Abstract
Patients with metastatic pancreatic ductal adenocarcinoma (PDAC) have an average survival of less than one year, underscoring the importance of evaluating novel targets with matched targeted agents. We recently identified that poly (ADP) ribose glycohydrolase (PARG) is a strong candidate target due to its dependence on the pro-oncogenic mRNA stability factor HuR (ELAVL1). Here, we evaluated PARG as a target in PDAC models using both genetic silencing of PARG and established small molecule PARG inhibitors, PDDX-001/004 (PARGi). Homologous repair-deficient cells compared to homologous repair-proficient cells were more sensitive to PARG inhibitors in vitro. In vivo, silencing of PARG significantly decreased tumor growth. PARGi synergized with DNA damaging agents (i.e., oxaliplatin and 5-FU), but not with PARP inhibitor therapy. Mechanistically, combined PARGi and oxaliplatin treatment led to persistence of detrimental PARylation, increased expression of cleaved caspase 3 and increased ү-H2AX foci. In summary, these data validate PARG as a relevant target in PDAC and establish current therapies that synergize with PARGi.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is estimated to become the second leading cause of cancer-related death in the US, with a five-year survival rate of only 9% [1, 2]. Despite recent advancements in treatment options for PDAC, the prognosis remains poor. In 2008, global genomic analyses on human pancreatic cancers revealed 12 core signaling pathways that were altered in the majority of PDAC [3–5],and subsequent sequencing studies have supported and expanded upon these initial findings [6, 7]. Initial results from the “Know Your Tumor Initiative” for PDAC patients, as well as several related publications [5–8], have identified certain patient populations with 27% highly actionable mutations. Actionable mutations commonly found were in DNA repair genes (BRCA1/2 or ATM; 8.4%) and cell cycle genes (CCND1/2/3 or CDK4/6; 8.1%) [6, 7]. Germline or sporadic mutations in homologous recombination genes found in PDAC and several others cancers compromise repair of damaged DNA, most likely helping to facilitate tumorigenesis [8–11]. Selectively targeting alternative DNA repair pathways in such tumors provides a putative “personalized” therapeutic strategy via a synthetic lethal approach [12, 13].
In patients carrying germline or somatic defects in homologous-repair (HR) genes, DNA repair is heavily dependent on poly (ADP-ribose) polymerases (e.g. PARP1) [7, 11]. Hence, these tumors are highly susceptible to PARP inhibitors (PARPi) [13–15]. Recent data are also emerging with the use of PARPi in the treatment of PDAC [16–20]. Many clinical trials are ongoing for locally advanced or metastatic PDAC, either with single agent PARPi (olaparib, veliparib, talazoparib, rucaparib) or in combination with standard chemotherapy, including gemcitabine, FOLFIRINOX and cisplatin [16–20]. A recent dose-escalation, Phase 1 trial with talazoparib as a single agent confirmed responses in PDAC patients [21]. Three PARPi are approved for patients with metastatic breast and ovarian cancer, and are being used routinely as standard of care [22, 23]. However, while PARPi can be very effective, resistance to PARPi is an evolving concern [24–26]. Additionally, it is becoming increasingly apparent that not all tumors that share phenotypic features of BRCA1/2 mutant tumors (BRCAness) respond to PARPi therapies. Besides conventional resistance mechanisms [26], recent studies from our group showed that PDAC cells develop resistance to DNA damaging agents through post-transcriptional upregulation of WEE1 and poly (ADP-ribose) glycohydrolase (PARG), mediated by the pro-oncogenic, RNA binding protein, HuR [27, 28].
While WEE1 has been explored as a target in PDAC by us and others [27, 29], PARG has been explored by others as a target in colon cancers, lung cancers and acute myeloid leukemia [30–32]. PARG is an enzyme that breaks down poly (ADP-ribose) (PAR) chains for a number of different cellular processes including: release and recycling of repair proteins, break repair resolution and replication fork progression. PAR chains are added by the PARP enzymes as covalent modifications to acceptor proteins, primarily in response to DNA damage by PARP1, and PARP2 [33, 34]. This reversible post-translational modification is critical for signaling and recruitment of repair factors, and has typically been targeted via PARPi to prevent or weaken the DNA damage response [28, 33–35]. Other studies have demonstrated that inhibiting PARG could: i) sensitize cells to radiation [36]; ii) slow down DNA repair and cause mitotic abnormalities [37]; and iii) synergize with silencing of BRCA1, BRCA2, PALB2, FAM175A (ABRAXAS) and BARD1 genes to increase DNA damage and stall replication forks [38, 39]. Finally, we recently showed that the pro-oncogenic factor, HuR, upregulates PARG in a cancer-specific manner [28], providing a strong rationale (i.e., a therapeutic window) to explore targeting PARG in PDAC cells. Thus, we hypothesized that targeting PARG could provide an alternative and complementary therapeutic strategy to treating PDAC [38, 40]. To date, no one has explored targeting PARG in both homologous repair- deficient and proficient (HR-D and HR-P) PDAC cells due to a lack of available specific and potent PARGi. Herein, we evaluate for the first time targeting PARG, through both genetic silencing and small molecule inhibitors [38, 41, 42] in pre-clinical models of PDAC.
Materials and Methods
Cell lines
Homologous repair proficient (HR-P):
MIA PaCa-2, PANC-1, and homologous repair deficient (HR-D): Hs 766T PDAC cells were obtained from ATCC (Manassas, VA). Normal HPNE cells were purchased from ATCC. DLD1.BRCA2 and RKO.FANCC isogenic colorectal cell lines were a generous gift from Dr. Scott Kern and were generated by targeted disruption of either BRCA2 or FANCC (Fanconi Anemia pathway) DNA repair genes by homologous recombination and provide excellent cancer models for studying drug sensitivity in a HR-deficient background [43, 44]. PDAC PDX derived cells were obtained from Dr. Golan [45] (Supplementary Table 1A). These cells were derived from pancreatic ascites or pleural effusion cancer cells from PDAC patients and represent clinically relevant models to study PARG as a target since many of these models recapitulate metastatic PDAC, and have been characterized for BRCA1/2 mutations and KRAS status. KPC BRCA2 WT and BRCA2 null cell lines were a kind gift from Dr. Kenneth P Olive (Columbia University, NY) [46, 47]. PDX cell lines were cultured in RPMI in a humidified incubator at 37°C and 5% CO2, as recommended. All other cell lines were cultured in DMEM medium supplemented with 10% FBS, 1% L-glutamine and 1% penicillin-streptomycin. All cell lines were STR authenticated, and were Mycoplasma-tested monthly, using PCR based mycoplasma detection kit (# MP0035, Sigma Aldrich, St. Louis, MO). We also evaluated cell lines for maintenance of a defining oncogenic driving mutation (i.e., Kras). Cells were passaged at least twice after thawing before experimental use.
Generation of doxycycline-inducible shPARG
The pCW-shPARG plasmid was prepared using Gibson cloning: A pCW-cas9 tetracycline inducible plasmid (# 50661, Addgene, Cambridge, MA) was enzyme digested with NheI and BamHI (New England BioLabs, Ipswich, MA) as per manufacturers’ specifications and run through a 0.75% agarose gel for subsequent purification with the QIAquick Gel Extraction Kit (Qiagen Inc., Germantown, MD) to extract the TET-inducible pCW vector backbone. An shPARG insert flanked by BamHI enzyme restriction sites and an upstream NheI restriction site was purchased from IDT (Skokie, IL), PCR amplified using designed cloning primers (Forward Primer: 5′-cagatcgcctggagaattggGGATCCGCTAGCGCCACC-3′ [17], Reverse Primer: 5′-aaggcgcaaccccaaccccgGGATCCCAAAAAGCGATCTTAGGAAAC-3′) and gel purified as detailed previously. The insert was then Gibson cloned into the vector using the NEBuilder® HiFi DNA Assembly Cloning Kit (# E5520S, New England BioLabs) at a vector and insert count of 0.05 picomols and 0.12 picomols, respectively. The newly formed plasmid was used to transform NEB 5-alpha competent cells (# C2988J, New England BioLabs). After a 24 hour incubation at 37°C, individual clones were then selected, cultured and sequenced for validation using sequencing primers (Primer 1: 5′-GGGCTGCCTTGGAAAAG-3′, Primer 2: 5′-CAGATCGCCTGGAGAATTG-3′).
Lentivirus production and generation of shPARG cell lines
A commercially available unique 29mer shRNA construct against human PARG in a lentiviral GFP vector (# TL310610, Origene, Rockville, MD) was packaged in HEK 293T cells using a lentiviral packaging kit (# TR30037, Origene). MIA PaCa-2 and PANC-1 cells were infected with virus particles with the addition of 1 μg/ml Polybrene (# TR-1003-G, Sigma Aldrich). Cells were then puromycin (# P8833, Sigma Aldrich) selected and validated for PARG mRNA and protein knockdown.
Modified small interfering RNA for transient knockdown
MIA PaCa-2 and PANC cells were transfected with custom-made modified control and PARG siRNA oligonucleotides (Genisphere LLC, Hatfield, PA; PARG sense 5’-UACCAGAGCAGUUUAGUAA-3’). Transfections with unmodified ON-TARGET plus Non-targeting Control siRNA (GE Dharmacon, #D-001810–01-05) and ON-TARGET plus PARG siRNA (GE Dharmacon, # LU-011488–00-0002) were used as controls. All transfections were performed using 30nM of oligonucleotides and Lipofectamine 2000 (Life Technologies, #11668) according to manufacturer’s instructions.
Drugs and inhibitors
Olaparib (PARPi # S1060) and oxaliplatin (# S1224) were obtained from Selleck Chemicals, Houston, TX. 5-FU (# F6627) was obtained from Sigma Aldrich. Cell active small molecule PARG inhibitors (PDDX-001/PDD00017273 and PDDX-002/PDD00017238) were synthesized as previously described [42, 48] and were utilized since they have been reported to demonstrate robust pharmacology against PARG with great potency and specificity, as compared to other known inhibitors of PARG. PDDX-004/PDD00017272 was synthesized in collaboration with Dr. Joseph Salvino from The Wistar Institute and this compound has the exact quinazolinedione core as PDDX-01 as published before [42]. Compounds were either re-suspended in dimethyl sulfoxide (DMSO) or water to stock concentrations of 10mM.
Cell survival and combination analyses
800 cells/well were plated in 96-well plates in 100 µL of cell culture medium, and treated with drugs after 24 hours at indicated concentrations. Percentage cell survival relative to control treatments was assessed after 5 days, by staining cells with Quant-iT Pico Green Reagent (Cat# P7581, Life Technologies Corp, Grand Island, NY) for 1 hour and measuring fluorescence using a fluorescence microplate reader (excitation ~480 nm, emission ~520 nm) [49]. IC50 values were determined through non-linear regression analysis.
Cell survival for siDDR experiments was analyzed by MTT assay. 1000 cells/well were plated in 96-well plates in 100 µL of cell culture medium, and treated with drugs after 24 hours at indicated concentrations. After 5 days, MTT reagent (5mg/ml in 1XPBS) was added to each well and plates were incubated for 4 hours at 37°C. Following incubation, media was removed and purple formazan crystals were dissolved in DMSO (100 µL). Absorbance was read at 570 nm and results were plotted as percentage relative survival.
Combination analyses were performed by plating cells in 96-well plates at 1000 cells/well. After 24 hours, cells were treated with PDDX-001 (dose range 1.5 µM-25 µM) and either olaparib (dose range 1.5 µM-25 µM), oxaliplatin (dose range 0.03 µM-1 µM), or 5-FU (dose range 0.75 µM-25 µM) in a 5X5 or 5X6 matrix. Each treatment was done in triplicate wells. Cells were treated for 5 days, and percentage cell survival relative to control treatments was assessed by staining cells with Quant-iT Pico Green Reagent. Each experiment was done at least three times. After Pico Green analysis, synergistic/antagonistic/additive combinations were analyzed by using Combenefit software which provides synergy distribution plots by comparing experimental data to mathematical models (e.g. bliss model) of dose responses for additive/independent combinations [50]. In brief, the software first reads each experimental dose response as a matrix of percentage of the control and each single agent effect is fitted with a dose response curve. Bliss Independence model then compares experimental combination dose response surface to the model-generated dose response, resulting in a synergy distribution plot. The Bliss Independence model calculates the expected effect of two drugs as follows: where A and B represent two different drugs [51]. If the actual effect is greater than that calculated with the Bliss Independence model this implies synergism = indicates additivity, and less than indicates antagonism. Combenefit determines synergy as the change in efficacy of a combination of drugs as compared to expected. So for instance if two drugs through the Bliss model are expected to kill 50 percent of cells but instead kill 75, this would be a synergy score of 50 as at this combination there is a 50% increase in the expected effectiveness of the combination. Drug matrix heat map 5X5 or 5X6 grid illustrating bliss index and percentage inhibition are shown for n=3.
Colony Formation analyses
Long-term colony formation assays were performed as previously described [52] and colonies were counted using Image J software.
RT-qPCR and mRNA expression analysis
Total RNA was extracted using the RNeasy mini kit (Qiagen Inc., Germantown, MD). cDNA was made using 1000 μg total RNA using Applied Biosystems High Capacity cDNA Reverse Transcriptase kit (Life Technologies Corp) and quantitative PCR (RT-qPCR) was performed using as previously described [49]. Relative quantification was performed using the 2-ΔΔCt method.
Immunoblot analysis
Cells were lysed in ice cold RIPA buffer (# sc-24948A, Santa Cruz Biotechnology Inc., Dallas, TX) supplemented with fresh protease inhibitors (# 78430, Life Technologies Corp), immunoblotted and membranes were scanned and quantitated using Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) as previously described [49]. Primary antibodies used were GAPDH (1:10,000; #2118, Cell Signaling Technology, Danvers, MA), PARP1 (1:1000; #sc-365315, Santa Cruz Biotechnology Inc.), PAR (1:1000; # 4335-MC-100, Trevigen, Gaithersburg, MD), PARG (# NBP2–46320, 1:1000; Novus, Littleton, CO), Cleaved Caspase-3 (1:1000), Caspase-3 (1:1000) and Histone H3 (# 9661, #9662 and #4499, Cell Signaling Technology) followed by LI-COR IRDye secondary antibodies.
Immunofluorescence
5,000 cells were plated onto coverslips in 24-well tissue culture plates and allowed to adhere for 24 hours. After drug treatment for indicated times and concentrations, coverslips were washed with 1X phosphate buffered saline (PBS), fixed (4% paraformaldehyde for 10 minutes RT), permeabilized (0.1% Triton-X), blocked and immunostained with γ-H2AX 1:500 (#05–636 Millipore Sigma, Burlington, MA) antibody overnight, 4°C, followed by secondary antibody (Alexa-488 F anti-mouse, #A-10680, Life Technologies Corp) for 1 hour after washing. After the final wash step, coverslips were mounted onto slides using DAPI ProLong Gold antifade mounting medium (#P36931, Life Technologies Corp). Slides were imaged with a Leica DM4B fluorescence microscope and foci were counted using Image J software [28, 53]. Percentage of cells expressing >10 γ-H2AX foci per treatment condition ± SD was calculated and plotted.
PAR ELISA
PARylation was analyzed in total protein lysates using HT Colorimetric PARP/Apoptosis Assay as previously described [28].
Apoptosis assays
Apoptosis was detected by western blot analysis for cleaved caspase-3 in drug treated cells at different time points [27].
Chromatin tethering
Treated cells were washed with ice-cold 1XPBS, gently scraped and collected in 1mL 1XPBS and pelleted by spinning at 400g for 5 minutes. Sequential fractionation was performed with ice-cold 0.1% Triton X-100 in cytoskeletal PIPES buffer as previously described [54]. The final pellet containing (chromatin-bound proteins) and total cell pellets were lysed in RIPA buffer. Equal amounts of protein were loaded onto wells and membranes were immunoblotted with primary and corresponding secondary antibodies. Histone H3 was used as a positive control and GAPDH as a negative control for the chromatin-bound fraction.
Xenograft Study
Doxycycline inducible shPARG knockdown model
Doxycycline-inducible shPARG cell lines were generated for MIA PaCa-2 cells. 3X106 cells/flank (MIA.shPARG) were then injected subcutaneously (100 uL) in 5–6 week-old Hsd:Athymic Nude-Foxn1nu female mice (Envigo RMS, Inc., Indianapolis, IN) (n=10 for each cell line). Cells were prepared in 80% DPBS and 20% Matrigel (#356237, Corning Life Sciences, Tewksbury, MA). Mice bearing established tumors (50–100 mm3, MIA.shPARG) were randomized into two treatment groups (n=5 for each group) and one group was administered doxycycline (DOX) chow (200 mg/kg, Bio-Serv, Flemington, NJ) to downregulate PARG expression.
Statistical considerations.
Mouse weights and tumor sizes were measured three times/week and tumor volumes were calculated using the formula: Volume = (LengthXWidth2)/2. Relative fold change in tumor volumes was plotted. Relative fold change represents fold change in tumor volume relative to tumor volume on day 1 of randomization of mice into dox/no dox groups. No significant loss in body weight was observed (<5%) for all groups. Animals were euthanized when average tumor volume reached 2000 mm3.
All mouse protocols were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical analyses were performed using the one or two sample t-test as indicated in figure legends. GraphPad Prism 7.04 software was used for analysis. Results are expressed as Mean ± standard error of mean (SEM), if not specifically indicated.
Results
PARG silencing inhibits PDAC tumor growth in vivo and in vitro
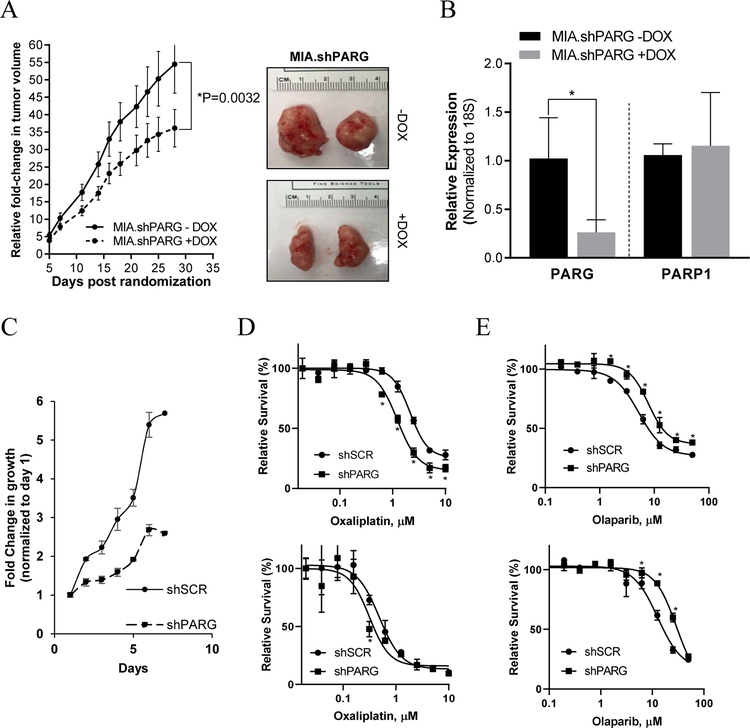
In vitro characterization of MIA.shPARG and Lenti.shPARG cell lines confirmed a decrease in mRNA and protein expression of PARG (Supplementary Figure 1A and B). For in vivo doxycycline induced shPARG knockdown model, 5–6 weeks old athymic nude female mice were injected with 100 µL of cell suspension subcutaneously in each flank, and mice were randomized into 2 groups of: i) no DOX and ii) DOX chow. In the DOX chow arm, knockdown of PARG significantly decreased tumor growth in MIA.shPARG (P≤0.05), compared with mice in the no DOX group (Figure 1A). At the conclusion of the study, tumors were harvested (Figure 1A) and validated for PARG mRNA knockdown. mRNA expression analysis shows PARG levels were significantly decreased in groups fed on DOX chow with no change in a negative control, PARP1 mRNA expression (Figure 1B). Lentivirus shPARG cell lines also show a decrease in growth rate as compared to a shScramble (control) cell line in vitro (Figure 1C).
Figure 1:
Knockdown of PARG with shPARG decreases PDAC tumor growth in vivo and in vitro. (A) Athymic nude mice bearing MIA.shPARG (left) xenografts were randomized into -DOX and +DOX arms and relative fold change in tumor volume was plotted. *P<0.05 using two-sample t-tests. And representative images (right) of tumor extracted after completion of in vivo study from MIA.shPARG mice (n=5 per group). (B) Relative mRNA expression of PARG and PARP in harvested tumors from MIA.shPARG (−/+ DOX) in vivo. Mean±SEM, n=3,*P =0.01 to 0.05.(C) In vitro fold change in growth of Lentiviral.shPARG PANC-1 cells compared to shScramble. (D) Lentiviral shPARG knockdown enhances sensitivity of cells to oxaliplatin and (E) decreases sensitivity to olaparib in PANC-1 (top) and MIA PaCa-2 (bottom) cells in vitro. Representative graphs from n=3 are shown, *P <0.05 using two-sample t-tests.
Silencing of PARG in the doxycycline-inducible MIA.shPARG or lentiviral shPARG models sensitized cells to oxaliplatin, as indicated by a decrease in relative cell survival compared to control arms (Figure 1D and Supplementary Figure 1C). Confirming previous reports, silencing of PARG decreased the sensitivity of cells to olaparib (Figure 1E) [55]. Collectively, these findings imply that PARG targeting mediates a reduction in PDAC cell survival and tumor growth in vivo.
PARG inhibitors decrease cell survival of cancer cell lines with genetically diverse backgrounds in vitro
Short term drug sensitivity assays
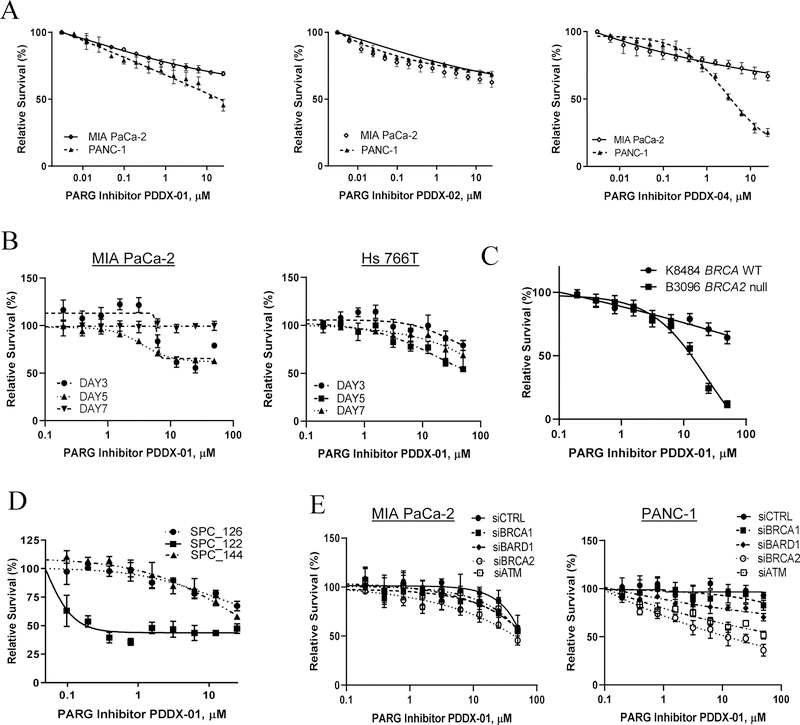
After establishing PARG as a therapeutic target from our in vivo studies, we assessed the efficacy of small molecule inhibitors of PARG (PARGi) [42] on cell survival of PDAC, PDX and KPC cell culture models (Figure 2A, B, C, and D). Both MIA PaCa-2 and PANC-1 HR proficient (HR-P) PDAC cells lines were treated with increasing doses of PARGis (PDDX-001, PDDX-002 and PDDX-004) and olaparib to determine IC50 concentrations. Hs 766T, PDAC PDX and KPC BRCA2 wild type and BRCA1/2 - null cell lines were treated with PDDX-01 and olaparib and cell survival was evaluated using a cell survival, Pico Green assay [49]. PARG inhibition was especially effective against cells with an HR deficiency (HR-D); Hs 766T, HR-D PDX cells (SPC_122) and KPC BRCA2-null cells were more sensitive to PARGi compared to their counterpart HR-P cells (Figure 2A, B, C, and D). Interestingly, in regards to our PDX lines, the cell line that responded the most to PARGi was SPC_122 (validated as BRCA1 mutant) and those that did not respond were SPC_144 (BRCA WT) and SPC_126 (BRCA 2 mutant). Of note, SPC_126 (Supplementary Table 1A) is obtained from a liver biopsy from a patient who became clinically resistant to therapy (i.e., previously exposed to PARPi/platinum therapy and recurred, and is also resistant to PARPi therapy in vitro and in vivo [45]. These data suggest that PARGi may not be the best therapeutic option in PARPi-resistant tumors.
Figure 2:
PARG inhibition is synthetic lethal with HR deficiency in PDAC cells. (A) Pico Green assays were performed after 5 days of treatment with PDDX-001, PDDX-002 and PDDX-004 to analyze relative cell survival in PDAC cells (MIA Paca-2, PANC-1). (B, C and D) Relative cell survival of MIA PaCa-2 and Hs 766T treated with PDDX-001 over 3, 5 and 7 days; KPC BRCA WT and KPC BRCA2 null cells as well as pancreatic cancer patient derived xenograft cell lines (SPC_126, SPC_122, SPC_144). (E) Synthetic lethality with PDDX-01 and siRNA DDR genes in MIA PaCa-2 and PANC-1 cells. Representative graphs from n=2 for (A) and n=3 for (B, C, D and E) are shown.
Additionally, we found that PARG is differentially expressed in PDAC and PDX models (Supplementary Figure 2A and Supplementary Table 1B). We observed that high PARG expressing cell lines responded better to PARGi treatment (SPC_122), as compared to cell lines with medium expression of PARG protein (MIA PaCa-2, PANC-1, SPC_144 and others). The “normal” epithelial HPNE cells with low levels of PARG protein expression (Supplementary Figure 2A and Supplementary Table 1B) did not respond to PARGi, supporting the possible therapeutic window we described previously [28]. We did find a perfect correlation (R2 =1) in PDX models, SPC_122 (high PARG expression/PARPi naïve-Supplementary Figure 2B) and SPC_126 (low PARG expression/PARPi resistant- Supplementary Figure 2B) PDX lines which corroborates previous published findings [55]. However, we did not find a correlation between PARG expression and PARPi response across PDAC lines (Supplementary Figure 2A and C) which suggests that defects in HR genes should still be the gold standard biomarker for PARPi therapy.
To analyze the spectrum of PARGi effects in isogenic models, and the possible therapeutic window between normal and somatic mutations found in a tumor, we also utilized genetically modified DLD1 (DLD1.BRCA2) and RKO (RKO.FANCC) colorectal cancer cells [43, 44]. We have previously tested these lines against various DNA damage agents and disruption of BRCA2 or FANCC renders them sensitive to oxaliplatin and PARPi [43]. Our results show that both BRCA2 and FANCC null cells required lower doses of PARGi to achieve IC50 when compared with isogenic parental cells (Supplementary Figure 2D).
To further characterize synthetic lethality with PARG inhibition and defects in genes involved in DNA damage response pathways in an isogenic model, we utilized HR-P PDAC cells, MIA PaCa-2 and PANC-1. Using siRNA oligos against BRCA1, BRCA2, ATM and BARD1, we compared drug sensitivities of olaparib and PDDX-01 in these knock-down conditions to a scramble siRNA control arm. In our screen, as expected, we found an increase in sensitivity to olaparib with BRCA1/BRAC2 knockdown, while only BRCA2 knockdown increased sensitivity to PDDX-01, demonstrating synthetic lethality of PDDX-01 and BRCA2 in PDAC cells (Figure 2E and Supplementary Figure 3A). Knockdown efficiencies of all the genes in both MIA PaCa-2 and PANC-1 cells are represented in Supplementary Figure 3B.
Drug combination analyses
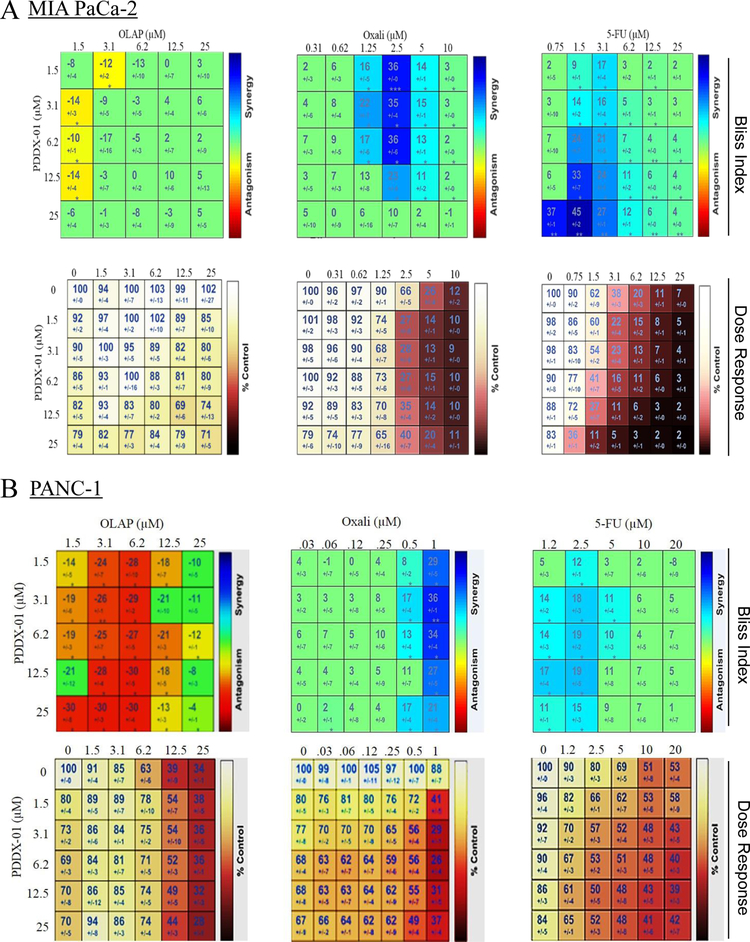
To test the efficacy of PARGi in combination with other commonly used DNA damaging agents and PARPi (olaparib) in PDAC cells, we utilized Combenefit software [50] to analyze synergy, additivity or antagonism between the combinations. The bliss index of combination analyses shows that PARGi (PDDX-001) did not synergize with olaparib and the combination is mostly antagonistic as seen in the bliss matrix for olaparib and PARGi in both MIA PaCa-2 and PANC-1 cells (Figure 3A and B). However, when PARGi was utilized with oxaliplatin or 5-fluorouracil (Figure 3A and B), inhibition of PARG synergized with these compounds, with some drug combinations being more synergistic than others. It is interesting to note that single agent PARGi, though not as effective in HR-P PDAC cells as HR-D cells, acts as a similar sensitizer to other DNA damage agents (oxaliplatin and 5-FU) in these cell lines. There was no synergy seen in HR-D cells HS 766T with either oxaliplatin/PARGi or 5-FU/PARGi combinations (Supplementary Figure 4A and B).
Figure 3:
PARGi synergizes with DNA damage agents and not with PARPi. Drug matrix heat map 5X5 (olaparib and 5-FU) and 6X5 (oxaliplatin) grid showing Bliss index and dose response as analyzed by Combenefit analysis for (A) MIA PaCa-2 and (B) PANC-1 cells. Combinations that are synergistic appear blue on the heat map.
Clonogenicity assays (Long term assays).
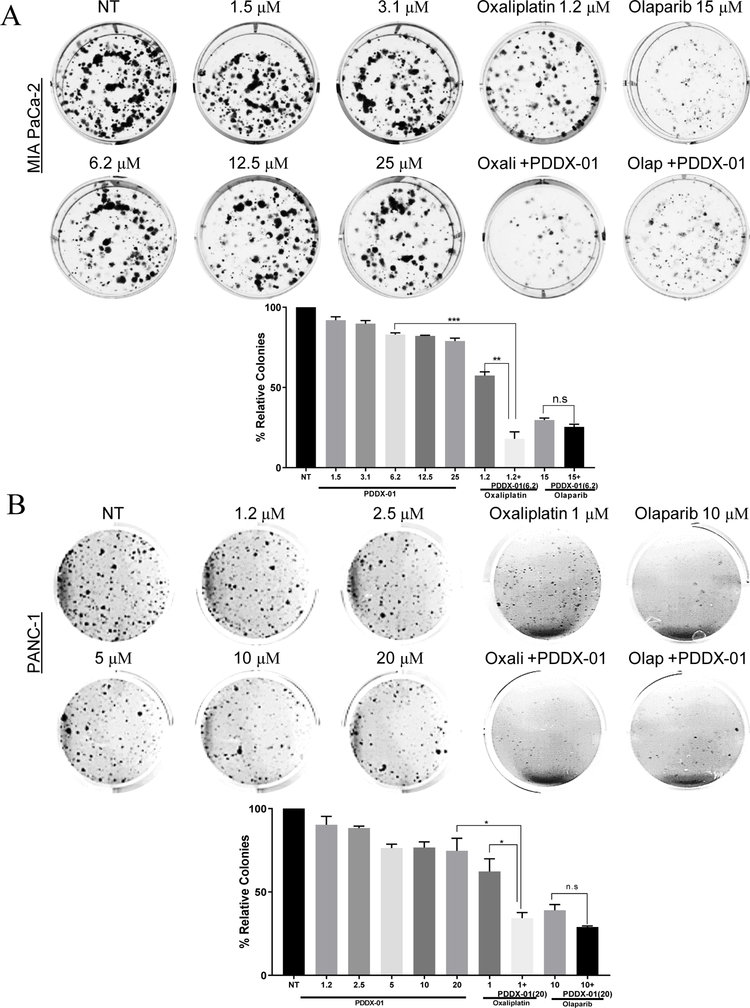
To understand the long term effects of PARGi on PDAC cells, we performed clonogenic assays. Both MIA PaCa-2 and PANC-1 cells were treated with PDDX-001 at increasing doses, and colonies were stained and counted after 14 days (Figure 4). PDDX-001 did not significantly reduce colony formation in either MIA PaCa-2 (Figure 4A) or PANC-1 cells (Figure 4B), confirming our results from short-term cell survival assays. From our combination analyses, we further tested the combination of PARGi and oxaliplatin/olaparib in both these cell lines. Interestingly, inhibition of PARG by PDDX-001 sensitized both MIA PaCa-2 and PANC-1 cells to oxaliplatin (Figure 4A and B), both of which are HR-P PDAC cell lines. Similar to the results from combination analyses, PDDX-001 did not synergize with olaparib in decreasing colony formation of PDAC cells. Together, these data indicate that PARGi has activity and efficacy against PDAC cells and could be utilized to increase the sensitivity of oxaliplatin and other DNA damage agents (like 5-FU) but not PARPi (olaparib) for the treatment of PDAC.
Figure 4:
PARGi decreases colony formation in PDAC cells in combination with oxaliplatin. (A) MIA PaCa-2 and (B) PANC-1 cell lines were treated with increasing doses of PDDX-001 (µM) alone and in combination with oxaliplatin (oxali) or olaparib (olap) and resulting colonies were fixed and stained with crystal violet solution. Representative images with graphs of relative colonies are shown. Mean ±SEM, n=3. ***P=0.0001 to 0.001, **P=0.001 to 0.01, *P=0.01 to 0.05, n.s= non-significant using two-sample t-tests.
PARGi affects PARylation dynamics of PDAC cells and regulates PARG activity
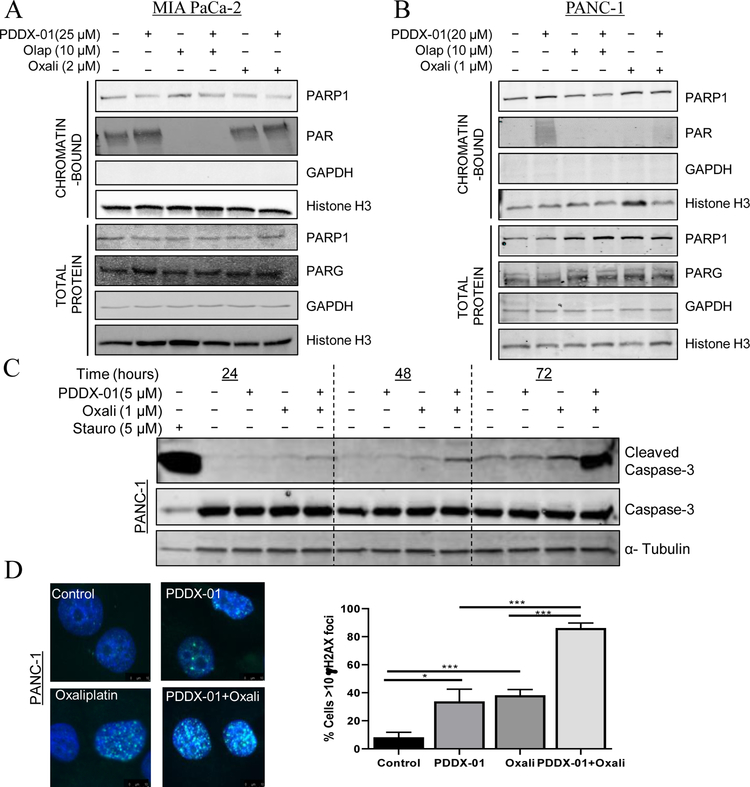
To further assess the mechanism of PARGi induced cytotoxicity and whether PARGi causes an increase in PAR accumulation in PDAC cells, we performed chromatin tethering assays [54]. We analyzed PARP and PAR protein expression in chromatin bound fractions after treatment with PARGi alone or in combination with oxaliplatin or olaparib in PDAC cell lines. GAPDH was used as a negative control and histone H3 expression was used as a positive control for chromatin fractions. PDDX-001 increased chromatin bound PAR expression in both MIA PaCa-2 and PANC-1 cells treated alone as shown in Figure 5A and B. We also observed a synergistic effect of PARGi with oxaliplatin as seen in Figures 3 and 4, which could not be attributed to synergistic increase in accumulated PAR with the combination (43). Contrary to this affect, and as previously shown [55], olaparib not only decreased relative PARylation as a consequence of inhibiting PARP1 activity alone, but also prevented PARGi induced increase in PAR (Figure 5A and B), implying that inhibition of PARGi does not benefit PARPi therapy, in agreement with previous results [55]
Figure 5:
PARGi induces PAR accumulation. (A) MIA PaCa-2 and (B) PANC-1 cells were treated with either PDDX-001, olaparib, oxaliplatin, alone or in combination for 24 hours and chromatin and whole cell fractions were isolated. Representative western blot images are shown from n=3. GAPDH and Histone H3 served as controls for whole cell fraction and chromatin fractions respectively. (C) PARGi in combination with oxaliplatin causes cleavage of caspase-3. PANC-1 cells were treated with PDDX-001, oxaliplatin or combination and apoptosis was assessed by western blot analysis for cleaved caspase-3 at 24, 48 and 72 hour time points. Representative image from n=3 is shown. (D) PARGi induces DNA damage in combination with oxaliplatin. γH2AX foci increases with the combination of PDDX-01 and oxaliplatin at 24 hour in PANC-1 cells. Representative graphs of cells with >10foci is shown. Mean±SD, n=3, *P<0.05, ***P<0.001
To evaluate whether PARG inhibitors affect relative PARylation, whole cell PARylation was assessed by ELISA assays [28]. All PARGi compounds significantly lead to persistence of whole-cell PARylation, as analyzed by ELISA assays (Supplementary Figure 5A) in both HR-P and HR-D PDAC cells. PARPi (olaparib) was used as the negative control, which has previously been shown to effectively prevent PAR polymer formation [28].
PARGi affects apoptosis and a marker of DNA damage response
In an effort to find the mechanism of synergy between PARGi and oxaliplatin in PDAC cells, we assessed apoptosis by cleaved caspase-3 expression in both MIA PaCa-2 and PANC-1 cells. Increase in cleaved caspase-3 with oxaliplatin was seen as early as 24 hours for both MIA PaCa-2 cells and PANC-1 cells (Figure 5C and Supplementary Figure 6A). This suggests that activation of caspases is a possible mechanism of synergistic cell death between PARGi and oxaliplatin.
Additionally, we analyzed gamma-H2AX expression as a marker of DNA damage and found a significant increase in gamma-H2AX foci in combination treatments across both the cell lines, indicating an increase in DNA damage (Figure 5D and Supplementary Figure 6B). Taken together, these results demonstrate that targeting PARG with DNA damage agents induces DNA damage and apoptosis in PDAC cells.
Discussion
Current standard of care chemotherapeutic regimens show only modest increases in overall survival of PDAC patients [56, 57]. Thus, there remains a desperate need to find targets and therapies, which eventually could be personalized for PDAC [2–7, 10, 58]. Recently, a successful phase 2 study of olaparib (PARPi) showed significant clinical activity in BRCA1/BRCA2 (HR-D) mutant pancreatic cancer patients previously treated with gemcitabine [19]. However, many studies have already started to emerge with PARPi resistance [24–26]. Previously, we published that PARPi treatment causes an increase in PARG protein through a novel post-transcriptional mechanism regulated by stress responsive RNA binding protein, HuR in PDAC [28]. Based on these data and the fact that PARG is part of the DNA repair process, we explored PARG as a therapeutic target in PDAC by first utilizing shRNA genomic approaches to silence PARG expression and activity, and then validating these findings through the use of established small molecule PARG inhibitors [38, 42].
The data presented herein also demonstrate that small molecule inhibitors of PARG decrease cell survival, proliferation and induce DNA damage in PDAC cells when used as monotherapy or in combination with DNA damage agents in vitro (Figures 2–5). IC50 values of PARGi values indicate that in various isogenic and non-isogenic PDAC cell models PARG inhibitors are more effective against HR-D cells when compared with HR-P cells, highlighting previous reports that PARG targeting may have beneficial synthetic lethal effects on cells with HR deficiencies [35–38, 59]. In sum, these data suggest PARGi should be explored as a novel synthetic lethal approach in PDAC tumors that share molecular features with BRCA - mutant PDAC subtypes.
Though our study shows that PARGi is more effective in HR-D PDAC cells, PARGi caused increased PARylation and a significant decrease in PARG activity in both HR-D and HR-P PDAC cells, without directly affecting protein levels of either PARG or PARP1. The increase in PARylation after PARGi emphasizes the specificity of PARGi and the importance of PARG as a primary PAR catabolizing enzyme [60]. This also suggests that PARG may be a more favorable target over other DNA repair proteins like PARP1, which has multiple family members [33, 41]. Although these inhibitors currently have limited bioavailability [42], our results support development of these lead compounds for more favorable bioavailable small molecule PARGis for effective targeting of PARG in vivo. In an ongoing effort, we have started investigating a 3DNA-nanocarrier delivery of modified siPARG to target PARG with a tumor specific targeting moiety in vivo (Supplementary Figure 7).
Combination treatments of PARGi with either oxaliplatin or 5-FU (components of FOLFIRINOX) in HR-P PDAC cell lines provide evidence that targeting PARG could synergize with current standard of care treatments for PDAC (Figure 3). Our results highlight that PARGi is antagonistic with olaparib (PARPi), as analyzed through bliss index of combination treatments (Figure 3). These results are in agreement with a recent study published by Gogola et al, that show loss of PARG as a resistance mechanism to PARPi therapy in murine mammary tumor cells and that PARP1 signaling is rescued by PARGi mediated PAR accumulation [55]. To understand the increased sensitivity of oxaliplatin in presence of PARGi, we looked at PAR levels in chromatin fractions. Accumulation of PAR polymers and further loss of NAD+ pools are reported to cause cytotoxicity [61–63]. From our results we did not find a synergistic increase in accumulation of PAR with the combination of oxaliplatin and PARGi (Figure 5A and B). However, we found an increase in DNA damage and elevated cleaved caspase-3 expression with the combination, both of which could implicate mechanisms behind synergy seen with oxaliplatin and PARGi (Figure 5C, D and Supplementary Figure 6A and B). Future studies will determine the precise mechanism of increased sensitivity of 5-FU or oxaliplatin with PARGi in an in vivo setting. Overall, these findings favor targeting PARG in combination with DNA damaging agents (e.g., oxaliplatin) and warrants further in vivo dose optimization and sequencing (i.e., timing) studies.
Taken together, we strongly believe this work provides the first evidence that acute targeting of PARG should be pursued as a treatment strategy for PDAC with both HR-proficient and deficient genetic backgrounds. More sophisticated, ongoing in vivo studies will evaluate next generation PARG targeting strategies (i.e., other small molecules and siPARG nanotherapy) in combination with other DNA damaging agents for the treatment of all pancreatic cancers.
Supplementary Material
Statement of Significance.
PARG is a potential target in pancreatic cancer, as a single agent anticancer therapy or in combination with current standard of care.
Acknowledgments
This work was supported by NIH-NCI R01 CA212600 (J.R. Brody), R37CA227865 (J.M.W and J.R.B support) and was also supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA056036 SKCC Core Grant (TJU). Additional work (JRB, MJP) is supported by a 2015 Pancreatic Cancer Action Network American Association for Cancer Research Acceleration Network Grant (15–90-25-BROD). Also the generous support of a Mary Halinski Fellowship (A.N) and the W. Kim Foster Pancreatic Cancer Research Endowment. KMS, IDW, DJO, DIJ and AMJ are supported by Cancer Research UK (Grant C480/A11411 and C5759/A17098). We would also like to thank Genisphere LLC, (Dr. Robert Getts, Hatfield, PA) for providing us with the modified siPARG. Mark Levine’s contributions to this work were made in memory of Ethel Levine.
Abbreviation list:
- PDAC
pancreatic ductal adenocarcinoma
- PAR
poly (ADP-ribose)
- PARP
poly (ADP-ribose) polymerase
- PARPi
PARP inhibitor
- PARG
poly (ADP-ribose) glycohydrolase
- PARGi
PARG inhibitor
- HR
homologous-repair
- HR-D
HR-deficient
- HR-P
HR-proficient
Footnotes
Conflict of interest statement: Genisphere, LLC provided the modified siPARG oligos described herein.
References
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2016. CA Cancer J Clin, 2016. 66(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Sheahan AV, et al. , Targeted therapies in the management of locally advanced and metastatic pancreatic cancer: a systematic review. Oncotarget, 2018. 9(30): p. 21613–21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, et al. , Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science, 2008. 321(5897): p. 1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey P, et al. , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 2016. 531(7592): p. 47–52. [DOI] [PubMed] [Google Scholar]
- 5.Waddell N, et al. , Whole genomes redefine the mutational landscape of pancreatic cancer. Nature, 2015. 518(7540): p. 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pishvaian MJ, et al. , Molecular Profiling of Pancreatic Cancer Patients: Initial Results from the Know Your Tumor Initiative. Clin Cancer Res, 2018. [DOI] [PubMed]
- 7.Pishvaian MJ and Brody JR, Therapeutic Implications of Molecular Subtyping for Pancreatic Cancer. Oncology (Williston Park), 2017. 31(3): p. 159–66, 168. [PubMed] [Google Scholar]
- 8.Aguirre AJ, et al. , Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov, 2018. [DOI] [PMC free article] [PubMed]
- 9.Chou A, et al. , Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med, 2013. 5(8): p. 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjoquist KM, et al. , Personalising pancreas cancer treatment: When tissue is the issue. World J Gastroenterol, 2014. 20(24): p. 7849–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank TS, et al. , Genomic profiling guides the choice of molecular targeted therapy of pancreatic cancer. Cancer Lett, 2015. 363(1): p. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papeo G, et al. , PARP inhibitors in cancer therapy: an update. Expert Opin Ther Pat, 2013. 23(4): p. 503–14. [DOI] [PubMed] [Google Scholar]
- 13.Kawahara N, et al. , Candidate synthetic lethality partners to PARP inhibitors in the treatment of ovarian clear cell cancer. Biomed Rep, 2017. 7(5): p. 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lumachi F, et al. , Proteomics as a Guide for Personalized Adjuvant Chemotherapy in Patients with Early Breast Cancer. Cancer Genomics Proteomics, 2015. 12(6): p. 385–90. [PubMed] [Google Scholar]
- 15.Weren RD, et al. , Novel BRCA1 and BRCA2 Tumor Test as Basis for Treatment Decisions and Referral for Genetic Counselling of Patients with Ovarian Carcinomas. Hum Mutat, 2017. 38(2): p. 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowery MA, et al. , Phase II trial of veliparib in patients with previously treated BRCA-mutated pancreas ductal adenocarcinoma. Eur J Cancer, 2018. 89: p. 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. FOLFIRI or Modified FOLFIRI and Veliparib as Second Line Therapy in Treating Patients With Metastatic Pancreatic Cancer.
- 18. Gemcitabine Hydrochloride and Cisplatin With or Without Veliparib or Veliparib Alone in Treating Patients With Locally Advanced or Metastatic Pancreatic Cancer.
- 19.Kaufman B, et al. , Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol, 2015. 33(3): p. 244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pishvaian MJ, et al. , A phase I/II study of the PARP inhibitor, ABT-888 plus 5-fluorouracil and oxaliplatin (modified FOLFOX-6) in patients with metastatic pancreatic cancer. Journal of Clinical Oncology, 2011. 29(15_suppl): p. TPS170-TPS170. [Google Scholar]
- 21.de Bono J, et al. , Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov, 2017. 7(6): p. 620–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh C, Targeted therapy for ovarian cancer: the rapidly evolving landscape of PARP inhibitor use. Minerva Ginecol, 2018. 70(2): p. 150–170. [DOI] [PubMed] [Google Scholar]
- 23.Keung MYT, Wu Y, and Vadgama JV, PARP Inhibitors as a Therapeutic Agent for Homologous Recombination Deficiency in Breast Cancers. J Clin Med, 2019. 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner NC and Ashworth A, Biomarkers of PARP inhibitor sensitivity. Breast Cancer Res Treat, 2011. 127(1): p. 283–6. [DOI] [PubMed] [Google Scholar]
- 25.Fojo T and Bates S, Mechanisms of resistance to PARP inhibitors--three and counting. Cancer Discov, 2013. 3(1): p. 20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pishvaian MJ, et al. , BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. Br J Cancer, 2017. 116(8): p. 1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal S, et al. , HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res, 2014. 74(4): p. 1128–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chand SN, et al. , Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors. Cancer Res, 2017. 77(18): p. 5011–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnak D, et al. , Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res, 2014. 20(19): p. 5085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotin LE, et al. , Ibrutinib synergizes with poly(ADP-ribose) glycohydrolase inhibitors to induce cell death in AML cells via a BTK-independent mechanism. Oncotarget, 2016. 7(3): p. 2765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauzee NJ, et al. , Silencing Poly (ADP-Ribose) glycohydrolase (PARG) expression inhibits growth of human colon cancer cells in vitro via PI3K/Akt/NFkappa-B pathway. Pathol Oncol Res, 2012. 18(2): p. 191–9. [DOI] [PubMed] [Google Scholar]
- 32.Nakadate Y, et al. , Silencing of poly(ADP-ribose) glycohydrolase sensitizes lung cancer cells to radiation through the abrogation of DNA damage checkpoint. Biochem Biophys Res Commun, 2013. 441(4): p. 793–8. [DOI] [PubMed] [Google Scholar]
- 33.Cortes U, et al. , Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol, 2004. 24(16): p. 7163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min W, et al. , Deletion of the nuclear isoform of poly(ADP-ribose) glycohydrolase (PARG) reveals its function in DNA repair, genomic stability and tumorigenesis. Carcinogenesis, 2010. 31(12): p. 2058–65. [DOI] [PubMed] [Google Scholar]
- 35.Koh DW, et al. , Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A, 2004. 101(51): p. 17699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breslin C, et al. , The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res, 2015. 43(14): p. 6934–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ame JC, et al. , Radiation-induced mitotic catastrophe in PARG-deficient cells. J Cell Sci, 2009. 122(Pt 12): p. 1990–2002. [DOI] [PubMed] [Google Scholar]
- 38.Gravells P, et al. , Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase. DNA Repair (Amst), 2017. 52: p. 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Toni EN, et al. , Inactivation of BRCA2 in human cancer cells identifies a subset of tumors with enhanced sensitivity towards death receptor-mediated apoptosis. Oncotarget, 2016. 7(8): p. 9477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh DW, Dawson VL, and Dawson TM, The road to survival goes through PARG. Cell Cycle, 2005. 4(3): p. 397–9. [DOI] [PubMed] [Google Scholar]
- 41.Gravells P, et al. , Radiosensitization with an inhibitor of poly(ADP-ribose) glycohydrolase: A comparison with the PARP1/2/3 inhibitor olaparib. DNA Repair (Amst), 2018. 61: p. 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James DI, et al. , First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib. ACS Chem Biol, 2016. 11(11): p. 3179–3190. [DOI] [PubMed] [Google Scholar]
- 43.Hucl T, et al. , A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res, 2008. 68(13): p. 5023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallmeier E, et al. , Targeted disruption of FANCC and FANCG in human cancer provides a preclinical model for specific therapeutic options. Gastroenterology, 2006. 130(7): p. 2145–54. [DOI] [PubMed] [Google Scholar]
- 45.Golan T, et al. , Recapitulating the clinical scenario of BRCA-associated pancreatic cancer in pre-clinical models. Int J Cancer, 2018. 143(1): p. 179–183. [DOI] [PubMed] [Google Scholar]
- 46.Olive KP, et al. , Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science, 2009. 324(5933): p. 1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreiber FS, et al. , Successful growth and characterization of mouse pancreatic ductal cells: functional properties of the Ki-RAS(G12V) oncogene. Gastroenterology, 2004. 127(1): p. 250–60. [DOI] [PubMed] [Google Scholar]
- 48.Waszkowycz B, et al. , Cell-active Small Molecule Inhibitors of the DNA-damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides. J Med Chem, 2018. [DOI] [PubMed]
- 49.Jimbo M, et al. , Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget, 2015. 6(29): p. 27312–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Veroli GY, et al. , Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics, 2016. 32(18): p. 2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roell KR, Reif DM, and Motsinger-Reif AA, An Introduction to Terminology and Methodology of Chemical Synergy-Perspectives from Across Disciplines. Front Pharmacol, 2017. 8: p. 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rafehi H, et al. , Clonogenic assay: adherent cells. J Vis Exp, 2011(49). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco FF, et al. , The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene, 2016. 35(19): p. 2529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sever-Chroneos Z, et al. , Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol Cell Biol, 2001. 21(12): p. 4032–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gogola E, et al. , Selective Loss of PARG Restores PARylation and Counteracts PARP Inhibitor-Mediated Synthetic Lethality. Cancer Cell, 2018. 33(6): p. 1078–1093 e12. [DOI] [PubMed] [Google Scholar]
- 56.Satyananda V, et al. , Advances in Translational Research and Clinical Care in Pancreatic Cancer: Where Are We Headed? Gastroenterol Res Pract, 2019. 2019: p. 7690528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. , Gemcitabine plus nab-paclitaxel for advanced pancreatic cancer after first-line FOLFIRINOX: single institution retrospective review of efficacy and toxicity. Exp Hematol Oncol, 2015. 4: p. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahib L, et al. , Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res, 2014. 74(11): p. 2913–21. [DOI] [PubMed] [Google Scholar]
- 59.Shirai H, et al. , PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis, 2013. 4: p. e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Sullivan J, et al. , Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun, 2019. 10(1): p. 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim MY, Zhang T, and Kraus WL, Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev, 2005. 19(17): p. 1951–67. [DOI] [PubMed] [Google Scholar]
- 62.Barkauskaite E, et al. , Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun, 2013. 4: p. 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez-Gonzalez R, et al. , Regulatory mechanisms of poly(ADP-ribose) polymerase. Mol Cell Biochem, 1999. 193(1–2): p. 19–22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.