathway, TPR-containing proteins have been shown to be crucial for protein translocation both into and out of the ER (Figure 3). ER post-translational translocation in yeast requires the Sec61 translocon channel and a complex of 4 additional proteins: Sec63, Sec62, Sec71 and Sec72 (Wu et al. 2018; Itskanov & Park 2019). Using a combined structural and biochemical approach, the role of Sec71/72 subcomplex was determined (Tripathi et al. 2017). The crystal structure reveals that Sec72 contains a TPR domain that has multiple chaperone interaction sites and is anchored to the ER membrane by Sec71 and Sec63. This TPR domain interacts with the C-terminus of Ssa1, cytosolic Hsp70, which binds to the concave inner surface of the TPR domain. Surprisingly it also interacts with Ssb1, a cytoplasmic Hsp70 that binds ribosome associated nascent polypeptide chains even though it lacks the C-terminal TPR-binding residues of Ssa1. Ssb1 instead interacts with the TPR domain through its ATPase domain along the backside (convex) of the TPRs allowing for translocation substrates to be recruited to the Sec71/72 complex both co- and post-translationally (Tripathi et al. 2017). Recent cryo-EM data reveals that Sec63 positions Sec71/72 for the capture of polypeptides associated with cytosolic Hsp70, an interaction mediated by the TPR domain of Sec72 (Figure 3–1) (Wu et al. 2018; Itskanov & Park 2019). While this post-translation translocation process has been worked out in yeast, a similar process is expected to occur in metazoans.
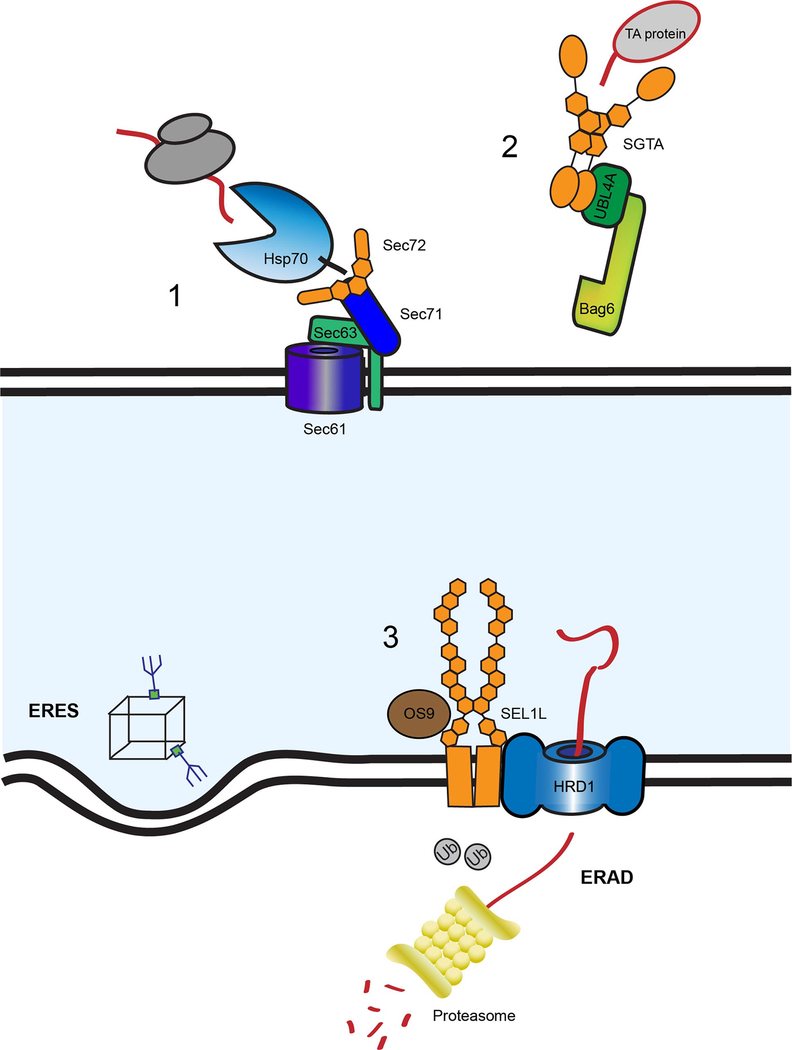
Figure 3. Sec72, SGTA and SEL1L are involved in ER protein translocation.
(1) Sec72 (orange) is anchored to the ER membrane by Sec71 (dark blue) and Sec63 (green) at the Sec61 (purple) translocon. The TPR domain, comprised of three TPR motifs (orange hexagons), binds the C-terminus of the yeast Hsp70 (blue), Ssa1, aiding in the recruitment of translated polypeptides (red) to the Sec61 translocon. (2) SGTA (orange) aids in post-translational translocation, specifically of tail-anchored proteins. SGTA is comprised of an N-terminal UBL-binding domain, which is also responsible for homo-dimerization, a middle TPR domain and C-terminal substrate binding domain that captures the tail-anchored proteins (light grey). SGTA is in a complex with UBL4A and Bag6 (dark and light green, respectively) and plays a role in molecular triage. This complex is responsible for the proper targeting of tail-anchored proteins to the ER membrane as well as segregating misfolded polypeptides and targeting them for degradation. As chaperones aid in this process, SGTA’s TPR domain is responsible for chaperone recruitment. The TPRs bind the C-termini of both Hsp70 and Hsp90 as well as proteasomal subunit, Rpn13. (3) As nascent polypeptides are queried for folding status, properly folded proteins are distinguished for export at ER exit sites (ERES) and misfolded proteins are targeted for degradation by ERAD. SEL1L (orange) is in complex with HRD1, the ER retro-translocon responsible for translocating misfolded proteins across the ER membrane for subsequent degradation by the proteasome (yellow) in the cytoplasm. SEL1L contains 11 SLR motifs (hexagons #1–11 from N to C-terminus), which are very similar to TPR motifs but with an extended consensus sequence length, that comprise the majority of the luminal portion of the protein. The SLR motifs nearest the membrane (#10–11) interact with a luminal portion of HRD1 and are necessary for interaction with ERAD factor, OS9 (brown). A SLR motif in the middle region (#9) is responsible for homo-oligomerization of SEL1L.
Another TPR-containing protein involved in a specialized post-translational translocation process is small glutamine-rich TPR-containing protein alpha (SGTA). SGTA acts as a co-chaperone to facilitate the targeting of tail-anchored membrane proteins to the ER membrane, as well as the sorting of membrane and secretory proteins that mislocalize to the cytosol for degradation (Martínez-Lumbreras et al. 2018). In combination with the BAG6 complex, SGTA helps to perform molecular triage to regulate the fate of tail-anchored membrane proteins and mislocalized secretory cargo by binding hydrophobic substrates and transferring selected protein clients to the BAG6 complex for sorting (Figure 3–2) (Shao et al. 2017). SGTA is comprised of an N-terminal dimerization domain, a central domain consisting of three TPR motifs and a C-terminal substrate binding motif (Dutta & Tan 2008; Chartron et al. 2011; Simon et al. 2013; Darby et al. 2014; Martínez-Lumbreras et al. 2018). The TPR domain interacts directly with Hsp70 and Hsp90 chaperones, as well as the proteasomal subunit, Rpn13 (Minami et al. 2010; Leznicki et al. 2015; Thapaliya et al. 2016). The TPR domain gives SGTA the co-chaperone-like ability to participate in both productive “on-pathway” folding, as well as protein degradation.
As proteins enter the secretory pathway, they are assessed by a number of ER resident quality control factors to determine if they are properly folded or require additional folding attempts (Ellgaard & Helenius 2003; Tannous et al. 2015). If a protein is deemed terminally misfolded, it is targeted for ERAD, which requires retro-translocation from the ER to the cytoplasm for polyubiquitination and subsequent degradation by the proteasome. SEL1L co-localizes with HRD1, one of the ER E3 ligases involved in ERAD (Gardner et al. 2000). It also associates with additional ERAD factors including OS-9 and GRP94 (the ER Hsp90) that have been shown to deliver mutant α1-antitrypsin to the HRD1-SEL1L ubiquitin complex for degradation, confirming its role in ERAD (Figure 3–3) (Christianson et al. 2008).
The recently solved structure of SEL1L provides insight into how it contributes to the ERAD process (Jeong et al. 2016). SEL1L is comprised of a fibronectin type II domain at the N-terminus followed by eleven SEL1L-like repeat (SLR) motifs and a C-terminal transmembrane domain (Figure 2). The SLR motif share a similar anti-parallel α-helical structure with the TPR motif, but the consensus sequence length is extended (36–44 amino acids compared to 34) (Mittl & Schneider-Brachert 2007). The SLR motifs make up the major portion of the luminal domain of SEL1L and are isolated into three clusters consisting of 4 SLRs (SLR-N(terminal) includes repeats #1–4), 5 SLRs (SLR-M(iddle) repeats #5–9) and 2 SLRs (SLR-C(terminal) repeats #10 and 11). SLR #9 of the SLR-M domain was shown to be required for the homo-oligomerization of SEL1L (Jeong et al. 2016). The SLR-C domain is responsible for mediating the interaction with a luminal portion of HRD1. Additionally, in yeast SLR-C has been shown to mediate interactions with Yos9 (OS-9 homologue) as truncation mutants in Hrd3p (the SEL1L homologue) lacking this region were no longer able to bind Yos9 (Figure 3–3) (Gauss et al. 2006). The N-terminal cluster contains 4 motifs and sequence analysis suggests that it may bind a specific partner or act as a classic co-chaperone (D’Andrea & Regan 2003). The SLR domains in SEL1L have been shown to support homo-oligomerization or complex formation with other ERAD components; however, the role of the N-terminal SLR domain is still unknown. Together, TPR and SLR-domain containing proteins aid in nucleating interactions at the ER membrane that support the translocation of proteins into and out of the ER lumen.
Molecular chaperone adapters and regulators
A main role for TPR-containing proteins in the cell is nucleating chaperone macromolecular complexes and regulating their activity (Caplan 2003; Brehme et al. 2014). Hsp70 and Hsp90 are major molecular chaperones in the eukaryotic cytosol, playing essential roles in protein quality control by preventing aggregation, catalyzing productive folding of newly synthesized proteins and promoting degradation of misfolded polypeptides (Hideaki & Yohtalou 1991; Hartl et al. 2011). Co-chaperones of Hsp70 and Hsp90 are characterized as non-client binding partners that participate in their function and regulation (Caplan 2003). Hsp70 and Hsp90 cooperate with cochaperones during the process of protein folding and require the help of co-chaperones containing TPR motifs for many of their functions.
One of the most well-studied TPR-containing proteins, Hsp70/Hsp90 Organizing Protein (HOP), mediates the spatial proximity of Hsp70 and Hsp90 by associating with both chaperones and facilitating the passage of cargo from one major chaperone to the other for efficient protein folding. HOP is a monomeric protein composed of nine TPR motifs, clustered into three distinct TPR domains (Odunuga et al. 2004; Yi et al. 2010). Both chaperone sequences end with the motif EEVD and this tetrapeptide is recognized by the HOP TPR1 and TPR2A domains for Hsp70 and Hsp90, respectively. Crystal structures of the TPR-peptide complexes show peptides spanning the groove in the concave surface of the TPR domains and binding is mediated by electrostatic interactions with the EEVD motif with the C-terminal aspartate acting as a two-carboxylate clamp (Figure 1B). The hydrophobic contacts between the peptide residues upstream of EEVD and the TPR domain backbone are critical for specificity (Scheufler et al. 2000). Studies of other TPR co-chaperones identified selective binding between chaperone-TPR pairs (Assimon et al. 2015). In vitro studies found that the co-chaperones HOP, CHIP and DNAJC7 bound both Hsp70 and Hsp90 with similar affinities but co-chaperones FKBP51 and 52 preferably bound Hsp90. The intrinsic affinity and post-translational modifications tune the interactions between the Hsp70 and Hsp90 proteins and their TPR co-chaperones. Phosphorylation of the Hsp70 or Hsp90 C-termini significantly decreased their binding affinity to CHIP. Many of the Hsp70 and Hsp90 co-chaperones contain functional domains outside of the TPR region that direct the localization or function of their requisite chaperone-TPR pairs, which could explain selective binding between these chaperone families and the distinct roles they play within the cell.
Although it is likely that more than 100 co-chaperones exist in mammals, it appears that of the ones identified and analysed, the proteins fall into two classes, those that contain a J-domain and those that contain TPR motifs (Caplan 2003). ERdj6 (also known as P58IPK) is an ER resident protein that contains both features (Figure 2). The crystal structure of ERdj6 revealed three N-terminal TPR domains, containing three TPRs each (similar to that found for HOP), and a C-terminal J-domain. Interestingly, ERdj6 can be found in complex with the ER Hsp70 paralog, BiP (Figure 4A) (Rutkowski et al. 2007). Evidence of its role in secretory protein maturation and quality control was provided when it was found to co-immunoprecipitate with a newly synthesized secretory protein in cells. A chaperone-like role for ERdj6 was further supported by the fact that protein maturation was stimulated upon its overexpression and knockout cells showed decreased protein synthesis under both normal and stressed conditions; and its expression was induced by ER stress (Yan et al. 2002; Rutkowski et al. 2007).
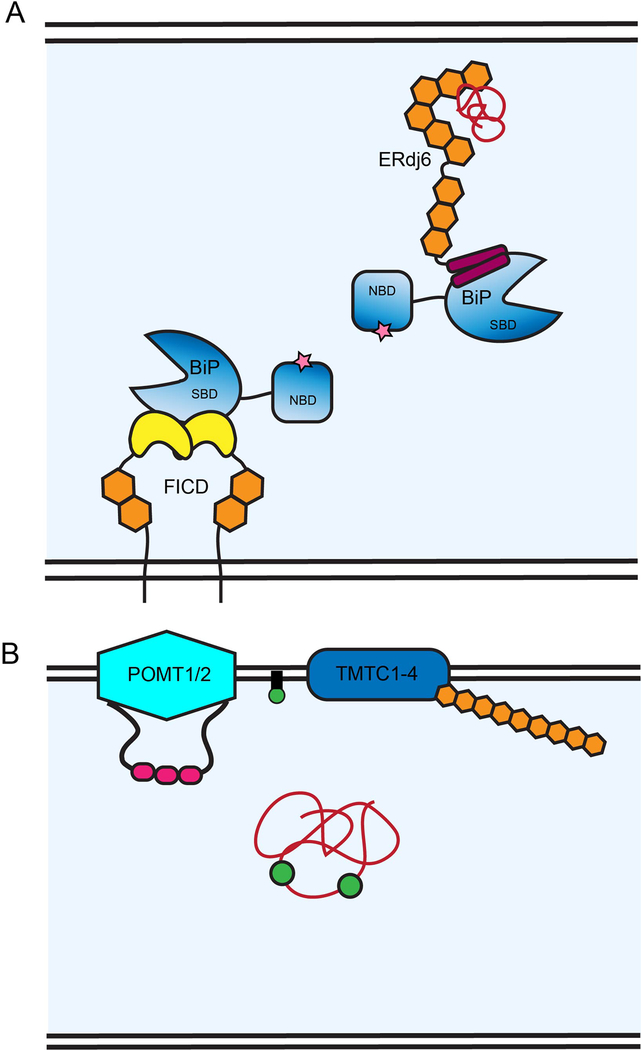
Figure 4. ER TPR co-chaperone, ERdj6, and protein modifiers, FICD and TMTC1–4 control protein homeostasis through interactions with BiP and glycosylation.
(A) ERdj6 has nine TPR motifs (orange hexagons) followed by a C-terminal J domain (magenta rectangles). The J-domain contains an Hsp70 interaction motif, HPD, and modulates the nucleotide binding activity of the ER Hsp70, BiP (gradient blue) by interacting with BiP on the substrate binding domain (SBD). The N-terminal TPR domain of ERdj6 can bind exposed hydrophobics on folding polypeptides (red) and potentially pass them off or sequester them for BiP bound at the J-domain. FICD is comprised of two N-terminal TPR motifs (orange hexagons) and a C-terminal FIC domain (yellow), which is responsible for regulating BiP chaperone activity by AMPylating BiP on its substrate binding domain (SBD), mimicking an ATP (pink star) bound state on the nucleotide binding domain (NBD). FICD forms a homodimer mediated through the FIC domain and AMPylates BiP under normal conditions to create an inactive BiP pool. As the UPR is activated, FICD de-AMPylates BiP so that it may engage unfolded substrate (red). (B) TMTC1–4 (blue and orange) are implicated in O-mannosylation. The TMTCs are composed of N-terminal hydrophobic domains (blue) embedding them in the ER membrane and 8–10 consecutive C-terminal TPR motifs (orange hexagons). POMT1 and 2 are the known protein O-mannosyl transferases of the ER. They are composed of a number of transmembrane domains represented by the light blue hexagon and contain three MIR domains (pink) in a luminal loop. The MIR domains are thought to recruit substrate to the membrane so that O-mannosyl transferases can transfer a mannose (green circle) from the dolichol-mannose precursor (black and green) in the membrane to the substrate (red).
Even though many of the cytosolic TPR co-chaperones interact with the C-terminal EEVD motif of Hsp70 or Hsp90, ERdj6 most likely binds to BiP via its C-terminal J-domain. Studies of classic Hsp chaperone systems in bacteria have shown that J-domain containing proteins have the ability to affect chaperone substrate binding by regulating nucleotide binding (Liberek et al. 1991). ERdj6 contains the critical Hsp70 binding tripeptide motif (HPD) in its C-terminal J-domain (Svärd et al. 2011). The N-terminal TPR cluster contains a conserved hydrophobic patch that may be involved in binding misfolded polypeptides. This conserved hydrophobic patch is 100 Å away from the BiP binding motif in the J-domain and structure-based mutagenesis for the conserved hydrophobic residues significantly reduced the molecular chaperone activity of ERdj6 (Figure 4A) (Tao et al. 2010). A flexible linker between the TPR subunits and the J-domain could allow movement suggesting that substrates bound to the hydrophobic surface/cleft in the TPR cluster could be brought to BiP bound at the C-terminal end (Svärd et al. 2011). ERdj6 constructs lacking the J-domain expressed in cells still bind misfolded substrate, suggesting that the N-terminal TPR motifs can bind polypeptides (Petrova et al. 2008).
As there has been estimated to be over 114 TPR containing proteins in the chaperome (Brehme et al. 2014), it is likely that there are TPR-containing proteins in addition to ERdj6 that contribute to the organization and regulation of the chaperone network in the ER. Given that many of the ER localized soluble folding factors possess the C-terminal ER retention motif, KDEL, it will be of special interest to determine if this sequence, in analogy to the EEVD motif of Hsp70 and 90 mediating HOP binding, directs binding to an ER adapter that might help support ER organization and retention.
Protein modifications
Protein modifications involve covalent additions to proteins that expands their function and regulation (Bürkle 2001; Truttmann & Ploegh 2017). Protein modifications play fundamental roles in regulating protein folding, targeting, interaction with ligands or protein partners, functional states and stability. TPR-containing proteins can contribute to protein regulation and function through post-translational modification in the ER by modifying chaperones with small molecules or by glycosylation (Schjoldager & Clausen 2012; Truttmann & Ploegh 2017) (Figure 4).
BiP is the ER member of the Hsp70 family and it relies on a number of partners, including J-domain co-chaperones (Hsp40 family members), nucleotide exchange factors, and signal transducers for its various activities (Dudek et al. 2009; Pobre et al. 2018). The activity of BiP is also modulated by AMPylation by the TPR-containing ER protein FICD (filamentation-induced by cyclic AMP domain containing protein) (Figure 4A) (Preissler et al. 2015; Preissler et al. 2016). AMPylation, also known as adenylation, is the process whereby adenosine monophosphate (AMP) from an ATP molecule is transferred to either a Tyr, Ser or Thr residue on the target protein. AMPylation of eukaryotic proteins is intimately related to the presence of FIC domains (Faber et al. 1998). FIC proteins are identifiable by their HXFX(D/E)(G/A)N(G/K)RXXR motif, which is the domain responsible for AMPylating substrate (Garcia-Pino et al. 2014). FICD, also known as Huntingtin interacting protein E (HYPE), is the only FIC-domain containing protein in the human genome and it also contains TPR motifs (Worby et al. 2009). BiP is a substrate of FICD (Sanyal et al. 2015). FICD expression was upregulated under stress conditions and its knockdown prevented induction of the unfolded protein response (UPR). BiP is AMPylated at residue Thr518 in the substrate binding domain leading to its inactivation (Preissler et al. 2015; Preissler et al. 2016). This modification is reversible as shown by the regulatory residue, Glu234, on FICD that switches the AMPylation/de-AMPylation reactivity (Preissler et al. 2017). When the cell is under normal homeostatic conditions, FICD AMPylates BiP to create an inactive BiP pool. As the UPR is activated, FICD de-AMPylates BiP so that it may engage unfolded substrate providing a rapid method to deploy active chaperones.
FICD is an ER resident type II membrane protein comprised of an N-terminal transmembrane (TM) domain followed by two TPR motifs, a linker region and a C-terminal FIC domain (Figure 2). The FIC domain exhibits well-conserved structure with Pfam canonical four α-helical FIC domain structure (Garcia-Pino et al. 2014). Dimerization of FICD can occur, leaving both of the FIC domains open for substrate binding and the TPR motifs available for protein-protein interactions; however, the interacting partners of the TPR domain have yet to be determined (Bunney et al. 2014).
Glycosylation, the covalent addition of carbohydrate molecules to a functional group on polypeptides, is one of the most abundant and diverse protein modifications observed in the cell (Schjoldager & Clausen 2012). Glycosylation can promote favorable folding energetics and lower aggregation propensity to help support proper protein folding (O’Connor & Imperiali 1996; Solá & Griebenow 2009; Price et al. 2012; Hebert et al. 2014). Two general types of glycosylation occur, N- and O-linked glycosylation, whereby saccharides are attached either to the amide group of an Asn residue (N-linked) or to the hydroxy of Ser or Thr residues (O-linked) (Ecker et al. 2003). Approximately one third of the mammalian proteome is targeted to the secretory pathway, and the majority of these proteins are glycosylated (Apweiler et al. 1999). While N-linked glycosylation is the most common modification in the ER, O-glycosylation also occurs here. Recently a family of membrane proteins containing long stretches of TPR domains was found to be involved in O-mannosylation in the ER.
O-linked glycosylation takes place in several parts of the cell. The main pathway is located in the Golgi, however O-mannosylation begins in the ER. It is catalysed by ER resident protein O-mannosyl transferases most commonly involving POMT1 and POMT2 (Figure 4B) (Ecker et al. 2003). These carbohydrate chain modifications can be used to monitor glycoprotein movement through the secretory pathway, acting as a maturation and quality control tag to indicate the status of the folding polypeptide in a manner similar to that previously observed for N-linked glycans (Hebert et al. 2014; Xu & Ng 2015).
A new class of TPR-containing proteins that localize to the ER has recently been revealed: transmembrane TPR-containing proteins (TMTC) 1–4 (Sunryd et al. 2014). TMTC1–4 contain homologous structural elements as their N-terminal halves are comprised of a number of hydrophobic domains and the C-terminal halves are composed mainly of TPR motifs (Figure 2). Although the structures for TMTC1–4 have yet to be determined, the predictive architecture could be compared to that of the POMT proteins and other O-linked transferases (Figure 4B). Both TMTC1–4 and POMT1/2 contain numerous hydrophobic segments anchoring them to the ER membrane. This positions the transferase close to the membrane to potentially enable the efficient transfer of the mannose from the dolichol-mannose precursor embedded in the membrane to the substrate. In order to attach, modify, or recognize glycoproteins, folding factors and machinery often possess multiple substrate recognition motifs or domains mediating specificity. In the POMT proteins, three MIR domains, named for the three proteins in which they occur (Mannosyltransferase, Inositol 1,4,5-triphosphate receptor (IP3R) and Ryanodine receptor), are present on a loop that faces the ER lumen and putatively bind substrates. In contrast, the TMTC proteins possess 8–10 consecutive TPR motifs in place of the MIR domains that potentially interact with a broad range of target substrates as observed for other O-linked transferases such as O-linked N-acetylglucosamine (GlcNAc) transferase (OGT).
OGT resides in the cytoplasm and catalyzes the transfer of GlcNAc from UDP-GlcNAc to Ser and Thr of cytoplasmic, nuclear and mitochondrial proteins including numerous transcription factors, tumor suppressors, kinases, phosphatases and histone-modifying proteins (Shafi et al., 2000; Yang et al., 2002). OGT contains an N-terminal domain comprised of 12.5 TPRs that mediates the recognition of a broad range of target proteins, as well as a C-terminal glycosyltransferase domain (Lubas et al. 1997; Iyer & Hart 2003). The crystal structure of the homodimeric TPR domain of OGT, containing 11.5 of the consecutive TPR motifs, displays an elongated superhelical structure (Figure 1C, left panel). The concave surface of the superhelix is lined by conserved Asn, in a manner reminiscent of the peptide binding site of importin-α, suggesting that this helical structure might help select and recruit substrates for modification (Figure 1C, right panel). Therefore, it would follow that the TPR domains of TMTC1–4 might also be used for substrate selection and positioning. TMTC1–4 have been implicated in the O-mannosylation of the cadherin and plexin family of proteins (Larsen et al. 2017). It will be of interest to determine if the TPR domains of TMTC1–4 can specifically bind the repeat domains of cadherins and plexins.
Though similar in composition, TMTC1–4 may play varying roles within the ER as they are associated with unique disease phenotypes. Interestingly, TMTC3 is implicated in two diseases that involve O-linked glycosylation. Biallelic mutations in TMTC3 are associated with recessive forms of cobblestone lissencephaly (COB), a severe brain malformation due to the over-migration of neurons and glial cells (Jerber et al. 2016). The cause of the over-migration defect is impaired interaction between glial limitans and the extracellular matrix (ECM), which is dependent upon glycosylated cell surface proteins making physical linkages between the cytoskeleton of glial cells and the ECM. Coincidentally, heterozygous variants of TMTC3 were also identified in periventricular nodular heterotopia, another disease resulting in defects in neuronal migration shown to cause brain malformations (Farhan et al. 2017). Expression analysis in patient-derived cells confirmed reduced transcript and protein levels of TMTC3. Neuron-specific knockdown of TMTC3 in flies resulted in increased susceptibility to induced seizures. This phenotype was rescued by neuron-specific expression of human TMTC3, suggesting a role for TMTC3 in synaptic cells and seizure biology.
A mutation in TMTC2 (Val381Ile) is linked to nonsyndromic sensorineural hearing loss (SNHL) (Runge et al. 2016). Exome sequencing, lineage and association analyses identified a fully penetrant sequence variant in the TMTC2 gene region that is associated with SNHL in a nine-family member cohort. This same variant was then found in a group of 363 unrelated individuals and associated with SNHL. TMTC4 was also linked to hearing loss as mice with TMTC4 inactivation in the cochlea caused acquired postnatal hearing loss (Li et al. 2018). After demonstrating a direct link between the more common noise-induced hearing loss (NIHL) and the UPR, mice with homozygous inactivation of both Tmtc4 and Chop had less hearing loss than knockout of Tmtc4 alone using inverse genetic complementation.
Post-translational modifications frequently affect protein function via changes in the protein structure and dynamics (Li et al. 2010). Post-translational modifications participate in a number of roles such as gene expression regulation, mediation of protein-protein interactions and, as demonstrated above, activation or deactivation of enzymatic activities or protein stability or destruction. As modulators of cell signalling and regulation, they perform a crucial role in maintaining cellular homeostasis. Diseases linked to post-translational modifications often involve mutations of post-translational target sites, however, as exhibited above, diseases can result from mutations in the protein modifiers themselves (Lazarus et al. 2011; Runge et al. 2016; Jerber et al. 2016; Li et al. 2018). Specifically, mutations in both the active sites as well as TPR region result in disease (Lazarus et al. 2011; Jerber et al. 2016).
Regulation of ER calcium homeostasis
Intracellular calcium is a universal second messenger in mammalian cells and chronic changes in Ca2+ signalling contribute to the pathogenesis of many diseases (Glaser et al. 2018). Ca2+ concentrations vary within the cell (10–100 nM) generally being lowest in the cytoplasm. One of the major functions of the smooth ER is calcium storage and it contains a host of factors dedicated to sensing and regulating intracellular calcium concentrations communicating with the cytoplasm, mitochondria and extracellular environment through the plasma membrane to maintain homeostasis (Lunz et al. 2019). The ER is also able to release stored Ca2+ through the ER resident IP3 and ryanodine receptors and upon depletion, the ER Ca2+ reuptake channel, SERCA, can refill the depleted stores. TPR-containing proteins have been shown to alter cytoplasmic calcium levels by interacting with and modulating the activity of SERCA (Sunryd et al. 2014).
TMTC1 and TMTC2 were identified and characterized as ER-localized, transmembrane proteins involved in calcium homeostasis (Sunryd et al. 2014). They interact with the calcium reuptake channel, SERCA2b, while TMTC2 also bound the carbohydrate binding chaperone calnexin. TMTC1 and 2 regulate the ability of SERCA to sequester Ca2+ back into the ER after stimulated Ca2+ release. Live cell Ca2+ measurements revealed that overexpression of either TMTC1 or TMTC2 caused a reduction of Ca2+ released from the ER following stimulation, whereas knockdown of either increased stimulated Ca2+ release. Additionally, TMTC2 is differentially expressed in patients receiving serotonin reuptake inhibitor (SSRIs) treatment for depression over the course of eight weeks (Madsen et al. 2018). TMTC2 was among other genes involved in calcium homeostasis that were shown to significantly increase with time and subsequent SSRI treatment.
SUMMARY
The ER utilizes protein-protein interaction motifs to efficiently execute various roles to maintain cellular homeostasis in a large single luminal space and a contiguous membrane. In particular, the presence of TPR motifs provides binding specificity for subsets of interactors while domains outside of the TPRs mediate activity. Thus far, ER TPR-containing proteins contribute to protein translocation both into and out of the ER membrane, post-translational modification and act as co-chaperones. Nine proteins, with tandem arrays from 2–10 sequential TPRs, have been identified that associate with or are localized within the ER, but more likely exist. Interestingly, the presence of TPR motifs has become increasingly easier to predict using algorithms (Letunic & Bork 2018), but the structures of the ER TPR-containing proteins are poorly understood, and oftentimes the function of the protein is identified prior to structure. Further studies using super resolution microscopy and proteomics are critical to provide a better understanding of the localization and identifying interacting partners of the ER TPR-containing proteins.
Given that the ER is involved in maintaining cellular protein, calcium and lipid homeostasis, it would be of interest to explore other families of repeat proteins, such as WD-40, ankyrin repeats or coiled-coil domains, to identify additional factors that contribute to ER morphology, organization and function. Coiled-coil domain containing proteins have previously been shown to contribute to ER membrane curvature and shape. The integral membrane protein Climp-63, which localizes exclusively in sheets, is thought to use its luminal coiled-coil domain to bridge between two apposed membranes (Klopfenstein et al. 2001). Additionally, the surface of ER sheets is kept flattened, likely by the sheet-enriched integral membrane proteins kinectin and p180, which possess a cytosolic coiled-coil domain that may form rod-like scaffolds (Shibata et al. 2010). WD-40 repeat containing protein, Sec13p, is involved in ER cargo export and ankyrin repeat containing ASB11 is a novel ER-associated ubiquitin ligase (Barlowe 1998; Andresen et al. 2014). Macromolecular complexes mediate many essential cellular processes. Key to understanding their mechanisms is knowing how these macromolecular assemblies are held together, localized and recruit substrates, properties that can be aided by critical repeat domains such as TPR motifs.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant GM086874 (to D. N. H.), and National Institutes of Health Chemistry-Biology Interface Program Training Grant T32GM008515 (to N. P. C.).
Footnotes
DISCLOSURE STATEMENT
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Alder NN, Shen Y, Brodsky JL, Hendershot LM, Johnson AE. 2005. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. The Journal of Cell Biology. 168:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T. 2011. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress and Chaperones. 16:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen CA, Smedegaard S, Sylvestersen KB, Svensson C, Iglesias-Gato D, Cazzamali G, Nielsen TK, Nielsen ML, Flores-Morales A. 2014. Protein Interaction Screening for the Ankyrin Repeats and Suppressor of Cytokine Signaling (SOCS) Box (ASB) Family Identify Asb11 as a Novel Endoplasmic Reticulum Resident Ubiquitin Ligase. J Biol Chem. 289:2043–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database11Dedicated to Prof. Akira Kobata and Prof. Harry Schachter on the occasion of their 65th birthdays. Biochimica et Biophysica Acta (BBA) - General Subjects. 1473:4–8. [DOI] [PubMed] [Google Scholar]
- Assimon VA, Southworth DR, Gestwicki JE. 2015. Specific binding of tetratricopeptide repeat (TPR) proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry. 54:7120–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C 1998. COPII and selective export from the endoplasmic reticulum. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1404:67–76. [DOI] [PubMed] [Google Scholar]
- Benyair R, Ogen-Shtern N, Lederkremer GZ. 2015. Glycan regulation of ER-associated degradation through compartmentalization. Semin Cell Dev Biol. 41:99–109. [DOI] [PubMed] [Google Scholar]
- Benyair R, Ogen-Shtern N, Mazkereth N, Shai B, Ehrlich M, Lederkremer GZ. 2015. Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Mol Biol Cell. 26:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G 1980. Intracellular protein topogenesis. PNAS. 77:1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. 2003. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 5:1051–1061. [DOI] [PubMed] [Google Scholar]
- Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, et al. 2014. A Chaperome Subnetwork Safeguards Proteostasis in Aging and Neurodegenerative Disease. Cell Reports. 9:1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik A, Stephens DJ. 2009. ER exit sites – Localization and control of COPII vesicle formation. FEBS Letters. 583:3796–3803. [DOI] [PubMed] [Google Scholar]
- Bunney TD, Cole AR, Broncel M, Esposito D, Tate EW, Katan M. 2014. Crystal Structure of the Human, FIC-Domain Containing Protein HYPE and Implications for Its Functions. Structure. 22:1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle A 2001. Posttranslational Modification In: Brenner S, Miller JH, editors. Encyclopedia of Genetics [Internet]. New York: Academic Press; [cited 2019 Jan 15]; p. 1533 Available from: http://www.sciencedirect.com/science/article/pii/B0122270800010223 [Google Scholar]
- Caplan AJ. 2003. What is a co-chaperone? Cell Stress Chaperones. 8:105–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro LG. 1964. Protein Synthesis, Storage, and Discharge in the Pancreatic Exocrine Cell: An Autoradiographic Study. The Journal of Cell Biology. 20:473–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Gonzalez GM, Clemons WM. 2011. A Structural Model of the Sgt2 Protein and Its Interactions with Chaperones and the Get4/Get5 Complex. J Biol Chem. 286:34325–34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Helenius J, Braakman I, Helenius A. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. PNAS. 92:6229–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. 2008. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nature Cell Biology. 10:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends in Biochemical Sciences. 28:655–662. [DOI] [PubMed] [Google Scholar]
- Daniels R, Kurowski B, Johnson AE, Hebert DN. 2003. N-Linked Glycans Direct the Cotranslational Folding Pathway of Influenza Hemagglutinin. Molecular Cell. 11:79–90. [DOI] [PubMed] [Google Scholar]
- Darby JF, Krysztofinska EM, Simpson PJ, Simon AC, Leznicki P, Sriskandarajah N, Bishop DS, Hale LR, Alfano C, Conte MR, et al. 2014. Solution Structure of the SGTA Dimerisation Domain and Investigation of Its Interactions with the Ubiquitin-Like Domains of BAG6 and UBL4A. PLOS ONE. 9:e113281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Benedix J, Cappel S, Greiner M, Jalal C, Müller L, Zimmermann R. 2009. Functions and pathologies of BiP and its interaction partners. Cell Mol Life Sci. 66:1556–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Tan Y-J. 2008. Structural and Functional Characterization of Human SGT and Its Interaction with Vpu of the Human Immunodeficiency Virus Type 1,. Biochemistry. 47:10123–10131. [DOI] [PubMed] [Google Scholar]
- Ecker M, Mrsa V, Hagen I, Deutzmann R, Strahl S, Tanner W. 2003. O- Mannosylation precedes and potentially controls the N- glycosylation of a yeast cell wall glycoprotein. EMBO reports. 4:628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. 2003. Quality control in the endoplasmic reticulum. Nature Reviews Molecular Cell Biology. 4:181–191. [DOI] [PubMed] [Google Scholar]
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. 1998. Huntingtin interacts with a family of WW domain proteins. Human molecular genetics. 7:1463–1474. [DOI] [PubMed] [Google Scholar]
- Fan ACY, Bhangoo MK, Young JC. 2006. Hsp90 Functions in the Targeting and Outer Membrane Translocation Steps of Tom70-mediated Mitochondrial Import. J Biol Chem. 281:33313–33324. [DOI] [PubMed] [Google Scholar]
- Farhan SMK, Nixon KCJ, Everest M, Edwards TN, Long S, Segal D, Knip MJ, Arts HH, Chakrabarti R, Wang J, et al. 2017. Identification of a novel synaptic protein, TMTC3, involved in periventricular nodular heterotopia with intellectual disability and epilepsy. Hum Mol Genet. 26:4278–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pino A, Zenkin N, Loris R. 2014. The many faces of Fic: structural and functional aspects of Fic enzymes. Trends in Biochemical Sciences. 39:121–129. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. 2000. Endoplasmic Reticulum Degradation Requires Lumen to Cytosol Signaling: Transmembrane Control of Hrd1p by Hrd3p. J Cell Biol. 151:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. 2006. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nature Cell Biology. 8:849. [DOI] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJC, Hallett M, et al. 2006. Quantitative Proteomics Analysis of the Secretory Pathway. Cell. 127:1265–1281. [DOI] [PubMed] [Google Scholar]
- Giorgi C, De Stefani D, Bononi A, Rizzuto R, Pinton P. 2009. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. The International Journal of Biochemistry & Cell Biology. 41:1817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Arnaud Sampaio VF, Lameu C, Ulrich H. 2018. Calcium signalling: A common target in neurological disorders and neurogenesis. Seminars in Cell & Developmental Biology [Internet]. [cited 2019 Jan 14]. Available from: http://www.sciencedirect.com/science/article/pii/S1084952118300685 [DOI] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. 2011. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 332:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove TZ, Cortajarena AL, Regan L. 2008. Ligand binding by repeat proteins: natural and designed. Current Opinion in Structural Biology. 18:507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Luzio JP, Chandler JA, Herman L. 1974. Localization of calcium in the smooth endoplasmic reticulum of rat isolated fat cells. J Cell Sci. 15:1–15. [DOI] [PubMed] [Google Scholar]
- Harada Y, Hua Li, Huilin Li, Lennarz WJ. 2009. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. PNAS. 106:6945–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness TA, Nargang FE, van der Klei I, Neupert W, Lill R 1994. A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. The Journal of Cell Biology. 124:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475:324. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su T-P. 2009. MAM: more than just a housekeeper. Trends in Cell Biology. 19:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Lamriben L, Powers ET, Kelly JW. 2014. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nature Chemical Biology. 10:902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideaki I, Yohtalou T. 1991. The stress (heat shock) proteins. International Journal of Biochemistry. 23:1185–1191. [DOI] [PubMed] [Google Scholar]
- Itskanov S, Park E. 2019. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science. 363:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SPN, Hart GW. 2003. Roles of the Tetratricopeptide Repeat Domain in O-GlcNAc Transferase Targeting and Protein Substrate Specificity. J Biol Chem. 278:24608–24616. [DOI] [PubMed] [Google Scholar]
- Javadi Y, Main ERG. 2009. Exploring the folding energy landscape of a series of designed consensus tetratricopeptide repeat proteins. PNAS. 106:17383–17388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D, Schekman R. 2011. COPII-mediated vesicle formation at a glance. Journal of Cell Science. 124:1–4. [DOI] [PubMed] [Google Scholar]
- Jeong H, Sim HJ, Song EK, Lee H, Ha SC, Jun Y, Park TJ, Lee C. 2016. Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway. Scientific Reports. 6:20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerber J, Zaki MS, Al-Aama JY, Rosti RO, Ben-Omran T, Dikoglu E, Silhavy JL, Caglar C, Musaev D, Albrecht B, et al. 2016. Biallelic Mutations in TMTC3, Encoding a Transmembrane and TPR-Containing Protein, Lead to Cobblestone Lissencephaly. Am J Hum Genet. 99:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez B, Ugwu F, Zhao R, Ortí L, Makhnevych T, Pineda-Lucena A, Houry WA. 2012. Structure of Minimal Tetratricopeptide Repeat Domain Protein Tah1 Reveals Mechanism of Its Interaction with Pih1 and Hsp90. J Biol Chem. 287:5698–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jínek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. 2004. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 11:1001–1007. [DOI] [PubMed] [Google Scholar]
- Johnson AE, van Waes MA. 1999. The Translocon: A Dynamic Gateway at the ER Membrane. Annual Review of Cell and Developmental Biology. 15:799–842. [DOI] [PubMed] [Google Scholar]
- Kajander T, Cortajarena AL, Mochrie S, Regan L. 2007. Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins. Acta Crystallographica Section D Biological Crystallography. 63:800–811. [DOI] [PubMed] [Google Scholar]
- Kalantari F, Bergeron JJM, Nilsson T. 2010. Biogenesis of lipid droplets – how cells get fatter. Molecular Membrane Biology. 27:462–468. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Nomura Y. 2003. ER signaling in unfolded protein response. Life Sciences. 74:199–205. [DOI] [PubMed] [Google Scholar]
- Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri H-P. 2001. Subdomain-Specific Localization of Climp-63 (P63) in the Endoplasmic Reticulum Is Mediated by Its Luminal α-Helical Segment. The Journal of Cell Biology. 153:1287–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR, Sitia R. 2000. Aggresomes and Russell bodies: Symptoms of cellular indigestion? EMBO Rep. 1:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamriben L, Graham JB, Adams BM, Hebert DN. 2016. N -Glycan-based ER Molecular Chaperone and Protein Quality Control System: The Calnexin Binding Cycle: The Calnexin Binding Cycle. Traffic. 17:308–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen ISB, Narimatsu Y, Joshi HJ, Siukstaite L, Harrison OJ, Brasch J, Goodman KM, Hansen L, Shapiro L, Honig B, et al. 2017. Discovery of an O-mannosylation pathway selectively serving cadherins and protocadherins. PNAS. 114:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. 2011. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 469:564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Korac-Prlic J, Kliza K, Husnjak K, Nyathi Y, Dikic I, High S. 2015. Binding of SGTA to Rpn13 selectively modulates protein quality control. J Cell Sci. 128:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Akil O, Rouse SL, McLaughlin CW, Matthews IR, Lustig LR, Chan DK, Sherr EH. 2018. Deletion of Tmtc4 activates the unfolded protein response and causes postnatal hearing loss. J Clin Invest. 128:5150–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Iakoucheva LM, Mooney SD, Radivojac P. 2010. Loss of Post-translatinal Modification sites in Disease. Pac Symp Biocomput.:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. PNAS. 88:2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. 1997. O-Linked GlcNAc Transferase Is a Conserved Nucleocytoplasmic Protein Containing Tetratricopeptide Repeats. J Biol Chem. 272:9316–9324. [DOI] [PubMed] [Google Scholar]
- Lunz V, Romanin C, Frischauf I. 2019. STIM1 activation of Orai1. Cell Calcium. 77:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynes EM, Simmen T. 2011. Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta. 1813:1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen MB, Kogelman LJA, Kadarmideen HN, Rasmussen HB. 2018. Systems genetics analysis of pharmacogenomics variation during antidepressant treatment. The Pharmacogenomics Journal. 18:144–152. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Regan L. 2005. Sequence variation in ligand binding sites in proteins. BMC Bioinformatics. 6:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, et al. 2018. Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control. BMC Biology. 16:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast FD, Fagarasanu A, Knoblach B, Rachubinski RA. 2010. Peroxisome Biogenesis: Something Old, Something New, Something Borrowed. Physiology [Internet]. [cited 2018 Dec 18]. Available from: https://www.physiology.org/doi/abs/10.1152/physiol.00025.2010 [DOI] [PubMed] [Google Scholar]
- Mazzarello P, Calligaro A, Vannini V, Muscatello U. 2003. The sarcoplasmic reticulum: its discovery and rediscovery. Nature Reviews Molecular Cell Biology. 4:69–74. [DOI] [PubMed] [Google Scholar]
- McGraw CF, Somlyo AV, Blaustein MP. 1980. Localization of calcium in presynaptic nerve terminals. An ultrastructural and electron microprobe analysis. The Journal of Cell Biology. 85:228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. 2010. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 190:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl PRE, Schneider-Brachert W. 2007. Sel1-like repeat proteins in signal transduction. Cellular Signalling. 19:20–31. [DOI] [PubMed] [Google Scholar]
- Molinari M, Helenius A. 1999. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature. 402:90–93. [DOI] [PubMed] [Google Scholar]
- Molinari M, Helenius A. 2000. Chaperone Selection During Glycoprotein Translocation into the Endoplasmic Reticulum. Science. 288:331–333. [DOI] [PubMed] [Google Scholar]
- Muto T, Obita T, Abe Y, Shodai T, Endo T, Kohda D. 2001. NMR identification of the Tom20 binding segment in mitochondrial presequences11Edited by M. F. Sumers. Journal of Molecular Biology. 306:137–143. [DOI] [PubMed] [Google Scholar]
- O’Connor SE, Imperiali B. 1996. Modulation of protein structure and function by asparagine-linked glycosylation. Chemistry & Biology. 3:803–812. [DOI] [PubMed] [Google Scholar]
- Odunuga OO, Longshaw VM, Blatch GL. 2004. Hop: more than an Hsp70/Hsp90 adaptor protein. BioEssays. 26:1058–1068. [DOI] [PubMed] [Google Scholar]
- Palade GE, Porter KR. 1954. STUDIES ON THE ENDOPLASMIC RETICULUM. J Exp Med. 100:641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE, Siekevitz P. 1956. Liver Microsomes: An Integrated Morphological and Biochemical Study. The Journal of Cell Biology. 2:171–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Rapoport TA. 2012. Mechanisms of Sec61/SecY-Mediated Protein Translocation Across Membranes. Annual Review of Biophysics. 41:21–40. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. 2008. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. The EMBO Journal. 27:2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. European Journal of Biochemistry. 268:2351–2361. [DOI] [PubMed] [Google Scholar]
- Pobre KFR, Poet GJ, Hendershot LM. 2018. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J Biol Chem.:jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rato C, Chen R, Antrobus R, Ding S, Fearnley IM, Ron D. 2015. AMPylation matches BiP activity to client protein load in the endoplasmic reticulum.Gilmore R, editor. eLife; 4:e12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rato C, Perera LA, Saudek V, Ron D. 2017. FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nature Structural & Molecular Biology. 24:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rohland L, Yan Y, Chen R, Read RJ, Ron D. 2016. AMPylation targets the rate-limiting step of BiP’s ATPase cycle for its functional inactivation. eLife [Internet]. [cited 2019 Jan 13]; 6 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5667935/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Culyba EK, Chen W, Murray AN, Hanson SR, Wong C-H, Powers ET, Kelly JW. 2012. N-glycosylation of enhanced aromatic sequons to increase glycoprotein stability. Peptide Science. 98:195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racapé M, Huyen J-PDV, Danger R, Giral M, Bleicher F, Foucher Y, Pallier A, Pilet P, Tafelmeyer P, Ashton-Chess J, et al. 2011. The Involvement of SMILE/TMTC3 in Endoplasmic Reticulum Stress Response. PLOS ONE. 6:e19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Stenmark H. 2015. ER-endosome contact sites: molecular compositions and functions. EMBO J. 34:1848–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. 1998. Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses. Science. 280:1763–1766. [DOI] [PubMed] [Google Scholar]
- Runge CL, Indap A, Zhou Y, Kent JW, King E, Erbe CB, Cole R, Littrell J, Merath K, James R, et al. 2016. Association of TMTC2 With Human Nonsyndromic Sensorineural Hearing Loss. JAMA Otolaryngol Head Neck Surg. 142:866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang S-W, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. 2007. The Role of p58IPK in Protecting the Stressed Endoplasmic Reticulum. Mol Biol Cell. 18:3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, Koller A, Mattoo S. 2015. A Novel Link Between Fic (Filamentation induced by cAMP)-mediated Adenylylation/AMPylation and the Unfolded Protein Response. J Biol Chem.:jbc.M114.618348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Ross CA, Villa A, Supattapone S, Pozzan T, Snyder SH, Meldolesi J. 1990. The inositol 1,4,5,-trisphosphate receptor in cerebellar Purkinje cells: quantitative immunogold labeling reveals concentration in an ER subcompartment. The Journal of Cell Biology. 111:615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer N, Chen J, Regan L. 2013. All Repeats are Not Equal: A Module-Based Approach to Guide Repeat Protein Design. J Mol Biol. 425:1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. 2000. Structure of TPR Domain–Peptide Complexes: Critical Elements in the Assembly of the Hsp70–Hsp90 Multichaperone Machine. Cell. 101:199–210. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT-BG, Clausen H. 2012. Site-specific protein O-glycosylation modulates proprotein processing — Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochimica et Biophysica Acta (BBA) - General Subjects. 1820:2079–2094. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Hebert DN. 2003. Protein Translocons: Multifunctional Mediators of Protein Translocation across Membranes. Cell. 112:491–505. [DOI] [PubMed] [Google Scholar]
- Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS. 2017. Mechanistic basis for a molecular triage reaction. Science. 355:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. 2005. Proteomic Analysis of Mammalian Oligosaccharyltransferase Reveals Multiple Subcomplexes that Contain Sec61, TRAP, and Two Potential New Subunits. Biochemistry. 44:5982–5992. [DOI] [PubMed] [Google Scholar]
- Simmen T, Lynes EM, Gesson K, Thomas G. 2010. Oxidative protein folding in the endoplasmic reticulum: Tight links to the mitochondria-associated membrane (MAM). Biochimica et Biophysica Acta (BBA) - Biomembranes. 1798:1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Simpson PJ, Goldstone RM, Krysztofinska EM, Murray JW, High S, Isaacson RL. 2013. Structure of the Sgt2/Get5 complex provides insights into GET-mediated targeting of tail-anchored membrane proteins. PNAS. 110:1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solá RJ, Griebenow K. 2009. Effects of glycosylation on the stability of protein pharmaceuticals. JPharmSci. 98:1223–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein O, Stein Y. 1967. Lipid Synthesis, Intracellular Transport, Storage, and Secretion. J Cell Biol. 33:319–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonça N, Liu X, Locasale JW, Simpson KW, Duhamel GE, et al. 2014. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. PNAS. 111:E582–E591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunryd JC, Cheon B, Graham JB, Giorda KM, Fissore RA, Hebert DN. 2014. TMTC1 and TMTC2 are novel endoplasmic reticulum TPR-containing adapter proteins involved in calcium homeostasis. J Biol Chem.:jbc.M114.554071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd M, Biterova EI, Bourhis J-M, Guy JE. 2011. The Crystal Structure of the Human Co-Chaperone P58IPK. PLOS ONE. 6:e22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Stukenbrok H, Metcalf A, Mignery GA, Sudhof TC, Volpe P, Camilli PD. 1992. Ca2+ stores in Purkinje neurons: endoplasmic reticulum subcompartments demonstrated by the heterogeneous distribution of the InsP3 receptor, Ca(2+)-ATPase, and calsequestrin. J Neurosci. 12:489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous A, Patel N, Tamura T, Hebert DN. 2015. Reglucosylation by UDP-glucose:glycoprotein glucosyltransferase 1 delays glycoprotein secretion but not degradation. Brodsky JL, editor. Molecular Biology of the Cell; 26:390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Petrova K, Ron D, Sha B. 2010. Crystal Structure of P58(IPK) TPR Fragment Reveals the Mechanism for its Molecular Chaperone Activity in UPR. Journal of Molecular Biology. 397:1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapaliya A, Nyathi Y, Martínez-Lumbreras S, Krysztofinska EM, Evans NJ, Terry IL, High S, Isaacson RL. 2016. SGTA interacts with the proteasomal ubiquitin receptor Rpn13 via a carboxylate clamp mechanism. Scientific Reports. 6:36622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Mandon EC, Gilmore R, Rapoport TA. 2017. Two alternative binding mechanisms connect the protein translocation Sec71/Sec72 complex with heat shock proteins. J Biol Chem.:jbc.M116.761122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann MC, Ploegh HL. 2017. rAMPing Up Stress Signaling: Protein AMPylation in Metazoans. Trends in Cell Biology. 27:608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veratti E 1961. Investigations of the Fine Structure of Striated Muscle Fiber. The Journal of Cell Biology. 10:1–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE. 2009. The Fic Domain: Regulation of Cell Signaling by Adenylylation. Molecular Cell. 34:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cabanos C, Rapoport TA. 2018. Structure of the post-translational protein translocation machinery of the ER membrane. Nature.:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sha B. 2006. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 13:589–593. [DOI] [PubMed] [Google Scholar]
- Xu C, Ng DTW. 2015. O-mannosylation: The other glycan player of ER quality control. Seminars in Cell & Developmental Biology. 41:129–134. [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. 2002. Control of PERK eIF2α kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. PNAS. 99:15920–15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Doudevski I, Regan L. 2010. HOP is a monomer: Investigation of the oligomeric state of the co-chaperone HOP. Protein Sci. 19:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. 2003. Molecular Chaperones Hsp90 and Hsp70 Deliver Preproteins to the Mitochondrial Import Receptor Tom70. Cell. 112:41–50. [DOI] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. 2012. Structural and Functional Discussion of the Tetra-Trico-Peptide Repeat, a Protein Interaction Module. Structure. 20:397–405. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu J. 2016. Shaping the Endoplasmic Reticulum into a Social Network. Trends in Cell Biology. 26:934–943. [DOI] [PubMed] [Google Scholar]